Abstract
The most frequent KIT mutations reported in core-binding factor acute myeloid leukemia are point mutations and insertions/deletions in exons 17 and 8. The vast majority of KIT mutation detection procedures are time-consuming, costly, or with a high lower limit of detection. High-resolution melting (HRM) is a gene scanning method that combines simplicity and rapid identification of genetic variants. We describe an HRM method for the simultaneous screening of exons 8 and 17 KIT mutations and report the results obtained in 69 core-binding factor acute myeloid leukemia patients. Mutation detection was compared with sequencing as the gold standard. The HRM method used high-resolution melting master reagents (Roche) and the LightCycler 480 (Roche) platform. HRM was reproducible, showed a lower limit of detection of 1%, and discriminated all patients with mutated KIT from controls without false positive or false negative results. Additionally, most of the mutations were differentiated from the other mutations. KIT mutations were present in 15.9% of patients, showing a higher incidence in inv(16) (25.8%) than in t(8;21) (7.9%). The presence of a KIT mutation was associated with a high white blood cell count, and adult patients with an exon 17 mutation had a higher incidence of relapse. These findings verify that HRM is a reliable, rapid, and sensitive method for KIT mutation screening. Furthermore, our study corroborates the unfavorable prognosis associated with exon 17 KIT mutations.
Activating mutations in tyrosine kinases, FLT3, KRAS, NRAS, and KIT, are found in more than 30% of patients with de novo acute myeloid leukemia (AML).1 These tyrosine kinase mutations confer a proliferative or survival advantage to blastic cells and may cooperate with mutated transcription factors such as CBF complex, RARa, and MLL, by interfering with differentiation and contributing to the AML phenotype.2
KIT gene encodes a transmembrane glycoprotein receptor that is member of the type III receptor tyrosine kinase family. The structure of this receptor consists of three domains: an extracellular domain composed of five immunoglobulin-like motives, a transmembrane and juxtamembrane domain, and an intracellular tyrosine kinase domain.3,4 After stem cell factor binding to the KIT receptor, the receptor dimerizes and becomes phosphorylated, activating the downstream signaling pathways implicated in cell proliferation, differentiation, and survival.4
Ligand-independent activation of KIT receptor may be achieved by gain-of-function mutations reported in AML as well as in several other malignancies. Activating KIT mutations are most frequently located within exon 17, which encodes the KIT activation loop in the kinase domain, and exon 8, which encodes a region of the extracellular domain of the KIT receptor.3,5,6 The most frequent KIT mutations reported are point mutations at codon 816 of exon 17 and insertions/deletions in exon 8.
KIT mutations have been reported to occur preferentially in AML with core-binding factor (CBF) rearrangements such as RUNX1-RUNX1T1 and CBFb-MYH11, caused by the t(8;21) and inv(16) or t(16;16), hereafter abbreviated as inv(16). According to different reports, the incidence of KIT mutations in these malignancies varies widely from 6% to 46%.5,6,7,8 Several studies have shown an adverse prognostic value for exon 17 KIT mutation in CBF AML3,5,6; however, the relevance of exon 8 mutations still remains unknown.
Various sensitive methods have been used to detect KIT mutations in CBF AML, such as polymerase chain reaction (PCR) followed by the screening of heteroduplex by high-performance liquid chromatography,3,5,9 sequencing,10 and real-time PCR using hybridization probes.6 However, these methods are expensive, time-consuming, or have a reduced sensitivity, not allowing the detection of these mutations in samples with low blast cell count.
The recent development of platforms and software for high-resolution melting (HRM) analysis11 has provided the introduction of this technology in the clinical laboratory. The great advantages of HRM (sensitivity, rapidity, simplicity, low cost, and high throughput) have justified the widespread use of these methods for detection of many small genetic variations.12,13,14 We report the results obtained in a group of 69 CBF AML patients by using a fast, sensitive, and reliable HRM method that allows the simultaneous screening of exons 8 and 17 KIT mutations.
Materials and Methods
Patients
The study includes 69 CBF AML patients recruited between 1998 and 2008: 55 adults [30 with t(8;21), 24 with inv(16), and one with t(16;16)] and 14 children [8 with t(8;21) and six with inv(16)], who were diagnosed in four Spanish institutions. The leukemia subtypes were established according to the morphological and cytochemical criteria of the French-American-British (FAB) classification.15 The presence of t(8;21) or inv(16) was detected by standard cytogenetics and the corresponding rearrangements, RUNX1-RUNX1T1 and CBFb-MYH11, by reverse transcription quantitative PCR (RQ-PCR).
Treatment
Patients received intensive chemotherapy following the PETHEMA LMA 96, LMA 99, and LMA 2006 protocols for adults in which induction therapy consisted of standard combinations of anthracycline plus cytarabine with or without etoposide. Children were treated according the SHOP 2001 protocol, which consisted of induction therapy with standard combinations of idarubicine plus cytarabine with or without etoposide.
The Institutional Ethics Committee for Clinical Research approved this study, and informed consent was obtained from all patients and conducted in accordance with the recommendations of the Declaration of Human Rights, the Conference of Helsinki, and institutional regulations.
Samples and DNA Extraction
KIT mutation study of exons 8 and 17 was performed on genomic DNA extracted from bone marrow samples collected during active disease (diagnosis and relapse). Genomic DNA was directly extracted from 500 μl of bone marrow using MagNA Pure LC DNA isolation kit large volume (Roche, Mannheim, Germany) automated in the MagNA Pure LC system (Roche). The quality and concentration of DNA extracted was assessed with a NanoDrop-1000 (NanoDrop Technologies Inc., Wilmington, DE).
High-Resolution Melting Analysis of KIT Mutations
All samples were tested in duplicate using LightCycler 480 high resolution melting master (Roche Diagnostics, Mannheim, Germany) x1 containing Taq, nucleotides, and the dye ResoLight. Two DNA normal controls and one positive mutated DNA sample for each exon were included in each run. The HRM assays were performed using the LightCycler 480 instrument (Roche Diagnostics, Roche Instrument Center AG, Rotkreuz, Switzerland).
Thirty nanograms of DNA were amplified in a 10-μl final volume containing 1X LightCycler 480 high resolution melting master (Roche), 0.5 μmol/L each primer pair (KIT F 5′-GCTGAGGTTTTCCAGCACTC-3′ and KIT R 5′-AATTGCAGTCCTTCCCCTCT-3′), and 2.0 mmol/L MgCl2 for exon 8 or 0.2 μmol/L of each primer pair (KIT S 5′-CAGCCAGAAATATCCTCCTTACT-3′ and KIT B 5′-TTGCAGGACTGTCAAGCAGAG-3′) and 3 mmol/L MgCl2 for exon 17. The expected size of the PCR product was 219 and 137 bp for exons 8 and 17, respectively.
The PCR program consists of an initial preheating at 95°C for 10 minutes to activate the Taq DNA polymerase, followed by 45 amplification cycles. Each cycle comprises an annealing step at 56°C for 13 seconds, an elongation step at 72°C for 14 seconds, and denaturation at 94°C for 10 seconds. The final melting program consists of three main steps beginning with a denaturation at 95°C for 1 minute, renaturation at 40°C for 1 minute, and melting from 60°C to 95°C, with a ramp of 0.02°C per second and 25 fluorescence acquisitions per degree centigrade.
Results were analyzed as fluorescence versus temperature graphs using Gene Scanning software version 1.5.0 (Roche Instrument Center, Rotkreuz, Switzerland). The melting curve analysis comprises three steps: normalization of melting curves, equaling to 100% the initial fluorescence and to 0% the fluorescence remnant after DNA dissociation; shifting of the temperature axis of the normalized melting curves to the point where the entire double-stranded DNA is completely denatured; and finally, the difference plot analyzing the differences in melting curve shape clustering the samples into groups based on the internal software calculation.12
Sequencing of PCR Products
The samples were directly sequenced from the LightCycler PCR amplification. The PCR products were mixed with ExoSap-IT (USB Corporation, Cleveland, OH) to remove the remaining primers and bidirectionally sequenced with forward and reverse primers using ABI PRISM terminator cycle sequencing kit version 1.1 (Applied Biosystems, Foster City, CA) on the ABI PRISM 3130 genetic analyzer (Applied Biosystems, Foster City, CA).
Statistical Analysis
The association of the presence of KIT mutation with the biological and demographic characteristics of leukemias was proved using the χ2 and Fisher's exact test. Unadjusted time-to-event analyses were performed using the Kaplan-Meier estimate16 and for comparisons the log-rank test.17 Disease-free survival (DFS) and relapse-free survival (RFS) were estimated from the date of complete remission. In the analysis of DFS, relapse and death in complete remission were considered uncensored events, whichever occurred first. For RFS, relapse in complete remission was considered the uncensored event. Patient follow-up was updated on June 30, 2008. All tests were two-sided, and a P value less than 0.05 was considered statistically significant. Computations were performed using the SPSS version 12.0 statistical package (Chicago, IL).
Results
Study of the High-Resolution Melting Method
Sixty-nine patients were screened for the presence of KIT mutations (67 at diagnosis and two relapses). The minigel electrophoresis of PCR products showed a single fragment with the expected size of 219 bp for exon 8 and 137 bp for exon 17. The melting analysis revealed the presence of a single peak with a mean melting temperature of 83.7°C and 79.5°C for exons 8 and 17, respectively. After normalization and temperature shift a clear difference between the group of wild-type DNA and the rest of the samples was observed. This difference was further enhanced with the difference plot, which establishes clusters of samples (Figure 1A and 1B). Hence, four positive samples in exon 8 and eight positive samples in exon 17 showed a clearly distinctive shape of the difference curve plot compared with those of the wild-type DNA clustered around baseline, thus allowing the distinction of these mutations. It should be noted that fluorescence curves obtained for the duplicates of each sample were practically superimposable either in the same assay or between assays.
Figure 1.
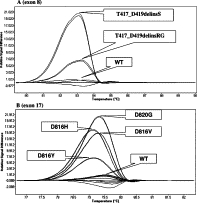
HRM method. Examples of difference plots obtained for wild-type (WT) DNA and different exon 8 (A) and exon 17 (B) KIT mutations.
Correspondence of HRM Results with Sequencing Studies
Patients' results were confirmed by sequencing directly from PCR products. Direct sequencing evidenced that all of the samples of exons 8 and 17 clustered around baseline as wild-type had normal DNA sequence, and the 11 patients classified in different clusters showed sequence variations/mutations. Considering the 69 samples analyzed by HRM, no false positive or negative results were found, yielding both sensitivity and specificity equal to 100%.
Characterization of the Mutations Detected
We identified three patients with insertion/deletions in exon 8, seven patients with point mutations in exon 17, and one patient who presented mutations in both exons. In exon 8 we found the mutations T417_D419delinsRG, T417_V422delinsSRIL, T417_D419delinsS and T417_ D419delinsG. Moreover, in exon 17 we found two D816V, two D816Y, two D816H, one D820G, and one N822K mutation (Table 1).
Table 1.
Cytogenetic and KIT Mutations Found in Exons 8 and 17
| Exon | No. of patients | Cytogenetics | Nucleotide change | Mutation description |
|---|---|---|---|---|
| 8 | 1 | inv (16) | del cttacgac ins ggggg | T417_D419delinsRG |
| 1 | inv (16) | del acttacgacaggctcg ins tcgaggatct | T417_V422delinsSRIL | |
| 1 | inv (16) | del ttacga ins gtc | T417_D419delinsS | |
| 1 | inv (16) | del ttacga ins cgg | T417_D419delinsG | |
| 17 | 2 | t (8;21), inv (16) | gac → cac | D816H |
| 2 | inv (16) | gac → gtc | D816V | |
| 2 | inv (16) | gac → tac | D816Y | |
| 1 | t (8;21) | gat → ggt | D820G | |
| 1 | t (8;21) | aat → aag | N822K | |
Lower Limit Detection of HRM
To determine the minimum detectable percentage of KIT mutated-cell DNA we serially diluted positive samples T417_D419delinsS for exon 8 and D820G for exon 17, which presented blast cell count close to 100% (1:10, 1:100, 1:300, 1:500, and 1:1000) in wild-type DNA. After performing the PCR assay we could still detect mutations for both exons up to dilution 1:100 (Figure 2A and 2B).
Figure 2.
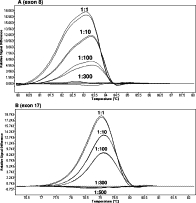
Lower limit detection of HRM assay for exons 8 and 17 KIT mutations. Differential plots obtained with the exon 8 mutation T417_D419delinsS (A) and exon 17 mutation D820G (B) serially diluted in wild-type DNA.
KIT Mutations and Biological and Demographic Characteristics of AML
The presence of KIT mutations was detected in 11 of 69 AML patients (15.9%; one infant and 10 adults); according to cytogenetic findings, three of 38 patients (7.9%) with t(8;21) and eight of 31 patients (25.8%) with inv(16) (P = 0.043). KIT mutations were associated with a high white blood cell count (P = 0.012). Thus, KIT mutated patients showed median values of white blood cells of 103*103/μl versus 20*103/μl of patients negative for KIT mutations. No association was found with age and gender.
KIT Mutations and Disease Outcome
Complete remission was achieved in 87% of patients. No association was found between KIT mutations and response to induction chemotherapy. The median follow-up of adult patients was 17 months (range, 1–88 months) with estimated probabilities of DFS and RFS of 55% and 58%, respectively.
The presence of KIT mutations in exons 8 or 17 did not influence DFS or RFS in adult patients. However, isolated exon 17 KIT mutations significantly influenced RFS (P = 0.04), although they did not modify DFS. Thus, the estimated 17-month RFS was 59% for non-mutated patients (n = 31), whereas it was 27% for patients with exon 17 mutated (n = 7) (Figure 3). With regard to the cytogenetic groups t(8;21) or inv (16), no differences were found in RFS or DFS.
Figure 3.
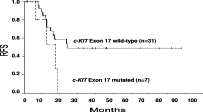
RFS in adult patients according to mutations in KIT exon 17.
Discussion
Our data support the finding that the HRM method here established allows the distinction of wild-type DNA from other DNA harboring KIT mutations in exons 8 and 17. Thus the differential plot clearly distinguishes the group of wild-type DNA samples from that with KIT mutations of AML patients. In this regard, the method is specific and sensitive enough for KIT mutation screening, since no false positive or negative results were found. Moreover, most of the mutations were well differentiated between them according to differential plot shapes, with the only exception of D816Y and D820G mutations that both were classified in the same group. In any case, all samples screened as positive for KIT mutations could be confirmed and characterized, in most cases by direct sequencing of PCR products of the HRM.13 However, in some cases we used locked nucleic acid oligonucleotides to sequence PCR products (data not shown)18,19,20 due to the high limit detection of direct sequencing.10,13 The method is reproducible enough, as it is supported by the practical identity of differential plots of duplicates.
The lower limit detection achieved with this method, 1% for exons 8 and 17 KIT mutations, is lower than that accomplished by most of the conventional methods. Furthermore, other relevant advantages of the method are rapidity, low cost, and high-throughput capability, allowing the simultaneous detection of 48 or 192 duplicate samples for the respective 96 or 384 multiwell plates. These characteristics give the HRM setup clear advantages over the methods reported to date,3,5,9,10,21 including classical real-time PCR methods with labeled probes that are expensive and only detect mutations in the fragment covered by the probes.6,22 Previously, Willmore et al23 used HRM for the screening of KIT gene mutations in gastrointestinal germ cell tumors. However, this HRM assay did not include exon 8, which is a hot spot in AML, and encompasses two steps, initial PCR in LightCycler capillary followed by HRM in an HR-1 instrument with the “drawback problem” of manipulations and time delay.
The incidence of KIT mutations in adult patients with CBF AML reported in our study, 18.2% in adults and 7.1% in children, was within the range of 6% to 48% reported for CBF AML.3,5,6,10,21,24,25,26,27,28,29,30 The wide variation in the incidence reported could be due to the size of series, demographic influences, sensitivity of the methods applied,3,6,10,13,22 and the exons studied.5,6,9,10,22
We found a higher incidence of KIT mutations in AML with inv(16) (25.8%) than in AML with t(8;21) (7.9%). This low incidence of mutations in t(8;21) patients has been previously reported by Lasa et al22 in a Spanish population and could be attributable to ethnic influences.
In accordance with other reports, the KIT mutations most frequently detected have been the point mutations at codon 816 in exon 17 within the KIT tyrosine kinase domain 2 and complex insertion/deletion mutations in exon 8 affecting the extracellular domain.3,5,30 Moreover, concordant with previous reports3,10,21 we found that all mutations in exon 17 of patients with inv(16) were located at codon D816, whereas in patients with t(8;21) translocation the mutations were scattered among different codons. D816 mutations produce ligand-independent activation of the KIT receptor, whereas exon 8 mutations cause hyperactivation of receptor in response to its natural ligand stem cell factor.26,31
The association of KIT mutations with higher levels of white blood cells found here has been reported in previous studies.3,5,24 This association may be explained by the activation of KIT caused by the mutation that would confer proliferative advantage to blast cells harboring these mutations.
In agreement with previous reports3,5 we found that adult patients harboring exon 17 mutations showed a higher relapse incidence than those patients lacking mutations, but this finding could not be verified for mutations in exon 8. However, the prognostic impact of KIT mutations remains controversial, and large series of mutated patients are needed to clarify this impact.6,8,9,22
In conclusion, we have set up a sensitive, specific, low-cost, high-throughput HRM method that allows the screening of exons 8 and 17 KIT mutations. The unfavorable prognosis conferred by KIT mutations together with a relative high incidence in CBF AML suggests that the study of these mutations should be included in the workup of this particular setting of AML. This subgroup of patients harboring KIT mutations would benefit from an aggressive up-front therapy and a future clinical trial with therapy based on tyrosine kinase inhibitors.
Footnotes
Supported in part by grants 06/0657 and RD06/0020/0031 from the Fondo de Investigación Sanitaria/Instituto de Salud Carlos III, Ministry of Health, Spain.
References
- 1.Omasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD, Payton JE, Westervelt P, DiPersio JF, Link DC, Walter MJ, Graubert TA, Watson M, Baty J, Heath S, Shannon WD, Nagarajan R, Bloomfield CD, Mardis ER, Wilson RK, Ley TJ. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukaemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renneville A, Roumier C, Biggio V, Nibourel O, Boissel N, Fenaux P, Preudhomme C. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22:915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- 3.Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW, Carroll AJ, Kolitz JE, Larson RA, Bloomfield CD, Cancer and Leukemia Group B Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;16) J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 4.Lennartsson J, Jelacic T, Linnekin D, Shivakrupa R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005;23:16–43. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 5.Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB, Colapietro P, Nichelatti M, Pezzetti L, Lunghi M, Cuneo A, Viola A, Ferrara F, Lazzarino M, Rodeghiero F, Pizzolo G, Larizza L, Morra E. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood. 2006;107:3463–3468. doi: 10.1182/blood-2005-09-3640. [DOI] [PubMed] [Google Scholar]
- 6.Schnittger S, Kohl TM, Haferlach T, Kern W, Hiddemann W, Spiekermann K, Schoch C. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107:1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 7.Shimada A, Taki T, Tabuchi K, Tawa A, Horibe K, Tsuchida M, Hanada R, Tsukimoto I, Hayashi Y. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): a study of the Japanese Childhood AML Cooperative Study Group. Blood. 2006;107:1806–1809. doi: 10.1182/blood-2005-08-3408. [DOI] [PubMed] [Google Scholar]
- 8.Care RS, Valk PJ, Goodeve AC, Abu-Duhier FM, Geertsma-Kleinekoort WM, Wilson GA, Gari MA, Peake IR, Löwenberg B, Reilly JT. Incidence and prognosis of c-KIT and FLT3 mutations in core binding factor (CBF) acute myeloid leukaemias. Br J Haematol. 2003;121:775–777. doi: 10.1046/j.1365-2141.2003.04362.x. [DOI] [PubMed] [Google Scholar]
- 9.Goemans BF, Zwaan CM, Miller M, Zimmermann M, Harlow A, Meshinchi S, Loonen AH, Hählen K, Reinhardt D, Creutzig U, Kaspers GJ, Heinrich MC. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19:1536–1542. doi: 10.1038/sj.leu.2403870. [DOI] [PubMed] [Google Scholar]
- 10.Wang YY, Zhou GB, Yin T, Chen B, Shi JY, Liang WX, Jin XL, You JH, Yang G, Shen ZX, Chen J, Xiong SM, Chen GQ, Xu F, Liu YW, Chen Z, Chen SJ. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci USA. 2005;102:1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 12.Vaughn CP, Elenitoba-Johnson KSJ. High-Resolution melting analysis for detection of internal tandem duplications. J Mol Diagn. 2004;6:211–216. doi: 10.1016/S1525-1578(10)60512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones AV, Cross NC, White HE, Green AR, Scot LM. Rapid identification of JAK2 exon 12 mutations using high resolution melting analysis. Haematologica. 2008;93:1560–1564. doi: 10.3324/haematol.12883. [DOI] [PubMed] [Google Scholar]
- 14.Rapado I, Grande S, Albizua E, Ayala R, Hernández JA, Gallardo M, Gilsanz F, Martinez-Lopez J. High resolution melting analysis for JAK2 exon 14 and exon 12 mutations: a diagnostic tool for myeloproliferative neoplasms. J Mol Diagn. 2009;11:155–161. doi: 10.2353/jmoldx.2009.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benett JM, Catovsky D, Daniel MT, Flandrin G, Galton DAG, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia: a report of the French-American-British Cooperative Group. Br J Haematol. 1976;33:451–458. [Google Scholar]
- 16.Kaplan EL, Meier P. Nonparametric estimations from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 17.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 18.Sotlar K, Escribano L, Landt O, Möhrle S, Herrero S, Torrelo A, Lass U, Horny HP, Bültmann B. One-Step detection of c-KIT Point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes. Am J Pathol. 2003;162:737–746. doi: 10.1016/S0002-9440(10)63870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallén M, Tomás E, Visakorpi T, Holli K, Mäenpää J. Endometrial K-ras mutations in postmenopausal breast cancer patients treated with adjuvant tamoxifen or toremifene. Cancer Chemother Pharmacol. 2005;55:343–346. doi: 10.1007/s00280-004-0923-x. [DOI] [PubMed] [Google Scholar]
- 20.Thiede C, Creutzig E, Illmer T, Schaich M, Heise V, Ehninger G, Landt O. Rapid and sensitive typing of NPM1 mutations using LNA-mediated PCR clamping. Leukemia. 2006;20:1897–1899. doi: 10.1038/sj.leu.2404367. [DOI] [PubMed] [Google Scholar]
- 21.Nanri T, Matsuno N, Kawakita T, Suzushima H, Kawano F, Mitsuya H, Asou N. Mutations in the receptor tyrosine kinase pathway are associated with clinical outcome in patients with acute myeloblastic leukemia harboring t(8;21)(q22;q22) Leukemia. 2005;19:1361–1366. doi: 10.1038/sj.leu.2403803. [DOI] [PubMed] [Google Scholar]
- 22.Lasa A, Carricondo MT, Carnicer MJ, Perea G, Aventín A, Nomdedeu JF. A new D816 c-KIT gene mutation in refractory AML1-ETO leukemia. Haematologica. 2006;91:1283–1284. [PubMed] [Google Scholar]
- 23.Willmore C, Holden JA, Zhou L, Tripp S, Wittwer CT, Layfield LJ. Detection of c-Kit-activating mutations in gastrointestinal stromal tumors by high-resolution amplicon melting. Am J Clin Pathol. 2004;122:206–216. doi: 10.1309/4E6U-YBY6-2N2F-CA6N. [DOI] [PubMed] [Google Scholar]
- 24.Boissel N, Leroy H, Brethon B, Philippe N, de Botton S, Auvrignon A, Raffoux E, Leblanc T, Thomas X, Hermine O, Quesnel B, Baruchel A, Leverger G, Dombret H, Preudhomme C. Acute Leukemia French Association (ALFA); Leucémies Aiguës Myéloblastiques de l'Enfant (LAME) Cooperative Groups: Incidence and prognostic impact of c-Kit, Flt3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML) Leukemia. 2006;20:965–970. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 25.Lin P, Chen L, Luthra R, Konoplev SN, Wang X, Medeiros LJ. Acute myeloid leukemia harboring t(8;21)(q22;q22): a heterogeneous disease with poor outcome in a subset of patients unrelated to secondary cytogenetic aberrations. Mod Pathol. 2008;21:1029–1036. doi: 10.1038/modpathol.2008.92. [DOI] [PubMed] [Google Scholar]
- 26.Kohl TM, Schnittger S, Ellwart JW, Hiddemann W, Spiekermann K. KIT exon 8 mutations associated with core-binding factor (CBF)-acute myeloid leukemia (AML) cause hyperactivation of the receptor in response to stem cell factor. Blood. 2005;105:3319–3321. doi: 10.1182/blood-2004-06-2068. [DOI] [PubMed] [Google Scholar]
- 27.Beghini A, Peterlongo P, Ripamonti CB, Larizza L, Cairoli R, Morra E, Mecucci C. C-kit mutations in core binding factor leukemias. Blood. 2000;95:726–727. [PubMed] [Google Scholar]
- 28.Ning ZQ, Li J, Arceci RJ. Activating mutations of c-kit at codon 816 confer drug resistance in human leukemia cells. Leukemia Lymphoma. 2001;41:513–522. doi: 10.3109/10428190109060342. [DOI] [PubMed] [Google Scholar]
- 29.Meshinchi S, Stirewalt DL, Alonzo TA, Zhang Q, Sweetser DA, Woods WG, Bernstein ID, Arceci RJ, Radich JP. Activating mutations of RTK/ras signal transduction pathway in pediatric acute myeloid leukemia. Blood. 2003;102:1474–1479. doi: 10.1182/blood-2003-01-0137. [DOI] [PubMed] [Google Scholar]
- 30.Beghini A, Ripamonti CB, Cairoli R, Cazzaniga G, Colapietro P, Elice F, Nadali G, Grillo G, Haas OA, Biondi A, Morra E, Larizza L. KIT activating mutations: incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplication. Haematologica. 2004;89:920–925. [PubMed] [Google Scholar]
- 31.Cammenga J, Horn S, Bergholz U, Sommer G, Besmer P, Fiedler W, Stocking C. Extracellular KIT receptor mutants, commonly found in core binding factor AML, are constitutively active and respond to imatinib mesylate. Blood. 2005;106:3958–3961. doi: 10.1182/blood-2005-02-0583. [DOI] [PubMed] [Google Scholar]


