Abstract
The recent release of the domestic dog genome provides us with an ideal opportunity to investigate dog-specific genomic features. In this study, we performed a systematic analysis of CpG islands (CGIs), which are often considered gene markers, in the dog genome. Relative to the human and mouse genomes, the dog genome has a remarkably large number of CGIs and high CGI density, which is contributed by its noncoding sequences. Surprisingly, the dog genome has fewer CGIs associated with the promoter regions of genes than the human or the mouse. Further examination of functional features of dog-human-mouse homologous genes suggests that the dog might have undergone a faster erosion rate of promoter-associated CGIs than the human or mouse. Some genetic or genomic factors such as local recombination rate and karyotype may be related to the unique dog CGI features.
Keywords: CpG islands, Dog, Promoter, Homologous genes, Domestication, Gene Ontology, Genome evolution, Essential genes, Housekeeping genes
Introduction
The dog has long been a subject of scientific curiosity because of its great diversity in both morphological (e.g., size, shape, coat color and texture) and behavioral traits [1, 2]. Although the dog genome is largely similar to the human genome [3, 4], it has much greater variance among its individual breeds [5]. This unique position in the mammalian phylogeny makes the dog genome suitable for evolutionary and comparative genomics studies [6, 7]. Moreover, the dog represents an important model organism because it has a large catalog of disease syndromes that are more similar to the human than any other laboratory or domestic species [8, 9]. Because of these important features, sequencing the dog genome (Canis familiaris) has been a high priority and its genome was recently completed [7]. This provides us an unprecedented opportunity to examine dog-specific features at the genome-wide level and compare it to other model genomes such as the human and mouse. As an example, a comparative genomics study suggested the euchromatic portion of the dog genome being ~18% smaller than the human genome and 6% smaller than the mouse genome, which could be explained by a lower rate of repeat insertions rather than a higher rate of deletions in the dog genome [8].
With more than twenty mammalian genomes having been sequenced thus far, a fundamental question is how the genomes have changed and what genetic factors have impacted sequence composition, size, function and complexity during the course of evolution. For instance, CpG dinucleotides are largely under-represented in most mammalian genomes, occurring only ~20–25% of their expected frequency overall [10–12]. This deficit of CpG dinucleotides is largely attributed to the high rate of deamination of methylated CpGs, which in turn accounts for approximately 80% of the total CpGs in mammalian genomes [13, 14]. Conversely, CpG islands (CGIs), which are clusters of CpGs in GC-rich regions, have nearly the expected frequency of CpGs [12]. CGIs are frequently located in the 5’ region of the genes and are considered as gene markers [15, 16]. Recent genome-wide investigation revealed that promoter-associated CGIs overall remained unmethylated [17], although a sizable fraction of them might be fully methylated in normal cells [17–20]. Methylation changes in promoter-associated CGIs have been found to cause transcriptional silencing and disruption of gene function [21]. In particular, many recent studies revealed that aberrant hypermethylation in promoter-associated CGIs of tumor suppressor genes may cause tumorigenesis [22]. Although CGIs have been used to estimate the number of genes in a genome [23, 24], our recent study revealed large variation on the number of CGIs and their density in mammalian genomes with comparable gene number [12]. Interestingly, the dog genome had the largest number of CGIs and the highest CGI density among the ten mammalian genomes we studied. The number of dog CGIs was nearly 3 times that in rodent genomes [12]. It has been commonly thought that rodents might have underwent a stronger process of CpG erosion to TpGs/CpAs by de novo methylation and that rodent CGIs had weaker selective constraint than humans [21, 23, 25]. However, it remains largely unknown whether the dog genome has a relative gain of CGIs to other mammalian genomes during evolution or it has still been under similar process of erosion.
To better understand the genome features of the dog and their relationship with the morphological and behavior traits, we performed a systematic investigation of CGIs in the dog genome. We examined the CGIs and their distribution in different genomic regions including promoter, 3’-, genic, intronic and intergenic regions and further compared them with those in the human and mouse genomes. To understand the functional implications of CGIs, we examined promoter-associated CGIs in the genes with different expression level (e.g., housekeeping vs. tissue specific genes) or functional importance (e.g., essential genes). We also examined the functional bias of genes that have likely lost CGIs in the dog lineage. This study provides detailed information of CGIs and their functional features in the dog genome and has important implications for mammalian genome evolution and gene function.
Results
Distribution and features of CGIs in the dog genome
We used Takai and Jones’ (2002) algorithm [26] to identify CGIs in the dog, human and mouse genomes (see Materials and Methods). Here, we first describe the distribution and features of CGIs in the dog genome. There were 58,327 CGIs in the dog genome, with an average length of 1102 bp, average GC content of 62.2%, and average ObsCpG/ExpCpG ratio of 0.753 (Table 1). Here, ObsCpG/ExpCpG ratio was measured by the ratio of the observed CpG dinucleotides over the expected CpGs in a sequence [16]. These dog CGIs had a total length of 64.3 Mb and accounted for 2.8% of the dog genome sequence. On average, we observed 25.2 CGIs per Mb in the dog genome; however, the standard deviation was high (±40.5 CGIs/Mb). When we examined CGIs in the non-repeat portion of the dog genome, we still found 53,102 CGIs, which accounted for 3.7% of the non-repeat portion of the dog genome (Table 1). This finding supports the assertion that Takai and Jones’s algorithm can effectively exclude the short repeats, especially Alu repeats [26]. Correspondingly, CGI density in the non-repeat portion of the dog genome (37.9 /Mb) is much higher than that (25.2 /Mb) of the whole genome.
Table 1.
Overview of CGIs in the whole genome and non-repeat regions
| Species | Number of CGIs |
Length (bp) |
GC content (%) |
ObsCpG/ ExpCpG |
CGIs covered (Mb) |
Genome size (Gb) |
CGI density/Mb (S.D.) |
|---|---|---|---|---|---|---|---|
| Whole genome | |||||||
| Dog | 58,327 | 1102 | 62.2 | 0.753 | 64.3 (2.8%a) | 2.31 | 25.2±40.5 |
| Human | 37,729 | 1090 | 62.0 | 0.743 | 41.1 (1.4%) | 2.85 | 13.2±16.8 |
| Mouse | 21,326 | 1044 | 60.9 | 0.752 | 22.2 (0.9%) | 2.61 | 8.2±8.4 |
| Non-repeat region | |||||||
| Dog | 53,102 | 975 | 59.0 | 0.791 | 51.8 (3.7%) | 1.40 | 37.9 |
| Human | 28,380 | 1098 | 59.4 | 0.798 | 31.1 (2.0%) | 1.52 | 18.7 |
| Mouse | 17,109 | 1048 | 58.7 | 0.789 | 18.0 (1.2%) | 1.52 | 11.3 |
Proportion of the total length of CGIs in the whole genome sequence.
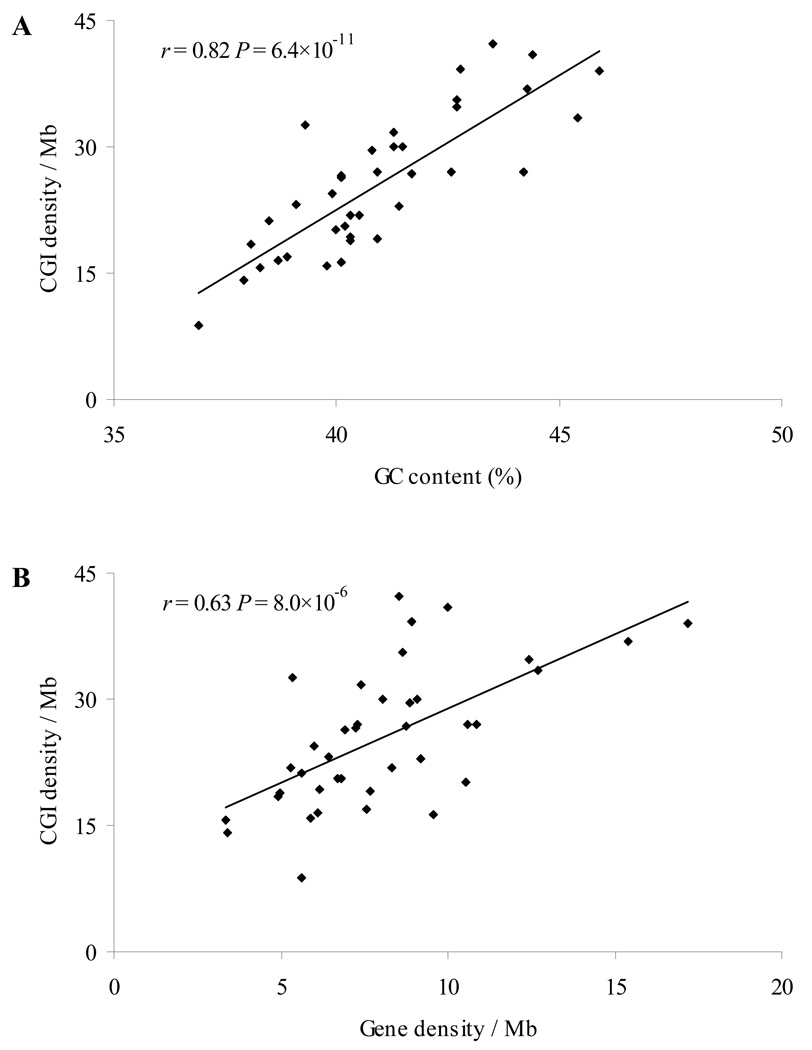
We further examined the distribution and features of CGIs on each dog chromosome. The results are shown in Supplementary Table S1. The number of CGIs and CGI density varied greatly. Chromosome 1, the largest autosome, had the largest number of CGIs (3636) while chromosome 32 had the smallest number of CGIs (342). Moreover, the highest CGI density was found on chromosome 28 (42.2 CGIs/Mb), which was 4.8 times the lowest CGI density found on chromosome 32 (8.8 CGIs/Mb). As expected, we observed a trend that larger chromosomes had more CGIs (linear regression, r= 0.76, P= 1.2 ×10−8, Supplementary Fig. S1A). The number of CGIs in a chromosome was significantly correlated with the number of genes in the chromosome (r= 0.86, P= 1.9 ×10−12, Supplementary Fig. S1B), supporting the notion that CGIs can function as gene markers. Moreover, CGI density in a dog chromosome was highly correlated with genomic factors such as GC content (r= 0.82, P= 6.4×10−11, Fig. 1A) and gene density (r= 0.63, P= 8.0×10−6, Fig. 1B), indicating that CGIs depend on both local genomic features and gene number.
Fig. 1.
Correlation between CGI density and genomic features on each chromosome in the dog genome. (A) CGI density versus GC content (%). (B) CGI density versus gene density (/Mb).
Comparison of CGIs in the dog, human and mouse genomes
The characteristics of CGIs in the dog genome were consistently stronger than those in the human and mouse genomes including average length (dog: 1102 bp; human: 1090 bp; and mouse: 1044 bp), average GC content (dog: 62.2%; human: 62.0%; and mouse: 60.9%) and average ObsCpG/ExpCpG ratio (dog: 0.753; human: 0.743; and mouse: 0.752). Dog CGIs covered a larger portion (2.8%) of the dog genome than human and mouse CGIs (human: 1.4% and mouse: 0.9%) (Table 1). Interestingly, when we compared CGIs in the non-repeat portion of the three genomes, the characteristics (length, GC content and ObsCpG/ExpCpG ratio) of dog CGIs became weaker than the corresponding ones of human CGIs (Table 1), even though the extent of decreasing the number of dog CGIs (58,327 to 53,102, 9.0%) is weaker than that of human CGIs (37,729 to 28,380, 24.8%) or mouse CGIs (21,326 to 17,109, 19.8%). Our further analysis indicated that short repeats such as SINEs may be more likely to be part of or more closely linked to the CGIs identified in the whole dog genome than in the human and mouse genomes. When repeats were excluded, those regions might still meet the criteria of CGIs – in the case of repeats closely linking to CGIs; or the original CGIs were broken into two – in the case of repeats inside of long CGIs; however, their characteristics, especially the length, weakened.
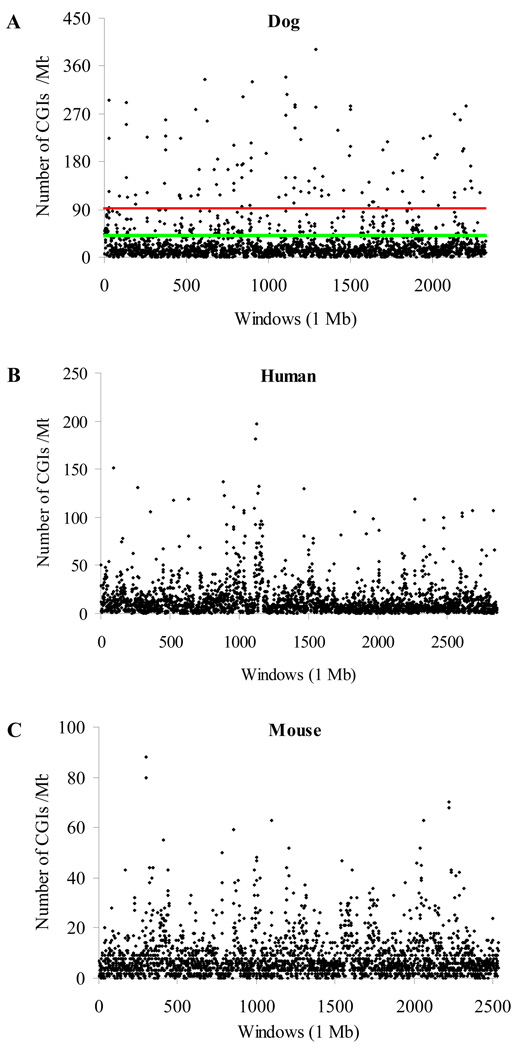
CGI density, measured by the number of CGIs per Mb in a sequence, is 25.2 per Mb in the dog genome. This is nearly twice that in the human genome (13.2 CGIs/Mb) and more than three times that in the mouse genome (8.2 CGIs/Mb). Figure 2 displays CGI density in each 1-Mb window in these three genomes. It reveals a strong variance of CGI density across each genome. For example, in the dog genome, the highest CGI density was 392 per Mb while there were 29 windows in which no CGI was identified. In the human genome, 99% of the windows had a CGI density that is equal to or below 92 per Mb; however, we observed 110 windows (4.7%) having CGI density higher than this threshold value in the dog genome. In the mouse genome, this threshold value decreased to 42 while we observed 313 dog windows (13.5%) above it (Fig. 2). Moreover, when CGI density measured by 1-Mb window was higher than 10 per Mb (e.g., 11–20 and 21–30 CGIs/Mb, Supplementary Fig. S2), we found that the proportion of dog CGIs was consistently higher than that of human or mouse CGIs. This is consistent with the previous finding that the dog has more fractions of windows with GC content over 50% [7].
Fig. 2.
Distribution of CGI density in 1-Mb window in the dog, human and mouse genomes. (A) The dog genome. (B) The human genome. (C) The mouse genome. The scale on the Y-axis is different. In Fig. 2A, a line indicates that 99% of the windows in the human (red line) or mouse genome (green line) had CGI density equal to or lower than that threshold value.
Distribution of CGIs in different genomic regions
As gene markers, CGIs that are associated with the promoter regions (i.e., promoter-associated CGIs) have been under greater scrutiny than other CGIs [15, 21, 27]. Here we first compared the features of promoter-associated CGIs in the dog, human and mouse genomes. The numbers of promoter-associated CGIs were 9825 (dog), 12,917 (human), and 10,595 (mouse), respectively. Interestingly, the difference in the number of promoter-associated CGIs is much smaller than that in the total number of CGIs among these three genomes. Surprisingly, the number of promoter-associated dog CGIs is even smaller than that of human or mouse CGIs. This might be due to the relatively poor gene annotations in the dog genome compared to the human or mouse genome. However, our examination of the 10,196 dog-human-mouse homologous genes found a lower frequency of promoter-associated CGIs in the dog genes than the human and mouse genes. Thus, the low prevalence of promoter-associated CGIs in the dog genome is unlikely caused by poor gene annotation (see results below).
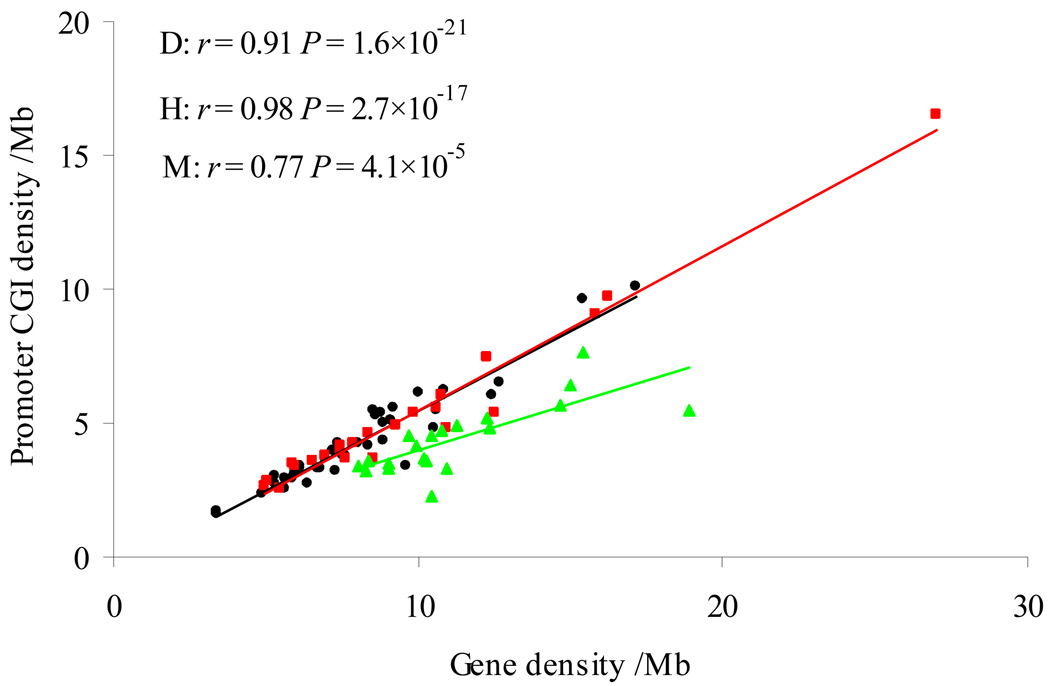
Moreover, the density of promoter-associated CGIs was very close between the dog (160.7 per Mb) and the human (156.8 per Mb), but both were higher than the mouse (118.4 per Mb) (Table 2). We found a highly significant correlation between the promoter-associated CGI density and gene density among the chromosomes in each of the three genomes. The correlation coefficient in the dog genome (r= 0.91, P= 1.6×10−21) is close to that in the human genome (r= 0.98, P= 2.7×10−17), but again both were higher than that in the mouse genome (r= 0.77, P= 4.1×10−5) (Fig. 3). While the lower CGI density in the mouse genome could be explained by its faster erosion rate than in the human genome [21, 25], the observations above indicated that the large difference in the number of CGIs among these three genomes was not because of the promoter-associated CGIs.
Table 2.
CGI density (/Mb) and ratio in genomic regions of three species
| Genome | Promoter a | 3’-region a | Genic | Intronic | Intergenic | |
|---|---|---|---|---|---|---|
| CGI density | ||||||
| Dog | 25.2 | 160.7 | 75.1 | 21.1 | 11.9 | 17.2 |
| Human | 13.2 | 156.8 | 43.1 | 10.6 | 5.9 | 5.2 |
| Mouse | 8.2 | 118.4 | 22.3 | 5.4 | 2.0 | 2.7 |
| Ratio | ||||||
| Dog/human | 1.9 | 1.0 | 1.7 | 2.0 | 2.0 | 3.3 |
| Dog/mouse | 3.1 | 1.4 | 3.4 | 3.9 | 6.0 | 6.4 |
Because the promoter and 3’ region is short, in this comparison, a CGI completely located within the promoter or 3' region was counted as 1 and partially located in the promoter or 3' region was counted as 0.5.
Fig. 3.
Correlation between promoter-associated CGI density and gene density in the dog (black), human (red), and mouse (green) genomes. Each dot represents one chromosome.
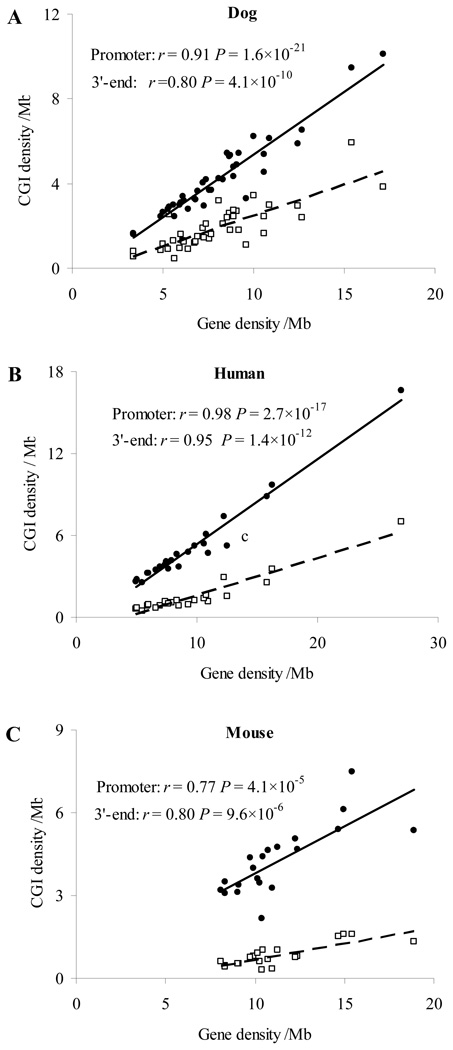
We next examined the distribution of CGIs in other genomic regions, including 3’-, genic, intronic and intergenic regions. CGI density in these regions, especially in the intronic and intergenic regions, was remarkably lower than that in the promoter regions (Table 2), supporting CGIs being an important gene feature in mammalian genomes. The CGI density in the 3’ region is moderately high, for example, 75.1 CGIs/Mb in the dog genome. Similar to promoter-associated CGIs, we found a significant correlation between 3’-region CGI density and gene density in each chromosome in all the three genomes (Fig. 4). In the 3’ region, the ratio of CGI density between the dog and human (1.7) or between the dog and mouse (3.4) was close to that in the whole genome (dog/human: 1.9; dog/mouse: 3.1, Table 2). However, the corresponding ratios in the noncoding regions (intergenic and intronic regions) were overall remarkably higher than those corresponding ratios at the whole genome level. For example, in the intergenic regions, the dog/human ratio was 3.3 and the dog/mouse ratio was 6.4 (Table 2). Our further comparison of the number of CGIs in different genomic regions among the three genomes showed the same pattern (Supplementary Fig. S3). For example, in the intergenic regions, the number of dog CGIs (28,324) was 6.4 times that of mouse CGIs (4401) and dog CGI density (17.2 per Mb) was also 6.4 times that of mouse CGI density (2.7 per Mb). This comparative analysis clearly indicated that the larger number of CGIs and higher CGI density in the dog genome than other mammalian genomes is mainly attributed to its abundance of CGIs in the noncoding regions, which are generally under weak or no selection.
Fig. 4.
Comparison of CGI density in the promoter and 3’- regions of genes in the dog, human and mouse genomes. (A) The dog genes. (B) The human genes. (C) The mouse genes. The scale is different on the Y-axis. Black dots: promoter regions; white dots: 3’ regions.
Comparison of CGIs in dog-human-mouse homologous genes
Because the level of gene annotations has varied among the three genomes [7, 28–30], we compared the CGIs in dog-human-mouse homologous genes. Homologous genes in general have been more reliably annotated and tend to be conserved. Thus, a comparative analysis of their gene features such as CGIs should provide us important insights on sequence, gene and genome evolution. We extracted a total of 10,196 dog-human-mouse homolog genes based on the NCBI HomoloGene database. Among them, 6418 genes (62.9%) had CGIs either present or absent in their promoter regions in all three genomes. Specifically, in a comparison of dog and human homologues, we found 5331 genes (52.3%) having promoter-associated CGIs present in both the genomes, 2013 genes (19.7%) without having promoter-associated CGIs in either genome, 717 genes (7.0%) having promoter-associated CGIs in the dog genome but not in the human genome, and 2135 genes (20.9%) having promoter-associated CGIs in the human genome but not in the dog genome (Supplementary Fig. S4). We named these four groups as D+H+, D-H-, D+H-, and D-H+, respectively. Here D+, H+, D- and H- denote the presence (+) or absence (−) of promoter-associated CGIs in the dog (D) or the human (H) genome. The number of D-H+ (2135) is three times that of D+H- (717). This implies that, although the dog genome has many more CGIs than the human genome, dog genes might be more vulnerable to lose CGIs in their promoter regions.
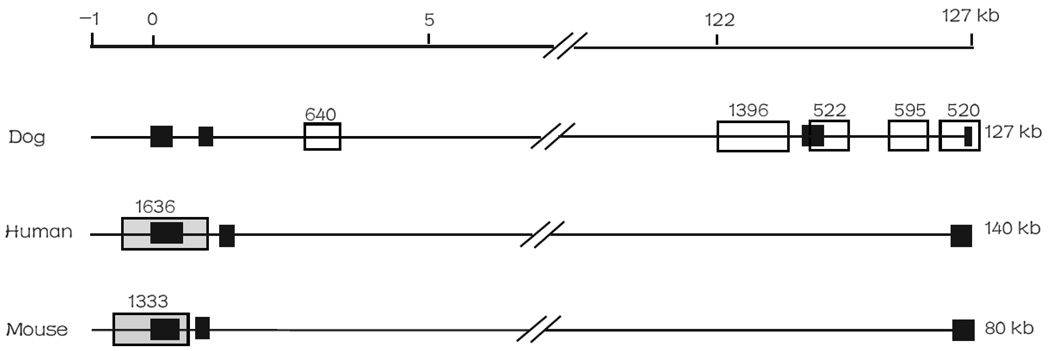
Here, we illustrated the opposite features of CGIs in the promoter versus other genomic regions by one example. We recently analyzed DTNBP1 (dystrobrevin-binding protein 1 gene) [31], one of the most studied and promising schizophrenia susceptibility genes. There was one CGI in the promoter region of the human and mouse DTNBP1, but none was detected in the promoter region of the dog DTNBP1 (Fig. 5). No unknown nucleotides “Ns” were found in these promoter regions, suggesting this difference is not because of low quality of the sequences. In the dog genic regions, there were five CGIs, three of which were completely located in its intronic regions and the other two were linked to its exons. In contrast, there was no CGI found in the genic regions of human or mouse DTNBP1 (Fig. 5).
Fig. 5.
Distribution of CGI(s) in dog, human and mouse DTNBP1 genes. On the top line, “0” represents the transcript start site. Grey box: promoter-associated CGI, white box: genic CGI, and black box: exons.
We next separated the dog and human homologous genes into different groups by their gene expression level or essential function (essential and non-essential genes) and then compared the prevalence of promoter-associated CGIs in these groups. The dog genes consistently showed a lesser presence of promoter-associated CGIs than human or mouse genes, regardless of essential genes or non-essential genes (Table 3) or housekeeping, widely-expressed, moderately-expressed, or narrowly expressed genes (Supplementary Table S2). For example, 62.7% of dog essential genes had promoter-associated CGIs; this compared to the 79.7% of human essential genes or 72.5% of mouse essential genes. Similarly, for the housekeeping genes, the presence of promoter-associated CGIs was 68.7% (dog), 86.1% (human) and 82.2% (mouse), respectively. Interestingly, it seems that the difference in the presence of promoter-associated CGIs between dog and human genes or between dog and mouse genes became greater when gene function became more important or gene expression level increased. For example, the frequency difference in the dog and human essential genes was 17.0% (79.7 – 62.7%), compared to the 9.7% (58.6 – 48.9%) in the dog and human non-essential genes (Table 3). Similarly, the frequency differences between the dog and human genes were 17.4% (housekeeping genes), 16.9% (widely expressed genes), 13.5% (moderately expressed genes), and 9.2% (narrowly expressed genes), respectively (Supplementary Table S2). Previous studies indicated that the rodent lineage had been under a faster erosion rate of CGIs [21, 23, 25]. The results here seem to suggest an even faster erosion rate of CGIs in the promoter regions of the dog genes, at least of these homologous genes.
Table 3.
Comparison of CpG islands in essential and non-essential genes
| Homologous genes |
Essential genes |
Non-essential genes |
||||
|---|---|---|---|---|---|---|
| Total | CGI+a (%) | Total | CGI+ a (%) | Total | CGI+a (%) | |
| Dog | 10,196 | 6048 (59.3) | 1263 | 792 (62.7) | 1049 | 513 (48.9) |
| Human | 10,196 | 7466 (73.2) | 1263 | 1007 (79.7) | 1049 | 615 (58.6) |
| Mouse | 10,196 | 6895 (67.6) | 1263 | 916 (72.5) | 1049 | 553 (52.7) |
For comparison purpose, only those genes that are homologous in the dog, human and mouse genomes were used.
Number of genes having promoter-associated CGIs.
Overrepresentation of Gene Ontology (GO) terms in D- genes
We examined functional biases of dog genes that have absence of CGIs in their promoter regions (D- genes as annotated above) by using GO terms. We first compared the GO terms that were annotated for the D+ genes (dog genes with promoter-associated CGIs) with those for the D- genes (dog genes without promoter-associated CGIs). We found 13 GO terms that are statistically enriched in the D- genes: receptor activity, signal transduction, plasma membrane, receptor binding, calcium ion binding, ion transport, peptidase activity, proteinaceous extracellular matrix, actin binding, lipid metabolic process, ion channel activity, protein kinase activity and phosphoprotein phosphatase activity (Supplementary Table S3).
Next, we examined functional biases of D- genes when their human homologs had CGIs, that is, GO terms that were overrepresented in the D-H+ genes relative to D+H+ genes. The results may imply which kinds of genes might have lost CGIs in their promoter regions during the course of dog lineage evolution. Table 4 shows 12 GO terms that are significantly enriched in the D-H+. Interestingly, 10 of the 12 GO terms were also detected in the D-versus D+ enrichment test (see above); they were: signal transduction, receptor activity, protein kinase activity, phosphoprotein phosphatase activity, ion transport, plasma membrane, calcium ion binding, ion channel activity, receptor binding and proteinaceous extracellular matrix. The substantial overlap of the enriched GO terms in the D- and D-H+ genes suggests that these genes tend to lose their promoter-associated CGIs in the dog lineage.
Table 4.
GO terms that are significantly overrepresented in the D-H+ genes compared to D+H+ genes
| GO code | GO term description a | D+ H+ (%) | D− H+ (%) | P-value b |
|---|---|---|---|---|
| GO:0007165 | B: signal transduction | 1171 (22.0) | 710 (33.3) | 4.3×10−24 |
| GO:0004872 | M: receptor activity | 483 (9.1) | 333 (15.6) | 4.0×10−16 |
| GO:0004672 | M: protein kinase activity | 385 (7.2) | 240 (11.3) | 1.9×10−8 |
| GO:0004721 | M: phosphoprotein phosphatase activity | 90 (1.7) | 79 (3.7) | 2.0×10−7 |
| GO:0006811 | B: ion transport | 395 (7.4) | 238 (11.2) | 2.1×10−7 |
| GO:0006464 | B: protein modification process | 899 (16.9) | 459 (21.5) | 3.2×10−6 |
| GO:0005886 | C: plasma membrane | 547 (10.3) | 299 (14.0) | 4.8×10−6 |
| GO:0005509 | M: calcium ion binding | 358 (6.7) | 210 (9.8) | 5.4×10−6 |
| GO:0005216 | M: ion channel activity | 204 (3.8) | 123 (5.8) | 2.9×10−4 |
| GO:0005102 | M: receptor binding | 170 (3.2) | 102 (4.8) | 1.2×10−3 |
| GO:0005578 | C: proteinaceous extracellular matrix | 87 (1.6) | 60 (2.8) | 1.3×10−3 |
| GO:0005622 | C: intracellular | 1050 (19.7) | 483 (22.6) | 5.1×10−3 |
Gene ontology organizing principles: Cellular component (C), biological process (B) and molecular function (M). Only those GO terms at the fourth level or lower were examined.
P values were calculated by χ2 test for 2×2 contingency table and only the P-values < 0.01 were used.
Discussion
In this study, we found a large number of CGIs and high CGI density in the dog genome but fewer CGIs associated with the promoter regions of dog genes. Several factors may influence the results. First, there are abundant repeats in mammalian genomes. One may wonder whether repeats have a major contribution to the large number of CGIs and their stronger characteristics in the dog genome, as some repetitive elements, such as Alu in primates, B1 in rodents, and SINEC in carnivores, have high GC content and might be potentially identified as CGIs [6, 26, 32]. Takai and Jones’ algorithm was developed to largely exclude the interference of Alu and SINEs in the human and mouse genomes [26]. The dog genome has some lineage-specific repeats such as SINEC_Cf subfamily (e.g., SINEC1A_CF, SINEC1B1_CF and SINEC1C1_CF, Repbase release 13.09) [6] and SINEs were found to be a major source of canine genomic diversity [33]. In this study, we examined the CGIs in non-repeat portions of the dog, human and mouse genomes. The CGI features (more CGIs in the dog genome) were essentially the same as what observed in the whole genomes (Table 1). Therefore, repeats should have limited influence on the observed large number of CGIs and high CGI density in the dog genome.
Second, quality of dog sequences may not be as good as the human or mouse sequences. Uncertain nucleotides (marked as “Ns”) may present in the promoter sequences. To assess whether such gaps in the promoter regions might have influenced our results, we examined the possible gap (“Ns”) in each 2-kb sequence that was defined as the promoter region of each gene. Among the 18,175 dog genes, 2469 had at least one N in the 2-kb promoter sequences and 2084 had >20 Ns (1% of the 2-kb sequence). Those 2084 genes had an average GC content 61.2% and ObsCpG/ExpCpG ratio 0.673 in their 2-kb promoter regions. Thus, the CGI searching algorithm seemed to work well with the uncertain nucleotides in the sequences. These genes had high frequency of CGIs associated with their promoter regions. For example, 84.6% of those 2084 genes had promoter-associated CGIs (Supplementary Table S4). In our comparison of dog-human-mouse homologous genes, the same conclusion of fewer promoter-associated CGIs in dog genes remained regardless whether these genes were excluded or not. For example, after excluding those genes with gaps >20 “Ns”, we had 8870 dog-human-mouse homologous genes. For those 8870 genes, we still observed fewer genes having promoter-associated CGIs in the dog genome (4933, 55.6%) than in the human (6311, 71.1%) or mouse (5836, 65.8%) genome (Supplementary Table S5).
Third, it is possible that the large number of CGIs and high CGI density in intergenic regions is partially attributed to the RNA genes that are not annotated as protein-coding genes. We extracted 2935 dog RNA genes from the Ensembl database (http://www.ensembl.org/), which has better annotations for RNA genes. These RNA genes have an average length of only 130 bp and have an average GC content of 44.3%. Only 6.8% of these RNA genes are associated with CGIs. The similar low frequency of CGIs around RNA genes was found in humans (3.3%) and mice (5.3%). Therefore, RNA genes unlikely have a major effect on the strong prevalence of CGIs in the intergenic regions.
Fourth, identification of CGIs depends on the computational algorithm. So far, multiple algorithms have been developed for identifying CGIs in a genome or a sequence. Most of these algorithms are based on three sequence parameters (length, GC content, and ObsCpG/ExpCpG) except for the CpGcluster, recently developed by Hackenberg et al. [34], that detects clusters of CpGs (i.e., CpGcluster) by statistical significance based on the physical distance between neighboring CpGs on a chromosome. Our recent evaluation suggested that Takai and Jones’s algorithm [26] performs more appropriately and CpGcluster strikingly inflates the number of islands [12, 35]. To examine the robustness of our results, we identified CpG clusters using the CpGcluster algorithm and found that the same conclusion still held. For example, CpG cluster density was 128.3 per Mb in the dog genome, remarkably higher than that in the human genome (69.4 per Mb) or in the mouse genome (44.4 per Mb) (Supplementary Table S6).
Some genetic and genomic factors may be related to the unique features in dog CGIs. First, recombination may drive the evolution of nucleotide compositions, elevating GC content [36]. An increase of GC content in a sequence may drive the sequence to meet CGI criteria or help to prevent it from loss of CGI characteristics caused by other genetic factors such as de novo methylation. Dog has more chromosomes (2n=78) than the human (2n=46) and the mouse genomes (2n=40). Given the similar genome size, more chromosomes in a genome result in an overall smaller size of chromosomes, thus, higher recombination rates. Indeed we previously observed a negative correlation between CGI density and chromosome size, a positive correlation between CGI density and recombination rate, and the trend of high CGI density towards the telomeric regions [12]. Similarly, the dog was recently estimated to have higher recombination rate (1.554 cM/Mb) than the human (1.133 cM/Mb) and the mouse genomes (0.523 cM/Mb) [37]. The higher recombination rate in the dog and human than the mouse is also likely because both the dog and human have more number of chromosome arms (dog NA=80, human NA=82) than the mouse (NA=40) [12]. Therefore, karyotype is likely related to the strong prevalence of dog CGI. Second, the high CGI density and strong variance in the dog genome are likely related to its local sequence variance and local recombination rates. The dog genome has strong variance of CGI density (SD: ±40.5 CGIs/Mb, Table 1 and Figure 2) and GC content [7]. More genomic fractions were found to have higher GC content (>50%) in the dog genome than the human and mouse genomes, although the average GC content is similar in the three genomes: 41.0 % (dog), 40.9% (human) and 41.7% (mouse) [7]. Taken together, these genomic factors support that recombination rate is one of the factors related to CGI features. A fine-scale map with local recombination rate in the dog genome is necessary for further confirmation.
Although the dog genome has a much larger number of CGIs and higher CGI density than the human or mouse genome, we found a smaller number of promoter-associated CGIs in the dog than in the human, or even in the mouse, which had the fewest CGIs among mammalian genomes [12]. The ratio of CGI density in the promoter regions between the dog and human (1.0) or between the dog and mouse (1.4) was substantially smaller than that in any other genomic region, especially intergenic region (dog/human: 3.3; dog/mouse: 6.4) (Table 2). Experimental and computational studies have supported that CGIs tend to vanish during the genome evolution by a mechanism of de novo methylation of their CpG dinucleotides, which subsequently change to TpGs or CpAs because of a very high methylation-dependent transition rate [21, 25, 38, 39]. Selective pressure on functional regions could protect CGIs from methylation because abnormal methylation in promoter-associated CGIs might result in serious diseases [22]. In the rodent genomes, their fewer CGIs and faster CGI loss rate were thought to be related to the weaker selective constraints [21, 23, 25]. It remains unknown whether the similar mechanism has acted in the dog genome. Recent studies revealed that domestication process might accumulate deleterious mutations and relax the selective constraints on both nuclear genes and mitochondrial DNA (mtDNA) [40–42]. It is possible that weak relaxation of selection during domestication weakened the protection of CGIs from erosion in the functional regions; however, such an effect is likely weak.
The domestic dog were separated from grey wolf in East Asia [43]. A comparative analysis of CGIs in the dog genome with the wolf or cat genomes will specify the roles of domestication in CGI evolution. At present, the wolf genome has not been sequenced yet. The cat genome assembly has only 1.9 × coverage and consists of large amount of gaps in sequences [44]. Because scanning CGI requires high quality sequence, minimum of 500 bp, and few gaps, at present it is not practical to perform a systematic comparison. For example, there were ~1000 “Ns” in the 2 kb promoter region of cat DTNBP1 gene. Although no CGI was detected in this sequence, we could not conclude that cat DTNBP1 is lack of CGI in its promoter region.
In conclusion, we systematically examined the CGIs in the dog genome and compared with those in the human and mouse genomes. Our results revealed a remarkably larger number of CGIs and much higher CGI density in the dog genome than in the human or mouse genome. Surprisingly, the dog had fewer promoter-associated CGIs than the human or the mouse, at least in the homologous genes we examined. This unique opposite feature in the dog genome is unlikely due to its poor gene annotation, dog-specific repeats, or RNA genes in the intergenic regions. We further revealed that the abundance of CGIs in the dog genome was largely contributed by the noncoding regions including intergenic and intronic regions. We discussed some genetic or genomic factors such as local recombination rate and karyotype that may be related to the unique features of CGIs in the dog genome. A further comparison with the genomes of wild animals and close-related domesticated cat shall provide us more insights on CGI and genome evolution, especially for the domesticated genomes.
Materials and methods
Genome sequences and gene annotations
The assembled dog (build 2), human (build 36) and mouse (build 37) genome sequences and their gene annotations were downloaded from the National Center for the Biotechnology Information (NCBI) (ftp://ftp.ncbi.nlm.nih.gov/genomes/). The genomic sequences with repeats being masked were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/). For data consistency, we downloaded the same version of each genome assembly from the NCBI and UCSC Genome Browser (dog: canFam2; human: hg18; mouse: mm9).
To compare the features of CGIs in different genomic regions, we applied the following criteria, as similarly shown in Jiang and Zhao [45]. For a gene, its promoter region was defined as an interval of −1500 to +500 bp around the transcriptional start site (TSS) or translational start site when its 5’ untranslated region (UTR) was not available, and correspondingly, its 3’ region was defined as −500 to +1500 bp around the transcriptional end site (TES) or translational end site when its 3’ UTR was not available. A genic region was defined from the TSS to TES. An intergenic region was a region that no gene was annotated and excluded any promoter or 3’ region that overlapped with it.
CGI identification and mapping to different genomic regions
CGIs were identified based on Takai and Jones’ (2002) algorithm [26]. The search criteria were: GC content ≥ 55%, ObsCpG/ExpCpG ≥ 0.65 and length ≥ 500 bp. These criteria can effectively identify CGIs associated with genes and exclude short interspersed repeats (e.g., Alu), which typically have a sequence length of 80–400 bp [26, 46]. Our recent comparative studies of different CGI identification algorithms [12, 47] indicated that Takai and Jones’ algorithm is most appropriate for CGI identification. For comparison purposes, we also used CpGcluster developed by Hackenberg et al. [34] to scan CGIs in the whole genome.
We compared the locations of CGIs with the coordinates of genomic regions (genes, intergenic regions, intronic regions, promoter regions, and 3’ regions). A CGI that was completely within a genic, intronic or intergenic region was defined as genic CGI, intronic CGI or intergenic CGI, respectively. Because promoter regions and 3’-regions were short (i.e., 2 kb), CGIs likely extended to their external sequences. We defined a CGI within or overlapping with the promoter or 3’ region as promoter-associated CGI or 3’-region CGI. However, when we compared CGI density among different genomic regions, we counted only 0.5 for a CGI that overlapped with a promoter or 3’ region and 1 for a CGI completely within a promoter or 3’ region. This is consistent with the calculation of CGI density in genic, intergenic and intronic regions, which required CGIs completely within these regions.
Homologous genes, essential genes and housekeeping genes
We retrieved 10,196 dog-human-mouse homologous genes from the NCBI HomoloGene database (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/, build 61). Because of the lack of essential and housekeeping gene data in the dog genome, we used essential genes and housekeeping genes from the mouse and human genomes and identified their homologous dog genes.
Essential genes were defined when the deletion of this gene resulted in either lethality before reproduction or sterility. We extracted the mouse essential genes processed by Liao and Zhang [48], who classified 2136 essential genes and 1736 non-essential genes. We obtained 1236 essential dog-human-mouse homologous genes and 1049 non-essential dog-human-mouse homologous genes.
Housekeeping genes were defined as those genes that were expressed ubiquitously. We used human expression data from the second version of Gene Expression Atlas [49] and considered a gene being expressed in a tissue when its average difference (AD) value was ≥ 200 [50]. We classified genes into four groups: 1) housekeeping genes that were expressed in all the tissues, 2) widely expressed genes that were expressed in more than 80% of the tissues, 3) moderately expressed genes that were expressed between 20% to 80% of the tissues, and 4) narrowly expressed genes (tissue specific genes) that were expressed in fewer than 20% of the tissues. Among the dog-human-mouse homologous genes, we had 1257 housekeeping genes, 3416 widely expressed genes, 2599 moderately expressed genes and 1357 narrowly expressed genes.
Gene Ontology (GO) annotations
We examined the features of genes whose promoter-associated CGIs might have been lost or gained. Among the dog-human homologues genes, we classified them based on their presence or absence of CGIs in the promoter regions: 1) D+H+, both the dog and human genes had CGIs; 2) D+H-, the dog gene had promoter-associated CGI(s) while the human gene had not; 3) D-H+, the dog gene had no CGI while the human gene had; 4) D-H-, neither the human nor the dog gene had CGI.
We obtained GO annotations of human genes from the EBI Gene Ontology Annotation (GOA) database (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/) [51]. To simplify the analysis, we used GO terms that were mapped to the goslim-generic subset (ftp://ftp.geneontology.org/pub/go/GO_slims/goslim_generic.obo). At present, there has been no GO term annotation for dog genes yet. Therefore, we used the GO terms for human genes to test the overrepresentation of GO terms in the D- or D-H+ groups. Because of a large number of GO terms and our tested genes, we restricted those GO terms at the fourth level or lower and used the cutoff P value 0.01.
Supplementary Material
Acknowledgments
We thank Dr. Jingchun Sun for her assistance in gene feature analysis. This project was supported by a NIH grant (LM009598) from the National Library of Medicine, an institutional research grant (IRG-73-001-31) from the American Cancer Society and the Thomas F. and Kate Miller Jeffress Memorial Trust research grant to Z. Zhao.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at xxx.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hare B, Brown M, Williamson C, Tomasello M. The domestication of social cognition in dogs. Science. 2002;298:1634–1636. doi: 10.1126/science.1072702. [DOI] [PubMed] [Google Scholar]
- 2.Wayne RK, Ostrander EA. Lessons learned from the dog genome. Trends Genet. 2007;23:557–567. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Yang F, et al. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- 4.Sargan DR, et al. Use of flow-sorted canine chromosomes in the assignment of canine linkage, radiation hybrid, and syntenic groups to chromosomes: refinement and verification of the comparative chromosome map for dog and human. Genomics. 2000;69:182–195. doi: 10.1006/geno.2000.6334. [DOI] [PubMed] [Google Scholar]
- 5.Parker HG, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 6.Kirkness EF, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- 7.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 8.Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1706–1716. doi: 10.1101/gr.3736605. [DOI] [PubMed] [Google Scholar]
- 9.Sargan DR. IDID: inherited diseases in dogs: web-based information for canine inherited disease genetics. Mamm. Genome. 2004;15:503–506. doi: 10.1007/s00335-004-3047-z. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Z, Zhang F. Sequence context analysis of 8.2 million single nucleotide polymorphisms in the human genome. Gene. 2006;366:316–324. doi: 10.1016/j.gene.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Z, Zhang F. Sequence context analysis in the mouse genome: single nucleotide polymorphisms and CpG island sequences. Genomics. 2006;87:68–74. doi: 10.1016/j.ygeno.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Han L, Su B, Li WH, Zhao Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008:R79. doi: 10.1186/gb-2008-9-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antequera F. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird AP. CpG Islands as Gene Markers in the Vertebrate Nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 16.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 17.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 18.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada Y, et al. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res. 2004;14:247–266. doi: 10.1101/gr.1351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang C, et al. Features and trend of loss of promoter-associated CpG islands in the human and mouse genomes. Mol. Biol. Evol. 2007;24:1991–2000. doi: 10.1093/molbev/msm128. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br. J. Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 25.Matsuo K, et al. Evidence for erosion of mouse CpG islands during mammalian evolution. Somat. Cell. Mol. Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- 26.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 28.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 29.Goodstadt L, Ponting CP. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clamp M, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo AY, et al. The dystrobrevin-binding protein 1 gene: features and networks. Mol. Psychiatry. 2009;14:18–29. doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bains W, Templesmith K. Similarity and divergence among rodent repetitive DNA-sequences. J. Mol. Evol. 1989;28:191–199. doi: 10.1007/BF02102476. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Kirkness EF. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackenberg M, et al. CpGcluster: a distance-based algorithm for CpG-island detection. BMC Bioinformatics. 2006;7:446. doi: 10.1186/1471-2105-7-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han L, Zhao Z. CpG islands or CpG clusters: how to identify functional GC-rich regions in a genome? BMC Bioinformatics. 2009;10:65. doi: 10.1186/1471-2105-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- 37.Dumont BL, Payseur BA. Evolution of the genomic rate of recombination in mammals. Evolution. 2008;62:276–294. doi: 10.1111/j.1558-5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Z, Jiang C. Methylation-dependent transition rates are dependent on local sequence lengths and genomic regions. Mol. Biol. Evol. 2007;24:23–25. doi: 10.1093/molbev/msl156. [DOI] [PubMed] [Google Scholar]
- 39.Fryxell KJ, Moon WJ. CpG mutation rates in the human genome are highly dependent on local GC content. Mol. Biol. Evol. 2005;22:650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- 40.Bjornerfeldt S, Webster MT, Vila C. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 2006;16:990–994. doi: 10.1101/gr.5117706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, et al. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 2006;22:126–131. doi: 10.1016/j.tig.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Cruz F, Vila C, Webster MT. The legacy of domestication: Accumulation of deleterious mutations in the dog genome. Mol. Biol. Evol. 2008;25:2331–2336. doi: 10.1093/molbev/msn177. [DOI] [PubMed] [Google Scholar]
- 43.Savolainen P, et al. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 44.Pontius JU, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang C, Zhao Z. Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms. Genomics. 2006;88:527–534. doi: 10.1016/j.ygeno.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Giraldez F, Andres O, Domingo-Roura X, Bosch M. Analyses of carnivore microsatellites and their intimate association with tRNA-derived SINEs. BMC Genomics. 2006;7:269. doi: 10.1186/1471-2164-7-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han L, Zhao Z. Comparative analysis of CpG islands in four fish genomes. Comp. Funct. Genomics. 2008:565631. doi: 10.1155/2008/565631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Su AI, Li WH. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol. Biol. Evol. 2005;22:2113–2118. doi: 10.1093/molbev/msi206. [DOI] [PubMed] [Google Scholar]
- 51.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.