Abstract
For many years, research on a suite of plant defense responses initiated when plants are exposed to general microbial elicitors was underappreciated, for a good reason: There has been no critical experimental demonstration of their importance in mediating plant resistance during pathogen infection. Today, these microbial elicitors are named pathogen/microbe - associated molecular patterns (PAMPs or MAMPs) and the plant responses called PAMP - triggered immunity (PTI). Recent studies provide an elegant explanation for the difficulty of demonstrating the role of PTI in plant disease resistance. It turns out that the important contribution of PTI to disease resistance is masked by pathogen virulence effectors that have evolved to suppress it.
Plants are exposed to myriads of potential microbial pathogens, but the world is still green. Why? Plants possess an innate immune system that efficiently detects and wards off potentially dangerous microbes(1–3). A first layer of this system is based on the amazingly sensitive perception of pathogen/microbe– associated molecular patterns (PAMPs or MAMPs) through pattern recognition receptors (PRR) at the plant’s cell surface (Fig. 1). For example, plants perceive bacterial flagellin through a PRR known as FLS2 (flagellin sensitive 2), a leucine - rich repeat receptor kinase (LRR - RK) located in the plasma membrane. Similarly mammals use through TLR5 to perceive bacterial flagellin, and mount multifaceted downstream immune responses (1, 4). The responses to flagellin and other MAMPs have been called PAMP-triggered immunity (PTI). Successful pathogens produce effectors to inhibit PTI, but plants, in turn, can perceive such effectors through additional receptors, typically nucleotide - binding leucine - rich repeat (NB - LRR) proteins, to mount a second layer of defense called effector-triggered immunity (ETI). Although the importance of ETI (formerly known as gene - for - gene resistance) in plant immunity is well established, only recently have we begun to appreciate a fundamental role of PTI in mediating plant - microbe interactions. In this Perspective, we highlight recent literature on PRR signaling and the ability of microbial pathogens to suppress PTI as a key virulence strategy.
Fig. 1.
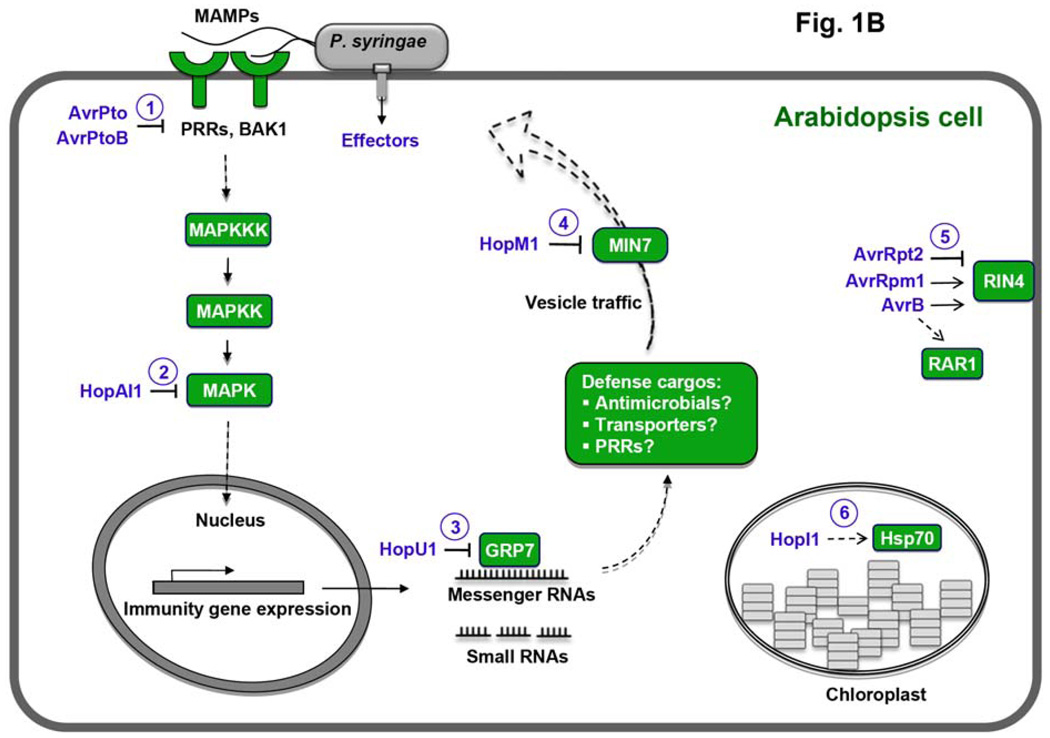
Concept of activation and suppression of PTI during pathogen infection. (A) An Arabidopsis plant showing disease symptoms (in the foreground; natural size) after infection by P. syringae bacteria (electron microscopy image in the background; magnification, 10,000x). (B) A conceptual diagram of PRR signaling and action of several P. syringae effectors for which the plant targets and immune suppression function have been characterized. Green and purple colors indicate plant targets and P. syringae effectors, respectively. Numbers in circles denote six steps targeted by effectors: MAMP perception (PRRs), the MAPK cascade (MPK3 and MPK6), RNA metabolism (GRP7), vesicle traffic (MIN7), regulators of PTI (RIN4 and RAR1), and chloroplast function (Hsp70) (22).
Perception of microbes through pattern recognition receptors
A hallmark of PRRs is their sensitivity and specificity: Plants possessing the appropriate PRRs perceive a specific MAMP at subnanomolar concentrations, whereas plants lacking the PRRs are completely blind to it. A given MAMP is recognized through a specific conserved epitope, such as the stretch of 22 amino acids (flg22) in the N - terminus of flagellin. Can pathogens avoid perception by the PRRs? It appears they can, but at a cost. Introduction of mutations into flagellin that make the molecule unrecognizable by FLS2 also render the microbe motionless and reduce its virulence (5). Thus, the specificity of the PRR appears to be focused exactly on a highly conserved domain of the MAMP that is functionally important to the microbe.
FLS2 homologues exist in all higher plants for which genomic information is available (1), and the rice homologue is functionally active as a flagellin receptor (6). Hence, flagellin perception through FLS2 homologues is evolutionarily old and conserved. Another well - characterized PRR of Arabidopsis, EFR (EF - TU receptor), perceives bacterial EF - Tu (elongation factor thermo unstable). Perception of this MAMP seems to be confined to the Brassicaceae, and is not found in other dicots or monocots, suggesting that EF - Tu perception is evolutionarily young. However, intriguingly, all plant genomes so far sequenced to contain homologues of the EFR gene with a comparable LRR - structure. The rice genome encodes about 40 such homologues (1). One of them, found in some rice cultivars, is the disease resistance gene Xa21, the protein product of which appears to recognize a quorum - sensing molecule of the rice pathogen Xanthomonas oryzae; thus, although the EFR - type PRRs show elements of conserved sequences among plants, they appear to recognize different MAMPs in different plant families (1).
One of the first steps in signaling of the FLS2 receptor is its interaction with BAK1, an LRR - RK known as the BRI1 - associated kinase (7, 9). This comes as a surprise, since BAK1 has previously been known, as its name says, as a co - receptor of the plant hormone brassinosteroid receptor BRI1. How can one and the same co - receptor function both in defense signaling and hormonal signaling? BRI1 and BAK1 are phosphorylated upon activation in hormonal signaling (9). Does this also occur in the FLS2 - BAK1 interaction, and does differential phosphorylation contribute to specificity of downstream responses? Is there competition between PTI and the brassinosteroid response for the co - receptor? What is the function of the four BAK1 homologues, the somatic embryogenesis - related kinases (SERKs), in Arabidopsis? Dysfunction of two of these, bak1 and serk4, sends Arabidopsis seedlings to death with symptoms of the hypersensitive response, a hallmark immune response of ETI (7, 9, 10). Despite the specificity and sensitivity of MAMP perception by PRRs, it has taken the scientific community a long time to accept that such systems could support plant disease resistance during pathogen infection. Opinion began to shift with the discovery that mutations in the fls2 receptor left Arabidopsis plants unusually susceptible to the bacterial pathogen Pseudomonas syringae (11). Even more convincing were observations that pathogens actively deploy virulence factors as a virulence strategy to suppress PTI.
Suppression of PTI by pathogen effectors
A variety of bacterial virulence factors, including the phytotoxin coronatine, extracellular polysaccharides, and proteinaceous effectors secreted through the type III secretion system (TTSS), suppress PTI (1–3). Most spectacularly, two secreted effectors, AvrPto and AvrPtoB (from P. syringae strain DC3000), physically interact with the kinase domains of FLS2, EFR, or BAK1 (12–14). Such physical interactions inhibit the kinase activity of PRRs (12) or interfere with formation of FLS2 - BAK1 complexes (13). Whereas AvrPto seems to have no homologues, AvrPtoB contains a C - terminal domain that resembles E3 ubiquitin ligase; ubiquitination by this domain initiates degradation of a tomato kinase (Fen) that is part of a unique and presumably ancient ETI pathway (15). The same domain also initiates degradation of PRRs, and thus its more important role may be in defeating PTI (14, 16). The ability of AvrPto and AvrPtoB to derail PRRs provides a satisfying explanation for previous discoveries that these effectors could suppress a variety of responses downstream of PTI, including callose deposition, activation of kinase cascades, and expression of MAMP - responsive proteins and small RNAs (17–19). Not all bacteria, indeed not even all strains of P. syringae, express AvrPto and AvrPtoB, suggesting that other strategies exist to inhibit PRR signaling. Indeed, HopAI1, present in many but not all P. syringae strains, is a phosphothreonine lyase that dephosphorylates MPK3 and MPK6 to terminate PRR signaling (20). Interestingly, another member of this effector family dephosphorylates kinases involved in mammalian innate immunity (21), showing that pathogens can apply the same mechanism of host immune modulation to both plants and mammals.
Pathogen effectors target more than PRRs or the MAPK cascade to suppress PTI. These effectors also attack processes directly downstream of PRR signaling and other consequent events (Fig. 1; 22). For example, the P. syringae effector HopU1 modifies several Arabidopsis RNA - binding proteins, including GRP7, by ADP - ribosylation. HopM1, another P. syringae effector, triggers degradation of the Arabidopsis MIN7 protein, which is a member of the ARF family of guanine nucleotide exchange factors involved in vesicle trafficking. Plants lacking either grp7 or min7 are abnormally susceptible to bacterial infection, implicating RNA metabolism and vesicle trafficking as part of the plant’s response to pathogens (22). The P. syringae effector HopI1 resides in the chloroplast, where its action (presumably through interaction with Hsp70 chaperones) suppresses accumulation of salicylic acid, a plant hormone key to defense responses; three other P. syringae effectors, AvrRpm1, AvrB, and AvrRpt2, interact with or modify proteins such as RIN4 and RAR1 that regulate pathogen- and effector-triggered immunity (22).
The many examples of physical associations between pathogen effectors and regulators of host immune responses have spurred a trendy notion that pathogen effectors can be used as molecular probes to identify unknown components of the plant innate immune system, including those involved in PTI. This is an exciting time for researchers in this area, especially since recent functional genomics studies suggest that bacteria, fungi, oomycetes, and nematodes that are pathogenic to plants could collectively deliver hundreds of virulence effectors into host cells. Identification of the plant targets of this vast repertoire of pathogen effectors will likely yield many new discoveries that could greatly impact our understanding of plant immunity, pathogenesis, and plant biology for years to come.
Concluding remarks
The past few years have witnessed significant paradigm - shifting advances in the field of plant - microbe interactions. Contributing to these advances are experimental demonstrations of a functional role of PRRs in plant disease resistance and the discovery that many bacterial virulence factors are involved in suppressing PRR signaling and PTI - associated immune responses. Nonetheless a major limitation of current research is the heavy reliance on information derived from essentially a single pathosystem, the interaction between the plant Arabidopsis and the bacterium P. syringae (Fig. 1). Thus our current understanding of plant-pathogen interactions is of a pioneering, but preliminary nature. It remains to be seen whether the conceptual framework emerging from the study of this pathosystem will translate to other plant - microbe interactions. The incredibly diverse interactions between plants and microbes suggest that other systems will involve many novel mechanisms, which are likely to refine or even challenge the current models. However, because all pathogens carry MAMPs that may be recognized by plants, yet all plants are still susceptible to virulent pathogens, it is certain that activation and suppression of PTI is a general principle underpinning plant - microbe interactions.
References and Notes
- 1.Boller T, Felix G. Ann. Rev. Plant Biol. 2009;60:379. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Jones JDG, Dangl JL. Nature. 2006;444:323. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.Clay NK, Adio AM, Denoux C, Ausubel FM. Science. 2009;323:95. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naito K, et al. Mol. Plant - Microbe Interact. 2008;21:1165. doi: 10.1094/MPMI-21-9-1165. [DOI] [PubMed] [Google Scholar]
- 6.Takai R, et al. Mol. Plant - Microbe Interact. 2008;21:1635. doi: 10.1094/MPMI-21-12-1635. [DOI] [PubMed] [Google Scholar]
- 7.Chinchilla D, et al. Nature. 2007;448:497. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 8.Heese A, et al. Proc. Nat. Acad. Sci. U.S.A. 2007;104:12217. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XF, et al. Dev. Cell. 2008;15:220. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 10.He K, et al. Curr. Biol. 2007;17:1109. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Zipfel C, et al. Nature. 2004;428:764. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 12.Xiang T, et al. Curr. Biol. 2008;18:74. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 13.Shan L, et al. Cell Host Microbe. 2008;4:17. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Göhre V, et al. Curr. Biol. 2008;231:824. [Google Scholar]
- 15.Rosebrock TR, et al. Nature. 2007;448:370. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimenez-Ibanez S. Curr. Biol. 2009;19:423. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 17.Hauck P, Thilmony R, He SY. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8577. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He P, et al. Cell. 2006;125:563. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 19.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Science. 2008;321:964. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, et al. Cell Host Microbe. 2007;1:175. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. Science. 2007;315:1000. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 22.Block A, Li G, Fu ZQ, Alfano JR. Curr. Opin. Plant Biol. 2008;11:396. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Research in the authors’ laboratories is supported by grants from Swiss National Science foundation to T.B. and U.S. NIH (R01AI060761), DOE (DE - FG02 - 91ER20021), NSF, USDA to S.Y.H.