Abstract
Human invariant natural killer T cells (iNKT cells) are a unique population of T cells that express an invariantly rearranged T-cell receptor (TCR) composed of TCRVα24 and TCRVβ11 chains which recognize glycosphingolipid antigens presented by the CD1d molecule. iNKT cells are absent in patients with X-linked lymphoproliferative disease (XLP) due to SH2D1A mutation, and are reported to be decreased in patients with XLP due to BIRC4 mutations. However, mice deficient in the BIRC4 gene product, X-linked Inhibitor of Apoptosis (XIAP), have normal iNKT cell populations. Because of this, we studied iNKT cell populations in 6 patients with XLP due to BIRC4 mutations, with comparison to 103 pediatric and adult normal control samples. We found that iNKT cells constitute 0.008%-1.176% of normal peripheral blood T cells, and that iNKT cell populations were normal or increased in patients with BIRC4 mutations. We conclude that XLP due to BIRC4 mutation is not associated with decreased populations of iNKT cells, and that XIAP is likely not a requirement for iNKT cell development.
Introduction
Human invariant natural killer T cells (iNKT cells) are a unique population of T cells that express an invariantly rearranged T-cell receptor (TCR) consisting of TCRVα24 and TCRVβ11 chains which recognize glycosphingolipid antigens presented by the CD1d molecule. iNKT cells are involved in a variety of immunoregulatory processes [see references 1 and 2 for recent reviews], and are known to be absent in patients with X-linked lymphoproliferative disease (XLP) due to SH2D1A mutation. Because the SH2D1A gene product, SLAM-associated protein (SAP), is a requirement for iNKT cell development in both humans and mice, iNKT cells are also absent in SAP knockout mice.[3,4]
Recently, the phenotype of XLP was found to be caused by mutations in a second gene, BIRC4, which encodes X-linked Inhibitor of Apoptosis (XIAP).[5] This disorder is sometimes referred to as XLP2. In the originally described cohort of patients, iNKT cell populations were reported to be decreased, and XIAP has been postulated to be involved with iNKT cell development or survival.[6] However, XIAP knock out mice have been reported to possess normal iNKT cell populations[5,7], a finding which we have also noted (R.A.M. and M.B.J., unpublished data). Because of this, we considered the possibility that the iNKT cell populations described in the original cohort of patients with XLP2 were decreased only when considered with regard to the small, age-matched control group that was used for comparison. We hypothesized that comparison to a large control group might reveal different results. Therefore, to more accurately evaluate the impact of BIRC4 mutation upon human iNKT cell populations, we studied peripheral blood iNKT cell populations of 6 patients with XLP2 and of 103 pediatric and adult normal controls. We observed that iNKT cell populations are not decreased in patients with XLP due to BIRC4 mutations.
Patients, Materials, and Methods
Patient and Normal Control Samples
Patients were evaluated at Cincinnati Children's Hospital Medical Center, and EDTA-anti-coagulated blood samples were taken after informed consent was obtained according to an Institutional Review Board (IRB)-approved research protocol. Patients 1-6 are 12, 4, 27, 15, 9, and 4 years of age, respectively. Patient 1 was studied while taking tacrolimus because of prior liver transplantation, and patients 2 and 5 were studied while taking prednisone and dexamethasone, respectively, for treatment of recurrent HLH-like disease. Patients 3, 4, and 6 were studied while healthy and free from any immunomodulatory therapy.
In order to establish an age-dependent normal whole blood iNKT cell range, EDTA-anti-coagulated pediatric control samples were obtained from the Diagnostic Immunology Laboratory at Cincinnati Children's Hospital Medical Center (CCHMC) which, through an agreement with the hospital, obtains pediatric control blood specimens from an outpatient clinic which are delivered within 24 hours to the laboratory. The samples were de-identified except for age and gender. Inclusion criteria for the pediatric control samples included normal CBC indices and white blood cell differential. EDTA-anti-coagulated adult control samples were obtained from healthy adult volunteers associated with the Cincinnati Children's Hospital Diagnostic Immunology Laboratory. All control samples were obtained according to IRB-approved policies, and held at room temperature until analysis.
Mutational Analysis of the BIRC4 Gene
Mutational analysis of the BIRC4 gene was performed as described elsewhere (manuscript accepted, Clinical Cytometry). We previously found that Patient 1 possesses an isolated deletion of exon 6. Patients 2 and 3 are maternally related, and have large deletions spanning exons 1-5. Patient 4 possesses a nonsense mutation, 997C>T, resulting in an early stop codon, Q333X. Patient 5 possesses a nonsense mutation in exon 1, 310C>T, resulting in Q104X. Patient 6 possesses a missense mutation in exon 1, 563G>A (G188E).
Flow Cytometric Identification of Invariant Natural Killer T-cell Populations
All patient and control anti-coagulated blood samples were incubated with fluorochrome-conjugated anti-CD3 (BD Biosciences), anti-TCRVα24, and anti-TCRVβ11 (Beckman Coulter) monoclonal antibodies to identify iNKT cells. 7 randomly selected control samples were also studied in parallel using anti-CD3 and PBS-57 loaded CD1d Tetramer (NIH Tetramer Core Facility, Atlanta, Georgia). Patient 4 was also studied with the PBS-57 loaded CD1d Tetramer. Erythrocytes were lysed by incubation in FACSLyse (BD Biosciences). Samples were then washed, fixed, and analyzed on a BD FACSCanto flow cytometer (BD Biosciences) using FACSDiva software (BD Biosciences). Data analysis was performed with either FACSDiva or FCS Express (De Novo Software) software. After gating on the lymphocyte population (identified by forward and side light scatter), invariant Natural Killer T-cell (iNKT cell) populations were defined as CD3+ T cells that either co-expressed TCRVα24 and TCRVβ11, or as CD3+ lymphocytes that recognized PBS-57 loaded CD1d tetramer. 50-1000 events were collected in the iNKT cell gate. Evaluation of background non-specific staining was performed using fluorochrome conjugated isotype control antibodies (BD Biosciences) or unloaded CD1d tetramer (NIH Tetramer Core Facility) as appropriate.
Statistical Analysis
The statistical analysis of control results was performed using Student's t-test after log-transformation of the data to account for the skewed data distribution. For comparison of the XIAP-deficient patient results to the control sample results, the data was again log-transformed, and the means of the transformed data were compared using a mixed effects model analysis. The mixed effects model analysis enables us to incorporate more than one observation per patient by assuming two sources of variances, between subject and within subject variances. Statistical significance was considered for p<0.05.
Results
Normal iNKT cell populations constitute 0.008%-1.176% of peripheral blood T cells
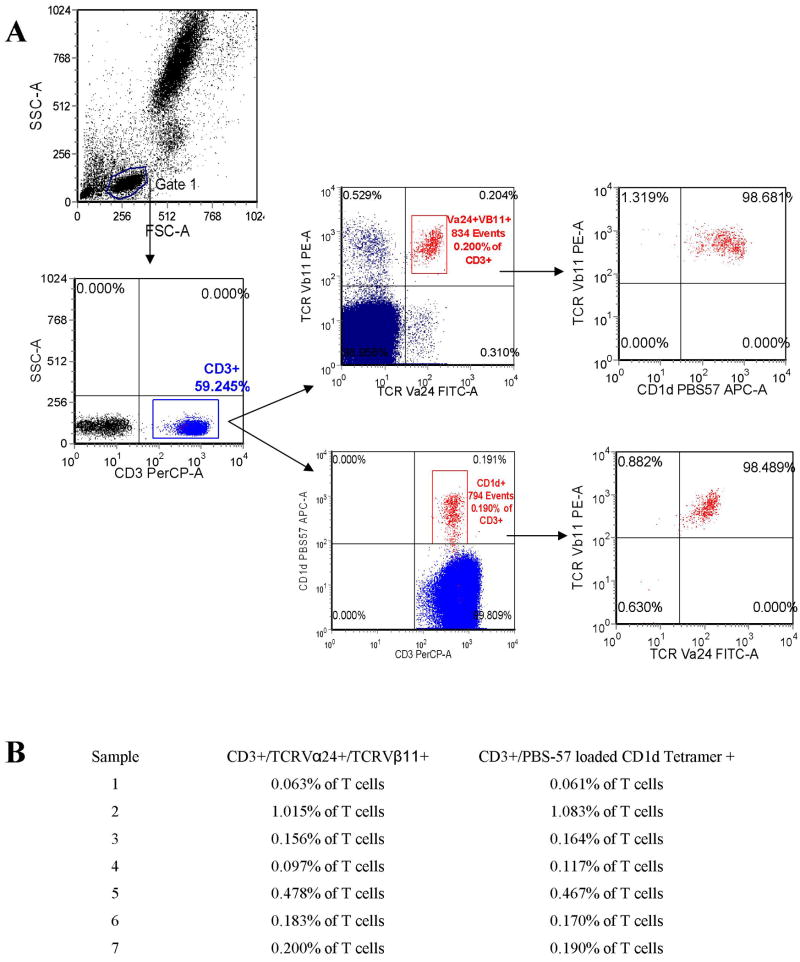
It has been previously shown that iNKT cells can be easily defined by CD3 expression and co-expression of TCRVα24 and TCRVβ11, with good correlation to staining performed using PBS-57 loaded CD1d tetramers.[8] (PBS-57 is a modified α-galactosyl ceramide that stains iNKT cells when loaded into CD1d tetramers comparably to KRN7000.[9]) We thus initially evaluated our ability to identify iNKT cells using this methodology. After gating on the lymphocyte population, we found that iNKT cell populations were easily identified and quantified based on CD3/TCRVα24/TCRVβ11 expression (Figure 1A). Seven randomly selected control samples were also studied using CD3 and PBS-57 loaded CD1d tetramer as a means to verify that our results were accurate. We found good correlation between methods (Figure 1A and 1B). Three of these samples were evaluated with simultaneous staining with CD3/TCRVα24/TCRVβ11/PBS-57 loaded CD1d tetramer. We observed that over 90% of tetramer + cells were also TCRVα24/TCRVβ11 +, and vice-versa (Figure 1A and data not shown).
Figure 1.
A. Sample scatter plots are shown illustrating the identification of whole blood iNKT cell populations. After gating on the lymphocyte population (identified by forward and side light scatter) and CD3+ lymphocytes (T cells), iNKT cells were identified and quantified in all control and patient samples by co-expression of TCRVα24 and TCRVβ11, and in selected samples also by recognition of PBS57-loaded CD1d tetramer. As shown here, simultaneous staining with TCRVα24 and TCRVβ11 and PBS57-loaded CD1d tetramer demonstrates that both methods identify the same population. B. iNKT cell populations were studied in 7 randomly selected control samples using both co-expression of TCRVα24 and TCRVβ11 and by recognition of PBS57-loaded CD1d tetramer to aid in the validation of each method. Results as listed.
Next, in order to create a meaningful normal pediatric iNKT cell reference range, and to verify a previously described adult normal range, we studied iNKT cell populations among 80 pediatric (ages 1-18 years) and 23 adult (age 19 and up) controls using CD3/TCRVα24/TCRVβ11 staining. The age range for the entire group was 1-71 years with a median age of 13 years. 55% of controls were female. We found that peripheral blood iNKT cells constituted between 0.003% and 2.21% of all T cells (defined as CD3+ lymphocytes). Given such a wide range, simple determination of normal range via calculation of mean +/- 2 standard deviations was not possible. Therefore, outliers constituting approximately 2.5% of samples at both the lower and upper ranges were discounted, creating a normal range of 0.008%-1.176% (Figure 2A). This range reflects a normal range previously described in a large adult control group (0.01-0.92% of T cells).[8] The median value was 0.046% of T cells (Figure 2B, shown with outliers removed and results rounded to include only 2 decimal places for ease of illustration).
Figure 2.
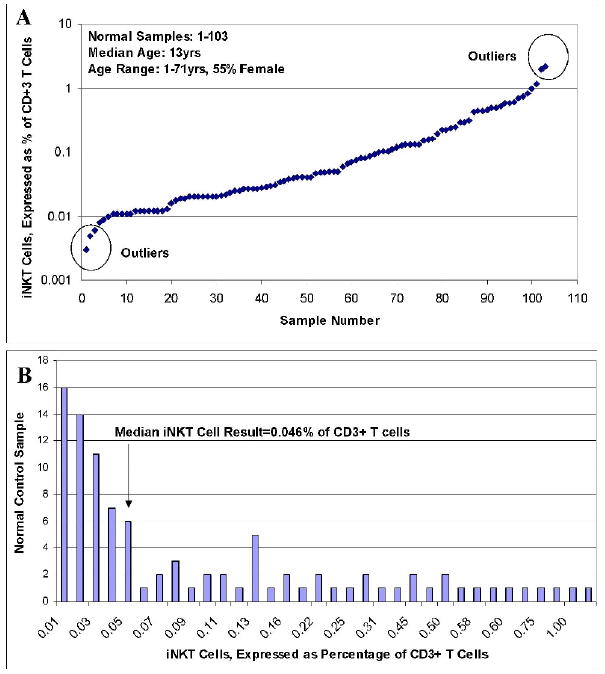
A. iNKT cells populations were identified and quantified among 80 pediatric and 23 adult control samples (based on co-expression of TCRVα24 and TCRVβ11 on CD3+ T cells). Results as shown. B. The data from A, rounded to 2 decimal places and shown by frequency of result after exclusion of outliers.
The control data was next log-transformed for further analysis (Figure 3Aa and 3Ab). Using the mean and standard deviation of the log-transformed data, we calculated a normal iNKT cell range of 0.003%-1.171%, verifying our accepted range above. iNKT cell populations were not found to be significantly linearly associated with age (Figure 3Ab) (p-value = 0.2352 from regression analysis, R2 = 0.0139), and iNKT cell populations were not noted to be significantly different between adults and children (Figure 3B) (p-value = 0.4982). iNKT cell populations were only marginally significantly different between genders (p-value = 0.0923) (Figure 3C).
Figure 3.
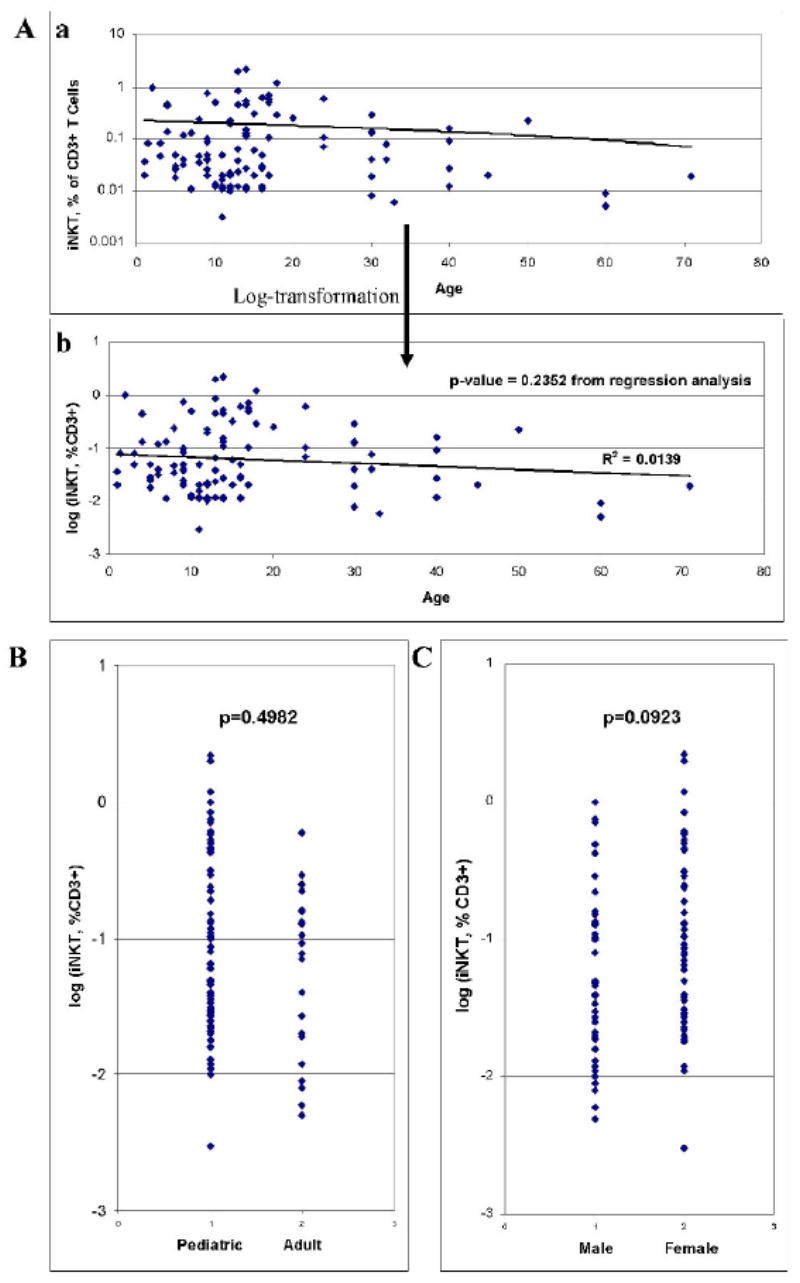
A. Results of iNKT cell analysis plotted in relation to age (a), and also shown after log-transformation (b). B. Results of iNKT cell control group analysis with comparison between pediatric (age 1-18) and adult (age 19 and up) groups. C. Results of iNKT cell control group analysis with comparison between genders.
With regard to absolute numbers of iNKT cells, complete blood count information was available for 88 of the control donors. Within this group, we observed a range of 0.08-35.43 iNKT cells per microliter of blood (80-35,430 iNKT cells per milliliter of blood). No statistically significant differences in absolute numbers of iNKT cell populations were noted based on age or gender (analysis not shown).
iNKT cells are not decreased in patients with XLP due to BIRC4 mutations
Before studying patients with BIRC4 mutations, we first verified our ability to differentiate very small populations of iNKT cells from absence of iNKT cells by studying patients with XLP due to SAP deficiency. As shown in Figure 4Ab, SAP deficient patients have an absence of iNKT cells which is easily distinguished from lower limit iNKT cell populations (Figure 4Aa). Following this, we studied iNKT cell populations in 6 patients from 5 unrelated families with XLP due to BIRC4 mutations. Patient 1 (Pt1) was studied 6 times over the course of 4 months. Patient 2 (Pt2) was studied 7 times over the course of 6 months. Patient 3 (Pt3) was studied twice during 1 month. Patient 4 (Pt4) was studied 3 times over the course of 6 months. Patient 5 (Pt5) was studied once. Patient 6 (Pt6) was studied twice during 1 month. All patients possessed iNKT cells (Figure 4B). When taking into account the percent and absolute number of peripheral blood iNKT cells, patients 1, 2, 3, 5, and 6 possessed essentially normal iNKT cell populations (Figure 5A and 5B). Interestingly, patient 4 repeatedly displayed mildly increased iNKT cell populations. To verify this finding, we studied this patient's iNKT cell population using PBS-57 loaded CD1d tetramer staining. We confirmed an increased iNKT cell population representing 2.956% of total T cells (result not shown), corresponding to an iNKT cell population of 2.853% of total T cells as measured by TCRVα24 and TCRVβ11 costain in parallel. To ensure that there was no statistically significant difference between the control and patient groups, the log-transformed data was compared (Figure 5C). Using a mixed effects model analysis, no significant difference was noted (p-value = 0.8562).
Figure 4.
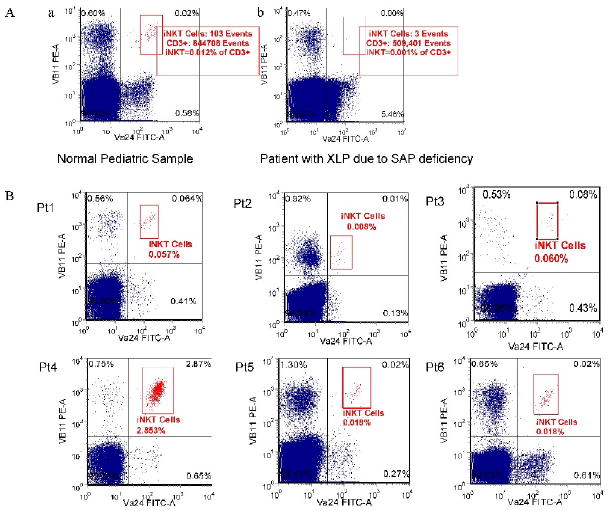
Aa. Sample scatter plot of the iNKT cell population in a control sample possessing low normal levels of iNKT cells (0.012%). Ab. Sample scatter plot of the (absence of) iNKT cell population in a patient with XLP due to SH2D1A mutation (0.001% or absence of iNKT cells). B. Sample scatter plots of iNKT cell populations among 6 patients with XLP due to BIRC4 mutation (designated as Pt1-6).
Figure 5.
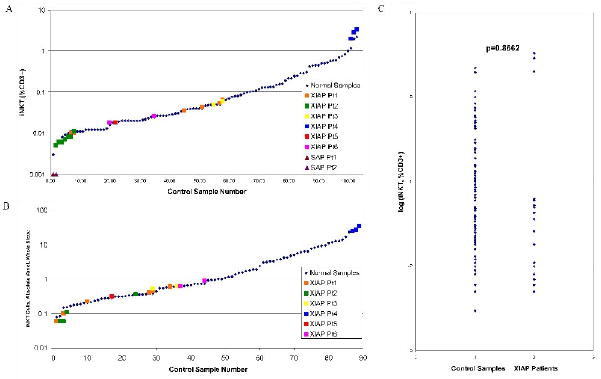
Results of iNKT cell analysis among patients with XLP due to BIRC4 mutation (designated as XIAP Pt1-6) and controls. Pt1 was studied 6 times over the course of 4 months. Pt2 was studied 7 times over the course of 6 months. Pt3 was studied twice during 1 month. Pt4 was studied 3 times over the course of 6 months. Pt5 was studied once. Pt6 was studied twice during 1 month. A. Patient results are shown overlaid onto graphical representation of the control samples, expressed as % of CD3+ T cells. Results from 2 patients with XLP due to SH2D1A mutation are also shown (SAP Pt1-2). B. Patient results are shown overlaid onto graphical representation of the control samples, expressed as number of iNKT cells per microliter of whole blood. C. Comparison of iNKT cell populations of patients (more than 1 result per patient) and control group (1 result per control sample). The log-transformed data are shown. Data were compared using a mixed effects model analysis (p-value = 0.8562).
Discussion
While iNKT cells are known to be absent in patients with XLP due to SH2D1A mutation (XLP1), the data that we present here demonstrates that iNKT cell populations are normal in patients with XLP due to BIRC4 mutation (XLP2). Our work also helps to define the normal range of iNKT cell populations in a large pediatric population, and reveals that the range is very broad and without significant differences across ages or between genders. Based on our work, we conclude that XIAP is not likely to be a requirement for iNKT cell development, and this is further supported by the previous observations that iNKT cells are normal in mice lacking XIAP. Additionally, analysis of iNKT cells does not appear to be useful as a screening tool for XLP2. Our work does not rule out the possibility that there are functional impairments of iNKT cells lacking XIAP, that there is a lack of a specific iNKT cell subpopulation (such as defined by CD4 expression, etc), or that there is a survival defect in the setting of activation or HLH. Further work is needed to investigate these possibilities.
Acknowledgments
The authors wish to thank the patients who have generously contributed samples for study. We thank the NIH Tetramer Core Facility for supplying the PBS-57 loaded CD1d tetramer. We thank Phil Shin, Barb Lawall, Sue Vergamini, and Kristi Smiley. We thank Lisa Madden, MD and Brenda Kitchen, MD for patient samples. This work was supported by grants from the Histiocytosis Association of America and NIH R03 1R03AI079797-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, Kronenberg M. Going both ways: Immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–88. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Medicine. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Ex Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigaud S, Fondanèche MC, Lambert N, Pasquier B, Mateo V, Soulas P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–4. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 6.Latour S. Natural Killer T cells and X-linked lymphoproliferative syndrome. Curr Opin Allergy Clin Immunol. 2007;7:510–14. doi: 10.1097/ACI.0b013e3282f1bad6. [DOI] [PubMed] [Google Scholar]
- 7.Bauler LD, Duckett CS, O'Riordan MX. XIAP regulates cytosol-specific immunity to Listeria infection. PLoS Pathog. 2008;4:e1000142. doi: 10.1371/journal.ppat.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody 6B11. Immunology. 2007;122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Goff RD, Zhou D, Mattner J, Sullivan BA, Khurana A, et al. A modified α-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods. 2006;312:34–9. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]