Abstract
Virion infectivity factor (Vif) is an HIV accessory protein that is essential for the infection of CD4+ T cells. Vif recruits a Cullin-5 (Cul5)-based ubiquitin ligase that targets a host cytidine deaminase, apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G), for proteasomal degradation. The Vif N-terminus binds APOBEC3G and the C-terminus interacts with the Cul5-based ubiquitin ligase machinery. Within the C-terminus, a highly conserved H108-X5-C114-X17–18-C133-X3–5-H139 (HCCH) motif binds zinc and is implicated in the Vif-Cul5 interaction. We have employed the biomimetic peptide HCCHp (HIV-1 Vif amino acids 101–142) in order to determine the zinc ligands and investigate the role of zinc binding in Cul5 recognition. Using CD spectroscopy, a competitive zinc binding assay, and a light scattering assay, we found that mutation of the conserved His and Cys residues in HCCHp had little effect on secondary structure, but reduced zinc-binding affinity and altered the aggregation properties of the peptides. X-ray absorption spectroscopy was used to study zinc coordination in wild type HCCHp. The data are consistent with S2N(imid)2 coordination and strongly suggest that His-108, Cys-114, Cys-133, and His-139 are zinc ligands. Mutation of one or both conserved Cys residues in HCCHp led to a decrease in Cys ligation, and an increase in the number of (N,O) ligands, with noninteger coordination numbers suggesting zinc site heterogeneity. A purified fragment of human Cul5 was found to inhibit zinc-induced aggregation of HCCHp, and pull-down experiments revealed that zinc binding to HCCHp increases the strength of the HCCHp-Cul5 interaction by eight-fold.
HIV-1 accessory proteins modulate the host cellular environment to permit viral replication and transmission (reviewed in (1–4)). The HIV-1 vif gene encodes virion infectivity factor (Vif),1 a 23-kDa basic protein of 192 amino acids. Vif is expressed late in the viral life cycle (5, 6), and is also packaged into virus particles (7–10). Deletions in vif reduce HIV infectivity (11–13) to levels that vary depending on the host cell type (reviewed in (4)). In nonpermissive cells, such as macrophage and lymphocytes, vif is essential for the production of infectious virus particles (10, 14, 15), while the infectivity of viruses produced from permissive cells, such as CEM or HeLa, does not depend upon vif (15–17). Sheehy et al. identified apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G) as the cellular factor responsible for the restriction of vif-deficient HIV-1 in nonpermissive cells (18).
APOBEC3G belongs to a family of zinc-dependent cytidine deaminases that includes APOBEC1 and activation-induced deaminase (19). APOBEC3G is packaged into virus particles and is delivered to target cells where it deaminates cytidines in the minus strand of viral reverse transcripts, causing hypermutation in the plus strand (20–22). Vif blocks incorporation of APOBEC3G into virus particles. This in part is due to Vif-mediated recruitment of a Cullin-5 (Cul5)-based E3 ubiquitin ligase (23–25). The Vif-E3 complex polyubiquitinates APOBEC3G leading to its recognition and hydrolysis by the 26S proteasome (24, 26–29).
Formation of the Vif-E3 ubiquitin ligase complex requires at least two conserved elements of Vif, the BC box motif (amino acids 144–150) and the HCCH motif (amino acids 108–139). The BC box in HIV-1 Vif (SLQYLAL) is similar to BC boxes found in suppressors of cytokine signaling proteins (30) and mediates interactions with Elongin C (23–25, 27, 28). The HCCH motif (H108-X5-C114-X17–18-C133-X3–5-H139) is a novel zinc-binding motif (31–33) that mediates interactions with Cul5 (31, 32). A GST-Vif fragment (amino acids 100–142) interacts with Cul5 (32), suggesting that this fragment of Vif folds into a biologically active conformation. Studies with the model peptide HCCHp (amino acids 101–142 of HIV-1 Vif) indicate that zinc binding to the HCCH motif is specific, reversible, and induces a conformational change that leads to the formation of protein aggregates, a phenomenon also observed for full-length Vif (33). A recent fluorescence and CD spectroscopic study of HCCHp reveals that zinc binding also increases tertiary packing, further supporting the concept that zinc binding and protein conformation are tightly coupled (34). Based on mutational analyses, the interface between Vif and Cul5 is believed to be hydrophobic and zinc binding is thought to position hydrophobic residues for optimal binding to Cul5 (32, 35, 36). This possible role of zinc in stabilizing the Vif-Cul5 hydrophobic interface may explain the ability of zinc to induce aggregation of Vif.
In this study, the model peptide HCCHp was employed to gain further insight into zinc coordination and Cul5 binding by the HCCH motif in Vif. Using CD spectroscopy and zinc binding assays, the effects of mutating the Cys and His residues in HCCHp on peptide secondary structure, zinc binding affinity, and zinc-induced aggregation were studied. All the HCCHp peptides displayed varying affinities for zinc and different aggregation properties, suggesting that in the absence of one or more of the conserved His/Cys ligands, alternate ligation could facilitate zinc binding. X-ray absorption spectroscopy (XAS) was used to characterize zinc coordination by wild type and mutant HCCHp peptides. XAS revealed S2N(imid)2 zinc coordination for wild type HCCHp. Mutation of one or both conserved Cys residues in HCCHp led to a decrease in Cys ligation and an increase in (N,O) ligation. The HCCH motif in Vif is a unique zinc-binding and protein-protein interaction domain. Interactions between the HCCH motif and Cul5 may provide a target for the development of new antiretroviral compounds to block HIV pathogenesis. It is assumed that the Vif-Cul5 interaction is mediated by zinc binding to the HCCH motif, but detailed analyses of zinc coordination and Cul5 recognition by the HCCH motif are lacking. Studies with purified human Cul5 demonstrated a direct and zinc-dependent interaction with HCCHp. Zinc enhanced the HCCHp-Cul5 interaction by eight-fold. While the involvement of additional Vif domains in Cul5 recognition cannot be ruled out, our data strongly suggest that the HCCH motif in its zinc-bound form is necessary and sufficient for Cul5 binding.
EXPERIMENTAL PROCEDURES
Peptide purification and manipulation
Unless stated otherwise, all chemicals were obtained in high purity from Sigma/Aldrich, St. Louis, MO. HCCHp is a 4822-Da peptide based on amino acids 101–142 of HIV-1HXB2 Vif, H2N-ELADQLIH108LYYFDC114FSDSAIRKALLGH127IVSPRC133EYQAGH139NKV-CO2H, and was synthesized by solid phase peptide synthesis using Fmoc protection (Bio-Synthesis Inc., Lewisville, TX). Crude peptides were solubilized and purified as described (33). Purified peptides were handled in an anaerobic glovebox (Coy Laboratory Products, Grass Lake, MI) and were dissolved in anaerobic buffers containing tris-carboxyethyl phosphine (TCEP). The molar absorption coefficients (ε280) used for determining the concentration of HCCHp and the mutant peptides were calculated as described (37). The absorbance (280 nm) of peptide samples prepared in 6M guanidine hydrochloride and 30 mM MOPS pH 7.0 was measured in triplicate. Peptide stock solutions were stored under nitrogen at −80 ºC.
Cloning, expression and purification of Cul5
A fragment of human cul5 encoding amino acids 1–384 was cloned into the BamHI/SalI sites of pGEX-5X-1 (GE Healthcare). This construct was used to transform BL21(DE3) E. coli cells. Bacteria were cultured in LB at 37 ºC until OD600 = 0.6. GST-Cul5 expression was induced by the addition of 1 mM IPTG and the cells were harvested after 4 hr. Cell pellets were resuspended in 50 mM HEPES pH 7.4, 150 mM NaCl, 200 µM TCEP and 0.1% Tween, supplemented with RNase/DNase (Pierce Biotechnology, Inc.) and protease inhibitors (Roche Applied Science), and lysed in a French pressure cell. The supernatant of the cell lysate was purified using a glutathione-agarose column (GSTPrep FF 16/10) according to the manufacturer’s instructions (GE Healthcare). Eluted protein was further purified using a Superdex-200 column equilibrated in 50 mM HEPES pH 7.4, 150 mM NaCl, 200 µM TCEP and 0.1% Tween and operated at a flow rate of 0.5 mL min−1. Based on densitometric analysis GST-Cul5 was determined to be ∼95% pure. Protein concentration was determined by the Bradford assay (Bio-Rad) using bovine serum albumin as a standard.
Far-UV circular dichroism spectroscopy
Protein samples were diluted to 15 µM in 10 mM MOPS pH 7.4, 20 mM NaCl, and 40 µM TCEP. Circular dichroic spectra were recorded on a JASCO J-815 spectropolarimeter with the bandwidth set at 1 nm. Peptide samples were analyzed at 20 ºC in quartz cuvettes of 1 mm path length. Each spectrum represents the background-corrected average of five successive scans. The mean residue molar ellipticity, [θ], was calculated according to the equation, [θ] = (θ × MR) / (10 × l × c). θ is the measured ellipticity in millidegrees, MR is the mean residue mass (molecular weight of the peptide divided by the number of amino acid residues), l is the optical path length in cm and c is the protein concentration in mg/mL. Deconvolution of far-UV CD spectra into secondary structural components (helix, strand, random coil) was performed using the K2d algorithm (38) from the suite of programs available at the online server DICHROWEB (http://dichroweb.cryst.bbk.ac.uk) (39, 40).
Light scattering assays
HCCHp was diluted to 15 µM in 200 mM MOPS pH 7.4, 80 mM NaCl, and 1.8 mM TCEP. Sample temperature was maintained at 20 ºC using a Peltier-controlled sample compartment. Zn2+ (1 molar equivalent), from a standard Zn(NO3)2 stock solution, was manually mixed with HCCHp and light scattering was measured using an ISS PC1 single-photon counting fluorometer (ISS, Champaign, IL) as previously described (33). Single exponential functions were fitted to the raw data (intensity vs. time) using Mathematica (Wolfram Research, Champaign, IL).
Light scattering assays involving Cul5
HCCHp (6.8 µM) was mixed with an equimolar amount of purified GST-Cul5 or GST in buffer containing 50 mM HEPES pH 7.4, 150 mM NaCl, 200 µM TCEP and 0.1% Tween and incubated for 15 min at room temperature. To this solution was added 1 molar equivalent Zn2+ and light scattering was measured as described above. In control experiments, HCCHp, GST, or GST-Cul5 were reacted with zinc under the same conditions.
Competitive zinc binding assay using 4-(2-pyridylazo)resorcinol
Zinc binding to HCCHp was measured by a competitive binding assay involving the metallochromic indicator 4-(2-pyridylazo)resorcinol (PAR), which binds Zn2+ to form a 2:1 complex with a strong absorbance at 500 nm (41). In the competition experiment, 3 µM PAR in 20 mM MOPS pH 7.4, 50 mM NaCl, 200 µM TCEP was titrated with Zn(NO3)2 to 85% saturation and then back titrated with wild type HCCHp or mutant peptides. Changes in absorbance were measured using a Lambda 25 UV/vis spectrophotometer (Perkin Elmer). For the purposes of data analysis, we assumed a competitive binding model in which zinc exists in three possible states: bound to PAR, bound to HCCHp, and free in solution (i.e., not bound to PAR or HCCHp).
The competitive binding data (A500) reflect the decrease in the concentration of the PAR2-Zn2+ complex caused by increasing the concentration of HCCHp. Using Mathematica (Wolfram Research, Inc) an equation relating [PAR2-Zn2+] to [HCCHp] was derived. This equation is a cubic with the form a[PAR2-Zn2+]3 + b[PAR2-Zn2+]2 + c[PAR2-Zn2+] – d = 0 (42). Solving this expression for [PAR2-Zn2+] yields three roots. Two of the roots are experimentally descriptive, depending on whether Kd1 > Kd2 or Kd1 < Kd2. Taking these two cases into account, the Mathematica algorithm evaluates the cubic expression at the same points for which A500 was measured. The experimental data are compared to the simulated data and the squared difference between these two values is calculated. These differences are measured across a wide range of Kd2 values and the value of Kd2 giving rise to the smallest difference is the best-fit value. The algorithm requires user inputs for the total concentration of PAR, the total concentration of Zn2+, and the dissociation constant for the PAR2-Zn2+ complex (Kd1), which was measured experimentally by titrating 3 µM PAR with Zn(NO3)2. Errors in Kd2 values were estimated by computationally determining the variance in Kd2 required to double the residual difference between the experimental and simulated data.
X-ray absorption spectroscopy (XAS) and extended X-ray absorption fine structure (EXAFS) of HCCHp-Zn2+ complexes
Peptides were HPLC purified, dried in vacuo, and dissolved in 100 mM MOPS pH 7.4, 80 mM NaCl, and 1.8 mM TCEP (Buffer A) to give a final peptide concentration of 160 µM. Peptides were reacted with 1 molar equivalent Zn(NO3)2 for 1 hr at 30 ºC in an anaerobic chamber. Samples were repeatedly centrifuged into a pellet and washed with Buffer A. After a final centrifugation step, the pellets were resuspended in 6 µL Buffer A, and 3 µL was transferred to a custom cuvette. The cuvette was immediately frozen in liquid N2.
Zn K-edge X-ray absorption spectra and EXAFS data were collected at 10 K at the Stanford Synchrotron Radiation Lightsource (SLAC National Accelerator Laboratory) beam line 9–3 with the SPEAR3 ring operating at 3.0 GeV, 80–100 mA. The harmonic rejection mirror was set for a nominal 13 keV cutoff and Si[220] crystals were fully tuned to provide a monochromatic X-ray beam that was apertured to 1 × 1 mm to illuminate the 3 µL samples. Data collection used an array of 30 intrinsic Ge fluorescence detector elements and 4–5 scans were averaged to provide the raw XAS data.
XAS data processing and reduction were carried out using the EXAFSPAK program suite (http://www-ssrl.slac.stanford.edu/exafspak.html). Internal energy calibration utilized an elemental Zn foil standard, assuming a first inflection point energy of 9660.7 eV and Zn EXAFS data were extracted using E0 of 9670 eV. Analysis of the EXAFS data also used EXAFSPAK and histidine imidazole multiple scattering contributions were calculated using feff 7.2 (43). To determine approximate numbers of imidazole ligands, Debye-Waller factors (i.e., the exp(– 2σas2k2) term in the EXAFS expression) for all imidazole multiple-scattering legs were fixed at the values that provided the best fit of a Zn(imid)4 model and only Zn-N(imid) coordination number was optimized (along with Zn-S(Cys) coordination number and a single ΔE0). Given the minor contribution of the Zn-N(imid) scattering to the overall EXAFS, the uncertainty in the resulting Zn-N(imid) coordination number is expected to be large (we estimate ± 25%). Zn-S(Cys) scattering dominates the EXAFS (when cysteines are present) so this coordination number should be more accurate (± 15%).
HCCHp pull-down by GST-Cul5
Purified GST-Cul5 (1.3 nmol, 92 µg) was incubated for 10 min at 4 ºC with 0.5 mL glutathione-agarose resin (GSTrap HP, GE Healthcare) equilibrated in 50 mM HEPES pH 7.4, 150 mM NaCl, and 200 µM TCEP (Buffer B). After centrifugation,the supernatant was discarded and the resin was washed with Buffer B. The supernatant was removed and purified HCCHp (10 nmol, 48 µg) was added to the resin in a total volume of 500 µL to give a final peptide concentration of 20 µM. Following 10 min of incubation, the HCCHp-resin slurry was split equally into two tubes. To one tube, 10 nmol Zn(NO3)2 (40 µM final concentration, 2 molar equivalents) was added, and to the other tube, an equal volume of Buffer B was added. After 10 minutes the tubes were centrifuged and the supernatants were discarded. Resin-bound proteins were washed six times with 1 mL aliquots of Buffer B. After the final centrifugation step, the supernatant was removed, and the resin was resuspended with 200 µL SDS-PAGE sample buffer (Invitrogen) plus 200 mM DTT. Samples were boiled for 10 min and 20 µL was loaded onto a Novex 4–12% Bis-Tris Gel. After electrophoresis the gel was stained overnight with SYPRO Ruby (Invitrogen). The gel was analyzed using a Fujifilm LAS-3000 luminescent image analyzer equipped with a blue LED (470 nm) and Y515 cutoff filter.
HCCHp and GST-Cul5 band intensities were normalized against the intensity of the 27-kDa triosephosphate isomerase marker band (New England Biolabs) and compared to normalized band intensities from HCCHp and GST-Cul5 standard curves.
RESULTS
Secondary structure of wild type and mutant HCCHp
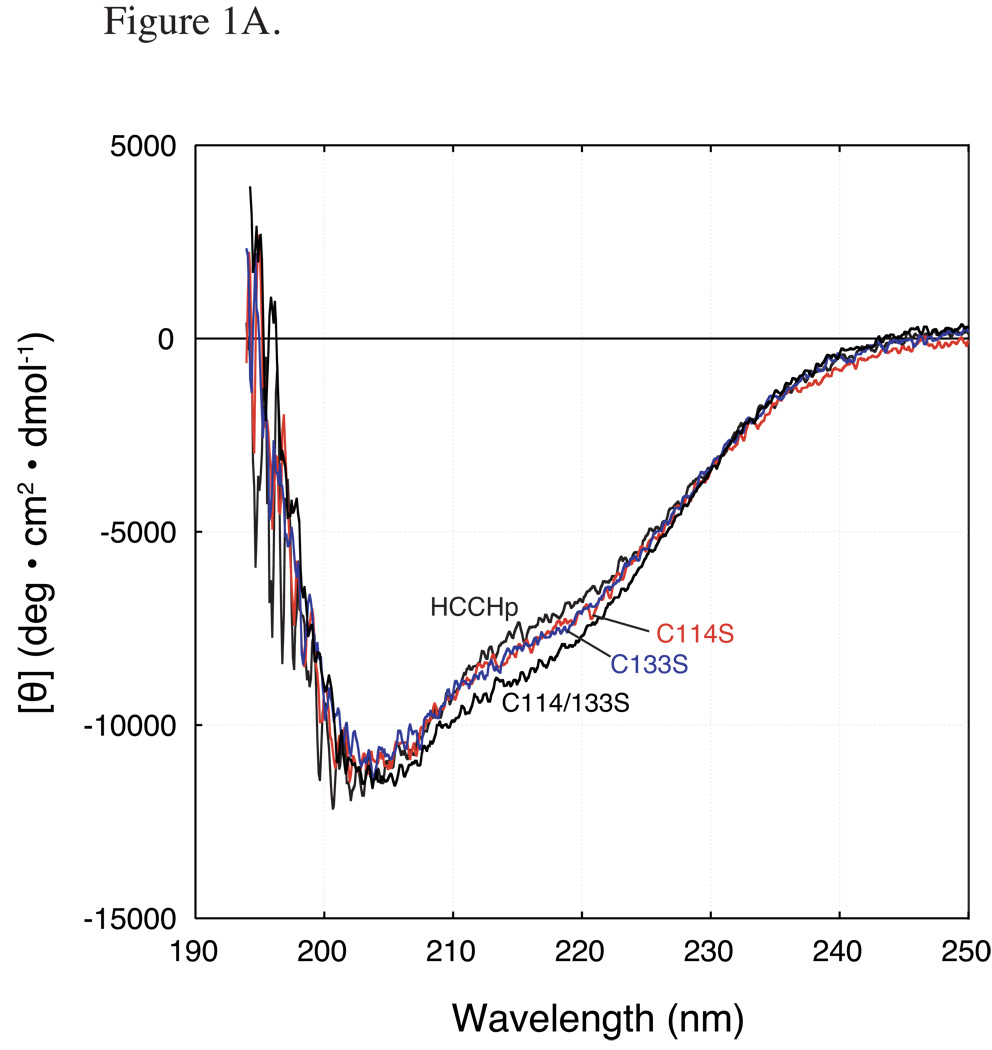
Wild type and mutant HCCHp peptides (15 µM) were analyzed by far-UV circular dichroism (CD) spectroscopy to determine the effects of Cys/His mutation on average secondary structural content. The K2d algorithm (38) was used to estimate percent α-helix, β-sheet, and random coil. Analysis of the far-UV CD spectrum for wild type HCCHp indicated that the peptide is 55% random coil, 12% α-helix and 33% β-sheet (Figure 1A). An earlier prediction of HCCHp secondary structure (36% random coil, 59% α-helix and 5% β-sheet) (33) differs significantly from our experimental results. However, the short length of the HCCHp peptide and sequence variability in the HCCH motif may affect the accuracy of the predicted secondary structure (44). The secondary structural content of the Cys mutant peptides, C114S, C133S and C114/133S, were very similar to wild type HCCHp (Figure 1A). RMSDs from the wild type spectrum were calculated to be 0%, 1.4% and 2.8% for the C114S, C133S and C114/133S mutants, respectively.
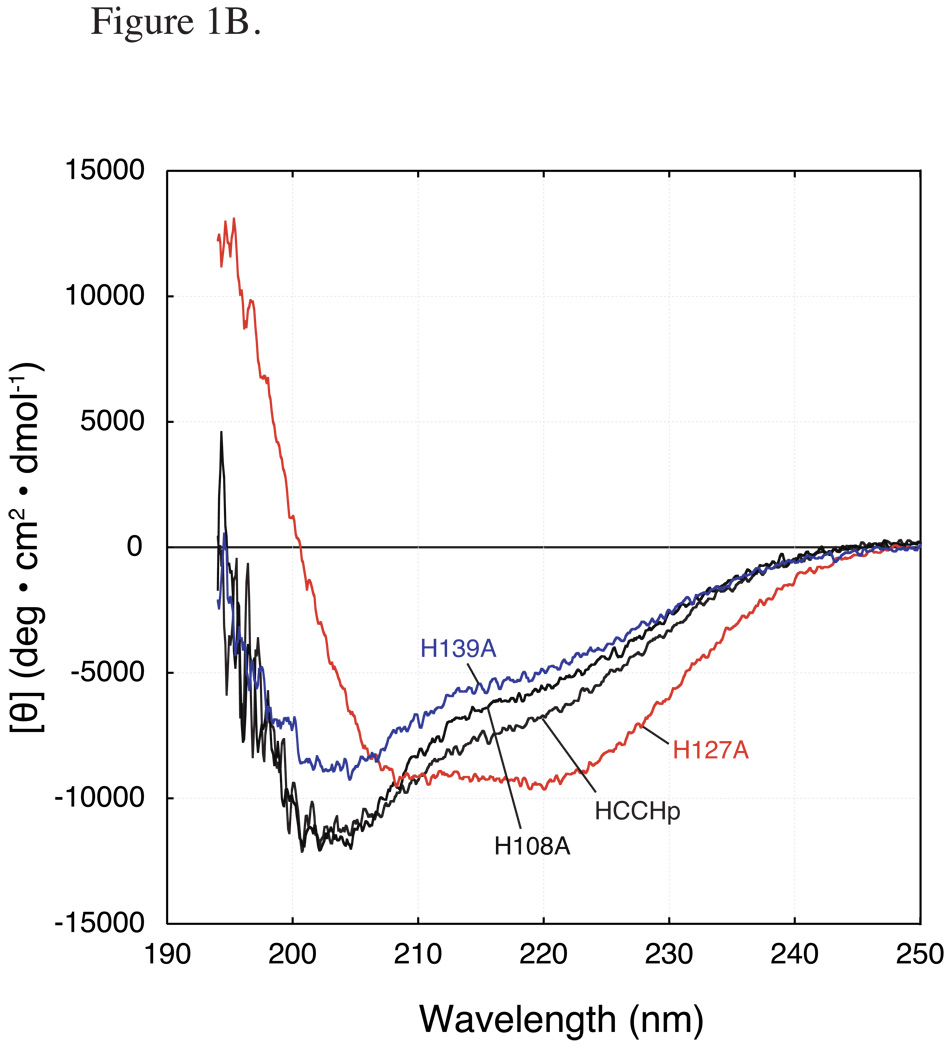
Figure 1.
Far-UV CD spectra of wild type and mutant HCCHp. Peptides were prepared at a final concentration of 15 µM. A CD spectra of wild type HCCHp and Cys mutants: C114S, C133S (dashed line), and C114/133S. B CD spectra of wild type HCCHp and His mutants: H108A, H127A, and H139A.
The HCCHp sequence (Experimental Procedures) contains three His residues (His-108, His-127 and His-139). The far-UV CD spectra of the H108A and H139A mutants were similar to wild type HCCHp although the values of [θ]222 were slightly smaller, indicating less helical content (Figure 1B). Component analysis of the CD spectra gave 10% helical content for the H108A and H139A mutants (compared to 12% for wild type HCCHp) and nearly identical fractions of β-sheet and random coil structure. Compared to the wild type spectrum, the overall RMSD in secondary structure for the H108A and H139A mutants were 3.6% and 2.8%. These deviations are small compared to that observed for the H127A mutant (30% RMSD) (Figure 1B). The CD spectrum of H127A revealed that Ala substitution at this position caused a dramatic increase in α-helical content from 12% in wild type to 31% in the mutant. This increase in α-helicity was offset by a nearly proportionate decrease in β-sheet content. According to a secondary structural prediction by PSI Pred (45), His-127 is immediately downstream from a helix that is terminated by Gly-126. Taken together, the data in Figure 1 suggest that mutation of the conserved Cys and His residues in HCCHp (His-108, Cys-114, Cys-133 and His-139) did not significantly affect the overall secondary structure of HCCHp.
Zinc binding properties of HCCHp
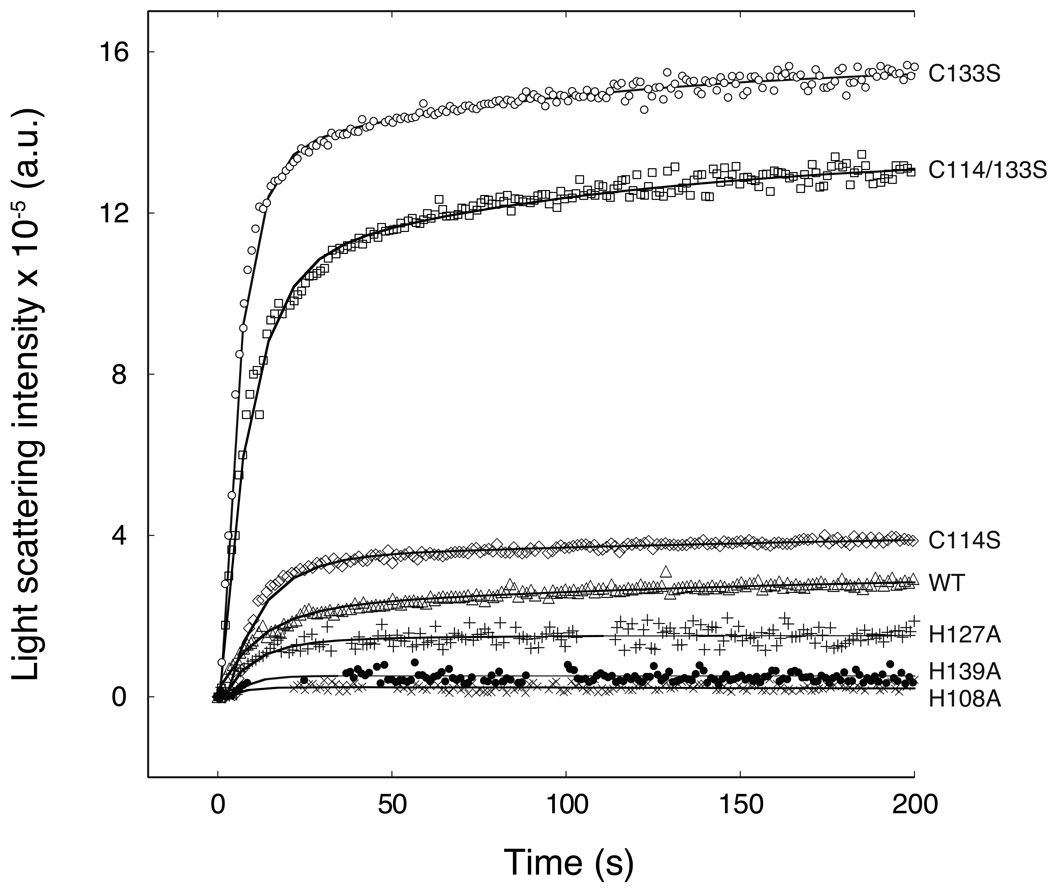
To study the role of the HCCHp His and Cys residues in zinc binding, two assays were employed. The first assay measures zinc-induced aggregation of HCCHp by light scattering. Zinc-induced aggregation is observed for full-length Vif as well as HCCHp, is reversible, and is preceded by an increase in β-sheet content (33). Wild type and mutant HCCHp peptides (15 µM) were reacted with 1 molar equivalent Zn2+ by manual mixing and the time-dependent increase in light scattering was measured (Figure 2). The data reveal that all of the peptides aggregated in the presence of zinc. However, the total change in scattered light intensity varied by more than two orders of magnitude. This change was smallest for H108A and increased in the order H108A < H139A < H127A < wild type < C114S < C114/133S < C133S. Since the light scattering intensity is a function of the average molecular mass and weight concentration of aggregated species, the data in Figure 2 could be explained if the mutants bound zinc to form aggregates with different size distributions or concentrations. However, the variations in light scattering intensity could also be explained if the zinc binding affinities of the mutant peptides differed significantly from that of wild type HCCHp.
Figure 2.
Zinc-induced light scattering by wild type and mutant HCCHp. Peptides (15 µM) were mixed with 1 molar equivalent zinc at t = 0 and light scattering intensity was measured over the course of 200 s. Data are shown for H108A (×), H139A (•), H127A (+), wild type (Δ), C114S (◇), C133S (○), and C114/133S (□). Solid lines represent single-exponential functions that were fitted to the data using Mathematica.
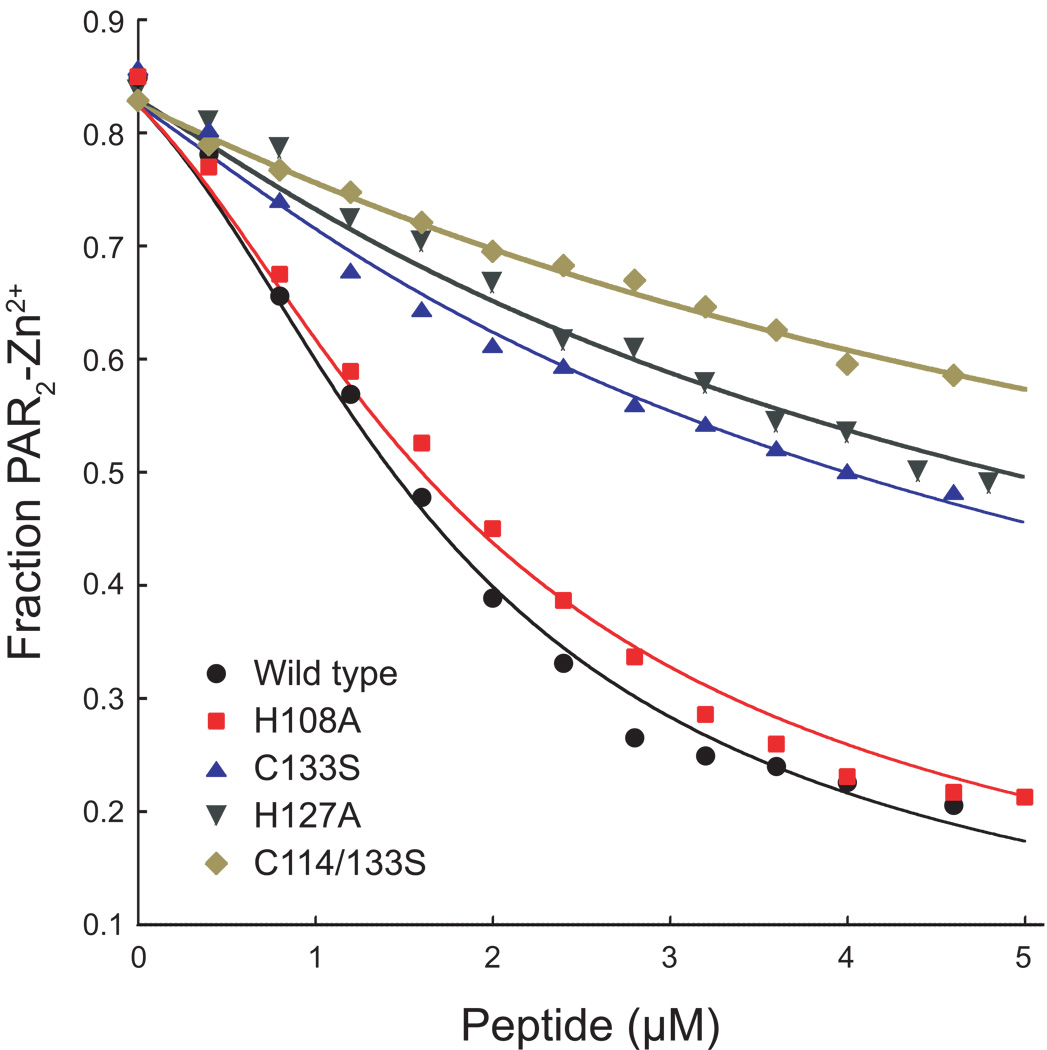
An assay based on the metallochromic ligand 4-(2-pyridylazo)resorcinol (PAR) was used to measure the zinc binding affinity of wild type and mutant HCCHp. PAR binds Zn2+ to form a complex that absorbs strongly at 500 nm (41). To measure the zinc binding affinity of HCCHp, a competitive binding assay was developed. PAR (3 µM) was titrated to 85% saturation with Zn2+ and back titrations were performed with wild type or mutant HCCHp. Figure 3 shows data from competitive titrations of the PAR2-Zn2+ complex with wild type and mutant HCCHp. Wild type HCCHp bound zinc more tightly than the mutants with the exception of the H139A and C114S peptides whose titration curves were visually indistinguishable from wild type HCCHp. For simplicity, the titration data for these mutants have been omitted from Figure 3. Titration data were analyzed using a curve-fitting algorithm written in Mathematica (Experimental Procedures). The results from curve fitting are shown in Table 1. Zinc bound to wild type HCCHp with a dissociation constant of 54 ± 10 nM. The Kd values for the C114S mutant (59 ± 15 nM) and H139A mutant (59 ± 17 nM) were within error of the wild type value and the Kd for the H108A mutant (74 ± 14 nM) was only marginally reduced compared to wild type HCCHp. This is in contrast to the C133S and C114/133S mutants, which bound zinc with dissociation constants of 370 ± 50 nM and 830 ± 70 nM, respectively. Interestingly, these mutants also showed the most dramatic elevation in zinc-induced light scattering compared to wild type HCCHp (Figure 2). The mutants that bound zinc more tightly (C114S, H139A and H108A) behaved more like wild type HCCHp in the light scattering assay. An exception is the H127A mutant, which displayed light scattering properties like those of the H139A mutant or wild type HCCHp (Figure 2), but bound to zinc much more weakly (Kd = 470 ± 130 nM) like the C133S and C114/133S mutants. It should be noted that the H127A mutant was significantly more structured than wild type HCCHp and all the other mutants (Figure 1). The data in Figure 2 and Figure 3 suggest that zinc coordination by HCCHp can occur using more than one unique set of ligands. The strongest zinc interaction occurs with wild type HCCHp. Mutations that weaken zinc binding affinity also appear to impact the aggregation pathway(s) as evidenced by gross changes in light scattering intensity (Figure 2).
Figure 3.
Competitive back titration of PAR2-Zn2+ with wild type and mutant HCCHp peptides. Peptides were titrated into a solution of PAR and Zn(NO3)2 and absorbance at 500 nm was monitored. Titration data were analyzed using a fitting algorithm written in Mathematica.
TABLE 1.
Best-fit HCCHp-zinc dissociation constants from curve-fitting analysis
| Peptide | Best-fit Kd (nM) |
|---|---|
| Wild type HCCHp | 54 ± 10 |
| C114S | 59 ± 15 |
| H139A | 59 ± 17 |
| H108A | 74 ± 14 |
| C133S | 370 ± 50 |
| H127A | 470 ± 130 |
| C114/133S | 830 ± 70 |
X-ray absorption spectroscopy and extended X-ray absorption fine structure (EXAFS) of HCCHp-Zn2+ complexes
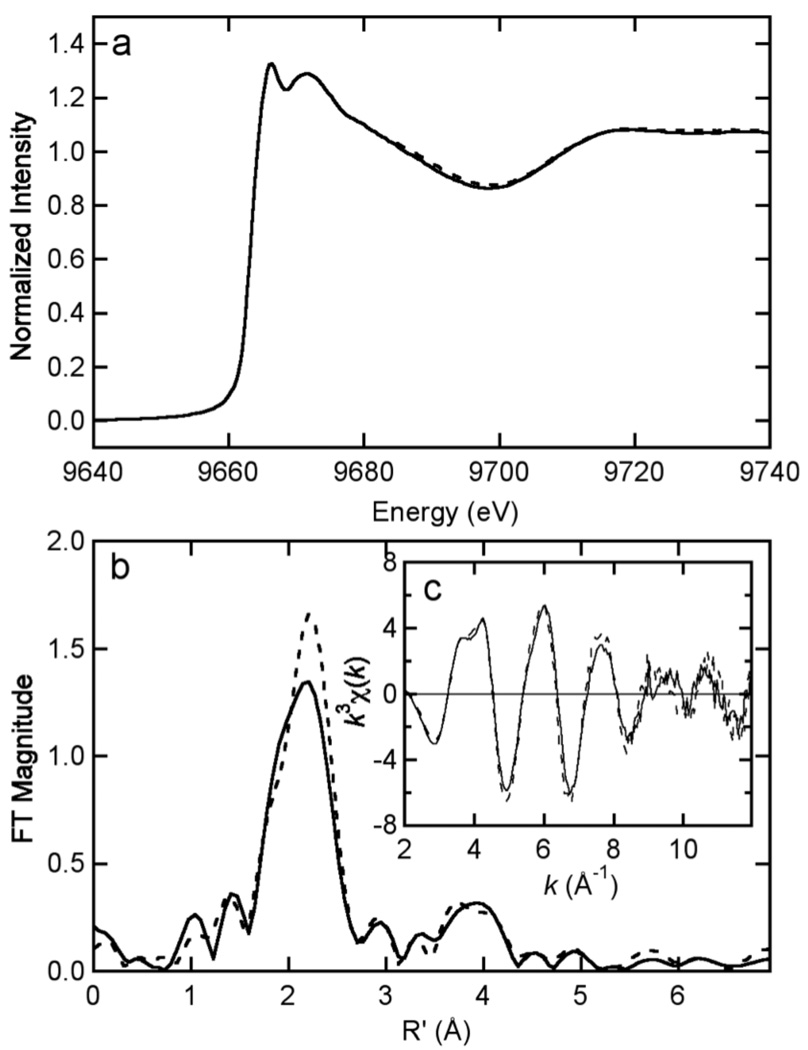
X-ray absorption spectroscopy (XAS) was used to examine the differences in zinc ligation by wild type HCCHp and the mutants H127A, C114S, C133S, and C114/133S.1 Peptides were reacted with 1 molar equivalent Zn2+ for 1 hr at 30 ºC under anaerobic conditions. Aggregates were repeatedly centrifuged into a pellet and washed with zinc-free buffer. A suspension of each aggregate was transferred to a cuvette and frozen in liquid N2. The Zn K-edge EXAFS data for wild type HCCHp and the H127A mutant are shown in Figure 4. The data reveal very similar, if not identical, zinc coordination by wild type HCCHp and the H127A mutant, leading to the conclusion that His127 is not required for zinc coordination. The first-shell peak in the Fourier transform data (centered at 2.2 Å, Figure 4B) is asymmetric and indicative of a mixture of S and N/O ligands. The peaks in the 3 – 4 Å range of the Fourier transform data arise from second shell scattering involving the ε2 ring nitrogen and carbons of coordinated imidazole and support a direct role of histidine in zinc coordination.
Figure 4.
Comparison of Zn K edge XAS data for wild type HCCHp (solid) and the H127A variant (dashed) in the edge (A) and EXAFS (C) regions. The Fourier transforms of the EXAFS (k = 2 – 12 Å–1k3 weighting, phase corrected based on S scattering) are shown in (B).
Models used to analyze the EXAFS data assumed site heterogeneity in zinc coordination. Such heterogeneity would result in average Zn-(N,O) or Zn-S coordination numbers that are non-integer, so the curve-fitting optimizations allowed coordination numbers for S and (N,O) ligands to float. Data analysis for wild type HCCHp and the H127A mutant revealed coordination numbers close to an S2N2 model (see Table 2 and Supplementary Table 2 for curve-fitting results). Data analysis gave histidine coordination numbers of 2.8 ± 0.7 and 2.4 ± 0.6 for wild type HCCHp and the H127A mutant, respectively. These coordination numbers were most sensitive to the Fourier transform data in the 3 – 4 Å range (Figure 4B), resulting in significant uncertainty. The Cys coordination numbers for wild type HCCHp (1.9 ± 0.3) and the H127A mutant (2.3 ± 0.3) are within error of the expected value of 2.0. Overall, the curve-fitting analyses for wild type HCCHp and the H127A mutant are consistent with predominant S2N(imid)2 zinc coordination.
TABLE 2.
Curve fitting results for Zn EXAFSa (plots of fits in Supplementary Table 2)
| Sample, filename (k range) | Fit | Shell | Ras | σas2 | ΔE0 | f'b |
|---|---|---|---|---|---|---|
| Δk3 χ | (Å) | (Å2) | (eV) | |||
| HIV Vif HCCHp (wt), | 1 | Zn-S1.90 | 2.31 | [0.0032]c | −1.22 | 0.075 |
| ZVC0A + ZVCGA | Zn-N2.79 | 2.02 | [0.0026] | [−1.22] | ||
| (k = 2 – 12 Å–1) | Zn-C2.79 | [3.00] | [0.0039] | [−1.22] | ||
| Δk3χ = 11.836 | Zn-C2.79 | [3.05] | [0.0039] | [−1.22] | ||
| Zn-N2.79 | [4.18] | [0.0034] | [−1.22] | |||
| Zn-C2.79 | [4.23] | [0.0034] | [−1.22] | |||
| HIV Vif [H127A]HCCHp | 2 | Zn-S2.32 | 2.30 | [0.0032] | −3.25 | 0.086 |
| ZVA0A + ZVAGA | Zn-N2.41 | 2.01 | [0.0026] | [−3.25] | ||
| (k = 2 – 12 Å–1) | Zn-C2.41 | [2.99] | [0.0039] | [−3.25] | ||
| Δk3χ = 12.142 | Zn-C2.41 | [3.04] | [0.0039] | [−3.25] | ||
| Zn-N2.41 | [4.17] | [0.0034] | [−3.25] | |||
| Zn-C2.41 | [4.22] | [0.0034] | [−3.25] | |||
| HIV Vif [C114S]HCCHp | 3 | Zn-S1.54 | 2.30 | [0.0032] | −1.24 | 0.112 |
| ZVSCA | Zn-N3.14 | 2.01 | [0.0026] | [−1.24] | ||
| (k = 2 – 12 Å–1) | Zn-C3.14 | [2.99] | [0.0039] | [−1.24] | ||
| Δk3χ = 11.836 | Zn-C3.14 | [3.04] | [0.0039] | [−1.24] | ||
| Zn-N3.14 | [4.17] | [0.0034] | [−1.24] | |||
| Zn-C3.14 | [4.22] | [0.0034] | [−1.24] | |||
| HIV Vif [C133S]HCCHp | 4 | Zn-S1.08 | 2.31 | [0.0032] | 0.34 | 0.100 |
| ZVCSA + ZVCSB | Zn-N3.39 | 2.02 | [0.0026] | [0.34] | ||
| (k = 2 – 12 Å–1) | Zn-C3.39 | [3.00] | [0.0039] | [0.34] | ||
| Δk3 χ = 10.860 | Zn-C3.39 | [3.05] | [0.0039] | [0.34] | ||
| Zn-N3.39 | [4.18] | [0.0034] | [0.34] | |||
| Zn-C3.39 | [4.23] | [0.0034] | [0.34] | |||
| HIV Vif [C114,133S]HCCHp | 5 | Zn-O1 | 2.08 | 0.0072 | −2.41 | 0.139 |
| ZVSSA + ZVSSB | Zn-N3 | 1.98 | [0.0026] | [−2.41] | ||
| (k = 2 – 12 Å–1) | Zn-C3 | [2.96] | [0.0039] | [−2.41] | ||
| Δk3χ = 12.514 | Zn-C3 | [3.01] | [0.0039] | [−2.41] | ||
| Zn-N3 | [4.14] | [0.0034] | [−2.41] | |||
| Zn-C3 | [4.19] | [0.0034] | [−2.41] | |||
Shell is the chemical unit defined for the multiple scattering calculation. Subscripts denote the number of scatterers per metal. Ras is the metal-scatterer distance. σas2 is a mean square deviation in Ras. ΔE0 is the shift in E0 for the theoretical scattering functions.
Numbers in square brackets were constrained to be either a multiple of the above value (σas2) or to maintain a constant difference from the above value (Ras, ΔE0). If there is no unbracketed value in that column, these were fixed to the specified value and not optimized.
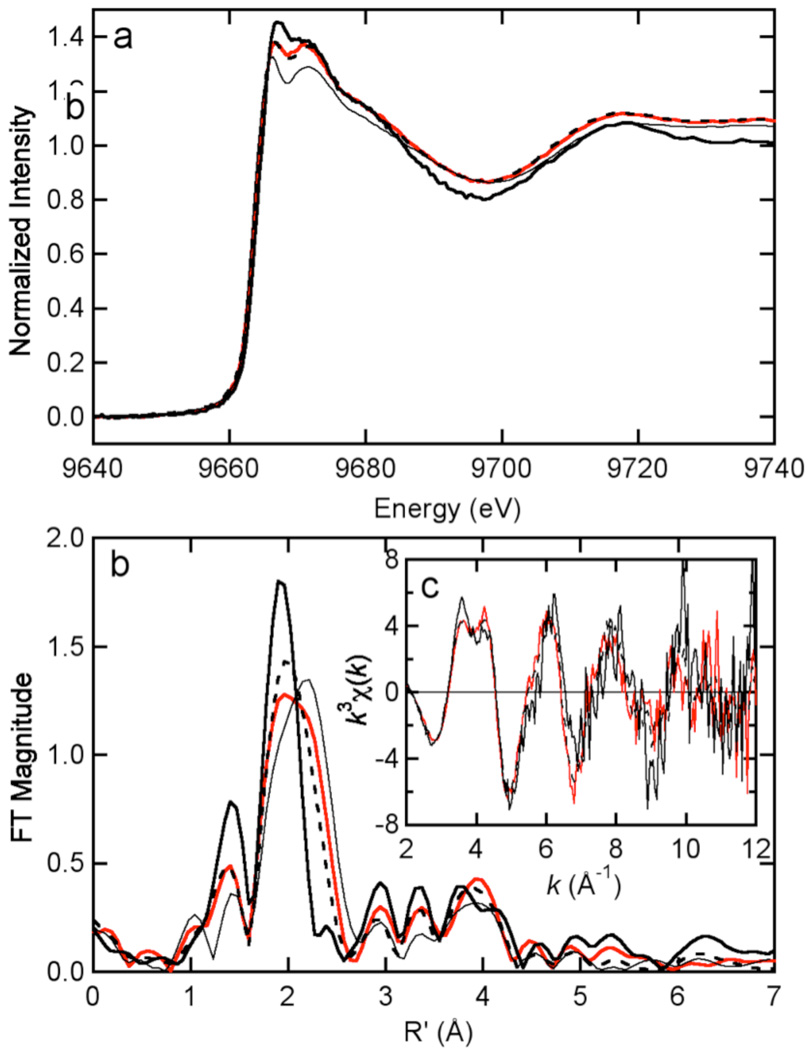
EXAFS data for the C114S mutant (Figure 5) revealed a decrease in S ligation (1.5 ± 0.3) and an increase in imidazole ligation (3.1 ± 0.7) compared to wild type HCCHp. The Cys coordination number was higher than expected for S1N3 coordination, suggesting possible heterogeneity in zinc coordination. For example, a mixture of S1N3 zinc sites and S2N2 zinc sites, the latter potentially formed by two HCCHp peptides each donating one Cys-(S) and one His-(N) ligand to zinc, could account for the higher than expected Cys coordination number. EXAFS data for the C133S mutant revealed numbers of Cys ligands (1.1 ± 0.2) and His ligands (3.39 ± 0.8) expected for predominant S1N3 coordination (Table 2). Although the error in curve fitting could explain the elevated His coordination number, this could also be caused by zinc coordination site heterogeneity. The FT data for the C114/133S mutant (Figure 5B) showed a symmetric first-shell peak centered at 2.0 Å and several peaks in the 3–4 Å range, consistent with strict (N,O) ligation and significant contribution from imidazole. Taken together the EXAFS data suggest that wild type HCCHp and the H127A mutant coordinate zinc using His-108, Cys-114, Cys-133 and His-139. Mutation of either Cys residue leads to a decrease in the number of S ligands and an increase in the number of (N,O) ligands, with noninteger coordination numbers suggesting zinc site heterogeneity. Replacement of one or both Cys residues in HCCHp with Ser may allow ligands in the same peptide molecule or from another peptide molecule to complete the zinc coordination sphere.
Figure 5.
Comparison of Zn K edge XAS data for wild type HCCHp (thin solid in a and b) with that for the C114S variant (red), the C133S variant (dashed bold), and the C114/133S variant (solid bold) in the edge (A) and EXAFS (C) regions. The Fourier transforms of the EXAFS (k = 2 – 12 Å–1k3 weighting, phase corrected based on S scattering) are shown in (B). Note that the wild type EXAFS data are not displayed in (C) for clarity.
Zinc-dependent interaction between HCCHp and Cul5
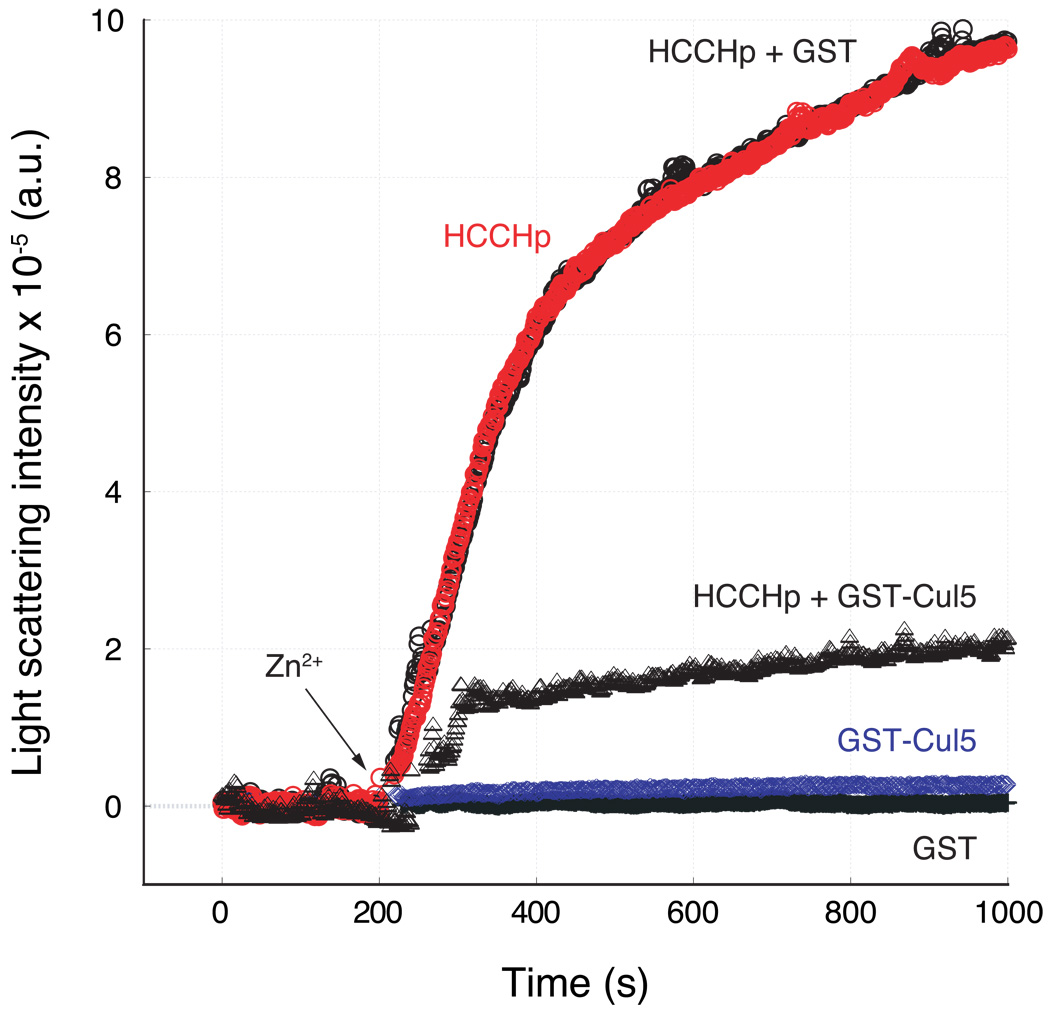
In the absence of zinc, HCCHp exists predominantly in a random-coil conformation. Zinc binding to HCCHp alters its conformation to one that is rich in β-sheet content, and leads to aggregation (33). If zinc binding to HCCHp exposes a hydrophobic surface that serves as a Cul5 recognition motif, we considered the possibility that aggregation and Cul5 binding might be mutually exclusive. We tested this hypothesis by determining the ability of Cul5 to block zinc-induced aggregation of HCCHp. A fragment of Cul5 containing three cullin repeats (helical bundles) was purified as a GST fusion to a purity of ∼95%. The CD spectrum of this Cul5 fragment was consistent with the alpha-helical fold of the cullin motif (Supplemental Data), suggesting proper protein folding. Equimolar amounts of HCCHp and GST-Cul5 were incubated together for 15 minutes. Following this incubation step, 1 molar equivalent of Zn2+ was added to the solution and light scattering was measured over time (Figure 6). GST-Cul5, but not GST, inhibited zinc-induced aggregation of HCCHp suggesting that the inhibitory effect is dependent on Cul5. Neither GST-Cul5 nor GST aggregated in the presence of zinc, strongly suggesting that light scattering was caused by zinc binding to HCCHp and not to either of these proteins. The data support the hypothesis that Cul5 binds to a hydrophobic surface in HCCHp that becomes exposed upon zinc binding. In the absence of Cul5 this hydrophobic surface drives the zinc-induced aggregation of HCCHp.
Figure 6.
Effect of Cul5 on zinc-induced aggregation of HCCHp. Zinc-induced light scattering of HCCHp was monitored in the absence of GST-Cul5 ( ), in the presence of GST-Cul5 (Δ), or in the presence of GST (○). As controls, zinc-induced aggregation of GST-Cul5 (
), in the presence of GST-Cul5 (Δ), or in the presence of GST (○). As controls, zinc-induced aggregation of GST-Cul5 ( ) or GST (+) was monitored.
) or GST (+) was monitored.
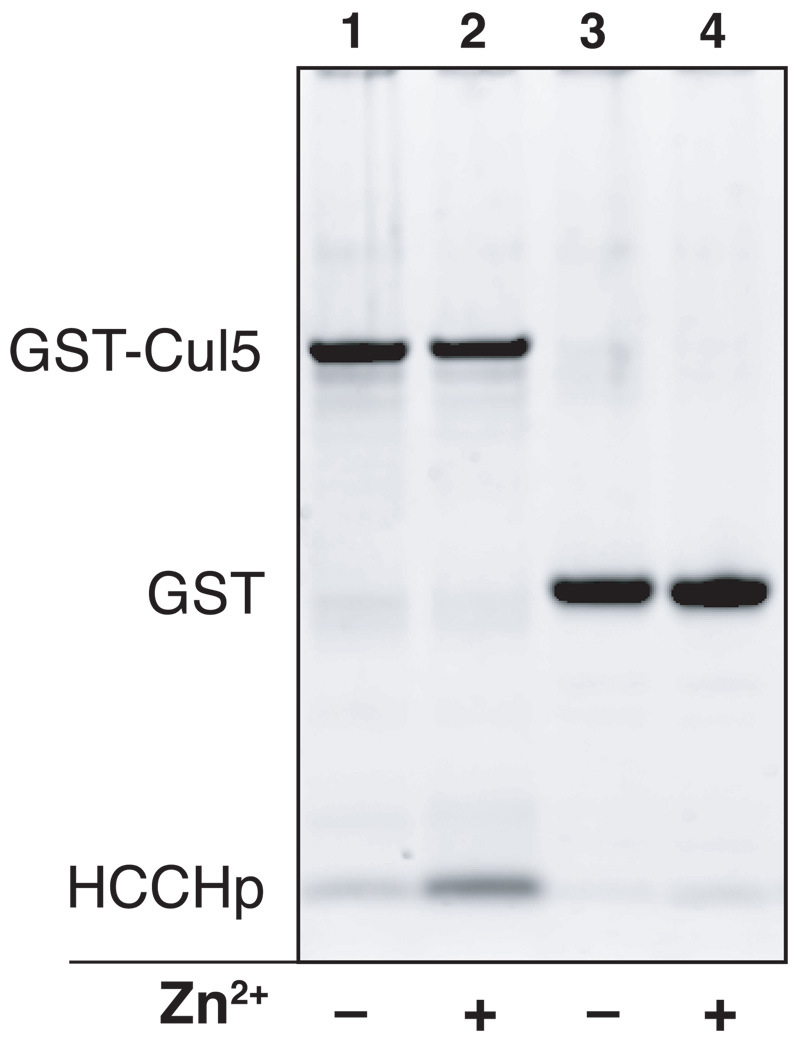
It has been demonstrated that a GST-Vif fusion protein containing amino acids 100–142 of Vif is sufficient to precipitate Cul5 expressed in a human cell line or from a cell-free reticulocyte system (32). However, a direct interaction between Vif and Cul5 has not been demonstrated and the zinc dependence of such an interaction has not been explored. In order to study the interaction between Cul5 and HCCHp, purified GST-Cul5 was immobilized on a glutathione matrix and used in an HCCHp pull-down experiment. The relative amount of HCCHp pulled down by Cul5 in the presence or absence of zinc was quantified by densitometric analysis following denaturing gel electrophoresis and protein staining with SYPRO Ruby. As can be seen from Figure 7, a small but detectable amount of HCCHp interacted with GST-Cul5 (lane 1), while very little peptide bound GST (lane 3). After normalizing for differences in GST-Cul5 and GST loading, the calculated amount of HCCHp associated with GST-Cul5 was at least 7-fold higher than the amount bound to GST, which was at the lower limit of detection. When zinc was included in the incubation step, the amount of HCCHp bound by GST-Cul5 increased 8.4-fold (lane 2), while the amount bound to GST did not increase significantly (lane 4). These data demonstrate a direct and zinc-dependent interaction between HCCHp and Cul5. To quantify the amount of HCCHp bound to GST-Cul5, the band intensities in lanes 1 and 2 in Figure 7 were compared to intensities from HCCHp and GST-Cul5 standards (Experimental Procedures). Based on this analysis the HCCHp:Cul5 molar ratio was estimated to be 2.0:1 in the presence of zinc. In the absence of zinc, the ratio was 0.24:1. Further studies to characterize the affinity and stoichiometry of the Vif-Cul5 interaction are under way.
Figure 7.
HCCHp pull-down experiment using purified GST-Cul5. Purified GST-Cul5 was immobilized on a glutathione sepharose column and used to pull down HCCHp in the absence or presence of zinc. Relative amounts of HCCHp pulled down were quantified by SYPRO Ruby staining and gel image analysis (Experimental Procedures).
DISCUSSION
To gain further insight into the structure and metal-binding properties of the HCCH motif in Vif, we studied the effects of mutating potential metal-binding residues on protein secondary structure, metal binding affinity, and ligand composition of the zinc binding site using the peptide HCCHp (amino acids 101–142 of HIV-1 Vif). The rationale for the use of HCCHp as a model peptide is supported by a previous study in which a similar Vif fragment (amino acids 100–142) was capable of interacting with Cul5, a component of the E3 ligase that targets APOBEC3G for proteasomal degradation (32). CD spectroscopy of wild type HCCHp and the mutant peptides H108A, C114S, H127A, C133S, H139A, and C114/133S revealed similar fractions of α-helix, β-sheet, and random coil, except the H127A peptide, which contained nearly three times the fraction of α-helix (Figure 1). The sequence of wild type HCCHp is derived from Vif expressed by the HXB2 strain of HIV-1. Inspection of Vif sequences from other HIV and SIV strains reveals that the occurrence of histidine at position 127 is not highly conserved. Vif sequences from other HIV-1 strains, including HIV-1NL4.3, contain Arg or Gln at position 127 while Vif sequences from strains of HIV-2 and SIV most frequently contain Glu at the corresponding position. The side chains of Glu, His, Gln, and Arg confer varying α-helix forming propensities (46), suggesting that HIV-1 Vif proteins may have secondary structures that differ significantly from their HIV-2 and SIV orthologs. The region immediately upstream from His-127 (IRKALLGH127) has been implicated in Cul5 binding (32, 35, 36). Thus, sequence variation may affect secondary structure(s) surrounding this putative Cul5 binding site and affect binding affinity.
XAS studies of HCCHp were performed to determine the zinc ligands in the HCCH motif. XAS is a direct method and is the sole spectroscopic technique that can be used to elucidate the molecular structure of Zn2+ binding sites in proteins. Vif contains two Cys residues that lie within the HCCH motif. The EXAFS data for wild type HCCHp indicate that there are two Cys(S) ligands in the primary Zn2+ coordination sphere, implicating both Cys-114 and Cys-133 as direct ligands to zinc (Figure 4 and Table 2). There are three His residues in HCCHp and the EXAFS shows that two (possibly three) are coordinated to zinc. However, mutation of the non-conserved His-127 did not significantly change the XAS spectrum, indicating that this residue does not contribute to the primary zinc coordination sphere. Taken together, the XAS/EXAFS data strongly suggest that the side chains of His-108, Cys-114, Cys-133, and His-139 in HCCHp coordinate zinc. Mutation of Cys-133 reduced zinc binding affinity by about 7-fold (Table 1 and Figure 3) consistent with the XAS data (Figure 4) that implicate this residue in zinc coordination. Mutation of His-108, Cys-114, and His-139, had a much smaller effect on zinc binding affinity, suggesting alternate modes of zinc coordination exist. Indeed, EXAFS of the single Cys mutants and the double Cys mutant revealed increasing Zn2+-N/O ligation and decreasing contribution from S ligands (Figure 5). This supports the hypothesis that these mutants coordinate zinc using alternate N/O ligands and provides further evidence that Cys-114 and Cys-133 coordinate zinc in wild type HCCHp.
In a previous study, point mutants of the HCCH motif in GST-Vif(100–142) were analyzed for their ability to bind to a zinc-charged matrix (32). The mutants studied (H108N and H139N) were severely compromised in their ability to bind to the matrix, although residual binding by the H108N mutant was apparent. All of the HCCHp mutants in this study bound zinc with affinities in the low-to-high nanomolar range and the zinc binding affinities of the H108A and H139A mutants were marginally reduced compared to wild type HCCHp (Table 1). These differences could be explained if immobilization of Vif on a zinc-charged surface alters the conformation and zinc-binding properties of the HCCH motif. Though the HCCHp mutants all retained significant zinc binding affinity, similar mutations in Vif (C114S, C133S, H108N, H139N) impair Cul5 binding, reduce Vif-mediated degradation of APOBEC3G, and attenuate HIV infectivity, suggesting that the targeted residues are required for normal Vif function (32). Mutation of one or more of the metal binding residues in HCCHp allows competing ligands in the same or another molecule to coordinate zinc. Such mutations would be expected to alter the zinc-bound conformation of Vif and disrupt interactions with the Cul5 E3 ubiquitin ligase necessary for function. Therefore, improper metal binding by Vif-HCCH mutants may provide a mechanism by which these mutations exert their toxic effect.
The data in Figure 7 demonstrate that the interaction between HCCHp and Cul5 is greatly enhanced by zinc binding. In the absence of zinc, HCCHp exists largely in a random-coil conformation (Figure 1), which does not bind tightly to Cul5. Zinc binding to HCCHp stabilizes a conformation that allows optimal binding to Cul5. Other studies have led to the general hypothesis that the interface between Vif and Cul5 is hydrophobic (32, 35, 36). Consistent with this hypothesis, zinc induced aggregation of HCCHp was specifically inhibited by Cul5 (Figure 6), suggesting that aggregation and Cul5 recognition are mediated by a common hydrophobic surface in Vif.
The interface between Vif and Cul5 represents a potential target for the development of antiretroviral compounds. Molecules designed to block the interaction of Vif with the E3 ubiquitin ligase machinery would be expected to increase levels of APOBEC3G, an innate retroviral restriction factor (47–49). Zinc binding to Vif is required for its E3 adaptor protein function. Therefore, removal of zinc from Vif is one way to disrupt its biological function. In support of this idea, a previous study demonstrated that micromolar levels of the chelator N,N,N′,N′-tetrakis-(2-pyridylmethyl) ethylenediamine inhibits Cul5 recruitment and APOBEC3G degradation (50). A second approach would be to disrupt the Vif-Cul5 interface by inhibiting the zinc-induced folding of the HCCH motif or by altering the zinc-bound conformation of the HCCH motif. HCCHp aggregation is induced by zinc binding and is inhibited by Cul5. Based on our results, molecules that disrupt the hydrophobic Vif-Cul5 interface would also be expected to block zinc-induced aggregation, and this could provide a new strategy for identifying inhibitors of the Vif-Cul5 interaction. Given the unique nature of the HCCH motif and the role of zinc binding in stabilizing the Vif(HCCH)-Cul5 interface, future studies should concentrate on the mechanistic and molecular details of this important virus-host interaction.
Supplementary Material
ACKNOWLEDGMENT
We greatly appreciate the human cul5 clone, which was a gift from Dr. Joan W. Conaway (Stowers Institute for Medical Research, Kansas City, Missouri 64110).
Footnotes
This work was supported by an intramural grant to E.L.M. and an NIH grant (GM42025) to R.A.S. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
Abbreviations: APOBEC3G, apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G; CD, circular dichroism; Cul5, Cullin-5; EXAFS, extended X-ray absorption fine structure; Fmoc, 9-fluorenylmethoxycarbonyl; GST, glutathione S-transferase; HEPES, N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; HPLC, high-performance liquid chromatography; IPTG, isopropyl-β-D-thiogalactopyranoside; MOPS, 3-N-morpholino propanesulfonic acid; TCEP, tris-carboxyethyl phosphine; Vif, virion infectivity factor; XAS, Xray absorption spectroscopy.
The H108A and H139A mutant peptides did not undergo zinc-induced aggregation and were not examined by XAS. However, EXAFS data for wild type HCCHp and the H127A mutant (Figure 4) are indicative of His imidazole coordination and strongly suggest that His-108 and His-139 coordinate zinc.
SUPPORTING INFORMATION
CD spectroscopic analysis of human Cul5(1–384) and detailed XAS/EXAFS curve-fitting results for all HCCHp samples. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Emerman M, Malim MH. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 2.Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Strebel K. Virus-host interactions: role of HIV proteins Vif, Tat, and Rev. Aids. 2003;17 Suppl 4:S25–S34. doi: 10.1097/00002030-200317004-00003. [DOI] [PubMed] [Google Scholar]
- 4.Strebel K. HIV accessory genes Vif and Vpu. Adv Pharmacol. 2007;55:199–232. doi: 10.1016/S1054-3589(07)55006-4. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz S, Felber BK, Pavlakis GN. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is Rev-dependent and regulated by splicing. Virology. 1991;183:677–686. doi: 10.1016/0042-6822(91)90996-o. [DOI] [PubMed] [Google Scholar]
- 6.Garrett ED, Tiley LS, Cullen BR. Rev activates expression of the human immunodeficiency virus type 1 vif and vpr gene products. J Virol. 1991;65:1653–1657. doi: 10.1128/jvi.65.3.1653-1657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karczewski MK, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Wu X, Newman M, Shaw GM, Hahn BH, Kappes JC. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borman AM, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 Vif- mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishi M, Nishino Y, Sumiya M, Ohki K, Kimura T, Goto T, Nakai M, Kakinuma M, Ikuta K. Cells surviving infection by human immunodeficiency virus type 1: vif or vpu mutants produce non-infectious or markedly less cytopathic viruses. J Gen Virol. 1992;73(Pt 1):77–87. doi: 10.1099/0022-1317-73-1-77. [DOI] [PubMed] [Google Scholar]
- 12.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV 'A' (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 13.Fisher AG, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo RC, Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 14.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai H, Shibata R, Sakuragi J, Sakuragi S, Kawamura M, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J Virol. 1993;67:1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan L, Peden K. Cell-free transmission of Vif mutants of HIV-1. Virology. 1992;190:19–29. doi: 10.1016/0042-6822(92)91188-z. [DOI] [PubMed] [Google Scholar]
- 18.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 19.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 20.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 21.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 25.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 27.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 28.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 29.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 30.Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- 31.Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc Natl Acad Sci U S A. 2005;102:11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 33.Paul I, Cui J, Maynard EL. Zinc binding to the HCCH motif of HIV-1 virion infectivity factor induces a conformational change that mediates protein-protein interactions. Proc Natl Acad Sci U S A. 2006;103:18475–18480. doi: 10.1073/pnas.0604150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giri K, Maynard EL. Conformational analysis of a peptide approximating the HCCH motif in HIV-1 Vif. Peptide Science. 2009 doi: 10.1002/bip.21209. Accepted. [DOI] [PubMed] [Google Scholar]
- 35.Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349:290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, Yu XF. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J Mol Biol. 2007;373:541–550. doi: 10.1016/j.jmb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 37.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 39.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 40.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt JB, Neece SH, Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- 42.Gatto GJ, Jr, Maynard EL, Guerrerio AL, Geisbrecht BV, Gould SJ, Berg JM. Correlating structure and affinity for PEX5:PTS1 complexes. Biochemistry. 2003;42:1660–1666. doi: 10.1021/bi027034z. [DOI] [PubMed] [Google Scholar]
- 43.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Real-space multiple-scattering calculation and interpretation of x-ray-absorption near-edge structure. Phys. Rev. B. 1998;58:7565–7576. [Google Scholar]
- 44.Wilson CL, Hubbard SJ, Doig AJ. A critical assessment of the secondary structure alpha-helices and their termini in proteins. Protein Eng. 2002;15:545–554. doi: 10.1093/protein/15.7.545. [DOI] [PubMed] [Google Scholar]
- 45.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 46.Van Holde KE, Johnson WC, Jr, Ho PS. Principles of physical biochemistry. New Jersey: Prentice-Hall, Inc; 1998. [Google Scholar]
- 47.Harris RS. Enhancing immunity to HIV through APOBEC. Nat Biotechnol. 2008;26:1089–1090. doi: 10.1038/nbt1008-1089. [DOI] [PubMed] [Google Scholar]
- 48.Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, Gabuzda D. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J Virol. 2007;81:13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nathans R, Cao H, Sharova N, Ali A, Sharkey M, Stranska R, Stevenson M, Rana TM. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Z, Ehrlich E, Luo K, Xiong Y, Yu XF. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. Faseb J. 2007;21:217–222. doi: 10.1096/fj.06-6773com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.