Abstract
Northwestern Europeans are among the tallest of human populations. The increase in body height in these people appears to have reached a plateau, suggesting the ubiquitous presence of an optimal environment in which genetic factors may have exerted a particularly strong influence on human growth. Therefore, we performed a genome-wide association study (GWAS) of body height using 2.2 million markers in 10 074 individuals from three Dutch and one German population-based cohorts. Upon genotyping, the 12 most significantly height-associated single nucleotide polymorphisms (SNPs) from this GWAS in 6912 additional individuals of Dutch and Swedish origin, a genetic variant (rs6717918) on chromosome 2q37.1 was found to be associated with height at a genome-wide significance level (Pcombined = 3.4 × 10−9). Notably, a second SNP (rs6718438) located ∼450 bp away and in strong LD (r2 = 0.77) with rs6717918 was previously found to be suggestive of a height association in 29 820 individuals of mainly northwestern European ancestry, and the over-expression of a nearby natriuretic peptide precursor type C (NPPC) gene, has been associated with overgrowth and skeletal anomalies. We also found a SNP (rs10472828) located on 5p14 near the natriuretic peptide receptor 3 (NPR3) gene, encoding a receptor of the NPPC ligand, to be associated with body height (Pcombined = 2.1 × 10−7). Taken together, these results suggest that variation in the C-type natriuretic peptide signaling pathway, involving the NPPC and NPR3 genes, plays an important role in determining human body height.
INTRODUCTION
Human body height has a heritability of at least 80% (1) but, in terms of its genetic complexity, it may still serve as a model for the architecture of human complex traits in general. Thus, recent genome-wide association studies (GWAS) have revealed that tens to hundreds of loci with small individual effects are likely to underlie the observed population variation in body height (1–9). What is more, taken together, the 44 height loci identified in the five first GWAS (2,3,5,8,9) were found to explain only 5% of this variation, with the most strongly associated single variant accounting for not more than 0.3% (1). The Genetic Investigation of Anthropometric Traits consortium is currently assembling a collection of at least 100 000 individuals from different GWAS worldwide, trying to achieve the power necessary to identify the hundreds or even thousands of genetic variants expected to influence human height and to jointly explain ∼15–20% of its population variation (1). However, it must be kept in mind that the genetic basis of body height may show regional differences owing to, for example, genetic heterogeneity or variable patterns of gene–environment interaction (7,10). Genetic associations with small effects therefore may have been obscured in instances where individuals of different origin were pooled in meta-analyses or used for the confirmation of findings made in other populations (2,3,5,6,11). Finally, different environments may result in different levels of heritability for complex traits, including height (12). We therefore decided to perform a GWAS of human stature that focused upon northwestern European individuals. In these people, adult height has potentially stabilized at a biologically determined maximum, suggesting environmental conditions that are more homogenous than in populations with an ongoing secular trend in height, such as the southern Europeans (13,14). Consequently, our study employed Dutch and German individuals in the stage 1 meta-analysis and Dutch and Swedish samples in the stage 2 meta-analysis. In addition to corroborating height associations from previous multi-regional studies, our data also revealed a region of height association that achieved genome-wide statistical significance for the first time.
RESULTS
Meta-analysis of height GWAS in the stage 1 data set (n = 10 074)
Genome-wide single nucleotide polymorphism (SNP) data from three Dutch and one northern German cohort, comprising a total of 10 074 individuals, were used to search for genetic variants associated with human height in stage 1 of the analysis (Table 1). These samples included individuals from the initial Rotterdam study cohort (RS-I; n = 5746), from an extension of the Rotterdam study (RS-II; n = 1891), the Erasmus Rucphen family study (ERF; n = 1473) and from the Kiel PopGen biobank study (PopGen-KIEL; n = 964). In each study, we performed an association analysis of up to 2 543 888 SNPs, including genotypes that were imputed with reference to the International HapMap Project CEU panel release 22. Genotype imputation is an approach to overcome the missing data problem in the analysis of data from different genotyping platforms. A recent pan-European study (15), including a subset of the samples used here, revealed that the HapMap CEU samples are genetically closest to western and northern Europeans. Therefore, the CEU data can be assumed to represent an appropriate basis for genotype imputation for the present study. Following quality control (QC), summary statistics from each study for the 2 228 850 remaining SNPs were subjected to the stage 1 meta-analysis. In agreement with previous recommendations (16), results were deemed significant at a genome-wide level if the locus-specific unadjusted P-value was smaller than 5 × 10−8 (17).
Table 1.
Characteristics of the study samples used for stage 1 and stage 2 meta-analyses
| Study | Population origin | Gender | Average age, years (SD) | Average height, cm (SD) |
|---|---|---|---|---|
| RS-I | Dutch | Male (n = 2372) | 68.13 (8.16) | 174.85 (6.76) |
| Female (n = 3374) | 70.32 (9.60) | 161.35 (6.57) | ||
| RS-II | Dutch | Male (n = 862) | 64.68 (7.82) | 175.49 (6.59) |
| Female (n = 1029) | 65.65 (8.86) | 162.36 (6.27) | ||
| PopGen-KIEL | German | Male (n = 506) | 51.27 (14.24) | 180.31 (7.45) |
| Female (n = 458) | 50.79 (14.92) | 167.16 (6.73) | ||
| ERF | Dutch | Male (n = 557) | 51.12 (15.73) | 174.00 (7.73) |
| Female (n = 916) | 49.99 (15.82) | 161.50 (7.01) | ||
| Stage 1 data set | n = 10 074 | |||
| LASA | Dutch | Male (n = 371) | 72.25 (6.49) | 173.35 (6.64) |
| Female (n = 381) | 72.56 (6.51) | 160.44 (6.27) | ||
| EPOS | Dutch | Female (n = 1698) | 50.01 (2.14) | 164.67 (6.10) |
| NTR/NESDA | Dutch | Male (n = 1211) | 46.09 (13.42) | 181.59 (7.17) |
| Female (n = 2311) | 42.65 (13.25) | 169.06 (6.37) | ||
| GOOD | Swedish | Male (n = 940) | 18.90 (0.56) | 181.38 (6.74) |
| Stage 2 data set | n = 6912 | |||
| Combined stage 1 and stage 2 data set | n = 16 986 | |||
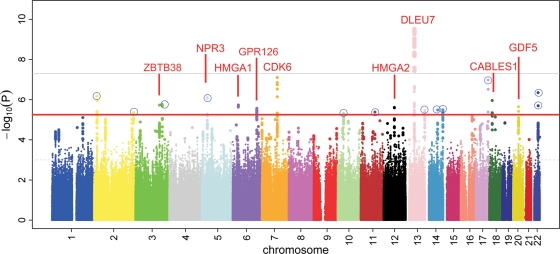
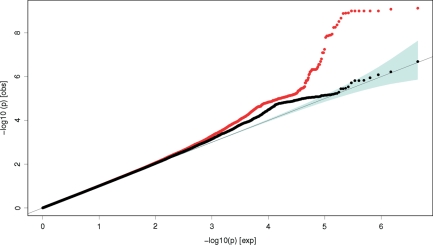
In the stage 1 data set, the strongest association with height was observed for rs3118905 (P = 3.1 × 10−10), thereby confirming a previously reported association of a locus near the DLEU7 gene (5). In addition, the previously identified associations with height in or around HMGA1, ZBTB38, CDK6, HGMA2, CABLES1, NPR3, GPR126 and GDF5 (2,3,5,18) were also replicated with P < 5 × 10−6 (Fig. 1). In total, 29 of 48 independent height loci totally known so far (2,3,5,6,8,9,18), signified by 38 of 57 SNPs in our study, attained nominal statistical significance (P < 0.05) in the stage 1 data set (Supplementary Material, Table S1). Inspection of the Quantile–quantile (Q–Q) plot (Fig. 2) of all SNPs included in the meta-analysis indicated an excess of significantly associated markers. Upon exclusion of all markers in LD with the most significant SNPs from the 48 known height loci (2,3,5,6,18), a deviation from the expected plot under the null hypothesis was still prevalent, suggesting the presence of additional, significantly associated loci in our data. After excluding all loci previously reported to be associated with human height, we ascertained 12 genetic variants with P < 5 × 10−6 (Fig. 2 and Supplementary Material, Table S2) for further analysis.
Figure 1.
Manhattan plot of the height association test results (log10(P)) for all SNPs in the stage 1 data set (n = 10 074). Red and grey lines are the suggestive (5 × 10−6) and genome-wide significance (5 × 10−8) P-value thresholds, respectively. Signals passing the suggestive threshold, and with the respective gene name given in red, are previously known regions of height association. Twelve loci for which SNPs were selected for the stage 2 analyses are emphasized by a blue circle.
Figure 2.
Quantile–quantile (Q–Q) plot of the height association test results (log10(P)) for all SNPs passing quality control (red line) in the stage 1 analysis including 10 074 subjects, excluding variants in 48 independent loci previously associated with height on a genome-wide level (black line). Depicted P-values were corrected for population stratification using the over inflation factor λGC = 1.049.
Combined meta-analysis of the 12 novel height-associated SNPs from stage 1 and stage 2 (n = 16 986)
The 12 putative height-associated SNPs newly identified in the stage 1 data set were next scrutinized in 4462 additional individuals of northern European ancestry (Table 1), namely 3522 Dutch from the Netherlands Twin Register study and the Netherlands Study of Depression and Anxiety (NTR/NESDA) and 940 Swedes from the Gothenburg Osteoporosis and Obesity Determinants (GOOD) study. Furthermore, de novo genotyping of the 12 SNPs was carried out in another 2450 individuals from two additional Dutch population-based studies: 752 participants of the Longitudinal Aging Study Amsterdam study (LASA) and 1698 participants of the European Prospective Osteoporosis Study (EPOS), thereby bringing the total number of Dutch, German and Swedish individuals in the combined stage 1 and stage 2 data set to 16 986.
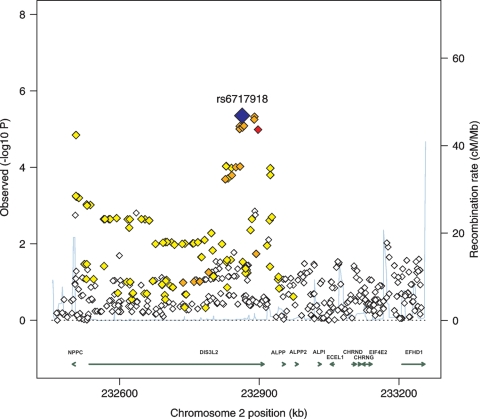
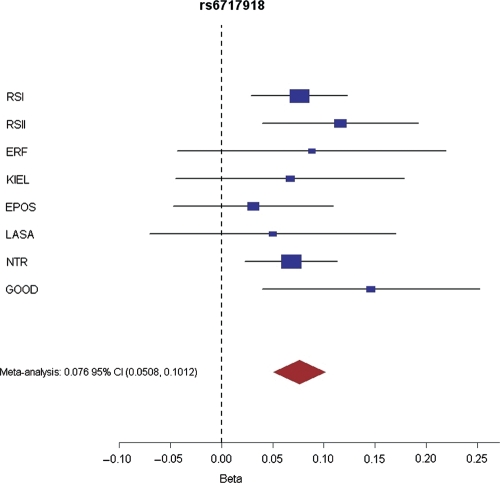
The combined analysis of all samples identified SNP rs6717918 on chromosome 2q37.1 as being associated with body height at a genome-wide significant level (P = 3.4 × 10−9). In the stage 1 data set, several SNPs in this region showed suggestive association with height and were only in relatively weak LD (0.2 < r2 ≤ 0.5) with rs6717918 (Fig. 3). The direction of the association between rs6717918 and height was consistent across all sub-samples, without significant evidence for any inter-study heterogeneity (Q-statistic P = 0.74) (Fig. 4). The T-allele of rs6717918 was associated with an increase in height by 0.44 cm per allele copy (Table 2). At the genome-wide significance level, two more loci, namely rs139909 mapping to the trinucleotide repeat containing 6B (TNRC6B) gene on chromosome 22 (Supplementary Material, Fig. S1) and rs10472828 near the natriuretic peptide receptor 3 (NPR3) gene on chromosome 5 (Supplementary Material, Fig. S2), showed suggestive evidence for an association with height in the combined data set (P < 5 × 10−7; Table 2).
Figure 3.
Local plot of the height association test results (log10(P)) around SNP rs6717918 (blue diamond). P-values are as obtained from the meta-analysis of the stage 1 data set. The combined P-value from the meta-analysis of stage 1 and stage 2 data sets equals 3.4 × 10−9 for rs6717918. The coloring of SNPs indicates the strength of LD with rs6717918, coded as red (strong, r2 > 0.8), orange (moderate, 0.5 < r2 ≤ 0.8), yellow (weak, 0.2 < r2 ≤ 0.5) or white (limited or none, r2 ≤ 0.2). The blue line depicts local recombination rates.
Figure 4.
Forrest plot for the most significant SNP (rs6717918) in the combined meta-analysis of both the stage 1 and the stage 2 data sets. Beta: increase in height per SNP allele. Blue squares represent effect size estimates (measured in standard deviations of height) and 95% CI for each study. The red diamond represents the summary effect size estimate.
Table 2.
Most significant height associations in the stage one, stage 2 and combined stage 1 and stage 2 meta-analyses
| SNP | Chr. | Position (bp)a | Genes | Effect. allele | Freq. effect. allele | Per-allele change in height in cm (SE)b | Stage 1 data P-value (n = 10 074) | Stage 2 data P-value (n = 6912) | Combined data P-value (n = 16 986) |
|---|---|---|---|---|---|---|---|---|---|
| rs6717918 | 2 | 232863344 | DISC3L2 ALPP NPPC | T | 0.78 | 0.44 (0.12) | 4.46 × 10−6 | 1.4 × 10−4 | 3.4 × 10−9 |
| rs139909 | 22 | 39027527 | TNRC6B | T | 0.68 | 0.25 (0.11) | 4.53 × 10−7 | 2.2 × 10−2 | 1.7 × 10−7 |
| rs10472828 | 5 | 32924575 | NPR3 | C | 0.56 | 0.22 (0.09) | 8.13 × 10−7 | 2.0 × 10−2 | 3.4 × 10−7 |
Only associations with an overall P < 5 × 10−7 are shown.
aPosition relative to Build 36.2.
bFrom the stage 2 data of the study only.
Next, we aimed at replicating the putative association of one or the other of the 12 SNPs in recently published GWAS results (2,3,5,6,8,9,18). We found additional evidence for an association with height only for the 2q37.1 region, using the Illumina data provided by Gudbjartsson et al. (2) for 25 174 Icelanders, 2876 Dutch and 1770 European Americans. However, all nine SNPs typed in the 2q37.1 region in the original study failed to attain genome-wide significance there. The strongest evidence for an association was obtained for rs749052 (P = 1.4 × 10−6). In a regional meta-analysis of their and our data SNP rs6718438, a proxy for rs6717918 in strong LD (r2 = 0.77) and only 456 bp away from it, was found to be the most significantly height-associated SNP (P = 8.4 × 10−12). Furthermore, all nine SNPs in the 2q27.1 region attained genome-wide significance (P < 4 × 10−8; Supplementary Material, Table S3 and Fig. S3). To determine whether the association with rs749052 observed by Gudbjartsson et al. (2) was independent of rs6718438 or whether it was due to LD between the two SNPs (r2 = 0.17), we conducted conditional association analyses of rs749052 controlling for the effect of rs6718438. A meta-analysis of the respective results for RS-I, RS-II and GOOD revealed that the height association of rs749052 remained nominally significant (P = 0.01). Taken together, the available data therefore provide conclusive evidence for an association between human body height and genetic variation at 2q37.1.
DISCUSSION
Northwestern Europeans are among the tallest of human populations (13). The average Dutch male, for example, is currently almost 20 cm taller than 150 years ago (13). Over the last 50 years, however, average human body height has increased much less in northern than in southern Europe (13,14). This suggests that height has approached a biologically determined maximum in the north, and that this leveling off has occurred against an optimal and comparatively homogeneous environmental background as regards growth-relevant factors (13,14). With an aim to identify new genetic variants determining human body height, we therefore conducted a two-stage meta-analysis of GWAS of 16 986 northwestern Europeans, comprising individuals of Dutch, German and Swedish origin. The first stage of this study not only confirmed the phenotype association of common genetic variants previously described as determinants of human stature in populations of various (mostly European) origins (Fig. 2, Supplementary Material, Table S1), but also pointed to additional putative height loci followed-up in the second-stage analysis using independent samples. Since GWAS are only suited to find phenotype associations with common genetic variants, however, we cannot exclude that rare alleles with large effects may also have contributed to the height variation observed in our samples. Similarly, environmental effects (e.g. diet) and gene–environment interaction effects may have contributed as well, but such considerations fell outside the scope of the current project.
In the combined stage 1 and stage 2 meta-analysis, we found for the first time that a locus at 2q37.1 is associated with human height at the genome-wide significance level. The strongest association was observed with intronic SNP rs6717918 in the DIS3 mitotic control homolog (Saccharomyces cerevisiae)-like 2 (DIS3L2) gene which, until very recently, had been considered a hypothetical protein-coding sequence only (MGC42174). Since there is no LD-based evidence for extensive recombination in the surrounding 500 kb region, however, it would still appear plausible that other genes in the vicinity of DIS3L2 may have contributed to the observed height association (Fig. 3), most notably the natriuretic peptide precursor type C (NPPC) gene ∼350 kb away from rs6717918 and only 6 kb away from another height-associated SNP, rs749052. In fact, we regard variation in the NPPC gene as the most likely cause of the height association observed with 2q37.1, represented by both rs6717918 and rs749052. The NPPC gene encodes the C-type natriuretic peptide (CNP), a molecule that regulates endochondral ossification of the cartilaginous growth plate and, hence, influences longitudinal bone growth (19,20). Recently, a balanced t(2;7) translocation has been reported in a patient with unusually high stature (>97th percentile), Marfanoid habitus and skeletal anomalies (19). The respective breakpoint on chromosome 2 was located halfway between rs6717918 and the NPPC gene and was shown to induce over-expression of CNP and consequent skeletal overgrowth. Furthermore, transgenic mice with CNP over-expression in osteoblasts exhibit a phenotype similar to the skeletal abnormalities of that patient (19). It has also been demonstrated that over-expression of CNP in chondrocytes can counteract dwarfism in a mouse model of achondroplasia (21). Taken together, these findings suggest that the association between human height and variation at 2q37.1, as observed in our study, reflects differential regulation of the NPPC gene expression with an impact on bone growth regulation and consequent body height.
Although not statistically significant at a genome-wide level in our study, the pronounced height association observed with rs10472828 on chromosome 5p14 deserves further attention. This is because rs10472828 is located only100 kb upstream of NPR3, a gene that encodes a receptor of the CNP ligand. Indeed, Soranzo et al. (18) recently found that this SNP is significantly associated with human height (P = 3.0 × 10−7) in a collection of British and Dutch individuals (which included the subset of the RS-I participants). When combining all our data with those provided by Soranzo et al. (18) in their Supplementary Material, Table S2, while excluding samples from the Rotterdam study (remaining n = 14 052), the combined P-value of 3.5 × 10−11 attains genome-wide significance. Furthermore, two SNPs near NPR3 (rs3811958 and rs13154066) showed a suggestive height association (P < 5 × 10−6) in the study by Gudbjartsson et al. (2), but these polymorphisms were not in strong LD with rs10472828. The NPR3 gene encodes one of three CNP receptors (NPR-C), and knock-out of NPR-C was found to result in significant skeletal overgrowth in mice (22). It has also been suggested that NPR-C may act as a clearance receptor modulating the effect of CNP (22,23). Recently, CNP-induced differentiation of osteoblasts was found to switch from NPR-B to NPR-C with aging in rat cells (24), thus implying an important role of NPR-C in the late stages of bone formation. The identification of SNPs near both the NPPC and the NPR3 genes as being strongly associated with human height clearly points to a prominent role of the CNP signaling pathway in the etiology of body height variation, at least in northwestern Europeans.
Meta-analyses of GWAS are not without limitations. False-positive associations due to multiple hypothesis testing or population stratification are inherent possibilities. Here, we minimized the impact of multiple testing by adopting a stringent genome-wide significance level. To alleviate the possible effects of population stratification, we adjusted all relevant test statistics by the inflation factor λGC (17) and by principal components (PC) derived from the multidimensional scaling analysis of identity-by-state distances between individuals (25). Furthermore, all studies included in our meta-analyses were confined to individuals of northwestern European descent and, consequently, the overall inflation factor of the stage 1 data set (λGC = 1.049) was low for a study of this size (26). This notwithstanding, the ERF samples exhibited a relative high inflation factor (λGC = 1.950) most likely due to intricate family relationships. We therefore conducted a sensitivity analysis, excluding the ERF samples, which revealed that the association between rs6717918 and height remained significant at the genome-wide level (P = 7.8 × 10−9). Taken together, multiple testing, population stratification and cryptic relatedness are therefore unlikely to have confounded our association findings.
In conclusion, we have unequivocally identified variation at 2q37.1 as being associated with human body height in northwestern Europeans. The fact that this locus has not been found in previous studies may either be due to chance (i.e. sampling variation, power differences etc.) or may be explicable in terms of a higher level of genetic and environmental heterogeneity in the other samples, compared with ours. Thus, sufficiently powered studies of additional, geographically confined populations are needed to clarify whether the observed height association of the 2q37.1 region we observed represents a region-specific effect or not. We further propose that the observed association is due to variation in the NPPC gene (encoding the CNP ligand), the most plausible functional candidate in the 2q37.1 region. The strong association observed with SNPs in the vicinity of the NPR3 gene on chromosome 5p14 (encoding the CNP receptor) lends additional support to the view that common variants in the CNP signaling pathway play a prominent role in the regulation of normal height variation in humans.
MATERIALS AND METHODS
Subjects
All studies were approved by the institutional ethics review committees of the respective organizations and all participants provided written informed consent. The RS-I is a prospective population-based cohort study of chronic disabling conditions in Dutch individuals aged 55 years or above (http://www.epib.nl/ergo.htm) (27,28). The RS-II is an extension of the RS-I, which started in 1999 and used the same inclusion criteria and design as the original cohort. In short, 3011 individuals (response rate 67%) who had turned 55 years of age or had moved into the study district of Ommoord, Rotterdam, since the start of the original study in 1990 was included in the extension cohort. The ERF study is a family-based study of a genetic isolate in the southwestern Netherlands to identify genetic risk factors for complex disorders (29). The PopGen-KIEL is a centralized platform for the recruitment and follow-up of probands for genetic epidemiological studies in Schleswig-Holstein, the most northern part of Germany. Since its establishment in 2003, PopGen-KIEL has assembled a collection of biomaterials, phenotypic and genotypic data from more than 60 000 individuals. This includes nearly 1000 controls for which height and genome-wide SNP genotype data were available for inclusion into the present study. The EPOS (30) is a cross-sectional study of 5896 women born between 1941 and 1947, and currently living in the city of Eindhoven, The Netherlands. DNA was available for 1798 of these women. The LASA study (31) is a population-based cohort study, including 919 individuals for whom DNA was available. NTR/NESDA: the two parent projects that supplied data are the Netherlands Study of Depression and Anxiety (NESDA) (32) and the Netherlands Twin Registry (NTR) (33). NESDA and NTR studies were approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Center, Amsterdam (IRB number IRB-2991 under Federal wide Assurance-3703; IRB/institute codes, NESDA 03-183; NTR 03-180). The sample consisted of 1777 NTR and 1763 NESDA participants. For NTR participants, longitudinal (1991–2004) survey and data on height were combined. Only one subject per family was selected. For NESDA participants, height was assessed during a visit to the clinic. The GOOD study was initiated to determine both environmental and genetic factors involved in the regulation of bone and fat mass. Male study subjects were randomly identified in the greater Gothenburg area in Sweden using national population registers, contacted by telephone and invited to participate (34). To be enrolled in the GOOD study, subjects had to be between 18 and 20 years of age. There were no other exclusion criteria, and 49% of the study candidates agreed to participate.
Genotyping and QC
The six GWAS were carried out using either the Illumina Infinium HumanHap550 Beadchip (RS-I and RS-II), the Illumina Infinium HumanHap610 (GOOD), the Illumina Infinium HumanHap300 (ERF), the Perlegen 600K (NTR/NESDA) or the Affymetrix Dual NspI/StyI GeneChip 2 × 250K (PopGen-KIEL). De novo genotyping of 12 SNPs in the LASA and EPOS samples was performed using Taqman allelic discrimination (Applied Biosystems Inc., Foster City, CA, USA) according to the manufacturer's protocols and QC standards (assay numbers and primer designs can be found in Supplementary Material). The following sample QC criteria were applied in the GWAS of RS-I, RS-II, PopGen-KIEL, ERF and GOOD: sample call rate ≥97.5%, gender mismatch with typed X-linked markers, evidence for DNA contamination in the samples using the mean of the autosomal heterozygosity >0.33, exclusion of duplicates or first-degree relatives identified using IBS probabilities, exclusion of outliers (three SD away from the population mean) using multi-dimensional scaling (MDS) analysis with four PC and exclusion of samples with missing height measurements. Complete information on genotyping protocols and QC measures for NTR/NESDA cohorts have been described elsewhere (35). The exclusion/filtering criteria for SNPs are described in Supplementary Material, Table S4.
Genotype imputation
Genotype imputation was used to evaluate the height association of one and the same SNP across samples typed on different genotyping platforms. Genotypes were imputed for all polymorphic SNPs (minor allele frequency >0.01) using either the MACH (36) or the IMPUTE (37) software, based upon phased autosomal chromosomes of the HapMap CEU Phase II panel (release 22, build 36), orientated on the positive strand. Imputation QC metrics from MACH and IMPUTE were used for filtering out SNPs with low-quality data. Detailed descriptions of the QC and imputation procedures are provided in Supplementary Material, Table S4.
GWAS
In each GWAS, the association between a SNP and height was assessed using sex-specific, age-standardized residuals that were analyzed under an additive (per allele) genetic model. To adjust for population substructure, we included as covariates in the regression analysis of RS-I, RS-II and PopGen-KIEL the four most important PC, derived from an MDS analysis of IBS distances using the PLINK (38) software. In the analysis of imputed genotypes, uncertainty in genotype prediction was accounted for by utilizing either the dosage information from MACH (36) or the genotype probabilities from IMPUTE (37). We carried out association testing for imputed SNPs using a linear regression framework as implemented in MACH2QTL (36), SNPTEST (37) and ProbABEL (39) (Supplementary Material, Table S4). A linear regression analysis (1 df) as implemented in PLINK (38) was performed for the genotype data in the EPOS and LASA samples, where genotyping had been carried out on Taqman assays.
Meta-analysis
The genomic control method (17), as implemented in METAL, was used to correct for any residual population stratification or relatedness not accounted for by the four most important PC. The estimated inflation factors were 1.089, 1.006, 1.000, 1.950, 1.086 and 1.030 for RS-I, RS-II, PopGen-KIEL, ERF, NTR-NESDA and GOOD, respectively. SNPs with a minor allele frequency<0.05, a MACH observed/expected allele dosage variance <0.05 or a SNPTEST proper_info <0.4 were excluded from the meta-analysis. A detailed description of each study is provided in Supplementary Material, Table S4. We obtained the combined results of 2 228 850 SNPs, pooling effect sizes by means of a fixed effects inverse variance meta-analysis as implemented in METAL. Estimated heterogeneity variance and forest plots were generated using the Rmeta R package. Regional association plots of the meta-analysis results were obtained with SNAP (40).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
The generation and management of GWAS genotype data for the Rotterdam Study (RS-I, RS-II) was supported by a Netherlands Organization for Scientific Research (NWO) Investments grant (nr. 175.010.2005.011, 911-03-012). This study was also supported by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/NWO project no. 050-060-810, the Netherlands Forensic Institute, and by a grant from the NGI/NWO within the framework of the Forensic Genomics Consortium Netherlands. The Rotterdam study is funded by the Erasmus University Medical Center, the Erasmus University Rotterdam, the Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports of the Netherlands, the European Commission (DG XII) and the Municipality of Rotterdam. The Erasmus Rucphen Family (ERF) study was supported by grants from the NWO, Erasmus MC and the Centre for Medical Systems Biology (CMSB). The PopGen-KIEL is directly funded by the Ministry of Science, Economy and Transport of Schleswig-Holstein, Germany. The GOOD study is funded by: The Swedish Research Council, The ALF grant at the Sahlgrenska University Hospital, Gothenburg, Sweden, the Lundberg Foundation, the Torsten and Ragnar Söderberg's Foundation and the Novo Nordisk Foundation. For NTR/NESDA, we acknowledge financial support from the NWO: twin-family database for behavior genetics and genomic studies (480-04-004), genetic basis of anxiety and depression (904-61-090); resolving cause and effect in the association between exercise and well-being (904-61-193); Center for Medical Systems Biology (NWO Genomics); Spinozapremie (SPI 56-464-14192); Centre for Neurogenomics and Cognitive Research (CNCR-VU); genome-wide analyses of European twin and population cohorts (EU/QLRT-2001-01254); Geestkracht program of ZonMW (10-000-1002); matching funds from universities and mental health care institutes involved in NESDA (GGZ Buitenamstel-Geestgronden, Rivierduinen, University Medical Center Groningen, GGZ Lentis, GGZ Friesland, GGZ Drenthe). Genotyping was funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and the analysis was supported by grants from GAIN and the NIMH (MH081802). Funding to pay the Open Access charge was provided by the Netherlands Genomics Initiative/Forensic Consortium Netherlands.
ACKNOWLEDGEMENTS
We thank Pascal Arp, Mila P. Jhamai, Dr Michael J. Moorhouse, Marijn Verkerk and Sander Bervoets for their help in creating the RS-I and RS-II GWAS database. We are grateful to all general practitioners for their support, to Petra Veraart for her help in verifying genealogy information of ERF, Jeannette Vergeer for the supervision of the ERF laboratory work and Peter Snijders for his help with the ERF data collection. The authors are grateful to all study participants, staff and the participating general practitioners and pharmacists.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Weedon M.N., Frayling T.M. Reaching new heights: insights into the genetics of human stature. Trends Genet. 2008;24:595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 3.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher P.M. Sizing up human height variation. Nat. Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 5.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson A., Marroni F., Hayward C., Franklin C.S., Kirichenko A.V., Jonasson I., Hicks A.A., Vitart V., Isaacs A., Axenovich T., et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet. 2009;18:373–380. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei S.F., Yang T.L., Tan L.J., Chen X.D., Guo Y., Guo Y.F., Zhang L., Liu X.G., Yan H., Pan F., et al. Genome-wide association scan for stature in Chinese: evidence for ethnic specific loci. Hum. Genet. 2009;125:1–9. doi: 10.1007/s00439-008-0590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanna S., Jackson A.U., Nagaraja R., Willer C.J., Chen W.M., Bonnycastle L.L., Shen H., Timpson N., Lettre G., Usala G., et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weedon M.N., Lettre G., Freathy R.M., Lindgren C.M., Voight B.F., Perry J.R., Elliott K.S., Hackett R., Guiducci C., Shields B., et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sammalisto S., Hiekkalinna T., Schwander K., Kardia S., Weder A.B., Rodriguez B.L., Doria A., Kelly J.A., Bruner G.R., Harley J.B., et al. Genome-wide linkage screen for stature and body mass index in 3.032 families: evidence for sex- and population-specific genetic effects. Eur. J. Hum. Genet. 2009;17:258–266. doi: 10.1038/ejhg.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei S.F., Tan L.J., Liu X.G., Wang L., Yan H., Guo Y.F., Liu Y.Z., Xiong D.H., Li J., Yang T.L., et al. Genome-wide association study identifies two novel loci containing FLNB and SBF2 genes underlying stature variation. Hum. Mol. Genet. 2008;18:1661–1669. doi: 10.1093/hmg/ddn405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silventoinen K., Kaprio J., Lahelma E., Koskenvuo M. Relative effect of genetic and environmental factors on body height: differences across birth cohorts among Finnish men and women. Am. J. Public Health. 2000;90:627–630. doi: 10.2105/ajph.90.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole T.J. The secular trend in human physical growth: a biological view. Econ. Hum. Biol. 2003;1:161–168. doi: 10.1016/S1570-677X(02)00033-3. [DOI] [PubMed] [Google Scholar]
- 14.Garcia J., Quintana-Domeque C. The evolution of adult height in Europe: a brief note. Econ. Hum. Biol. 2007;5:340–349. doi: 10.1016/j.ehb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Lao O., Lu T.T., Nothnagel M., Junge O., Freitag-Wolf S., Caliebe A., Balascakova M., Bertranpetit J., Bindoff L.A., Comas D., et al. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 16.Pe'er I., Yelensky R., Altshuler D., Daly M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 17.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J.B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bocciardi R., Giorda R., Buttgereit J., Gimelli S., Divizia M.T., Beri S., Garofalo S., Tavella S., Lerone M., Zuffardi O., et al. Overexpression of the C-type natriuretic peptide (CNP) is associated with overgrowth and bone anomalies in an individual with balanced t(2;7) translocation. Hum. Mutat. 2007;28:724–731. doi: 10.1002/humu.20511. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert S. Paraxial and Intermediate Mesoderm. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 21.Yasoda A., Komatsu Y., Chusho H., Miyazawa T., Ozasa A., Miura M., Kurihara T., Rogi T., Tanaka S., Suda M., et al. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat. Med. 2004;10:80–86. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- 22.Matsukawa N., Grzesik W.J., Takahashi N., Pandey K.N., Pang S., Yamauchi M., Smithies O. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc. Natl Acad. Sci. USA. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pejchalova K., Krejci P., Wilcox W.R. C-natriuretic peptide: an important regulator of cartilage. Mol. Genet. Metab. 2007;92:210–215. doi: 10.1016/j.ymgme.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Kaneki H., Kurokawa M., Ide H. The receptor attributable to C-type natriuretic peptide-induced differentiation of osteoblasts is switched from type B- to type C-natriuretic peptide receptor with aging. J. Cell. Biochem. 2008;103:753–764. doi: 10.1002/jcb.21448. [DOI] [PubMed] [Google Scholar]
- 25.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 26.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofman A., Breteler M.M., van Duijn C.M., Krestin G.P., Pols H.A., Stricker B.H., Tiemeier H., Uitterlinden A.G., Vingerling J.R., Witteman J.C. The Rotterdam study: objectives and design update. Eur. J. Epidemiol. 2007;22:819–829. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofman A., Grobbee D.E., de Jong P.T., van den Ouweland F.A. Determinants of disease and disability in the elderly: the Rotterdam elderly study. Eur. J. Epidemiol. 1991;7:403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- 29.Aulchenko Y.S., Heutink P., Mackay I., Bertoli-Avella A.M., Pullen J., Vaessen N., Rademaker T.A., Sandkuijl L.A., Cardon L., Oostra B., et al. Linkage disequilibrium in young genetically isolated Dutch population. Eur. J. Hum. Genet. 2004;12:527–534. doi: 10.1038/sj.ejhg.5201188. [DOI] [PubMed] [Google Scholar]
- 30.Smeets-Goevaers C.G., Lesusink G.L., Papapoulos S.E., Maartens L.W., Keyzer J.J., Weerdenburg J.P., Beijers L.M., Zwinderman A.H., Knottnerus J.A., Pols H.A., et al. The prevalence of low bone mineral density in Dutch perimenopausal women: the Eindhoven perimenopausal osteoporosis study. Osteoporos. Int. 1998;8:404–409. doi: 10.1007/s001980050083. [DOI] [PubMed] [Google Scholar]
- 31.Knipscheer C.P., Dykstra P.A., van Tilburg T.G., de Jong-Gierveld J. Living arrangements and social networks of elders. A selection of findings from a NESTOR-Study Leefvormen en sociale netwerken van ouderen. Een selectie van bevindingen uit een NESTOR-Studie. Tijdschr. Gerontol. Geriatr. 1998;29:110–119. [PubMed] [Google Scholar]
- 32.Penninx B.W., Beekman A.T., Smit J.H., Zitman F.G., Nolen W.A., Spinhoven P., Cuijpers P., De Jong P.J., Van Marwijk H.W., Assendelft W.J., et al. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int. J. Methods Psychiatr. Res. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boomsma D.I., de Geus E.J., Vink J.M., Stubbe J.H., Distel M.A., Hottenga J.J., Posthuma D., van Beijsterveldt T.C., Hudziak J.J., Bartels M., et al. Netherlands Twin Register: from twins to twin families. Twin Res. Hum. Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- 34.Lorentzon M., Swanson C., Andersson N., Mellstrom D., Ohlsson C. Free testosterone is a positive, whereas free estradiol is a negative, predictor of cortical bone size in young Swedish men: the GOOD study. J. Bone Miner. Res. 2005;20:1334–1341. doi: 10.1359/JBMR.050404. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan P.F., de Geus E.J., Willemsen G., James M.R., Smit J.H., Zandbelt T., Arolt V., Baune B.T., Blackwood D., Cichon S., et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol. Psychiatry. 2008;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Abecasis G.R. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 37.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics (Oxford, England) 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 40.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics (Oxford, England) 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.