Abstract
RECQL4 is a human RecQ helicase which is mutated in approximately two-thirds of individuals with Rothmund–Thomson syndrome (RTS), a disease characterized at the cellular level by chromosomal instability. BLM and WRN are also human RecQ helicases, which are mutated in Bloom and Werner's syndrome, respectively, and associated with chromosomal instability as well as premature aging. Here we show that primary RTS and RECQL4 siRNA knockdown human fibroblasts accumulate more H2O2-induced DNA strand breaks than control cells, suggesting that RECQL4 may stimulate repair of H2O2-induced DNA damage. RTS primary fibroblasts also accumulate more XRCC1 foci than control cells in response to endogenous or induced oxidative stress and have a high basal level of endogenous formamidopyrimidines. In cells treated with H2O2, RECQL4 co-localizes with APE1, and FEN1, key participants in base excision repair. Biochemical experiments indicate that RECQL4 specifically stimulates the apurinic endonuclease activity of APE1, the DNA strand displacement activity of DNA polymerase β, and incision of a 1- or 10-nucleotide flap DNA substrate by Flap Endonuclease I. Additionally, RTS cells display an upregulation of BER pathway genes and fail to respond like normal cells to oxidative stress. The data herein support a model in which RECQL4 regulates both directly and indirectly base excision repair capacity.
INTRODUCTION
Rothmund–Thomson syndrome (RTS) is a rare, autosomal recessive disorder associated with a characteristic skin rash (poikiloderma) that begins in infancy, small stature, skeletal dysplasia, radial ray defects, sparse hair and eyebrows and occasional cataract formation. Approximately one-third of individuals with RTS develop osteosarcoma at a median age of 11.5 years. Other malignancies are rarely seen but can include cutaneous squamous cell carcinoma and myelodysplasia (1). A high incidence of chromosome abnormalities, mainly mosaic trisomies and isochromosomes, are found in RTS cells and are assumed to underlie the cancer predisposition of these patients (1,2).
Two-thirds of RTS individuals have a RECQL4 mutation (and 90% of these patients have a mutation in both alleles) (3). Thus far, all genotyped RTS individuals who develop osteosarcoma carry a mutation in RECQL4 (3). RECQL4 mutations also cause RAPADILINO (Radial and patellar aplasia) (4) and Baller-Gerold (bilateral radial aplasia and craniosynostosis) syndromes (5). The genomic structure of the RECQL4 gene is unusual in that it has short introns (<100 bp) that are spliced inefficiently (6–8). Consequently, mutations in both the introns and exons of RECQL4 can cause RTS.
RecQ family helicases unwind DNA in a 3′ to 5′ direction in an ATP hydrolysis-dependent manner (9). RECQL4 shares homology to the highly conserved central helicase domain of RecQ helicases, including the Walker A-box motif, which is partially responsible for ATP binding and hydrolysis. However, RECQL4 does not share homology to two other conserved RecQ motifs, RecQ C and HRDC, which are present in human BLM and WRN RecQ helicases. It is unclear whether RECQL4 possesses helicase activity (10,11). Previously it was reported that RECQL4 lacked helicase and translocase activity (10). However, a recent report suggests that RECQL4 has two domains capable of DNA unwinding, one in the N-terminus and another within the helicase domain (11). The N-terminal region of RECQL4, which is not conserved in other RecQ helicases, has weak homology to yeast Sld2/DRC1, which is essential for the initiation of replication in yeast (12,13). Xu and Liu (11) suggest that RECQL4's helicase activity was masked by its more robust DNA annealing activity.
The function of RECQL4 and the cellular pathways in which it is involved remains poorly understood. However, it has been proposed that RECQL4 may play similar biological roles as human BLM and WRN. BLM- and WRN-deficient cells show genomic instability and increased sensitivity to ionizing radiation, reduced DNA repair synthesis, delayed S phase and hypersensitivity to agents that interfere with DNA replication (10,12,14). The cellular phenotype of RTS cells appears to be somewhat variable, which may reflect the genetic heterogeneity of these cells. Some RTS cell lines show lower survival and reduced DNA repair synthesis in response to radiation-induced DNA damage, while some RTS cell lines are not hypersensitive to radiation (15–19). Similarly, some RTS cells are proficient in nucleotide excision repair (20,21), while other RTS cells are hypersensitive to UV irradiation (22). Several RTS cell lines are not hypersensitive to bifunctional alkylating agents (18), and one RTS cell line was reported to undergo irreversible growth arrest after treatment with H2O2 (23); however, another study reported that several RTS cell lines are not hypersensitive to H2O2 (24). Embryonic fibroblasts from a mouse with a targeted deletion in RECQL4's helicase domain (exon 13) are not hypersensitive to UV or γ-irradiation (25). The heterogeneity in the response of RTS cell lines to DNA damage could be due, in part, to the fact that not all RTS cell lines have a defect in RECQL4. One-third of RTS patients have a mutation in an unknown gene product (3).
Interestingly, RECQL4 is present in both the nucleus and cytoplasm of human cells (26), while BLM and WRN are localized exclusively to the nucleus. In the nucleus, RECQL4 is associated with pro-myelocytic leukemia (PML) bodies, and with DNA double-strand break (DSB)-induced Rad51 foci (27). RECQL4 also accumulates in nucleoli, in cells treated with H2O2 or streptonigrin (28). The association of RECQL4 with PML bodies and Rad51 foci suggests that RECQL4 may play a role in DSB repair via homologous recombination (29). However, RECQL4 also interacts with PARP-1, and localization of RECQL4 to nucleoli is inhibited by a PARP-1 inhibitor (28). This observation, and the sensitivity of some RTS cells to oxidative DNA damage, as discussed earlier, suggests a possible role for RECQL4 in base excision repair (BER).
BER is the major mechanism for repairing oxidative DNA lesions. BER appears to be essential for viability in higher animals, because knockout mice for APE1/HAP1 (30), DNA polymerase β (POL β) (31), Flap Endonuclease I (FEN1) (32), XRCC1 (33), DNA ligase I (34) and DNA ligase III die in embryonic stages (35). There are two BER sub-pathways: short-patch BER, which involves a one nucleotide repair patch, and long-patch BER, which involves a 2–7 nucleotide repair patch. The initial step in BER is recognition and excision of base lesions a DNA glycosylase (36). DNA glycosylases catalyze hydrolysis of the N-glycosylic bond, releasing the damaged base to form an abasic (or apurinic/apyrimidinic; AP) site. The efficient repair of abasic sites is critical, because they are mutagenic lesions. In BER, an incision is made immediately 5′ to the AP site by apurinic endonuclease 1 (APE1), leaving a 5′ terminal deoxyribose phosphate (dRP) group (37). For short patch BER, DNA polymerase β (POL β) is recruited to the site to fill-in the single nucleotide gap and to remove the dRP group (38). The nick is then sealed by the DNA ligase III/XRCC1 complex (39).
Some DNA lesions inhibit the lyase activity of POL β (e.g. a reduced dRP group), and under these conditions strand displacement DNA synthesis is required. This is performed through the incorporation of multiple nucleotides (typically 2–7) by long patch BER (40). Strand displacement DNA synthesis is mediated by a proliferating cellular nuclear antigen-dependent polymerase, POL ε or δ (or by POL β), and replication factor C (41). Flap endonuclease, FEN1, then cleaves off the resulting flap structure, leaving a nick to be sealed by DNA ligase I (42).
Single strand break (SSB) repair is another subpathway of BER. SSBs can be generated directly by DNA damaging agents or during processing of base damage during BER. XRCC1 foci formation is a standard method for monitoring SSB formation (43,44). In situations of non-conventional SSB termini (e.g., 3′-phosphate), additional enzymes, including APE1, PNK, TDP1 or Aprataxin, are required to regenerate a 3′-OH or 5′-P terminus prior to SSB repair (43,44). This study examines a possible role for RECQL4 in several aspects of BER.
RESULTS
Defective repair of H2O2-induced DNA lesions in RTS and RECQL4 knockdown cells
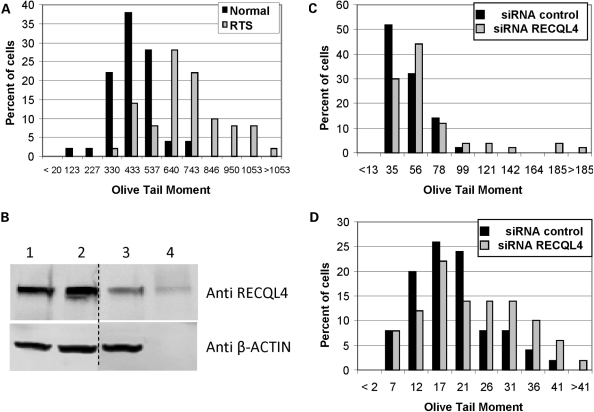
We have previously compared the normal human primary MRC5 cells to WRN deficient cells using the comet assay after H202 (45) and therefore elected to use the MRC5 cells in our analysis here. The primary RTS cell line, AG05013, was derived from a 10-yr-old male and carries two different frame shift mutations in RECQL4 (46,47). Based on Kitao et al. (47), one allele has a frame shift mutation in the RecQ helicase domain and in the other allele, the mutation is situated just beyond the RecQ helicase domain at amino acid 830. Neither allele gives rise to the expression of stable RECQL4 truncated proteins (26). These RTS cells along with the normal human fibroblasts, MRC5, were exposed to H2O2 and the number of Fpg-sensitive DNA lesions was estimated using the comet assay. The cells were treated with H2O2 (500 µM) for ∼30 min (15 min in culture medium on ice and 15 min in agarose suspension), lysed and treated with E. coli Fpg prior to analysis (Fpg converts oxidative DNA lesions to SSBs). Comet tail length and Olive tail moment (OTM) were calculated from images of ∼50 cells as described in Materials and Methods. Mean OTM values were calculated using the data shown in Fig. 1A, and they were significantly different for the AG05013 RTS (OTM 650 ± 26) and MRC5 normal (OTM 395 ± 17) cells (two-tailed Student's t-test P-value 1.5 E−11) (Fig. 1A). The above results suggest that RTS cells are hypersensitive to H2O2-induced DNA damage.
Figure 1.
Comet assay of RTS and RECQL4 knockdown cells treated with H2O2. (A) Comet assay for RTS fibroblasts. Exponentially growing normal, MRC5, or RTS, AG5013, cells were treated with 500 µM H2O2 for 15 min on ice, and comet assay was performed as described in Materials and Methods. (B) Western blot analysis of U2OS cells treated with RECQL4 siRNA. Blot was probed with the indicated antibody. Western blot of U2OS whole cell extracts showing the knockdown effects of RECQL4 siRNA: (1) No treatment; (2) Negative control siRNA; (3) RECQL4 siRNA; (4) Purified RECQL4 protein (∼5 ng). (C) Comet assay for RECQL4 knockdown U2OS cells treated with H2O2. RECQL4 knockdown U2OS cells were treated with 500 µM H2O2 for 15 min and comet assay was performed as described in Materials and Methods. (D) Comet assay for RECQL4 knockdown cells with 3 h recovery after treatment with H2O2. Same as in (C), except cells were incubated for 3 h in fresh medium lacking H2O2 before comet assay was performed. Comet assay data was analyzed using Komet 5.5 software. Shown is the percentage of cells with the observed Olive Tail moment values. In order to graphically represent the OTMs, the data were binned into 10–12 segments and, unless otherwise noted, the median of the bin is displayed on the X-axis.
This observation was confirmed using U2OS osteosarcoma cells transiently transfected with siRNA to RECQL4 that has a 60–90% knockdown efficiency (Fig. 1B). As observed in H2O2-treated RTS cells, RECQL4 knockdown U2OS cells had a significantly higher mean OTM (52.5) than control cells (38.1) (two-tailed P-value 0.007; Fig. 1C). Defective repair of H2O2-induced DNA damage was also observed in RECQL4 knockdown cells 3 h after removal of H2O2. Thus, a longer tail length (P-value 0.003; data not shown) and a trend towards a higher mean OTM (two-tailed P-value 0.07; Fig. 1D) was observed in RECQL4-knockdown cells when cells were given a 3 h recovery period after treatment with H2O2 before comet assay was performed. These data suggest that the RECQL4-deficiency is associated with a defect in the repair H2O2-induced DNA damage.
Increased number of XRCC1 foci in RTS cells
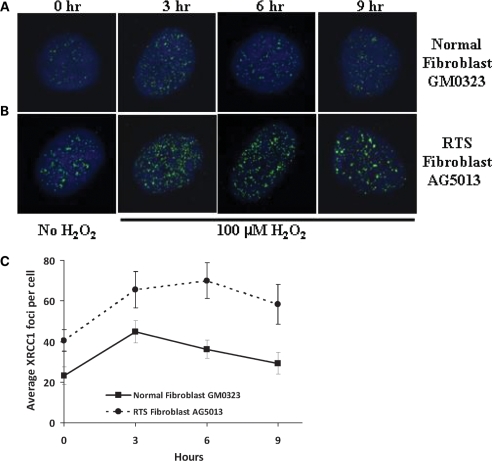
XRCC1 foci are important markers of SSBs in vivo (44), and they can be readily visualized using immunofluorescence and confocal microscopy. XRCC1 foci formation was monitored after exposure to 100 µM H2O2 in primary normal GM00323 and RTS fibroblasts AG05013 fibroblasts. XRCC1 foci were measured without treatment (0 h) and after treatment plus 3, 6 or 9 h following exposure to 100 µM H2O2 (Fig. 2A). The average number of foci per cell observed is shown in Fig. 2B. The images show that both basal and H2O2-induced XRCC1 foci are more abundant in RTS cells than in normal cells.
Figure 2.
XRCC1 foci in RTS cells treated with H2O2. The GM00323 normal (A) and AG5013 RTS fibroblast (B) cell lines were incubated in the absence (0 h) or presence of 100 µM H2O2 for 15 min, and then fixed 3, 6 or 9 h after H2O2 treatment. Cells were stained with rabbit anti-XRCC1 antibody as described in Materials and Methods. Images were captured with a Nikon TE2000 microscope at 600× magnification with five laser imaging modules and a CCD camera (Hamamatsu). (C) Average number of foci observed per cell, minimum number of cells analyzed for each time point was 10 cells. Significant P-values between the normal and RTS cells were 0.024, 0.044, 0.0031 and 0.001 for the 0, 3, 6 and 9 h points, respectively.
High endogenous level of FapyG and 8-oxoG in RTS cells
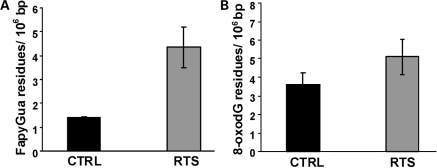
The relative abundance of 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 8-oxoG was measured in primary normal GM00323 and AG05013 RTS cells. DNA was isolated from the cells, digested with P1 nuclease and treated with alkaline phosphatase, then analyzed by GC/MS. The results showed a significant ∼3-fold higher level of FapyG (P= 0.03) in the RTS cells than in the normal cells (Fig. 3). In contrast, the 8-oxoG levels were not statistically significantly changed.
Figure 3.
Endogenous levels of FapyG and 8-oxoG in RTS cells. DNA was isolated from normal control, GM00323 and RTS fibroblasts, AG5013, digested with P1 nuclease and alkaline phosphatase, then analyzed by GC-MS for FapyG (A) or 8-oxoG (B) as described in Materials and Methods. Values indicated are modified residues per 106 bases.
Interactions between RECQL4 and BER proteins
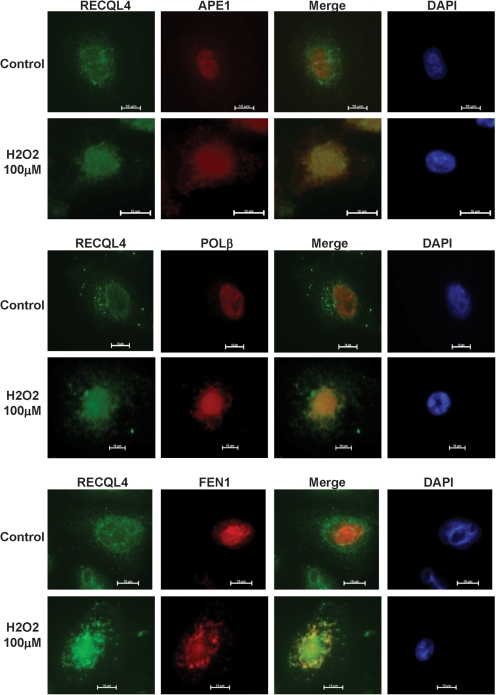
The above results suggest that RECQL4 may play a role in the response to H2O2 -induced DNA damage, which is largely repaired by BER. Thus, possible physical interactions between RECQL4 and the three core BER proteins APE1, POL β and FEN1, were examined in HeLa cells using an immunohistochemical approach. The results show that RECQL4 is present in the cytoplasm and nucleus and that it re-localizes to the nucleolus in response to oxidative stress (Fig. 4), which is consistent with previous findings (28). Furthermore, RECQL4 grossly co-localizes with APE1, and FEN1 in the nucleus in H2O2-treated cells (Fig. 4). In the case of POL β, there was a concomitant increase in the nuclear protein expression levels following H2O2 treatment, but we were unable to resolve clear foci. We also find a co-localization of RECQL4 to mitochondria (data not shown). These results are consistent with the possibility that RECQL4 participates in BER.
Figure 4.
Immunohistochemical localization of RECQL4, APE1, POL β and FEN1 in HeLa cells treated with H2O2. HeLa cells were incubated in the presence or absence of 100 µM H2O2 for 30 min and then fixed. Cells were stained for DAPI, RECQL4 and APE1, POL β or FEN1, as described in Materials and Methods.
RECQL4 stimulates APE1 endonuclease activity
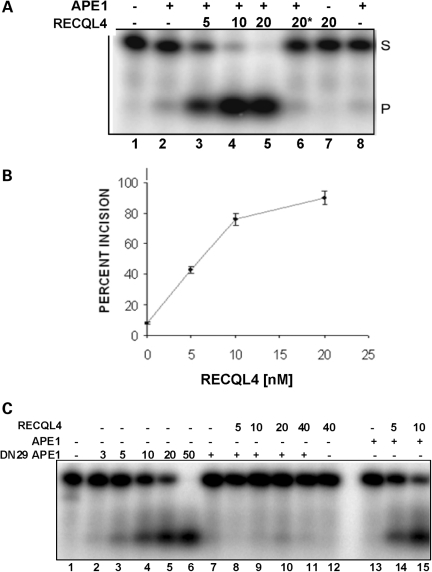
APE1 is the major BER AP endonuclease. We examined whether RECQL4 stimulates APE1 incision activity in vitro, using a 34 bp dsDNA oligonucleotide substrate with a tetrahydrofuran abasic site analog at position 16. In the presence of ∼0.05 nM APE1, which incises ∼10% of input DNA substrate under the conditions tested, RECQL4 (5 nM) stimulated APE1 endonuclease activity ∼4-fold, with higher stimulation at higher concentrations of RECQL4 (Fig. 5A and B). No stimulation was observed with heat-inactivated RECQL4 (Fig. 5A, lane 6), and RECQL4 had no intrinsic AP site incision activity (Fig. 5A, lane 7).
Figure 5.
Effect of RECQL4 on APE1 endonuclease activity. (A) APE1 incision assays were carried out using a 34 bp dsDNA substrate containing a tetrahydrofuran at position 16. APE1 (20 pg; ∼55 fM) was added as indicated before addition of the indicated amount of RECQL4. Asterisk indicates heat inactivation. (B) APE incision assays were quantified and average percent incision was calculated using data from three experiments. (C) AP incision assays were carried out with DN29 APE1 in the presence or absence of RECQL4. Plus symbol indicates addition of 2.5 pg DN29 APE1 or 20 pg of APE1, prior to addition of the indicated amount of RECQL4.
Previous studies show that the N-terminal 35 amino acids of APE1 are required for its interaction with WRN, but are not required for APE1 endonuclease activity (48). Therefore, a mutant of APE1 lacking the first 29 amino acids (DN29 APE1) was tested for its ability to interact with and be stimulated by RECQL4. The results show that RECQL4 does not stimulate DN29 APE1 (Fig. 5C, lanes 8–11), suggesting that RECQL4 may interact physically with the N-terminal region of APE1 to activate its endonuclease function.
RECQL4 modulates POL β catalytic activity
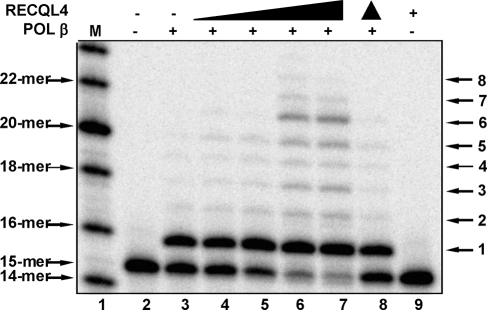
POL β is the primary DNA polymerase for BER. Thus, we also examined whether RECQL4 stimulates POL β nucleotide insertion and strand displacement DNA synthesis activities in vitro using a 34 bp gapped dsDNA substrate. The results show that RECQL4 stimulates primer extension and strand displacement DNA synthesis by POL β in a dose-dependent manner (Fig. 6). Heat-inactivated RECQL4 did not stimulate POL β and RECQL4 has no intrinsic nucleotide insertion or DNA strand displacement activity (Fig. 6, lanes 8–9).
Figure 6.
Effect of RECQL4 on POL β DNA synthesis activity. Increasing amounts of RECQL4 protein were added to reaction mixtures containing 12.5 nM DNA and 1 nM POL β where indicated. RECQL4 concentrations were 0, 0.75, 1.5, 3 and 6 nM. Filled triangle indicates heat inactivation. Assays were performed as described in Materials and Methods.
RECQL4 stimulates FEN1 incision activity
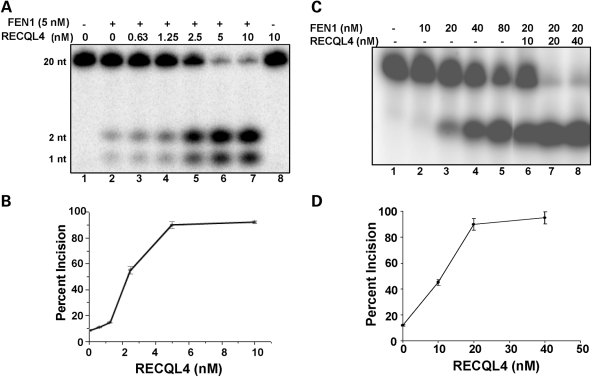
FEN1 endonuclease is required to remove DNA flaps (ssDNA 5′-protruding ends) generated by DNA strand displacement synthesis during long patch BER. Here, we examined whether RECQL4 stimulates FEN1 incision activity in vitro using a DNA substrate with a 1- or 10-nucleotide flap (Fig. 7). The results show that RECQL4 stimulates FEN1 incision activity on both DNA substrates in a dose-dependent manner. RECQL4 (2.5 nM) stimulated FEN1 (5 nM) incision of the 1-nucleotide flap DNA substrate ∼6-fold (Fig. 7A and B), and RECQL4 (10 nM) stimulated FEN1 (20 nM) incision of the 10-nucleotide flap ∼4.3-fold (Fig. 7C and D). RECQL4 has no intrinsic flap endonuclease activity (Fig. 7A, lane 8).
Figure 7.
Effect of RECQL4 on FEN1 flap incision activity. FEN1 incision activity was measured using a 1-nucleotide (A and B) or 10-nucleotide (C and D) flap DNA substrate as described in Materials and Methods. The indicated amount of RECQL4 was added. (B and D) FEN1 incision assays were quantified and mean ± standard error was calculated using data from at least two experiments.
Microarray analysis
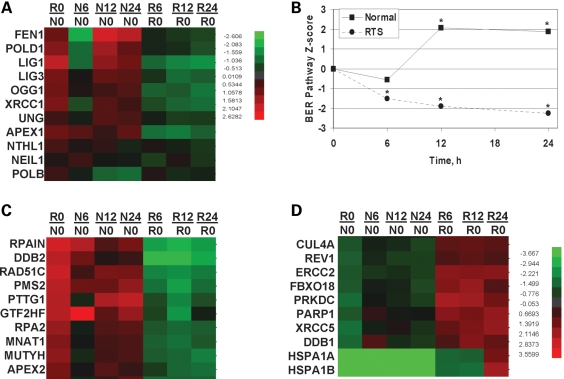
Three normal, GM00323, GM00969, GM01864 and three RTS, AG18371, AG05013, AG17524, cell lines were assessed for their ability to respond to oxidative stress induced by treatment with 25 µM menadione for 1 h. Cells were harvested immediately or 6, 12 or 24 h after treatment. As we had done previously (49), the mRNAs were pooled from the three samples in an attempt to minimize inter-individual variability. Pooled mRNAs were hybridized to the 23 k gene Illumina's Sentrix HumanRef-8 v2 Expression BeadChips (Illumina) three times and the results were consistent. While we recognize that many of the proteins below have roles in other DNA metabolic processes, we elected to focus on those genes whose gene products play a major role in the repair of oxidative DNA damage through either the short or long patch BER pathway. Specifically, OGG1, NTH1, NEIL1, UNG, APEX1, POLB, POLD, FEN1, LIG1, LIG3 AND XRCC1 gene expression profiles were evaluated in normal (N) and RTS (R) cells (Fig. 8). Gene expression profile changes for the individual genes are depicted in the heat map and are clustered as a function of time after treatment (Fig. 8A). Initially, the RTS cells over-express the BER genes relative to normal cells (no treatment RTS/no treatment normal, R0/N0, lane, Fig. 8A). However, after menadione treatment the RTS cells show a general trend to down-regulate the BER genes. Gene set analysis, as opposed to individual gene profile analysis, has been shown to be more sensitive at identifying significant biological changes (50). By the gene set analysis approach, at time zero, the RTS cells up-regulate BER genes relative to normal cells and this is highly statistically significant, BER pathway Z-score R0/N0 2.6 and P-value 3.6 × 10−6. For the pathway analysis, if the P-value was <0.05 then the pathway changes were considered statistically significant. For each cell line, the N0 and R0 ratios were set to zero and the response of the cells to menadione was plotted (Fig. 8B). After treatment, the normal cell lines up-regulated the BER genes while the RTS cell lines fail to respond in a similar manner (Fig. 8B). Six hours after menadione the data is not statistically significant, P-value 0.19, however, at both the 12 and 24 h time points there is a statistically significant difference between the normal and RTS cell lines, P-values 0.018 and 0.006 for the 12 and 24 h time points, respectively. Thus, the RTS cells display an elevated level of BER genes without treatment and failed to respond appropriately upon oxidative stress. These data also support our proposal that the RTS cells are experiencing endogenous DNA damage that needs to be repaired by BER and furthermore, that the RTS cells fail to respond appropriately after oxidative stress.
Figure 8.
BER pathway analysis of microarray data. (A) Clustering of RNA expression patterns for select BER genes observed after no treatment (R0/N0) or after treatment with 25 µM menadione and harvested at 6, 12 or 24 h later. (B) Relative BER pathway gene expression changes for 11 BER proteins, OGG1, NTH1, NEIL1, UNG, APEX1, POLB, POLD, FEN1, LIG1, LIG3 and XRCC1, were analyzed and are displayed. The relative RTS versus normal signal is plotted as a function of the time when the samples were harvested. All points were statistically significant (*) except the 6 h point in the normal cells. A P-value of 0.05 was considered statistically significant. (C and D) Cluster analysis of the top 10 most up- or down-regulated DNA repair genes after menadione treatment. R0/N0, ratio of z-scores of RTS to normal cells. Nx/N0 or Rx/N0, ratio of z-scores of normal (N) or RTS (R) relative to z-score at time zero (N0 or R0), where x represents the time of harvest after menadione treatment, 6, 12 or 24 h.
In addition to the BER genes listed earlier, we also analyzed expression patterns of all DNA repair genes. The DNA repair gene set (176 genes) was derived from the Broad Research Institute at MIT and the gene expression profile pattern for each DNA repair gene is shown in Supplementary Material, Fig. S1 and summarized in Supplementary Material, Table S1. As can be seen in the heat map, not all DNA repair genes are dysregulated. The relative gene expression profiles between the RTS and normal cells are shown in the R0/N0 lane (Supplementary Material, Figs 1 and 8C and D). Note many genes are up-regulated (red) in RTS cells at time zero relative to the normal controls, R0/N0 lane. Ten of the top genes whose profiles change the most after menadione treatment, either up- or down-regulated, are shown in Fig. 8C and D, respectively. The heat maps were clustered based on time after menadione treatment and normalized back to the expression level of each gene at time zero for the respective cells. These results demonstrate that the RTS cells do not show a general decline of all gene expression profiles but rather that the cells are capable of up-regulating some DNA repair genes after stress (Fig. 8D).
DISCUSSION
This study shows that RTS and RECQL4 knockdown cells accumulate higher levels of H2O2-induced DNA damage than control cells (Figs 1 and 2). RTS cells also have a higher level of basal and H2O2-induced XRCC1 foci (Fig. 2) and have higher basal levels of FapyG and 8-oxoG (Fig. 3), suggesting that a RECQL4 deficiency is associated with defective repair of oxidative DNA damage and may exhibit a hyper-oxidation phenotype. While survival assays after H2O2 treatment showed variability, the RTS cells tended to be more sensitive to H2O2 than normal cells (data not shown). However, RTS cell line AG05013 was more sensitive to another oxidative damaging agent, menadione (Supplementary Material, Fig. S2). Interestingly, WRN-deficient cells also accumulate specific oxidative DNA damage (51) and have a pro-oxidant phenotype (52,53). Furthermore, WRN cells are not hypersensitive to γ-irradiation, which generates oxidative damage and double strand breaks (54).
Additional support for the involvement of RECQL4 in the repair of oxidative DNA damage is provided by the fact that RECQL4 stimulates the enzymatic functions of three BER proteins, APE1, POL β and FEN1 in vitro (Figs 5–7) and shows gross co-localization with APE1 and FEN1 in the nucleus of cells after oxidative DNA damage (Fig. 4). Specifically, we found that the N-terminal 29 amino acids of APE1 are required for the interaction with RECQL4 (Fig. 5C); although WRN also interacts with this region of APE1, WRN does not stimulate APE1 incision activity in vitro (48). In total, RECQL4 may promote repair of oxidative DNA damage by activating BER. To our knowledge, this is the first report to describe functional protein interactions between RECQL4 and BER proteins.
Approximately one-third of RTS patients develop osteosarcomas at a median age of 11.5 years (1) and all genotyped RTS patients with osteosarcoma have a RECQL4 mutation (3). Both WS and RTS patients suffer from a high incidence of sarcomas, whereas BS patients display a broader spectrum of cancers, similar to what is seen in the general population at a later stage in life. The observation that both RECQL4 and WRN affect BER protein function might suggest that BER needs tight regulation in order to avoid malignancy.
APE1 is somewhat overexpressed in osteosarcomas (55), as well as colon, prostate, cervical and ovarian cancer (56), and is linked to radio- and chemoresistance during cancer therapy (57,58). In our gene expression profiling, APE1 (a.k.a. APEX1) was up regulated in RTS cells relative to normal cells (Fig. 8A). The etiology and mechanism of APE1 overexpression in these cells is not understood. Conversely, siRNA knockdown of APE1 can inhibit repair of abasic sites, inhibit proliferation and induce apoptosis in several cancer cell lines (59). APE1 knockdown also inhibits growth of ovarian tumor xenografts (60) and causes hypersensitivity to H2O2 in osteosarcoma cells (55). Inhibitors of APE1 cause hypersensitivity to H2O2 in some cancer cell lines (61). Since RECQL4 stimulates APE1 incision activity at abasic sites in vitro (Fig. 5), RECQL4 could potentially be explored as a novel target for modulating APE1 activity. Perhaps such compounds could have therapeutic efficacy against osteosarcomas or other cancers. Clearly, additional studies are needed to understand how APE1 and RECQL4 contribute to the development of osteosarcomas in individuals with RTS (23) and other cancers.
RECQL4, like WRN and BLM (48,62), stimulates strand displacement DNA synthesis by POL β (Fig. 6). WRN's stimulation of POL β's activity was dependent on an active WRN helicase (48,62). Furthermore, we showed that a deletion mutant of WRN which contained just the helicase domain was sufficient for POL β stimulation (48,62). While RECQL4 has been shown not to have helicase activity under normal helicase buffer conditions (10), it can have helicase activity under specialized conditions including the presence of large excess of single stranded DNA (11). In our experiments, there is only a slight excess of ssDNA, and so we consider it likely that helicase activity of RECQL4 is not required for POL β strand displacement synthesis. Xu and Liu (11) stated that the C-terminal domain of RECQL4 had a mild inhibitory effect on the SFII helicase domain and perhaps upon interaction with POL β, RECQL4's helicase could be transactivated. Thus, while RECQL4 does not appear to have helicase activity under the conditions of our experimentation, we cannot exclude the possibility that some activity is present.
RECQL4 (Fig. 7), WRN and BLM (63,64) stimulate FEN1 catalyzed incision of 1- and 10-nucleotide 5′-flap structures. WRN and BLM have similar affinities for FEN1 and the protein regions responsible for their interaction with FEN1 have been mapped (64). Although RECQL4 shares sequence similarity to these regions of WRN and BLM (63), RECQL4 does not co-immunoprecipitate with FEN1 from HeLa cells (data not shown). However, a direct protein interaction was observed when purified proteins were used (Supplementary Material, Fig. S3). Thus, RECQL4 could potentially stimulate FEN1 in vivo in one of the several pathways in which it acts, including BER, homologous recombination, lagging-strand DNA replication, re-initiation of stalled replications forks or telomere stability (42,65,66). Previously, we demonstrated that WRN's FEN1 stimulation was independent of WRN helicase activity (63). Since the FEN1 incision assays were done in the absence of ATP, RECQL4's FEN1 stimulation is not dependent upon an active helicase, as observed with WRN.
Finally, given the significant role that BER plays in response to both endogenous and exogenous oxidative damage, normal and RTS cell lines were assessed for their ability to respond to oxidative stress produced by 25 µM menadione. We elected to focus on those genes whose gene products play a major role in the repair of oxidative damage through the BER pathway, including those for short and long patch BER. The gene expression profiles for OGG1, NTH1, NEIL1, UNG, APEX1, POLB, POLD, FEN1, LIG1, LIG3 and XRCC1 were assessed 6, 12 or 24 h after the 1 h menadione treatment (Fig. 8). The analysis revealed that the RTS cells displayed an up-regulation of BER genes without stress treatment and failed to respond like the normal cell lines following oxidative stress. RTS cells showed a general down-regulation of BER genes. A previous microarray analysis using MCF7 breast cancer cells and menadione reported that genes for FEN1 and uracil DNA glycosylase were down-regulated in response to 25 µM menadione (67). In normal cells, we observed the same phenomenon for FEN1 at the 12 and 24 h points; however, the RTS cells showed little or no change in FEN1 expression after menadione treatment (Fig. 8A). In contrast, our normal and RTS cells displayed a differential response with respect to UNG, and the overall Z-ratio changes for UNG in both the normal and RTS cells were <1.5. The Z-ratio changes were not >1.5 for several of the BER pathway genes and thus the reason we elected to do the pathway analysis rather than the individual gene expression profiling. The gene set enrichment analysis strategy used here has been shown to improve the analysis of minimally changed gene expression profiles (50). The assumption behind this type of analysis is that the statistical significance of co-regulated genes will be greater than that for individual genes in a given set. As discussed earlier, in untreated normal and RTS cells there is a highly statistically significant difference in the expression of the BER pathway genes, P-value 3.6 × 10−6. It seems likely that the RTS cells are attempting to compensate for the loss of RECQL4 and are experiencing more endogenous stress.
This paper represents the first biochemical analysis of RECQL4 that describes three functional protein interactions that predict a potential role for RECQL4 in modulating core BER proteins. Due to the elevated levels of XRCC1 foci, and specific modulation of POL β and FEN1, it is likely that RTS cells have a defect in SSB repair. Altered BER regulation seems to be a re-occurring theme among the human RecQ helicase family, because both WRN and BLM share many of the same functional protein interactions and modulate core BER proteins (54). It may be interesting in future studies to evaluate whether RECQ1 and RECQ5 also possess the capacity to regulate the BER process.
MATERIALS AND METHODS
Cell culture
Primary cell lines were obtained from Coriell Cell Repositories and maintained at 37°C and 5% CO2. Normal fibroblast cell lines used were MRC5, GM00323, GM01864 and GM00969. RTS cell lines were: AG05013, AG18371 and AG17524. All lines were initially grown in Eagle's Minimum Essential Medium with Earle's salts and non-essential amino acids with 15% fetal bovine serum (not inactivated) and 1% penicillin–streptomycin. Normal cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin or AmnioMAX II Complete media (Invitrogen, CA, USA) while RTS cells were maintained in AmnioMAX II Complete media. All primary cells were grown in AmnioMAX II Complete media prior to and during the experiments. U2OS and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.
Comet assay
The comet assay was performed essentially as described in Von Kobbe et al. (45). Briefly, comet assays were performed with MRC5 (wild type) and AG05013 (RTS) cells. Cells were treated with 500 µM H2O2 for 15 min and analyzed immediately or washed and incubated in fresh medium at 37°C and 5% CO2 for 3 h. Cells were washed with PBS and 300 µl of mincing solution [1× HBSS (Ca2+, Mg2+ free), 20 mm EDTA, 10% DMSO) was added. Cells were scraped, collected in a 1.5 ml tube, pelleted and resuspended in 50 µl of the supernatant. Ten microliters of the cell suspension was mixed with 75 µl 0.5% low melting point agarose (Fisher Scientific, Fair Lawn, NJ) in 1× PBS and pipetted onto a slide coated with 1% agarose (IBI, Shelton, CT) and spread by coverslip. The slide was placed on a pre-chilled aluminum tray for 5 min, the cover slip removed and an additional 75 µl of low melting point agarose was added and spread. The slide was chilled again for 5 min. Slides were then placed in pre-chilled lysing solution (2.5 M NaCl, 100 mm EDTA, 10 mm Trizma base, pH 10.0) overnight at 4°C. Slides were washed three times in 0.4 M Tris (pH 7.5; Sigma) and once in 1× FLARE buffer (Trevigen, Inc.). Cells were treated with formamidopyrimidine-DNA glycosylase (Fpg) in FPG FLARE reaction buffer (1× FLARE, 1× BSA, Trevigen, Inc.) or buffer alone for 1 h at 37°C. Slides were incubated in alkali solution (pH 12.1) for 30 min and then placed in a horizontal electrophoresis chamber filled with pre-chilled 1× TBE and run for 30 min at 35 V. DNA was stained with ethidium bromide and viewed on a Zeiss Axiovert 200 M fluorescence microscope (Zeiss, Thornwood NY). Olive tail moment and comet tail length were determined using Komet 5.5 software (Kinetic Imaging, Durham, NC) using the following conventions: Tail length=Tail Extent; Olive tail moment is defined as the percent of DNA in the tail multiplied by the distance between the means of the head and tail distributions or (Tailmean− Headmean) × Tail%DNA/100.
siRNA method
Four RECQL4 siRNAs were purchased from Dharmacon and screened for knockdown efficiency. 5′-CAAUACAGCUUACCGUACCAUU-3′ RECQL4 siRNA achieved ∼70% knockdown of RECQL4, and was selected for use in subsequent experiments. U2OS cells were transfected with negative control siRNA (Silencer Negative Control#1, Ambion) or RECQL4 siRNA (100 nM) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol.
Measurement of XRCC1 foci
XRCC1 foci were measured in two control cell lines (GM00323) and two RTS cell lines (AG05013). Approximately 25 000 primary fibroblasts were seeded on slides, grown overnight and then treated with 100 mm H2O2 for 15 min. Cells were washed and incubated for 0 (control), 3, 6 or 9 h, and then immediately fixed and stained with rabbit polyclonal XRCC1 antibody (Santa Cruz, 1:200) and Alexa Donkey anti-rabbit 488 secondary antibody. Ten images representing about 15–20 cells per well were taken using a Nikon TE2000 spinning disk microscope with five laser imaging modules and a CCD camera (Hamamatsu). The data were analyzed using Volocity version 4.3.1 build 6 (Improvision).
Quantification of FapyG and 8-OHdG
DNA was prepared from wild-type control GM00323 cells and AG05013 RTS cells. Purified DNA was dissolved in water and samples were blinded. DNA quality and concentration were determined from the UV spectrum (200 and 350 nm). DNA (50 µg) was treated with 2 µg E. coli Fpg, ethanol precipitated and the supernatant was lyophilized, trimethylsilylated and analyzed by GC/MS as described previously (68,69). Samples were supplemented with purified homogeneous 8-OHG-13C2,15N2, FapyA-13C,15N2 and FapyG-13C,15N2 as internal standards. Selected-ion monitoring was used to quantify trimethylsilylated 8-OHG and FapyG, and isotope-labeled internal standards (70).
Colocalization
HeLa cells were grown overnight on chambered slides to 70% confluency then treated with 100 µM H2O2 for 30 min in serum-free medium. Cells were washed with DMEM, incubated at 37°C for 5.5 h in DMEM with 10% FBS and antibiotics, washed in PBS containing 0.04% Triton X-100 and fixed in PBS with 4% formaldehyde for 10 min at room temperature. Cells were washed in PBS with 0.04% Triton X-100 once for 2 min, permeabilized in PBS containing 0.2% Triton X-100 for 10 min, washed with PBS for 2 min and placed in blocking solution (2% BSA, PBS) for 1 h at room temperature. Primary antibodies APE1 (abcam abs722, 1:1000), FEN1 (abcam ab462, 1:500), POL β (Life Sciences, 1:500), RECQL4 (Santa Cruz sc-16925, 1:200) were added in blocking buffer for 1 h at room temperature. Cells were washed four times with PBS for 5 min and then incubated with appropriate secondary antibodies (Invitrogen, Alexa Fluor 594 donkey anti-mouse (A21203) or Alexa Fluor 488 donkey anti-goat (A11055), 1:1000) in blocking buffer for 1 h at room temperature. Cells were washed five times for 2 min with PBS. Slides were mounted with Prolong Gold with DAPI and cover slips were added. Samples were cured in the dark for 24 h at room temperature and visualized using a Zeiss Axiovert 200 M fluorescence microscope.
DNA and protein
Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). RECQL4 protein was purified as described (10). APE1, truncated D29 APE1, POL β and FEN1 were purified as described (71). An expression construct for DN29 APE1 was generated as follows: the human APE1 coding sequence was PCR amplified from the pET-APE plasmid DNA (71) to remove the first 29 amino acids residues of the N-terminus using PCR primers TRUNCATEAPE29, 5′-CAT GCC ATG GCA AAG AAA AAT GAC AAA GA-3′, and BAM3′APE1, 5′-CGG GAT CCT CAC AGT GCT AGG TA-3′. The PCR product was subsequently digested with NcoI and BamHI, and subcloned into the corresponding restriction sites of pET11d (Novagen). DN29 APE1 was expressed as described previously (71), with minor modifications due to different elution profile during S cation exchange chromatography. All proteins were judged to be greater than 95% pure as judged by SDS–PAGE analysis.
APE1 incision assay
The DNA substrate was prepared by annealing the following oligomers: 34G, 5′GTA CCC GGG GAT CCG TAC GGC GCA TCA GCT GCA G 3′ and 34F, 5′ CTG CAG CTG ATGCGC FGT ACG GAT CCC CGG GTA C 3′. 34F contained the base analog tetrahydrofuran (designated as F) and was 5′- 32P end-labeled. Reaction mixtures (10 µl) were incubated on ice in buffer containing 50 mm HEPES-KOH pH 7.4, 50 mm KCl, 10 mm MgCl2, 0.05% Triton X-100, 100 mg/ml BSA and 5% glycerol. APE1 (∼55 fM) followed by 100 fmol DNA substrate were added, incubated on ice for 3 min, and then RECQL4 was added and samples were incubated at 37°C for 10 min. The reaction was stopped by the addition of 10 µl of formamide buffer [95% (v/v) formamide, 20 mm EDTA, 0.1% (w/v) bromophenol blue and xylene cyanol]. Samples were analyzed by electrophoresis on a 20% TBE-urea denaturing, polyacrylamide gel. At least three experiments were performed; data are presented as mean ± standard error.
POL β DNA synthesis assay
POL β DNA polymerase activity was assayed by measuring nucleotide insertion into a gapped DNA substrate made by annealing an upstream primer, 5′CTG CAG CTG ATG CGC 3′, and a downstream primer, 5′GTA CGG ATC CCC GGG TAC 3′, to a complementary 34-bp oligonucleotide, 5′GTA CCC GGG GAT CCG TAC GGC GCA TCA GCT GCA G 3′. The 5′-end of the upstream primer was radiolabeled with 32P. The reaction was performed in buffer containing 50 mm HEPES-KOH, pH 7.5, 20 mm KCl, 2 mm DTT, 4 mm ATP, 10 mm MgCl2 and 20 µM dNTP. The indicated amounts of POL β were incubated with 12.5 nM DNA substrate in a final volume of 10 µl. The reaction mixtures was assembled on ice, transferred to 37°C and incubated for 25 min. The reaction was stopped by adding 10 µl formamide loading buffer, heating to 95°C for 5 min, and samples were analyzed on a 15% TBE-Urea denaturing, polyacrylamide gel. The experiment was repeated three times.
FEN1 incision assay
DNA substrates were constructed by annealing an upstream primer (5′CTG CAG CTG ATG CGC 3′) and a downstream oligonucleotide, either flap1, 5′ACG TAC GGA TCC CCG GGT AC 3′ or flap10, 5′GGT AGG TAA ACG TAC GGA TCC CCG GGT AC 3′ to a complementary strand (5′GTA CCC GGG GAT CCG TAC GGC GCA TCA GCT GCA G-3′). The downstream oligonucleotide was labeled on the 5′ end with 32P. Assays were performed with 1 nM DNA substrate and the indicated amount of protein in a final volume of 10 µl containing 20 mm HEPES, pH 7.5, 50 mm KCl, 0.5 mm DTT, 5 mm MgCl2, 0.05% Triton X-100, 100 µg/ml BSA and 5% glycerol. Reactions were carried out at 37°C for 15 min and stopped by adding 10 µl formamide loading buffer and heating at 90°C for 5 min. Samples were analyzed on a 20% TBE-Urea denaturing, polyacrylamide gel. Experiments were repeated 2–3 times; results are presented as mean ± standard error.
Microarray analysis
Normal, GM00323, GM00969, GM01864, and RTS fibroblast cells, AG18371, AG05013, AG17524, were grown in GIBCO AmnioMAX II complete medium (Invitrogen). At ∼70% confluency, the cells were treated with 25 µM menadione, prepared in media, for 1 h then washed once with 5 ml PBS. Conditioned media, 5 ml, was returned to the plates then the cells were cultured in a 37°C incubator until they were harvested. Cells were harvested after 6, 12 or 24 h. The control cells were manipulated as the experimental except no drug was added.
Total RNA was extracted from the normal and RTS fibroblast cells using QIAGEN RNeasy Mini Kit according to the manufacturer's protocol. The quantity of recovered RNA was assessed using the NanoDrop ND-1000 Spectrophotometer. The quality of the RNA samples was assessed using an Agilent BioAnalyzer (Agilent Technologies) and was equivalent across all samples analyzed. As we did before (49), in order to reduce the inter-individual variability, RNA from the three normal cell lines were pooled and likewise for the three RTS cells. These pooled RNAs were then used for the microarray experiment.
A 0.5 µg aliquot of total RNA from the pooled normal or RTS cell lines was labeled using the Illumina Total Prep RNA Amplification kit (Ambion). A total of 0.85 µg of biotin-labeled cRNA was hybridized for 16 h to the 23 k gene Illumina's Sentrix HumanRef-8 v2 Expression BeadChips (Illumina). The arrays were washed, blocked and then hybridized. Biotinylated cRNA was detected with streptavidin-Cy3 and quantitated using Illumina's BeadStation 500GX Genetic Analysis Systems scanner. Image processing and data extraction were performed using BeadStudio ver. 15 (Illumina).
Microarray data were analyzed using DIANE 6.0, a spread sheet-based microarray analysis program. Raw microarray data were subjected to Z normalization and tested for significant changes. Genes were determined to be differentially expressed after calculating the Z-ratio and false discovery rate. Individual genes with P-value <0.001, Z-ratio >2 and false discovery rate <0.3 were considered significantly changed. Ingenuity pathways analysis (IPA) (Ingenuity System) was used to identify biological networks with the greatest number of differentially expressed genes. For each network or pathway, probability scores were calculated using the right-tailed Fisher's exact test. For the pathway analysis, a P-value of <0.05 was considered statistically significant.
RECQL4:FEN1 interaction
HeLa cells, 1 × 106, were transfected with 5 µg of pCMV23-3 × FlagRECQL4. This vector has been described previously (27). Twenty-four hours after transfection, the cells were lysed with RIPA buffer supplemented with 500 mm NaCl. The lysates were rotated for 4 h at 4°C, vigorously shaken then centrifuged at 16 000g for 20 min at 4°C. The clarified lysate was subjected to Flag antibody immunoprecipitation. After an overnight incubation, the beads were washed five times with 1 ml of RIPA buffer supplemented with 500 mm NaCl. The beads were washed twice with RIPA buffer then purified FEN1 (1 µg) was added. The proteins were allowed to incubate together for 4 h with rotation, and then the beads were washed with 1 ml of RIPA buffer supplemented with 300 mm NaCl five times. Rabbit anti-Flag (Sigma) and rabbit anti-FEN1 (Trevigen) were used to visualize the individual proteins by western blotting.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the Intramural Research Program of the National Institute of Health, National Institute on Aging and by National Institute of Health Research Grant RO1ES-015632. Annual reports Z01 AG000726-16 and Z01 AG000727. E.S. was supported, in part, by a grant from the Polish Ministry of Science and Higher Education N303 391436.
ACKNOWLEDGEMENTS
We thank members of the Laboratory of Molecular Gerontology, NIA-NIH (Baltimore, MD) especially Regina Knight and Cynthia Kasmer for their technical support, Meltem Muftuoglu and Miral Dizdaroglu for technical assistance with the endogenous lesion quantification, Robert Wersto, Joe Chrest and Coung Nguyen for cell sorting. Additionally, we would like to thank Dr Scott Maynard and Dr Robert Maul for critically reading this manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wang L.L., Levy M.L., Lewis R.A., Chintagumpala M.M., Lev D., Rogers M., Plon S.E. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am. J. Med. Genet. 2001;102:11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Ying K.L., Oizumi J., Curry C.J. Rothmund-Thomson syndrome associated with trisomy 8 mosaicism. J. Med. Genet. 1990;27:258–260. doi: 10.1136/jmg.27.4.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L.L., Gannavarapu A., Kozinetz C.A., Levy M.L., Lewis R.A., Chintagumpala M.M., Ruiz-Maldanado R., Contreras-Ruiz J., Cunniff C., Erickson R.P., et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J. Natl Cancer Inst. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 4.Siitonen H.A., Kopra O., Kaariainen H., Haravuori H., Winter R.M., Saamanen A.M., Peltonen L., Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 5.Van Maldergem L., Siitonen H.A., Jalkh N., Chouery E., De Roy M., Delague V., Muenke M., Jabs E.W., Cai J., Wang L.L., et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beghini A., Castorina P., Roversi G., Modiano P., Larizza L. RNA processing defects of the helicase gene RECQL4 in a compound heterozygous Rothmund-Thomson patient. Am. J. Med. Genet. A. 2003;120:395–399. doi: 10.1002/ajmg.a.20154. [DOI] [PubMed] [Google Scholar]
- 7.Wang L.L., Worley K., Gannavarapu A., Chintagumpala M.M., Levy M.L., Plon S.E. Intron-size constraint as a mutational mechanism in Rothmund-Thomson syndrome. Am. J. Hum. Genet. 2002;71:165–167. doi: 10.1086/341234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balraj P., Concannon P., Jamal R., Beghini A., Hoe T.S., Khoo A.S., Volpi L. An unusual mutation in RECQ4 gene leading to Rothmund-Thomson syndrome. Mutat. Res. 2002;508:99–105. doi: 10.1016/s0027-5107(02)00189-6. [DOI] [PubMed] [Google Scholar]
- 9.Soultanas P., Wigley D.B. Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci. 2001;26:47–54. doi: 10.1016/s0968-0004(00)01734-5. [DOI] [PubMed] [Google Scholar]
- 10.Macris M.A., Krejci L., Bussen W., Shimamoto A., Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Xu X., Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachrati C.Z., Hickson I.D. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachrati C.Z., Hickson I.D. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 14.Hickson I.D. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 15.Smith P.J., Paterson M.C. Enhanced radiosensitivity and defective DNA repair in cultured fibroblasts derived from Rothmund Thomson syndrome patients. Mutat. Res. 1982;94:213–228. doi: 10.1016/0027-5107(82)90183-x. [DOI] [PubMed] [Google Scholar]
- 16.Kerr B., Ashcroft G.S., Scott D., Horan M.A., Ferguson M.W., Donnai D. Rothmund-Thomson syndrome: two case reports show heterogeneous cutaneous abnormalities, an association with genetically programmed ageing changes, and increased chromosomal radiosensitivity. J. Med. Genet. 1996;33:928–934. doi: 10.1136/jmg.33.11.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinya A., Nishigori C., Moriwaki S., Takebe H., Kubota M., Ogino A., Imamura S. A case of Rothmund-Thomson syndrome with reduced DNA repair capacity. Arch. Dermatol. 1993;129:332–336. [PubMed] [Google Scholar]
- 18.Lindor N.M., Furuichi Y., Kitao S., Shimamoto A., Arndt C., Jalal S. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet. 2000;90:223–228. doi: 10.1002/(sici)1096-8628(20000131)90:3<223::aid-ajmg7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 19.Jin W., Liu H., Zhang Y., Otta S.K., Plon S.E., Wang L.L. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum. Genet. 2008;123:643–653. doi: 10.1007/s00439-008-0518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miozzo M., Castorina P., Riva P., Dalpra L., Fuhrman Conti A.M., Volpi L., Hoe T.S., Khoo A., Wiegant J., Rosenberg C., et al. Chromosomal instability in fibroblasts and mesenchymal tumors from 2 sibs with Rothmund-Thomson syndrome. Int. J. Cancer. 1998;77:504–510. doi: 10.1002/(sici)1097-0215(19980812)77:4<504::aid-ijc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 21.Grant S.G., Wenger S.L., Latimer J.J., Thull D., Burke L.W. Analysis of genomic instability using multiple assays in a patient with Rothmund-Thomson syndrome. Clin. Genet. 2000;58:209–215. doi: 10.1034/j.1399-0004.2000.580308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan W., Luo J. RecQ4 facilitates UV-induced DNA damage repair through interaction with nucleotide excision repair factor XPA. J. Biol. Chem. 2008;283:29037–29044. doi: 10.1074/jbc.M801928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werner S.R., Prahalad A.K., Yang J., Hock J.M. RECQL4-deficient cells are hypersensitive to oxidative stress/damage: Insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem. Biophys. Res. Commun. 2006;345:403–409. doi: 10.1016/j.bbrc.2006.04.093. [DOI] [PubMed] [Google Scholar]
- 24.Cabral R.E., Queille S., Bodemer C., de Prost Y., Neto J.B., Sarasin A., Daya-Grosjean L. Identification of new RECQL4 mutations in Caucasian Rothmund-Thomson patients and analysis of sensitivity to a wide range of genotoxic agents. Mutat. Res. 2008;643:41–47. doi: 10.1016/j.mrfmmm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Hoki Y., Araki R., Fujimori A., Ohhata T., Koseki H., Fukumura R., Nakamura M., Takahashi H., Noda Y., Kito S., et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- 26.Yin J., Kwon Y.T., Varshavsky A., Wang W. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]
- 27.Petkovic M., Dietschy T., Freire R., Jiao R., Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 28.Woo L.L., Futami K., Shimamoto A., Furuichi Y., Frank K.M. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp. Cell. Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 29.Bischof O., Kim S.H., Irving J., Beresten S., Ellis N.A., Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xanthoudakis S., Smeyne R.J., Wallace J.D., Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabelof D.C., Guo Z., Raffoul J.J., Sobol R.W., Wilson S.H., Richardson A., Heydari A.R. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res. 2003;63:5799–5807. [PubMed] [Google Scholar]
- 32.Larsen E., Gran C., Saether B.E., Seeberg E., Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tebbs R.S., Flannery M.L., Meneses J.J., Hartmann A., Tucker J.D., Thompson L.H., Cleaver J.E., Pedersen R.A. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 34.Bentley D., Selfridge J., Millar J.K., Samuel K., Hole N., Ansell J.D., Melton D.W. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 35.Puebla-Osorio N., Lacey D.B., Alt F.W., Zhu C. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol. Cell. Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krokan H.E., Standal R., Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demple B., Sung J.S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Wilson S.H. Mammalian base excision repair and DNA polymerase beta. Mutat. Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 39.Tomkinson A.E., Chen L., Dong Z., Leppard J.B., Levin D.S., Mackey Z.B., Motycka T.A. Completion of base excision repair by mammalian DNA ligases. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- 40.Fortini P., Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Fan J., Wilson D.M., III Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic. Biol. Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Shen B., Singh P., Liu R., Qiu J., Zheng L., Finger L.D., Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 43.Caldecott K.W. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Wilson D.M., III Processing of nonconventional DNA strand break ends. Environ. Mol. Mutagen. 2007;48:772–782. doi: 10.1002/em.20346. [DOI] [PubMed] [Google Scholar]
- 45.Von Kobbe C., May A., Grandori C., Bohr V.A. Werner syndrome cells escape hydrogen peroxide-induced cell proliferation arrest. FASEB J. 2004;18:1970–1972. doi: 10.1096/fj.04-1895fje. [DOI] [PubMed] [Google Scholar]
- 46.Kitao S., Ohsugi I., Ichikawa K., Goto M., Furuichi Y., Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 47.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 48.Ahn B., Harrigan J.A., Indig F.E., Wilson D.M., III, Bohr V.A. Regulation of WRN helicase activity in human base excision repair. J. Biol. Chem. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 49.Kyng K.J., May A., Kolvraa S., Bohr V.A. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc. Natl Acad. Sci. USA. 2003;100:12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S.Y., Volsky D.J. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das A., Boldogh I., Lee J.W., Harrigan J.A., Hegde M.L., Piotrowski J., de Souza Pinto N., Ramos W., Greenberg M.M., Hazra T.K., et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 52.Oliver C.N., Ahn B.W., Moerman E.J., Goldstein S., Stadtman E.R. Age-related changes in oxidized proteins. J. Biol. Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 53.Pagano G., Zatterale A., Degan P., d'Ischia M., Kelly F.J., Pallardo F.V., Calzone R., Castello G., Dunster C., Giudice A., et al. In vivo prooxidant state in Werner syndrome (WS): results from three WS patients and two WS heterozygotes. Free Radic. Res. 2005;39:529–533. doi: 10.1080/10715760500092683. [DOI] [PubMed] [Google Scholar]
- 54.Bohr V.A. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D., Luo M., Kelley M.R. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 56.Evans A.R., Limp-Foster M., Kelley M.R. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 57.Herring C.J., West C.M., Wilks D.P., Davidson S.E., Hunter R.D., Berry P., Forster G., MacKinnon J., Rafferty J.A., Elder R.H., et al. Levels of the DNA repair enzyme human apurinic/apyrimidinic endonuclease (APE1, APEX, Ref-1) are associated with the intrinsic radiosensitivity of cervical cancers. Br. J. Cancer. 1998;78:1128–1133. doi: 10.1038/bjc.1998.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koukourakis M.I., Giatromanolaki A., Kakolyris S., Sivridis E., Georgoulias V., Funtzilas G., Hickson I.D., Gatter K.C., Harris A.L. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int. J. Radiat. Oncol. Biol. Phys. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 59.Fung H., Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 60.Fishel M.L., He Y., Reed A.M., Chin-Sinex H., Hutchins G.D., Mendonca M.S., Kelley M.R. Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair (Amst) 2008;7:177–186. doi: 10.1016/j.dnarep.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madhusudan S., Smart F., Shrimpton P., Parsons J.L., Gardiner L., Houlbrook S., Talbot D.C., Hammonds T., Freemont P.A., Sternberg M.J., et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrigan J.A., Opresko P.L., von Kobbe C., Kedar P.S., Prasad R., Wilson S.H., Bohr V.A. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J. Biol. Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 63.Brosh R.M., Jr, von Kobbe C., Sommers J.A., Karmakar P., Opresko P.L., Piotrowski J., Dianova I., Dianov G.L., Bohr V.A. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma S., Sommers J.A., Gary R.K., Friedrich-Heineken E., Hubscher U., Brosh R.M., Jr The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA. Nucleic Acids Res. 2005;33:6769–6781. doi: 10.1093/nar/gki1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y., Kao H.I., Bambara R.A. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 66.Saharia A., Guittat L., Crocker S., Lim A., Steffen M., Kulkarni S., Stewart S.A. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chuang Y.Y., Chen Y., Gadisetti Chandramouli V.R., Cook J.A., Coffin D., Tsai M.H., DeGraff W., Yan H., Zhao S., et al. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62:6246–6254. [PubMed] [Google Scholar]
- 68.Boiteux S., Gajewski E., Laval J., Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 69.Reddy P., Jaruga P., O'Connor T., Rodriguez H., Dizdaroglu M. Overexpression and rapid purification of Escherichia coli formamidopyrimidine-DNA glycosylase. Protein Expr. Purif. 2004;34:126–133. doi: 10.1016/j.pep.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 70.Dizdaroglu M. Quantitative determination of oxidative base damage in DNA by stable isotope-dilution mass spectrometry. FEBS Lett. 1993;315:1–6. doi: 10.1016/0014-5793(93)81120-o. [DOI] [PubMed] [Google Scholar]
- 71.Erzberger J.P., Barsky D., Scharer O.D., Colvin M.E., Wilson D.M., III Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.