Abstract
Regulatory T (Treg) cells play a central role in maintaining immune homeostasis. However, little is known about the stability of Treg cells in vivo. In this study, we demonstrate that a significant percentage of cells exhibited transient or unstable Foxp3 expression. These exFoxp3+ T cells express an activated-memory T cell phenotype, and produced inflammatory cytokines. Moreover, exFoxp3 cell numbers increased in inflamed tissues under autoimmune conditions. Adoptive transfer of autoreactive exFoxp3 cells led to the rapid-onset of diabetes. Finally, T cell receptor repertoire analyses suggested that exFoxp3 cells develop from both natural and adaptive Treg cells. Thus, the generation of potentially autoreactive effector T cells as a consequence of Foxp3 instability has important implications for understanding autoimmune disease pathogenesis.
INTRODUCTION
Autoimmunity represents a collection of over 80 heterogeneous disorders often controlled by complex genetic and environmental factors. The pathogenesis is thought to result from a breakdown in the mechanisms controlling central and/or peripheral tolerance, which ultimately culminates in autoimmune disease. Thus, the path to effective therapies likely requires an understanding of the mechanisms that underlie the breakdown of tolerance normally maintained by regulatory T (Treg) cells. Treg are a specialized CD4+ T cell lineage that plays a central role in the preservation of self-tolerance and whose dysfunction has been implicated in autoimmune diseases including type I diabetes(T1D)1–3. The cells are characterized as a small subset of CD4+ T cells with a unique phenotypic fingerprint driven by the expression of the forkhead transcription factor, Foxp3(http://www.signaling-gateway.org/molecule/query?afcsid=A002750)4. Foxp3 is essential for the suppressive activity of Treg, and Foxp3 deficiency leads to a multi-organ autoimmune disease as can be observed in the ‘scurfy’ mouse and in human immune dysregulation, polyendocrinopathy, enteropathy X–linked syndrome (IPEX) patients5–7. Moreover, ectopic Foxp3 expression in non-Treg cells confers suppressor function in vitro and in vivo4. Finally, there are two distinct subsets of Treg in peripheral lymphoid organs – natural Treg (nTreg) that develop in the thymus following high affinity self-antigen recognition, and adaptive Treg (aTreg) that develop from conventional T (Tconv) cells as a consequence of peripheral antigen exposure8. aTreg play a complementary role in maintaining peripheral tolerance, particularly for the self-antigens not expressed in the thymus.
Although the importance of Treg in the control of autoimmunity is now well established, little is known about the stability of Treg in vivo. Gavin et al. showed that most Treg retain high Foxp3 expression following adoptive transfer in a non-pathogenic setting and suggested that Foxp3 expression serves as a binary mode whose stability is controlled by Foxp3 itself through a positive feedback loop9,10. However, several studies have suggested that Treg isolated from inflammatory sites express reduced amounts of Foxp3 possibly increasing susceptibility to autoimmunity1,3. Supporting this idea, the inflammatory cytokine IL-6 synergizes with IL-1 to downregulate Foxp3 expression through a Stat3-dependent pathway11 and microRNA-deficient Treg cells can lose Foxp3 expression12. In addition, peripheral Treg, particularly the CD4+CD25− Foxp3+ subset, were unstable in a lymphopenic setting, and those unstable Treg can convert to follicular T helper cells and promote germinal center formation in mouse Peyer’s Patches. Together, these findings suggest that peripheral Treg can become unstable under certain experimental conditions.
In this study, we examined the stability of Treg using genetic lineage tracing of Foxp3+ Treg under both homeostatic and autoimmune conditions in unmanipulated mice. Foxp3-GFP-Cre BAC transgenic mice12 were crossed to Rosa26-loxP-Stop-loxP-YFP (R26-YFP) reporter mice13, allowing us to track Foxp3 induction and down-regulation concurrently. We observed that a significant percentage of cells that at some point expressed Foxp3 had down-regulated or completely lost Foxp3 expression in healthy animals; these exFoxp3 cells expressed an activated-memory phenotype and produced inflammatory cytokines. Of note, these exFoxp3 cells, which increased significantly in percentage in the autoimmune diabetes setting, could transfer diabetes. Finally, variable T cell receptor-α (TCRα) variable segment analyses revealed that the exFoxp3 cells shared ontogeny with Foxp3+ Treg as well as Tconv cells, suggesting that under some conditions both natural and adaptive Treg become unstable in the periphery and may promote autoimmunity.
Results
Foxp3+ T cells in Foxp3-GFP-Crex R26-YFP mice
We recently reported the creation of a BAC-transgenic mouse expressing a GFP-Cre fusion protein under the control of the Foxp3 promoter (Foxp3-GFP-Cre)12. Characterization of these mice demonstrated a Treg cell developmental profile similar to that previously observed in a Foxp3-GFP knock-in reporter strain14. Importantly, endogenous Foxp3 expression was not changed in BAC Tg cells (data not shown). These mice were crossed with a Rosa26-loxP-Stop-loxP-YFP (R26-YFP) reporter mouse13. Thus, in the resultant Foxp3-GFP-Crex R26-YFP transgenic mice, Foxp3-GFP-Cre+ Treg excise the floxed-stop cassette to drive constitutive YFP transcription off the R26 promoter, permanently marking the Foxp3+ T cells and their progeny. We noted that a few GFP+YFP+ thymocytes were observed in CD4+CD8+ double positive (DP) and CD8 single positive (SP) thymocytes suggesting that Foxp3 transcription was induced under certain conditions of self-antigen exposure during positive selection and CD8 T cell maturation (Fig. 1A). The majority of the Foxp3+GFP+ and/or YFP+ cells were present within the CD4 SP thymocyte subset (Fig. 1A). Using heat shock antigen (CD24) expression as an indicator of thymocyte maturity, it was possible to map the development of the lineage (Fig. 1B). Few, if any GFP+YFP+ cells were observed among the immature, CD4 SP CD24bright thymocytes. However, as the cells matured (indicated by reduced CD24 expression) a population of GFP+ cells became evident (Fig. 1A). These cells had low YFP fluorescence due to a delay between Foxp3 transcription, read out as GFP expression, and the loxP-Stop-loxP excision in the R26-YFP transgene. Thus, the GFP+YFPlo/− population represented cells that had upregulated Foxp3 most recently. YFP+ cells emerged as Treg further matured, as indicated by further reduced CD24 expression, such that the most mature cells expressed high amounts of both GFP and YFP. Thus, lineage tracing of Foxp3 promoter activity suggested that the majority of Foxp3 transcription is initiated after CD4SP thymocyte maturation. This stage of thymocyte maturation has been linked to negative selection, supporting previous suggestions that Foxp3+ Treg develop as an “escape” mechanism during negative selection following self-antigen exposure15,16. Finally, the CD4 SP cells included a small, but detectable cell population of GFP−YFP+ cells (Fig. 1A). This population, which constituted about 10% of the YFP+ cells in the thymus, appeared to develop in the mature CD4+ T cells since all the cells were CD24low, which was consistent with having derived from TCR-triggered antigen recognition. It is important to note that we did not observe any YFP expression among B cells or other non-T cells in these animals consistent with a high fidelity of Cre activity in this BAC Tg and lack of stochastic expression of YFP in other tissues.
Fig. 1. Development of Foxp3+ T cells inFoxp3-GFP-Cre × R26-YFPtransgenic mice.
(A)Thymocytes were isolated, stained for CD4 and CD8, and GFP and YFP expression was analyzed by flow cytometry. Thymocytes were gated as CD4+CD8+ double positive (DP) and individual CD4 or CD8 single positive (SP) populations. (B) The histograms show CD24 expression on the CD4SP GFP and YFP populations gated as shown in panel A. (C) GFP and YFP expression by gated CD4+ T cells from lymph nodes (LN) and spleen. (D) Methylation analysis of the Foxp3 locus. The methylation status of the CpG motifs of the TSDR of sub-cloned from purified Tconv, Tregs and exFoxp3 cells. Data is the average of data generated in3 independent experiments. (E) Graphs show the indicated GFP and YFP populations as a percentage of CD4+ T cells in spleen, LN, liver and Peyer’s patch of Foxp3-GFP-CrexR26-YFP mice. Each point represents an individual mouse. Experiments were performed with 6 -9 week old mice and are representative of 4 (A, B) and 20 (C) independent experiments.
Next, we examined the presence of different subsets in the periphery. As previously described12, the GFP-Cre construct in the BAC transgenic mice was highly efficient. About 10% of peripheral CD4+ T cells were GFP+YFP+ cells, whereas very few (0.4%)GFP+ YFPlo T cells which most likely represent newly generated aTreg as a consequence of peripheral induction of Foxp3 were seen (Fig. 1C). Interestingly, analysis of the peripheral spleen and lymph node (LN) tissues revealed that the GFP−YFP+ population accounted for up-to 20% of the peripheral YFP+ cells (Fig. 1C). To directly address the stability of the Foxp3 locus in this novel population, we examined the methylation status of the Treg-specific demethylated region (TSDR) of the Foxp3 gene in the various T cell subsets17. Methylation of CpG islands in this region of the Foxp3 locus is a primary control mechanism of Foxp3 expression in mouse CD4+ T cells, as 90% of the CpGs in the examined intron of Foxp3− naive CD4+ T cells were >90% methylated, whereas the CpGs in this intron were fully un-methylated in thymus-derived nTreg and in vivo generated aTreg18–20. Bisulphite sequence analysis confirmed previous findings that 100% of FACS sort-purified conventional CD4+ GFP−YFP− T cells (termed Tconv) cells had >85% of the CpGs methylated whereas <15% of the CpGs were methylated in 90% of the Treg (CD4+ GFP+YFP+ T cells) (Fig. 1D and supplementary Fig. 1). In contrast, FACS-purified GFP−YFP+ cells had a varied pattern of methylation status. Only 74% of the DNA strands had >85% of the CpGs in the TSDR methylated, which correlated positively with Foxp3 expression (supplementary Fig. 1); 11% of the clones had >85% of the CpG islands un-methylated and, most interestingly, 13% had partial methylation, with 15–85% of the CpGs being un-methylated (Fig. 1D). Interestingly, there appeared to be a random pattern of partial methylation in the GFP−YFP+ cells (supplementary Fig. 1), suggesting that factors which controlled the spontaneous loss of Foxp3 expression led to re-methylation of the Foxp3 locus in some cells. Alternatively, it was possible that a subset of GFP−YFP+ cells had never fully demethylated the locus even though they had expressed sufficient Foxp3 to turn on the Cre enzyme. In this regard, this methylation phenotype is reminiscent of what has been observed in in vitro TGFβ-induced Tregs (iTreg)18.
Analysis of spleen, LN, liver and Peyer’s Patches of multiple mice showed that there were similar proportions of the various YFP subsets throughout peripheral lymphoid compartments (Fig. 1E). Approximately 15% of the YFP+ cells were negative for Foxp3 staining (Fig. 2A), which correlated with the proportion of YFP+ cells that lacked GFP expression (Fig. 1C). These results suggested that a population of T cells existed in both the thymus and various lymphoid compartments that had expressed Foxp3 at one stage but ceased active translation of Foxp3 protein. We herein refer to the GFP−YFP+ cells as exFoxp3 cells.
Fig. 2. CD4+ YFP+ Foxp3− cells have a non-Treg surface phenotype.
(A) Lymph node and spleen CD4+ GFP+ T cells were FACS purified from Foxp3-GFP-Cre mice and CD4+ YFP+ T cells sort-purified from Foxp3-GFP-Crex R26-YFPmice. Purified populations were analyzed for CD127, CD25, and GITR and Foxp3 by intracellular staining. (B) As in (A), analysis of FR4, CTLA-4 and CD103 expression and Foxp3 by intracellular staining. Histograms show YFP− Tconv (thick line), YFP+ Foxp3+ Treg (thin line), and YFP+ Foxp3− (filled histogram). Representative plots from 6–9 week old mice of 3 experiments.
Next, we examined the phenotype of the exFoxp3 cells in the periphery. The Foxp3−YFP+ cells were uniformly CD25−, GITRlow and CD127high, differing markedly from the Foxp3+ YFP+ Tregs (Fig. 2A). Analyses of other cell surface molecules revealed that the exFoxp3 cells had a mixed phenotype with heterogeneous expression of signature Treg markers including folate receptor 4 (FR4), CTLA-4 and CD103 (Fig. 2B). Previous Foxp3 knockout studies showed that ablation of Foxp3 in vivo and ex vivo resulted in the loss of the Treg phenotypic signature; thus, the significant alterations in expression of CD25, GITR, CD127 and other surface markers in the GFP−YFP+ cells suggested a similar instability had occurred under homeostatic conditions.
ExFoxp3 cells have an activated-memory phenotype
Interestingly, the exFoxp3 cells did not represent a dead end, terminally differentiated Treg but rather a cell with plasticity that could develop an activated-memory cell phenotype, with heterogeneous CD62L expression and high expression of CD44 (Fig. 3A). To directly assess if the exFoxp3 cells were effector-memory T cells, YFP+ cells were sorted, stimulated with PMA and ionomycin and examined for intracellular cytokine production (Fig. 3B–D). A substantial percentage of the exFoxp3 cells produced interferon-γ (IFNγ), supporting the hypothesis that these were indeed effector-memory T cells. Previous studies had suggested a unique relationship between Treg and Th17 cell differentiation based on transcription factor plasticity during T cell differentiation, particularly in gut-associated lymphoid tissue11,21. Therefore, we examined the production of IL-17 by exFoxp3 cells isolated from gut-associated lymphoid tissue. A mean of 22.4% of exFoxp3 cells in Peyer’s Patches produced IL-17A, compared with a mean of 13.2% producing IFN-γ (Fig. 3C, D). This contrasted with other lymphoid tissues analyzed, where in the exFoxp3 cells had a T helper type 1 (TH1)-biased phenotype. For instance, in the flow cytometric analysis depicted in Fig. 3C, & D, means of 32%, 31.7% and 11.2% of exFoxp3 cells isolated from the liver, spleen, and LN, respectively, produced IFN-γ, but a much lower percentage of exFoxp3 cells in these tissues produced IL-17A. Together, these results suggest that exFoxp3 populations contain effector-memory T cells with distinct cytokine producing capability depending on their microenvironment.
Fig. 3. CD4+ YFP+ Foxp3− cells have a non-Treg, memory cell surface phenotype and produce IFN-γ and IL-17.
Analysis of peripheral lymphoid organ resident CD4+ GFP+YFP+, CD4+ GFP YFP+, and CD4+GFP−YFP− cells. (A) Spleen CD4+ populations were gated and analyzed for CD62L and CD44 expression. (B) Spleen CD4+ YFP+ and CD4+ YFP− T cells were purified then stimulated with PMA, ionomycin and monensin for 4 h; cells were then stained for intracellular Foxp3 and IFN-γ. (C,D) Graphs show the percent of cells in each population that produce IFN-γ (C) or IL-17 (D). Each point represents an individual mouse. Experiments were performed on 6 – 9 week old mice and are representative of 5 (A, B) independent experiments.
Autoimmune microenvironment favors loss of Foxp3
Decreased Foxp3 expression in Tregs has been reported in various autoimmune diseases including type 1 diabetes in NOD mice 1,3. To test the hypothesis that the reduction of Foxp3 expression under autoimmune setting shifted the balance of Treg to exFoxp3, we crossed the Foxp3-GFP-Cre × R26-YFPmice into the NOD strain, a spontaneous autoimmune diabetes model which shares many similarities to autoimmune T1D in humans. Our previous studies demonstrated a decreased Treg cell:T effector (Teff)cell ratio in the inflamed islets, which is tightly associated with disease progression1. Therefore, we examined the proportion of CD4+Foxp3+Tregs in aging, pre-diabetic Foxp3-GFP-Cre × R26-YFPNOD mice. Infiltrates were prepared from the pancreases of 16–18 week old mice and the abundance of the various GFP and/or YFP-expressing populations in these infiltrates were compared with those in draining pancreatic LN (PLN) and non-draining inguinal LN (ILN). Additionally, we analyzed the expression of markers of Treg and T memory-effector cells. We observed a significant decrease in the relative percentage of CD4+ GFP+YFP+ Treg within the CD4+ YFP+ T cell population in the pancreas as compared with PLN or ILN lymph nodes (data not shown). In contrast, the percentage of GFP−YFP+ exFoxp3 cells was increased specifically in the pancreas (average 32.7%), as compared to ILN and PLN lymph nodes (average 17.8% and 22.8%, respectively) (Fig. 4A). Interestingly, exFoxp3 cells isolated from the pancreas expressed the lowest amount of CD25andthe highest amount of CD127 compared with exFoxp3 cells from the other compartments (Fig. 4B, C), and produced IFN-γ (data not shown); these observations suggest that the autoimmune microenvironment altered T cell phenotypes and promoted pathogenicity. In addition, these results suggested that the appearance of the exFoxp3 cells was not simply a consequence of stochastic regulation of Foxp3 but rather a consequence of antigen recognition in the inflamed setting.
Fig. 4. The autoimmune microenvironment favors loss of Foxp3.
(A)Percent of GFP−YFP+ exFoxp3 cells among CD4+YFP+ cells in the pancreas, inguinal (ILN) and pancreatic (PLN) lymph nodes of NOD mice. Data points represent individual mice. (B,C) CD25 (B) orCD127 (C) expression on exFoxp3 cells isolated from each site, expressed as mean fluorescence intensity (MFI). Six 16–18-week old mice were analyzed in 2 independent experiments. (D) Development of Treg and exFoxp3 cells in BDC2.5 TCR Tg mice. Analysis of GFP and YFP expression in thymocytes, LN cells and splenocytes of 6–9 week-old BDC2.5 TCR Tg+ and control TCR Tg− mice. Plots are gated on live thymocytes and enriched CD4+ T cells from LN and spleen. Representative of 3 mice. (E) CD4+ YFP+ T cells purified from pancreatic LN (PLN) of BDC2.5.TCR Tg or non-Tg control mice were analyzed for Vβ4 (BDC2.5 TCR chain) and Foxp3 expression. Plots are representative of 3 mice, and each point on the graph represents the percentage of YFP+ FoxP3− cells in individual mice.
To directly address this hypothesis, the Foxp3-GFP-Cre × R26-YFPmice were further crossed with BDC2.5 TCR Tg+ mice22 whose TCR transgenic CD4+ T cells recognize a pancreatic islet autoantigen; this system allowed investigation of whether the expression of a pancreatic antigen-specific TCR would change the percentage of exFoxp3 cells and their pathogenic potential. The proportion of thymic CD4 SP Tconv and exFoxp3 cells were similar in control non-Tg Foxp3-GFP-Cre × R26-YFPand BDC2.5 TCR Tg Foxp3-GFP-Cre × R26-YFP mice, although there were generally fewer thymic Treg cells in the latter animals (Fig. 4D). However, there was up-to 2-fold more exFoxp3 cells in the spleen and PLNs of the BDC2.5 TCR Tg mice as compared with control littermates (Fig. 4D, E); this increase is similar to the percentage found in the pancreas of conventional NOD mice. Together these data suggest that strong self-antigen engagement, especially in the inflammatory setting, can promote the generation of exFoxp3 cells.
ExFoxp3 cells are pathogenic
To further test the hypothesis that established Treg can be unstable and potentially pathogenic in an autoimmune setting, Treg were sorted from Foxp3-GFP-Cre × R26-YFPBDC2.5 mice and transferred into T cell-deficient NOD.Tcra−/− mice (Fig. 5A and supplementary Fig. 2). Analyses of recipient mice 4 weeks after transfer of nTreg showed that on average a third of the cells (35.7%) had down-regulated Foxp3 based on the absence of GFP or Foxp3 protein. Importantly, these cells took on an effector-memory phenotype with on average 28.4% of the GFP−YFP+ Tg+ cells producing IFN-γ after PMA and ionomycin stimulation (Fig. 5A). A similar experiment was performed by transferring GFP−YFP− Tg+ cells into immunodeficient mice. In this study, about 0.5% of the transferred cells—all of which expressed the Vβ4 chain of the BDC2.5 TCRTg—turned on YFP in the pancreas, indicative of transient Foxp3 expression (Fig. 5B). Together, these results suggest that the exFoxp3 cells can be generated from nTreg instability and/or abortive aTreg induction dependent on self-antigen interaction. However, the aTreg pathway did not seem to efficiently generate exFoxp3 cells in vivo after adoptive transfer.
Fig. 5. exFoxp3 cells develop from Treg upon adoptive transfer.
(A)GFP+YFP+ GITRhigh BDC2.5 TCR Tg purified cells (left dot plot) were transferred into NOD Tcra−/− mice. After four weeks splenic YFP+ cells were purified, exposed to PMA, ionomycin and monensin for 3–4 hours and stained for intracellular Foxp3 and IFN-γ (right dot plot). Representative of 4 mice. (B) GFP−YFP− cells purified from inguinal LNs (ILN) of BDC2.5 TCR Tg+ mice were transferred into NOD Rag2−/− mice. Seven days after transfer, pancreatic mononuclear cells of acutely diabetic NOD Rag2−/− mice that received CD4+ BDC2.5 TCR Tg+ YFP− cells, or of control NOD Rag2−/− mice that did not receive cells were analyzed by flow cytometry. CD4+ Vβ4+ cells were gated and analyzed for YFP expression. Representative of 3 mice.
To investigate the functional consequences of the generated exFoxp3 cells, the various subsets, including the exFoxp3, Tconv, and Treg cells, were sorted from CD4-enriched LN and spleen of Foxp3-GFP-Crex R26-YFP BDC2.5 mice and expanded each population in vitro using anti-CD3-plus anti-CD28-coated beads and IL-2 (Fig. 6A)23. The various T cell subsets were stable throughout the 7–9 days in culture as both the GFP+YFP+ Treg and GFP−YFP+ exFoxp3 remained uniformly CD4+ and YFP+ and continued to express their respective high and low amounts of GFP. Intracellular Foxp3 staining showed that less than 2% of the expanded GFP−YFP+ T cells were Foxp3+; although the majority of expanded GFP+YFP+ cells maintained expression of Foxp3 about 20 % lost Foxp3 expression (Fig. 6B). Individually expanded BDC2.5 TCR Tg+ Treg, exFoxp3 and Tconv cells were then transferred into NOD Rag2−/− mice and followed for autoimmune diabetes development. Mice that received GFP+ YFP+ Treg had normal blood glucose concentrations (<250mg/dl) throughout study follow-up. In contrast, exFoxp3 cells, like Tconv cells, induced rapid is let destruction and development of diabetes (Fig. 6C, D). Over 95% of the TCRTg CD4+Vβ4+ cells expressed YFP indicating the transferred exFoxp3 cells were responsible for disease pathogenesis (Supplementary Fig. 3). These results suggested that exFoxp3 BDC2.5 cells function as effector T cells upon recognition of pancreatic self-antigen, and induced rapid and severe T1D.
Fig. 6. exFoxp3 cells are pathogenic.
(A)CD4+ T cells from Foxp3-GFP-Cre × R26-YFPBDC2.5 mice were sorted to >95% purity into GFP+YFP+ and GFP−YFP+ cells. The percentage of Foxp3+ within these sorted populations was determined using intracellular staining (pre-culture). (B) Purified cells were cultured withanti-CD3 and anti-CD28 coated beads + IL-2for 6–8days, and were stained by flow cytometry (post-culture). Representative plots from 3 experiments.(C) 5 × 105 expanded GFP−YFP−, GFP+YFP+ or GFP−YFP+ cells were transferred into NOD Rag2−/− mice, and blood glucose concentrations were recorded from 8 days later and onwards. Diabetes is considered when blood glucose exceeds 250 mg/dl. Each point represents an individual mouse. (D) Hematoxylin and eosin staining of pancreas sections from NOD Rag2−/− mice that did not receive cells, or that received GFP−YFP+ cells 9 days prior as described in (B). Representative of 4 mice.
The precursors of exFoxp3 are heterogeneous
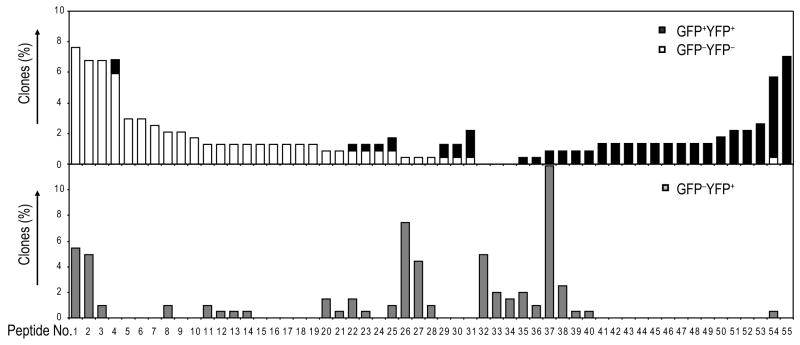
It was critical to address the ontogeny of the exFoxp3 cells directly since it remained unclear in an unmanipulated setting whether the exFoxp3 cells arose from aborted Foxp3+ aTreg that had converted from conventional T cells in the periphery or resulted from the loss of Foxp3 expression in bonafide CD4+Foxp3+ nTreg cells. Previous studies have suggested that rearrangement of endogenous TCRα chains occurs selectively within nTreg in the BDC2.5 mouse24 and these mixed TCR pairs result in a unique T cell repertoire in the bonafide Treg subset. Thus, we isolated the various CD4+ T cell subsets from Foxp3-GFP-Cre × R26-YFPBDC2.5 mice, and amplified, cloned and sequenced a region encompassing the CDR3 through the constant region of the Vα2 TCR in these cells. Of 384 clones per subset, 227 Treg, 202 exFoxp3, and 236 Tconv cells had productive V-and J-gene rearrangements (supplementary Table 1). As previously reported, the endogenous TCR Vα2 repertoire of Treg and Tconv cells from BDC2.5 TCR transgenic mice was distinct, as only 12.8% of the GFP+YFP+ Treg CDR3 amino acid (a.a.) sequences were found in the GFP−YFP− Tconv cells (Fig. 7). The exFoxp3 cells shared CDR3 amino acid sequences with both Treg and Tconv, overlapping 23.8% and 35.6%, respectively (Fig. 7, Table 1). Among the 12 most frequent CDR3 amino acid sequences found in the GFP−YFP+ cells (Table 1), 4 peptide sequences have been described in previous studies; peptide sequence CAAANSGTYQRF was observed to be dominant in Tconv cells whereas CAASALRTGSWQLIF, CAASGAGGYKVVF and CAATHNNNRIFF were uniquely found in Treg (supplementary Table 2)24. Given the broadness of the TCR repertoire, the number of sequenced clones from exFoxp3 cells was probably still relatively low, however, significant overlap of TCR repertoire with both Treg and Tconv cells suggested exFoxp3 cells are heterogeneous, deriving in part from established Treg that lost Foxp3 expression, and, in part from Foxp3− conventional T cells that expressed, then lost, Foxp3 following TCR engagement. Since all the Tconv cells expressed a transgenic BDC2.5 TCR, it is tempting to speculate that BDC2.5 TCR and an unknown islet antigen interaction actually leads to development of at least some of the exFoxp3 cells.
Fig. 7. exFoxp3 cells can develop from both nTregs and aTregs.
CD4+ GFP−YFP−, GFP−YFP+ and GFP+YFP+ T cells were sorted from BDC2.5 TCR Tg mice, cDNA was amplified with Vα2-specific primers and the amplicons were subcloned and sequenced. The frequency of each unique CDR3 amino acid sequence in Tconv (un-filled bar), exFoxp3 (grey bar) and Treg cells (black bar). Data is one mouse representative of 2 analysed.
Table 1.
Most frequent CDR3 amino acid sequences found in the GFP− YFP+ cells.
| CDR3 AA seq | J gene | GFP−YFP− (236)* | GFP+YFP+ (227)* | GFP− YFP+ (202)* |
|---|---|---|---|---|
| CAARPRHNVLYF | TRAJ21*01 | 0.0% | 0.9% | 11.9% |
| CAASAGGGRALIF | TRAJ15*01 | 0.4% | 0.0% | 7.4% |
| CAAKKGYNVLYF | TRAJ21*01 | 7.6% | 0.0% | 5.4% |
| CAAANSGTYQRF | TRAJ13*01 | 6.8% | 0.0% | 5.0% |
| CAAKAHLGFASALTF | TRAJ35*02 | 0.0% | 0.0% | 5.0% |
| CAAKPGYNKLTF | TRAJ11*01 | 0.4% | 0.0% | 4.5% |
| CAASALRTGSWQLIF | TRAJ22*01 | 0.0% | 0.9% | 2.5% |
| CAASGAGGYKVVF | TRAJ12*01 | 0.0% | 0.4% | 2.0% |
| CAATHNNNRIFF | TRAJ31*02 | 0.0% | 0.0% | 2.0% |
| CAAKSGGSNYKLTF | TRAJ53*01 | 0.8% | 0.4% | 1.5% |
| CAAKKGYKLTF | TRAJ9*01 | 0.8% | 0.0% | 1.5% |
| CAATGNSAGNKLTF | TRAJ17*01 | 0.0% | 0.0% | 1.5% |
Total number of sequence is noted.
Discussion
There have been controversies regarding the stability of Treg in the periphery. It was widely believed that Treg represented a highly stable lineage in which few if any cells lost Foxp3 under normal homeostatic conditions. Recently, Hori and colleagues reported a small subset of Treg, namely, a CD25−Foxp3+ subset, was unstable and rapidly lost Foxp3 expression upon transfer into a lymphopenic host. Other studies have also suggested that Treg can be unstable; however, these studies have been limited to in vitro cultures or in vivo transfers to lymphopenic recipients. In this study, we have utilized a Foxp3 reporter lineage marker system to demonstrate unequivocally that a subset of Foxp3-expressing cells indeed lose Foxp3 expression. The generation of the exFoxp3 cells is accelerated during autoimmune T1D progression. These cells develop an effector-memory phenotype, produce pathogenic cytokines and can trigger the development of autoimmunity. Thus, Foxp3 expression is dynamically regulated under normal physiologic conditions.
The paradigm in immunology is that autoimmunity is precipitated by an imbalance of pathogenic T cells and Foxp3+ regulatory T cells25,26. One dogma suggests that autoimmunity stems from a defect in Treg that “allows” pathogenic cells to escape regulation and mediate disease27. Based on our observations, we propose an alternative possibility, namely, that Foxp3 instability can lead to the generation of pathogenic effector-memory T cells that promote autoimmunity. The efficiency of the Treg system is a consequence of Treg cell expression of a TCR repertoire biased towards self-recognition16, 28,29. Thus, the frequency of precursors capable of recognizing self proteins is very high, and this facilitates efficient suppression. Moreover, Treg have multiple functional properties that target dendritic cells including both cell contact-dependent and soluble cytokine-specific suppressive activities25. These functional attributes allow this small subset to suppress local immunity through a combination of bystander suppression and infectious tolerance. However, these same attributes can backfire when a subset of the Treg lose Foxp3 expression and turn into autoreactive effector cells. In the pancreas of NOD mice, the balance was shifted from Foxp3+ Treg to exFoxp3 cells. exFoxp3 cells showed very high expression ofCD127, which is believed to be a direct Foxp3 target and represents a key marker of memory T cell subsets10,30.
Although the detailed molecular mechanism of exFoxp3 generation still needs to be clarified, it is possible that during an autoimmune pathological T cell response, a functional deficiency of IL-2 signaling in the Treg in the affected tissues may lead to a break in the positive feedback loop that controls Foxp3 stability such that self reactive Treg may convert to effectors with high diabetic potential. Consistent with this notion, we observed a specific enrichment of CD4+Foxp3+CD25− cells in the inflamed islet, which represents an unstable environment that favors the development of exFoxp3 cells, either through transient induction of Foxp3 in Tconv cells or loss of Foxp3 transcription in Treg. Moreover, IL-2 treatment can efficiently recover the CD25 expression of intra-islet Treg and protect mice from developing autoimmune diabetes 1. Furthermore, recently we demonstrated that Treg cells depend on Dicer and hence microRNAs (miRNAs) for proper function, and that exFoxp3 cells are largely increased in Foxp3-specific Dicer KO mice12. More recently, Rudensky and colleagues demonstrated that miR-155 modulates IL-2 signaling through targeting SOCS-131, and that miR-155-deficient Treg cells needed higher IL-2 concentrations in vitro to achieve STAT5 phosphorylation comparable to that seen in wild-type cells. Hence it is possible that miRNAs are involved in maintaining high Foxp3 expression in Treg cells.
The lineage tracing through analysis of the endogenous Vα TCR usage in BDC2.5 TCR transgenic cell subsets revealed that a significant percentage of exFoxp3 cells shared ontogeny with both Foxp3+ Treg and Tconv lineage. It is likely that the same inflammatory autoimmune setting actually destabilizes both nTreg and aTreg. aTreg share multiple characteristics with nTreg including the fully un-methylated the TSDR in Foxp3 locus17, similar cytokine expression and the Treg mRNA expression fingerprint. aTreg play a complementary role to nTreg cells, particularly in response to self-antigens not expressed in the thymus. Thus, it is possible that under inflammatory conditions, aTreg development is aborted allowing the self-reactive cells to develop directly into effector-memory T cells and promote autoimmune disease. Interestingly, Lochner et al reported a surprising finding that a transient Foxp3 expression actually favored the differentiation of IL-17 producing RORγt+ T cells32. We speculate that in all cases, the molecular mechanism of destabilized Foxp3 involves epigenetic changes of the Foxp3 locus in the inflamed site. In this regard, one study suggested that the IL-6 signaling pathway can re-methylate a critical Foxp3 CpG motif and suppress Foxp3 expression33. Recent studies suggest that subsets of Tregs preferentially and specifically suppress distinct T helper subset-mediated immune pathology. For instance, Treg-specific IRF-4 ablation led to impaired TH2 suppression but normal TH1 suppression34. On the other hand, Koch et al. reported that a subset of Tbet-expressing Treg suppress TH1-type inflammation35. Since Treg and Tconv are closely related and share a mRNA and miRNA signature 36, it is possible that some of this phenomenon is explained by Treg/exTreg pairs that are equipped with similar chemokine receptors to migrate to the same tissues and/or that some of the effectors are derived directly from Treg in the inflamed tissues.
Finally, the plasticity of Foxp3 expression in the inflamed setting may also have implications in infectious diseases where early inflammatory cytokines induced by the innate immune response may not only disable Treg but enhance immunity by creating a pathogenic autoreactive T cell repertoire locally in the infected tissues. This possibility may explain the commonly observed phenomenon of episodes of infections triggering autoimmune disease37. Thus, a better understanding of the extracellular and intracellular signals that maintain or destabilize Foxp3 expression may have important therapeutic implications in a variety of disease settings ranging from autoimmunity to cancer and infectious disease.
Methods
Mice
NOD.BDC2.5, Foxp3-GFP-Cre and Rosa26-loxP-Stop-loxP-YFP reporter mice have been described 12,13,22. All mice were housed and bred under specific pathogen-free conditions at the University of California, San Francisco Animal Barrier Facility. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
Antibodies
Labeled antibodies specific for CD4 (RM4-5), CD8 (Ly-2), CD25 (PC61), CD44 (pgp-1), GITR (DTA-1), CD62L (MEL14), CD103 (M290), CD127 (A7R34), CD152 (UC10-4F10), Foxp3 (FJK-16s), FR4 (eBio12A5), Vβ4 (KT4), IL-17A (eBio17B7), IFN-γ (XMG1.2) and specific isotype controls were purchased from BD PharMingen or eBioscience. Anti-CD24 (clone 91) was from Southern Biotechnology Associates.
Flow cytometry
For cytokine analyses, cells were incubated with 0.5 μM ionomycin, 10 ng/ml phorbol myristate acetate (PMA) and 3 μM monensin at 37°C for 3–4 hours before being fixed and permeabilized (Foxp3 Staining Kit, eBioscience) and stained for intracellular proteins. Stained cells were analyzed with a FACSCalibur and CELLQuest software or an LSRII flow cytometer and FACSDiva (BD Biosciences, San Jose, CA). T cells were sorted using a MoFlo cytometer high speed cell sorter (DakoCytomation) or FACSAria (BD Biosciences). 510/20-nm filter (495LP Dichroic mirror) and 560/40-nm filter (535LP Dichroic mirror) were used to discriminate the GFP from the YFP signal.
In vitro expansion and in vivo transfer of T cells
Sorted T cells were stimulated with anti-CD3 and anti-CD28Dynabeads (Invitrogen) at a 1:1 cell:bead ratio, supplemented with 2,000 IU/ml rhIL-2 in complete medium, which consisted of 10% heat-inactivated fetal bovine serum (Biosource International), nonessential amino acids, 0.5 mM sodium pyruvate, 5 mM Hepes, 1 mM glutaMax I (all from Invitrogen), and 55 μM β-mercaptoethanol (Sigma-Aldrich) in DMEM base. The cultures were monitored daily and maintained at 0.7–1 × 106/ml by diluting with IL-2-supplemented complete medium for 7–9 d. At the end of the culture, the anti-CD3 and anti-CD28 beads were removed by centrifugation on Ficoll. 0.5–1 × 106 cells were transferred i.v. to NOD Rag2−/− or NOD.Tcra−/− mice. Blood glucose concentrations were determined by an Accu-Chek glucose meter (Roche).
Foxp3 methylation analysis
Sort purified cells were subject to bisulphite sequence analysis as performed 18. Methylation specific PCR primer sequences were forward 5′-TATTTTTTTGGGTTTTGGGATATTA-3′, reverse 5′-AACCAACCAACTTCCTACACTATCTAT-3′.
TCR repertoire analysis
YFP−GFP−, YFP+GFP−, and YFP+GFP+ Vα2 TCR clones (384 each) were sequenced by McLab. A python script was used to extract the full length sequences from ab1 files, which were then trimmed to include only the PCR amplified TCR Vα2 domains. The sequences were submitted by batch to the IMGT website for V-gene and J-gene analysis (www.imgt.org). Of the original 384 per subset clones, 227 Treg, 202 YFP+GFP−, and 236 Tconv cells had productive V-and J-gene rearrangements. A separate script was used to compile occurrences of each oligonucleotide and CDR3 peptide sequence within each cell population, and to compare occurrences among the three populations.
Isolation of lymphocytes from pancreas
The pancreas was excised independent of lymph nodes and incubated in 1mg/mL collagenase P (Roche), 20μg/mL DNase, 0.2% BSA in RPMI for 30 minutes at 37°C. The digested tissue was strained through a 20uM cell strainer (BD Biosciences) and washed once with RPMI containing DNase. Lymphocytes were separated from low density pancreatic cells by centrifugation through a 40%/60% percoll gradient. Lymphocytes were washed and stained for flow cytometric analysis.
Supplementary Material
Acknowledgments
The authors would like to thank C.J. McArthur, M. Lee, N. Grewal, S. Jiang, J. Beilke and S. Zhu for technical assistance and N. Martinier and D. Fuentes for expert animal husbandry. We thank A. Abbas, Q. Tang, M. Anderson, R. Locksley and members of the Bluestone laboratory for helpful discussions and reading the manuscript. This work was supported by National Institutes of Health Grants P01 AI35297 and U19 AI056388 (to JAB), P30 DK63720 (for core support). XZ was supported by an ADA Mentor-Based Award. LTJ was supported by a fellowship for prospective researchers from the Swiss National Science Foundation (PBBSB-118644), the Roche Research Foundation and the Novartis Foundation formerly Ciba-Geigy Jubilee Foundation. CP was supported by NIGMS grant # 1 R25 GM56847.
References
- 1.Tang Q, et al. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–97. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bluestone JA, Tang Q, Sedwick CE. T regulatory cells in autoimmune diabetes: past challenges, future prospects. J Clin Immunol. 2008;28:677–84. doi: 10.1007/s10875-008-9242-z. [DOI] [PubMed] [Google Scholar]
- 3.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–70. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 4.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 5.Bennett CL, Ochs HD. IPEX is a unique X-linked syndrome characterized by immune dysfunction, polyendocrinopathy, enteropathy, and a variety of autoimmune phenomena. Curr Opin Pediatr. 2001;13:533–8. doi: 10.1097/00008480-200112000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–1. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 7.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 8.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nat Rev Immunol. 2003;3:253–7. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 9.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 10.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–84. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 11.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–91. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–41. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Kishimoto H, Sprent J. The thymus and negative selection. Immunol Res. 2000;21:315–23. doi: 10.1385/IR:21:2-3:315. [DOI] [PubMed] [Google Scholar]
- 16.Jordan MS, et al. Thymicselection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–6. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 17.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol. 2009;9:83–9. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 18.Floess S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–51. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polansky JK, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–63. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 23.Tang Q, et al. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J Exp Med. 2004;199:1455–65. doi: 10.1084/jem.20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J Exp Med. 2007;204:2039–45. doi: 10.1084/jem.20070822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Q, Bluestone JA. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat Immunol. 2008;9:239–44. doi: 10.1038/ni1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomon B, et al. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–40. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 27.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–10. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh CS, Rudensky AY. The role of TCR specificity in naturally arising CD25+ CD4+ regulatory T cell biology. Curr Top Microbiol Immunol. 2005;293:25–42. doi: 10.1007/3-540-27702-1_2. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–11. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lochner M, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+T cells. J Exp Med. 2008;205:1381–93. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–73. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–6. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009 doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–27. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose NR. The adjuvant effect in infection and autoimmunity. Clin Rev Allergy Immunol. 2008;34:279–82. doi: 10.1007/s12016-007-8049-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.