Abstract
The INFINITI HPV-QUAD assay is a commercially available genotyping platform for human papillomavirus (HPV) that uses multiplex PCR, followed by automated processing for primer extension, hybridization, and detection. The analytical performance of the HPV-QUAD assay was evaluated using liquid cervical cytology specimens, and the results were compared with those results obtained using the digene High-Risk HPV hc2 Test (HC2). The specimen types included Surepath and PreservCyt transport media, as well as residual SurePath and HC2 transport media from the HC2 assay. The overall concordance of positive and negative results following the resolution of indeterminate and intermediate results was 83% among the 197 specimens tested. HC2 positive (+) and HPV-QUAD negative (−) results were noted in 24 specimens that were shown by real-time PCR and sequence analysis to contain no HPV, HPV types that were cross-reactive in the HC2 assay, or low virus levels. Conversely, HC2 (−) and HPV-QUAD (+) results were noted in four specimens and were subsequently attributed to cross-contamination. The most common HPV types to be identified in this study were HPV16, HPV18, HPV52/58, and HPV39/56. We show that the HPV-QUAD assay is a user friendly, automated system for the identification of distinct HPV genotypes. Based on its analytical performance, future studies with this platform are warranted to assess its clinical utility for HPV detection and genotyping.
Specific HPV genotypes have been associated with an increased risk of progression to cervical precancer and cancer.1,2,3 In the United States, 70% of cervical cancers are caused by HPV types 16 and 18,4 with HPV16 more commonly associated with squamous cell carcinoma and HPV18 with adeno- and adenosquamous- carcinoma.5 Infection with HPV types 16 and 18 has been also associated with a higher risk for progression to carcinoma in situ III or greater.2,3,6,7,8 Additional evidence suggests that persistent infection with the same HPV carcinogenic type may confer increased risk for high grade lesions.9,10,11
Although the clinical utility of detecting and monitoring the persistence of carcinogenic HPV types continues to evolve, commercial methods for genotyping are becoming available. The biological relevance of HPV genotyping for routine use has not been well studied by prospective clinical trials with long term follow-up to determine clinical outcomes. However, commercial genotyping methods may provide clinicians information about infections with specific HPV types that may be helpful in certain clinical contexts. In this study, the analytical performance of one commercially available assay for determining HPV genotype is presented and compared with the FDA-approved digene Hybrid Capture 2 High-Risk HPV DNA Test (HC2) assay.
Materials and Methods
Cervical Specimens
Cervical specimens submitted to ARUP Laboratories for routine HPV DNA testing by the digene Hybrid Capture 2 (HC2) High Risk (HR) HPV DNA Test (Qiagen, Valencia, CA) were de-identified according to an approved protocol by the Institutional Review Board. Cervical specimens were submitted in either HC2 DNA cervical collection brush and specimen transport medium (STM, Qiagen, Valencia, CA), SurePath transport media (BD Franklin Lakes, NJ), or PreservCyt transport media for ThinPrep (Cytyc, Marlborough, MA). Residual material from original specimens following HC2 HPV testing was available for direct genotyping of specimens with the SurePath and PreservCyt media, but not for the HC2 STM specimens. Denatured material was available for all specimen types. Selected specimens were enriched for HC2 positive results but were otherwise blinded during analysis.
HC2 HPV DNA Assay
The HC2 assay was performed with a pool of RNA probes to HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 in the Infectious Disease Clinical Laboratory at ARUP Laboratories by the manufacturer's protocol. Results were reported based on the ratio of the specimen relative light unit divided by the established cutoff. Specimens with relative light unit/cutoff ratios greater than 2.5 were reported as positive for high risk HPV. Any specimens with relative light unit/cutoff ratios less than 1.0 were reported as negative for high risk HPV. When the ratio was >1.0 and <2.5 the specimen was considered equivocal. In the testing algorithm at ARUP Laboratories, an equivocal specimen was repeated two times and reported with the result (either positive, negative, or equivocal) that repeated two out of the three tests.
DNA Preparation
DNA was extracted from all specimen types using the QIAamp MinElute Media Kit (Qiagen) as described in the package insert. Briefly, the liquid remaining from the original SurePath and PreservCyt cervical specimens was vortexed, and 1 ml was removed and centrifuged at 14,000 rpm for 5 minutes to pellet cellular material. A volume of 750 μl of the supernatant was removed and discarded. To the remaining 250 μl, 80 μl of Buffer ATL and 20 μl Proteinase K were added and the specimens were vortexed and incubated for 30 minutes in a 56°C heat block. The only material available for the HC2 STM specimens was the denatured material remaining after processing through the HC2 HPV test. DNA from the denatured HC2 STM specimens, as well as from denatured SurePath specimens, was extracted by processing 200 μl of the denatured specimens. As for the pelleted specimens, 80 μl of Buffer ATL and 20 μl Proteinase K were added and the specimens were vortexed and incubated for 30 minutes in a 56°C heat block. Buffer AL containing carrier RNA was added at a volume of 250 μl to each tube and the specimens were incubated for 15 minutes in a 70°C heat block. Following incubation, 300 μl of ethanol was added and the tubes were held at room temperature for 5 minutes. The lysed solutions were added to the MinElute columns and pulled through by vacuum application. Following rinses with Buffer AW2 and ethanol, the DNA was eluted into a clean tube with centrifugation after the addition of 120 μl or 50 μl Buffer AVE to the original and denatured specimens respectively. The DNA was stored at −20°C. The DNA concentrations of the extracted samples ranged from 6.8 to 308 ng/uL based on A280 nm measurements.
HPV-QUAD Genotyping
The AutoGenomics HPV-QUAD assays targets the E1 gene of the HPV genome. Multiplex PCR for five individual HPV types (16, 18, 31, 33, and 45), five combinations of HPV types (35/68, 39/56, 58/52, 59/51, and 6/11), and a β globin internal control was performed on extracted DNA using Platinum TaqDNA Polymerase (Invitrogen, Carlsbad, CA) and an amplification mix provided with the INFINITI HPV-QUAD Assay (AutoGenomics, Carlsbad, CA). The reaction volume included 5 μl of DNA, 9.75 μl of amplification mix, and 0.25 μl of Platinum Taq (5 U/μL). Amplification was done in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) according to the AutoGenomics protocol. Following amplification of products of <300 bp in length, the PCR plate was placed on the INFINITI Analyzer with the appropriate Intellipac reagent module, BioFilmChip microarrays, buffers, and disposables. Inventory and worklists were generated and the instrument run was started. The INFINITI Analyzer automatically added labeled nucleotides, tagged primers, and other reagents for allele specific primer extension, which was done with a 1 minute hold at 95°C, followed by 40 cycles of 56°C for 20 seconds, and 95°C for 20 seconds. The instrument then added hybridization buffer and moved the solutions to the microarray chips, which were placed in an on-board hybridization chamber. The microarray chips were incubated at 39°C for 90 minutes to allow the labeled and tagged allele specific primer extension product to hybridize to the microarray chips. The chips were washed, dried, and then moved to an optics station for detection. HPV genotype results were based on the ratio of the fluorescence signal of the HPV genotype to the fluorescence signal of the nonspecific oligonucleotide background control spots. If the ratio was ≥2.0 the specimen was positive for the particular HPV type(s), if the ratio was <2.0 and the β globin control ratio was ≥1.6, the specimen was negative for the particular HPV type(s). Specimens with HPV ratios <2.0 and β globin ratio <1.6 were repeated and reported as indeterminate if there was no change. Specimens with HPV ratios >1.0 and <2.0 and β globin ratio ≥1.6 were considered intermediate and the extracted nucleic acid was submitted for repeat testing one time by PCR and INFINITI analysis and the second result was reported.
Real-Time PCR and Sequence Analysis
Real-time PCR was performed using MY09 and MY11 consensus primers.12 Amplification was done in 10 μl PCR mixture containing 20 mmol/L Tris (pH8.4), 50 mmol/L KCl, 250 μg/ml bovine serum albumin, 3.5 mmol/L MgCl2, 200 μmol/L each dATP, dCTP, and dGTP, and 600 μmol/L dUTP (Roche Applied Science, Indianapolis, IN), 1× LCGreen PLUS (Idaho Technology, Salt Lake City, UT), 0.01 U AmpErase uracil N-glycosylase (Applied Biosystems, Foster City, CA), 0.5 U Platinum TaqDNA Polymerase (Invitrogen, Carlsbad, CA), 0.25 μmol/L each primer, and 1 μl DNA specimen. PCR was initiated with a 10 minutes hold at 37°C for contamination control by uracil N-glycosylase and a 10 minutes hold at 95°C for polymerase activation. Thermal cycling was done in a LightCycler 1.5 Instrument (Roche) for 50 cycles with denaturation at 95°C for 10 s, annealing at 57°C for 30 s, and extension at 72°C for 30 seconds. All transition rates were 20°C/s with fluorescence acquisition at the extension step. Melting analysis was done by heating to 94°C and cooling to 65°C at 20°C/s then melting at 0.05°C/s with continuous acquisition of fluorescence until 95°C. Sequencing was done with MY consensus primers following removal of excess reagents using the MinElute PCR Purification Kit (Qiagen, Valencia, CA) following the manufacturer protocol. Specimens were sequenced using dye terminator sequencing chemistry at the ARUP sequencing facility on a 3730 DNA Analyzer (Applied BioSystems).
Statistical Analysis
Agreement between the HC2 and the HPV HPV-QUAD was analyzed with the Kappa statistic. All calculations were performed using SAS software, version 9.1 (Cary, NC).
Results
Forty-seven PreservCyt, 56 SurePath, 41 denatured SurePath, and 53 denatured HC2 STM specimens were tested for a total of 197 specimens. Results from HC2 HR HPV testing were available for all specimens; no cytology results were available. HC2 results for the selected 197 cervical specimens analyzed in this study are presented in Table 1 and included 134 HC2 positive, 58 HC2 negative, and 5 HC2 equivocal specimens. For a frame of reference, the unselected spectrum of HC2 results from patient specimens at ARUP Laboratories generally consists of 70% negative, 27% positive, and 3% equivocal (unpublished data).
Table 1.
Hybrid Capture 2 Results for Cytology Specimens Tested
| Number of specimens |
||||
|---|---|---|---|---|
| HC2* Result: | Positive | Negative | Equivocal | Total |
| RLU/CO†: | >2.5 | <1.0 | >1.0 <2.5 | |
| SurePath transport media | 38 | 15 | 3 | 56 |
| PreservCyt transport media | 33 | 13 | 1 | 47 |
| Denatured cervical collection brush | 33 | 19 | 1 | 53 |
| Denatured SurePath transport media | 30 | 11 | 0 | 41 |
| Total | 134 | 58 | 5 | 197 |
HC2, Hybrid Capture 2 High Risk HPV.
RLU/CO, relative light units/cutoff.
HPV-QUAD Genotyping Results
On initial testing by the HPV-QUAD assay, 95 of 134 HC2 positive specimens were reported with at least one genotype, 34 were negative for any HPV genotype, and 5 were indeterminate. Of 58 negative HC2 specimens, 52 were reported by HPV-QUAD as negative, 2 were reported positive, and 4 were indeterminate. Five HC2 equivocal specimens were HPV-QUAD positive in two cases, negative in two cases, and indeterminate in one. The overall agreement after initial genotyping was 95 positive and 52 negative for 75% concordance (147/197).
Indeterminate Specimens
A total of 10 specimens were reported as indeterminate by HPV-QUAD assay after the first analysis (10/197 = 5%), indicating possible specimen inadequacy or PCR inhibition. When the indeterminate specimens were repeated, 9 of 10 indeterminate specimens were resolved. Based on this repeat testing, the final agreement between HC2 and HPV-QUAD assays was 98 positive and 55 negative for 78% concordance (153/197). The only specimen that remained indeterminate was also equivocal in the HC2 assay.
Intermediate Genotyping Ratios
Results with intermediate ratios on HPV-QUAD were reported as negative. Seventeen specimens that were HC2 positive and HPV-QUAD negative had intermediate HPV-QUAD ratios for one or more HPV types. Additionally, one HC2 negative/HPV-QUAD negative specimen had an intermediate ratio. Assuming that the signal intensity is related to the amount of virus present, the specimens with ratios intermediate between 1.0 and 2.0 may contain low amounts of HPV. When the 18 specimens with intermediate ratios were repeated, 13 were identified as containing HPV types that were consistent with genotypes initially detected with intermediate ratios, 4 were reported negative, and 1 was indeterminate. These data are presented in Table 2.
Table 2.
Repeat HPV-QUAD Results for Specimens Reported as Negative (<2.0) but with Intermediate Signal Ratios (>1.0 <2.0)
| Specimens with positive repeat results | ||||||
|---|---|---|---|---|---|---|
| Run 2 |
Run 3 |
|||||
| Specimen # | HPV type(s) | Run 1* Ratio | Ratio | Result | Ratio | Result |
| 1 | 52/58 | 1.3 | 2.6 | pos | ||
| 2 | 31 | 1.0 | 10.4 | pos | ||
| 3 | 35/68 | 1.4 | 3.3 | pos | ||
| 4 | 35/68 | 1.6 | 2.2 | pos | ||
| 5 | 6/11 | 1.9 | 2.5 | pos | ||
| 6 | 35/68 | 1.6 | 2.2 | pos | ||
| 7 | 52/58 | 1.7 | 4.5 | pos | ||
| 8 | 52/58 | 1.6 | 2.6 | pos | ||
| 9 | 31 | 1.5 | 4.4 | pos | ||
| 10 | 39/56 | 1.9 | 1.9 | neg | ||
| 51/59 | 1.2 | 2.1 | pos | |||
| 52/58 | 1.7 | 2.5 | pos | |||
| 11† | 18 | 0.0 | 8.6 | pos | 0.0 | neg |
| 51/59 | 1.5 | 7.2 | pos | 3.1 | pos | |
| 12 | 16 | 0.4 | 3.4 | pos | ||
| 39/56 | 1.1 | 11.6 | pos | |||
| 13 | 16 | 1.0 | 4.1 | pos | ||
| 18 | 1.8 | 0.0 | neg | |||
| 39/56 | 1.2 | 3.4 | pos | |||
| Specimens with negative repeat results | ||||
|---|---|---|---|---|
| Run 2 |
||||
| Specimen # | HPV type(s) | Run 1 Ratio | Ratio | Result |
| 14 | 35/68 | 1.1 | 1.0 | neg |
| 15 | 51/59 | 1.3 | 1.7 | neg |
| 16 | 39/56 | 1.4 | 0.4 | neg |
| 17 | 6/11 | 1.4 | 1.7 | neg |
| 18‡ | 18 | 1.6 | 1.9 | neg/IND |
All specimens were positive in the HC2 assay, except Specimen 13.
All Specimens in Run 1 were reported as negative with HPV ratios <2.0.
The result for Specimen 11on Run 2 indicated the presence of HPV18. This was likely contamination and was not present in Run 3.
Specimen 18 was indeterminate (IND) on Run 2 (insufficient beta globin amplification).
HPV Genotype Results for All Specimen Types
Following the repeat testing of indeterminate specimens and those with intermediate ratios, the concordance between HC2 and HPV-QUAD analysis for all 197 specimens was 83% (kappa 0.65 [95% CI = 0.55−0.76]). On subset analysis, the results for the cervical collection brush specimens showed 87% agreement (kappa 0.73 [95% CI = 0.56−0.91]), SurePath transport media specimens had 82% agreement (kappa 0.67 [95% CI = 0.48−0.86]), denatured SurePath transport media specimens had 85% agreement (kappa 0.68 [95% CI = 0.46−0.91]), and PreservCyt specimens had a 77% agreement (kappa 0.52 [95% CI = 0.29−0.75]). There was no significant difference in agreement between these subsets using the kappa statistics.
Resolution of HC2 Positive/HPV-QUAD Negative Specimens
A total of 24 specimens were positive in the HC2 assay and negative in the HPV-QUAD assay. Following repeat testing, five of the specimens clearly remained negative in the HPV-QUAD assay. These specimens were evaluated in a real-time PCR using MY consensus primers. None of the specimens amplified well by PCR, all had crossing points of >40 cycles and had small or no melting peaks. Despite poor amplification, three of the specimens were submitted for sequencing with MY primers, and no HPV was identified.
Real-time PCR was performed on 12 other HC2 positive/HPV-QUAD negative specimens. Definite negative results were reported in the HPV-QUAD assay for 10 of the specimens and two specimens had low levels of HPV6/11 as evidenced by HPV ratios >1.0 and <2.0. These 12 specimens amplified well with MY primers and were submitted for sequencing. Among these specimens that were negative in the HPV-QUAD assay, HPV type 53 was identified by sequencing in three and HPV type 66 was identified in five specimens. One of the specimens with low level HPV6/11 contained HPV type 11 and one contained HPV type 53. The remaining specimens contained HPV types 64, 70, and 82. The HC2 package insert states that the assay cross-reacts with HPV types 53 and 66, and cross-reactivity to HPV types 11, 64, and 70 has been reported.13,14 None of these HPV types are targets in the either the HC2 assay or the HPV-QUAD assay. When the 17 discordant specimens that were shown to be false positive in the HC2 assay were re-assigned as negative, there was agreement between 109 HPV positive and 71 HPV negative specimens for an overall agreement of 91%.
False negative HPV-QUAD results were noted in seven specimens. Real-time PCR amplification and sequence analysis of these specimens was performed and sequencing identified HPV type 16 in one specimen, HPV 35 in two specimens, HPV 56 in three specimens, and HPV 59 in one specimen. When these specimens were repeated in the HPV-QUAD assay, there was evidence, based on HPV ratios >1.0 and <2.0, of low levels of HPV in four of the specimens. In each case, the HPV types with intermediate ratios were confirmed by sequence analysis. A summary of the results for all HPV-QUAD negative/HC2 positive specimens is presented in Table 3.
Table 3.
Resolution of Hybrid Capture 2 (HC2) Positive/HPV-QUAD Negative Specimens
| Likely false positive HC2 | Cross-reactive HC2 | Cross-reactive HC2 | False negative HPV-QUAD | False negative HPV-QUAD low HPV ratios* | |
|---|---|---|---|---|---|
| HPV-QUAD result: (# specimens) | Negative (5) | Negative (10) | HPV6/11† (2) | Negative (3) | HPV39/56 (1) HPV35/68 (2) HPV51/59 (1) |
| Real-time PCR‡ Cp: | >40 | 26.1 to 38.1 | 25.6 to 21.2 | 37.1 to 40.5 | 29.0 to 39.6 |
| Tm: | None | 79.8 to 81.8°C | 81.3 to 81.2°C | 78.5 to 79.4°C | 78.1 to 79.8°C |
| Sequencing result: | No HPV | HPV53 (2) | HPV53 | HPV16 | HPV56 |
| HPV66 (5) | HPV11 | HPV56 (2) | HPV35 (2) | ||
| HPV70 | HPV59 | ||||
| HPV64 | |||||
| HPV82 |
HPV ratios (R) were 39/56 r = 0.73, 35/68 r = 1.22, 35/68 r = 1.11, 51/59 r = 1.31.
HPV6/11 are low risk HPV types.
Real-time PCR and sequencing performed using MY consensus primers as described in the Materials and Methods. Cp, crossing point; Tm, melting temperature.
Resolution of HC2 Negative/HPV-QUAD Positive Specimens
A total of four specimens were reported as negative in the HC2 assay and positive in the HPV-QUAD assay. One specimen was first reported as HPV18, but was negative on a subsequent run. There was no amplification of this specimen in the real-time PCR assay using MY consensus primers. This specimen was located next to another specimen that was a strong positive for HPV18. A second specimen had HPV16 and HPV39/56 ratios that were >1.0 but <2.0, and repeated positive for HPV16 and HPV39/56. Following a third repeat, the specimen was negative, but with an HPV16 ratio of 1.37. This specimen did not amplify in the real-time PCR assay and was surrounded by specimens that were positive for HPV16 in each run. The third specimen was positive for HPV18 and HPV39/56 in two runs. This specimen amplified in the real-time PCR and sequencing identified HPV62, considered a non-oncogenic HPV type. The fourth specimen was positive for HPV16 then negative in two repeat runs. This specimen amplified poorly in the real-time PCR but was shown to contain HPV18 by sequencing. In each case the specimens were processed near other HPV positive specimens and cross-contamination during set-up was likely. Cross-contamination from instrument processing is less likely since the instrument processes in a linear manner and the contamination noted was random.
HPV Types Identified in the HPV-QUAD Assay
There were 109 specimens that were identified in the HPV-QUAD assay with at least one HPV type. Specific single infections were noted in 33 specimens with HPV16 (33/109, 30%), 6 specimens with HPV18 (6/109, 5%), 8 specimens with HPV31 (8/109, 7%), 1 specimen with HPV33 (1/109, 1%), and 5 specimens with HPV45 (5/109, 5%). Since there was no differentiation of HPV types when HPV35/68, HPV39/56, HPV58/52, HPV59/51, and HPV6/11 were reported, mixed infections were difficult to specifically identify. There were 18 specimens reported with at least 2 HPV types and 2 specimens reported, with at least 3 types for a total of 20 specimens with mixed infections (20/109, 18%). The total distribution of HPV types identified in this study is presented in Table 4. Specific single HPV types were identified in 53 specimens, 2 specific HPV types were identified in 5 specimens, at least 1 HPV type was identified in 36 specimens, at least 2 HPV types were noted in 13 specimens, and at least 3 HPV types in 2 specimens.
Table 4.
Distribution of HPV Types Identified in 109 Specimens in the HPV-QUAD Assay
| HPV type(s) identified | Number of specimens | HPV type(s) identified | Number of specimens |
|---|---|---|---|
| 16 | 33 | 16, 18 | 1 |
| 52/58 | 15 | 16, 45 | 1 |
| 39/56 | 12 | 18, 31 | 1 |
| 31 | 8 | 16, 35/68 | 1 |
| 18 | 6 | 16, 6/11 | 1 |
| 45 | 5 | 18, 51/59 | 1 |
| 35/68 | 5 | 33, 35/68 | 1 |
| 51/59 | 4 | 39/56, 52/58 | 1 |
| 16, 39/56 | 3 | 51/59, 52/58 | 1 |
| 16, 31 | 2 | 16, 18, 39/56 | 1 |
| 16, 52/58 | 2 | 35/68, 39/56, 51/59 | 1 |
| 18, 52/58 | 2 | 6/11 | 0 |
| 33 | 1 |
Reproducibility of HPV Genotype Calls
Thirty-seven of the 109 specimens identified as containing HPV genotypes were repeated at least one time. Twenty-eight of the specimens repeated with exactly the same genotype, five specimens repeated with the same genotype(s) plus an additional genotype, and four specimens lost one genotype when repeated. The variable genotypes were likely due to low level HPV as evidenced by ratios generally between 1.0 and 2.0 (see Supplementary Table S1 at http://jmd.amjpathol.org).
Reproducibility of Signal and Ratio Values
Controls routinely analyzed in each run during this study included HeLa 229 cells (ATCC number CCL-2.1) containing HPV18, CaSki cells (ATCC number CRL-1550) containing HPV16, and a negative PreservCyt specimen. The reproducibility of the signals and ratios reported over 14 separate runs was assessed by determining the minimum, lower quartile (Q1), median (Q2), upper quartile (Q3), and maximum values of the data. The box plots for the signal and ratio values generated from these data are presented in Figure 1A–B.
Figure 1.
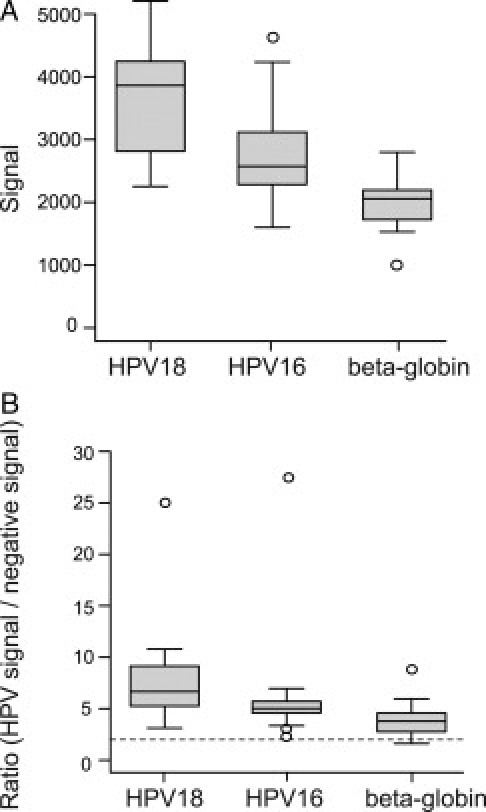
Signal and ratio values for HPV controls. Box plot data for signals (A) and HPV/negative ratios (B) obtained over 14 runs for control samples containing HPV18, HPV16, and β globin. In (B) the dotted line is the ratio cutoff level of 2. The boxes cover the 25th to 75th percentiles, the lines in the boxes represent the median values and the whiskers contain approximately 95% of the data. Open circles are outliers.
Dilution Studies
HeLa 229 (HPV18) and Ca Ski (HPV16) cells were diluted in pooled PreservCyt specimens that tested negative in the HC2 assay. The HeLa cells were assumed to contain approximately 30 HPV18 DNA copies per cell and the CaSki cells approximately 600 HPV16 DNA copies per cell.15,16 Nucleic acid extraction of the dilutions was performed as described in the Materials and Methods, and HPV genotype analysis was done in replicate in the HPV-QUAD assay. HPV18 was identified in 3/3 replicates at the level estimated to be 3000 HPV18 DNA copies/PCR. No HPV18 was detected at DNA levels below 3,000 copies/PCR. HPV16 was identified in 3/3 replicates at the level estimated to be 300 HPV16 DNA copies/PCR. No HPV16 was detected at DNA levels below 300 copies/PCR.
Discussion
The AutoGenomics INFINITI HPV-QUAD Assay is a commercially available method for the detection and identification of HPV genotypes in cervical specimens. We demonstrated an overall agreement of 83% (kappa 0.65 [95%CI = 0.55−0.76]) between the HC2 and HPV-QUAD assays for 197 specimens. Additionally, determination of specific genotypes was successfully performed with the HPV QUAD using multiple types of specimens such as SurePath or PreservCyt specimen transport media. We found that denatured material that remained after processing for the HC2 assay from cervical collection brush STM and SurePath specimens is also a suitable specimen for the HPV-QUAD genotyping assay. The ability to use residual denatured material from the HC2 assay is an important benefit for the clinical laboratory since it improves workflow by allowing reflex testing directly from the HC2 assay. Finally, use of residual material from HC2 testing circumvents the problem of insufficient quantities of original specimen for further testing.
The observed disagreement between the HC2 and HPV-QUAD assays in terms of positive and negative results was not a surprising finding since the two assays are based on different chemistries. The HC2 assay uses solid phase antibody capture of RNA:DNA hybrids and amplified chemiluminescent signal detection, while the HPV-QUAD assay relies on target DNA amplification by PCR, allele specific primer extension, and signal detection of labeled products on a microarray. These distinct methods can contribute to differences in both analytical sensitivity and specificity. The use of multiple probes in the HC2 assay may provide additional sensitivity at the cost of reduced specificity and increased cross-reactivity, potentially causing false-positive results. Exponential amplification afforded by PCR with the HPV-QUAD assay can improve sensitivity, but unexpected HPV sequence variations may affect primer and probe binding, and thus contribute to HPV-QUAD negative and HC2 positive results. Sensitivity can also depend on the volume of specimen processed and analyzed. Larger specimen volumes and adequate cellularity improve sensitivity. Additionally, nucleic acid purification and ensuring stability of specimens are important variables affecting assay performance. In this study, the HC2 assay processes a larger volume of original specimen but no purification is done; sample processing for the HPV-QUAD assay includes both concentration and purification. A limitation of this study was that specimen storage was longer before HPV-QUAD testing (∼1month) than for HC2 testing. Specimen processing differences and storage conditions could have contributed to observed differences in HC2 and HPV-QUAD results.
The number of indeterminate and intermediate results for the HPV QUAD assay may have occurred due to compromise of HPV DNA from specimen storage (ie, not performed on fresh specimens) and poor quality of original specimen from insufficient cellularity. Nine of 10 indeterminate specimens and 13 of 18 specimens with intermediate ratios >1.0 and <2.0, were resolved by repeat testing. A testing algorithm that includes repeat testing for specimens with indeterminate and intermediate results will increase the number of reportable results. It is expected that the analysis of newly acquired specimens will lower the number of indeterminate and intermediate results.
We found four specimens that were HC2 negative and HPV-QUAD positive and suspect that these discrepancies were due to technical errors during processing since most of them involved HPV types 16 and 18. Repeat analysis in the HPV-QUAD assay and real-time PCR support that these specimens were negative for HPV. The pattern of cross-contamination did not suggest an instrumentation related issue, but rather emphasizes the need for careful handling of specimens and reagents during nucleic acid extraction and PCR setup. Conversely, 24 specimens were negative in the HPV-QUAD assay and positive in the HC2 assay. Real-time PCR and sequencing indicated that five of the specimens contained no detectable HPV DNA and were likely false positive in the HC2 assay. An additional 12 specimens were shown to contain HPV types that are known to cross-react in the HC2 assay and 7 specimens contained low levels of HPV DNA.
The automation conferred with the INFINITI system provides a reliable and convenient platform for walk-away analysis in the clinical laboratory. The assay chemistry is robust and provides reproducible genotyping results. Further understanding of the clinical implications of HPV genotyping will require well-designed and controlled studies with supplemental patient results and outcomes. With the availability of the INFINITI assay and other HPV genotyping assays, the appropriate studies can be done. It is anticipated that genotyping will enhance patient care and improve outcomes when used as a reflex and adjunct test in some clinical situations.
Acknowledgements
We thank Andrew Wilson for help with statistical analyses.
Footnotes
Supported by the ARUP Institute for Clinical and Experimental Pathology and AutoGenomics, Inc., Carlsbad, California (reagents).
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Supplementary data
References
- 1.Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, Clifford GM. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–632. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 2.Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, Rush BB, Glass AG, Schiffman M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific hpv testing in clinical practice. J Nat Cancer Inst. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- 3.Castle PE, Solomon D, Schiffman M, Wheeler CM. Human papillomavirus type 16 infections and 2-year absolute risk of cervical precancer in women with equivocal or mild cytologic abnormalities. J Nat Cancer Inst. 2005;97:1066–1071. doi: 10.1093/jnci/dji186. [DOI] [PubMed] [Google Scholar]
- 4.Wallin K-L, Wiklund F, Angstrom T, Bergman F, Stendahl U, Wadell G, Hallmans G, Dillner J. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N Engl J Med. 1999;341:1633–1638. doi: 10.1056/NEJM199911253412201. [DOI] [PubMed] [Google Scholar]
- 5.Clifford GM, Rana RK, Franceschi S, Smith JS, Gough G, Pimenta JM. Human papillomavirus genotype distribution in low-grade cervical lesions: comparison by geographic region and with cervical cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1157–1164. doi: 10.1158/1055-9965.EPI-04-0812. [DOI] [PubMed] [Google Scholar]
- 6.Berkhof J, Bulkmans NW, Bleeker MC, Bulk S, Snijders PJ, Voorhorst FJ, Meijer CJ. Human papillomavirus type-specific 18-month risk of high-grade cervical intraepithelial neoplasia in women with a normal or borderline/mildly dyskaryotic smear. Cancer Epidemiol Biomarkers Prev. 2006;15:1268–1273. doi: 10.1158/1055-9965.EPI-05-0764. [DOI] [PubMed] [Google Scholar]
- 7.Bulkmans NW, Bleeker MC, Berkhof J, Voorhorst FJ, Snijders PJ, Meijer CJ. Prevalence of types 16 and 33 is increased in high-risk human papillomavirus positive women with cervical intraepithelial neoplasia grade 2 or worse. Int J Cancer. 2005;117:177–181. doi: 10.1002/ijc.21210. [DOI] [PubMed] [Google Scholar]
- 8.Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castle PE. Invited commentary: is monitoring of human papillomavirus infection for viral persistence ready for use in cervical cancer screening? Am J Epidemiol. 2008;168:138–144. doi: 10.1093/aje/kwn037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kjaer SK, van den Brule AJC, Paull G, Svare EI, Sherman ME, Thomsen BL, Suntum M, Bock JE, Poll PA, Meijer CJ. Type specific persistence of high risk human papillomavirus (HPV) as indicator of high grade cervical squamous intraepithelial lesions in young women: population based prospective follow up study. BMJ. 2002;325:572–579. doi: 10.1136/bmj.325.7364.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koshiol J, Lindsay L, Pimenta JM, Poole C, Jenkins D, Smith JS. Persistent human papillomavirus infection and cervical neoplasia: a systematic review and meta-analysis. Am J Epidemiol. 2008;168:123–137. doi: 10.1093/aje/kwn036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, Burk RD. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35:1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castle PE, Lorincz AT, Scott DR, Sherman ME, Glass AG, Rush BB, Wacholder S, Burk RD, Manos MM, Schussler JE, Macomber P, Schiffman M. Comparison between prototype hybrid capture 3 and hybrid capture 2 human papillomavirus dna assays for detection of high-grade cervical intraepithelial neoplasia and cancer. J Clin Microbiol. 2003;41:4022–4030. doi: 10.1128/JCM.41.9.4022-4030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castle PE, Schiffman M, Burk RD, Wacholder S, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Lorincz A. Restricted cross-reactivity of hybrid capture 2 with nononcogenic human papillomavirus types. Cancer Epidemiol Biomarkers Prev. 2002;11:1394–1399. [PubMed] [Google Scholar]
- 15.Yee C, Krishnan-Hewlett I, Baker C, Schlegel R, Howley P. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–363. [PMC free article] [PubMed] [Google Scholar]
- 16.Walboomers J, Jacobs M, Manos M, Bosch F, Kummer J, Shah K, Snijders P, Peto J, Meijer C, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


