Abstract
The relationship of mucoviscosity-associated (magA) and/or regulator of mucoid phenotype (rmpA) genes to the Klebsiella pneumoniae hypermucoviscosity (HMV) phenotype has been reported. We previously demonstrated that rmpA+ K. pneumoniae can cause serious disease in African green monkeys and isolated rmpA+ and magA+ HMV K. pneumoniae from other species of non-human primates. To rapidly screen African green monkeys/non-human primates for these infections, we developed three real-time PCR assays. The first was K. pneumoniae-specific, targeting the khe gene, while the others targeted rmpA and magA. Primer Express 2 was used with the three K. pneumoniae genes to generate sequence-specific TaqMan/TaqMan-Minor Groove Binder assays. Oral/rectal swabs and necropsy samples were collected; swabs were used for routine culture and DNA extraction. K. pneumoniae colonies were identified on the Vitek 2 with DNA tested using the K. pneumoniae-specific assays. Testing of 45 African green monkeys resulted in 19 khe+ samples from 14 animals with none positive for either rmpA or magA. Of these 19 khe+ samples, five were culture-positive, but none were HMV “string test”-positive. Subsequent testing of 307 non-human primates resulted in 64 HMV K. pneumoniae isolates of which 42 were rmpA+ and 15 were magA+. Non-human primate testing at the U.S. Army Medical Research Institute of Infectious Diseases demonstrated the ability to screen both live and necropsied animals for K. pneumoniae by culture and real-time PCR to determine HMV genotype.
Klebsiella pneumoniae is a Gram-negative, facultative anaerobic, non-motile bacillus in the family Enterobacteriaceae. It is a common cause of a broad range of infections, including septicemia, pneumonia, urinary tract infections, and meningitis.1 A distinctive clinical syndrome of invasive K. pneumoniae has been recognized in humans, primarily in Asia, and more recently in the United States.2,3,4 This invasive syndrome is characterized by primary bacteremic liver abscesses and is frequently associated with complications such as meningitis, endophthalmitis, lung abscess, or fasciitis.5
These invasive strains of K. pneumoniae have been highly associated with the hypermucoviscosity (HMV) phenotype. A strain is characterized as having the HMV phenotype when a standard bacteriological loop is passed through a colony and a mucoviscous string forms that is greater than 5 mm (ie, positive “string test”).6,7 The HMV phenotype confers resistance to serum complement and to phagocytosis by white blood cells.8,6 The two most commonly studied genes associated with the HMV phenotype in K. pneumoniae are the regulator of mucoid phenotype (rmpA) and mucoviscosity associated gene (magA).9
In 2005, seven African green monkeys (AGMs) from the research colony at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) succumbed to multisystemic abscesses due to invasive HMV K. pneumoniae.10 After this initial occurrence, screening of non-human primates (NHPs) for HMV K. pneumoniae was instituted during semiannual exams by oral culture. In March 2007, one AGM was found to be positive for HMV K. pneumoniae but was asymptomatic. Due to concerns for the health of the NHPs, as well as questions as to how these infections might affect the outcome of studies being done on the NHPs, a group of 45 AGMs that were exposed to this one case were screened by oral cultures and real-time PCR for the presence of HMV K. pneumoniae. Additionally, in March 2008, one Rhesus macaque tested positive via oral culture for HMV K. pneumoniae during its semiannual examination. As a result, immediate screening of the entire colony via oral and rectal culture was initiated.
Rapid detection of HMV K. pneumoniae in an NHP colony can allow for early isolation and treatment to prevent further spread of the organism. Molecular methods are sensitive and specific and allow for this early detection. While other PCR assays have been described in the literature, they are all standard PCR. Real-time PCR methods are faster and further ensure specificity with the use of probes.
Three rapid real-time TaqMan and TaqMan-Minor Groove Binder (MGB) PCR assays were developed to detect HMV K. pneumoniae. One assay is species-specific for K. pneumoniae, detecting the Klebsiella hemolysin gene (khe). The other two assays detect genes that are highly associated with the HMV phenotype, rmpA and magA.8,6
Materials and Methods
Positive Control DNA
The K. pneumoniae DNA used to develop and optimize the khe assay was ATCC 700721. The invasive strain of K. pneumoniae with the hypermucoid phenotype that was used for the rmpA assay was Kp V513, named for the AGM from which it was originally isolated.10 The K. pneumoniae DNA used to develop and optimize the magA assay were 06X-03044 and 06X-03046, which were human isolates obtained from the Department of Laboratory Medicine, University of Washington.
PCR Assay Development and Optimization
Primer Express 2 (PE2) (Applied Biosystems, Foster City, CA) was used with the K. pneumoniae khe gene (Accession: AF293352.1)11 to generate primers and a TaqMan probe specific for the sequence. The available magA genes (Accession: AB117611, AB085741, AB198423, AB355924 and AY762939)6,12,13,14 were aligned and PE2 was used to design an assay only to the homologous sequence regions. The available rmpA genes (Accession: AB289644, AB289642, AB298504, AP006726, AY059957, AY059958, AY378100, and X17518)12,15,16,17 were aligned and PE2 was used in an attempt to design a single assay to detect the homologous regions, however this was not successful. A new assay design program, AlleleID 7.0 (Premierbiosoft, Palo Alto, CA), was also tried without success. The rmpA gene from the NHP isolate10 was then sequenced (data not shown). Because the NHP rmpA gene most closely aligned with the AB289644 and AB298504 sequences, an NHP rmpA-specific assay was designed using the homologous sequences and PE2. The PCR assays were optimized on both the Idaho Technology, Inc. Ruggedized Advanced Pathogen Identification Device (R.A.P.I.D.) System (Salt Lake City, UT) and the Roche LightCycler 2.0 Real-Time PCR System (Indianapolis, IN) according to a Diagnostic Systems Division, USAMRIID standard protocol. Briefly, probe concentrations were standardized by diluting the probes so that fluorescence background was 10 to 30 on the R.A.P.I.D. System with a gain setting of “16.” MgCl2 and primer concentrations were optimized sequentially, as follows: MgCl2 was optimized in 1 mmol/L increments from 3 mmol/L to 7 mmol/L, and primers were optimized symmetrically in 0.1 μmol/L increments from 0.5 μmol/L to 1.0 μmol/L. The combinations that exhibited the earliest CT value and generated the highest end point fluorescence were chosen as the optimal conditions for each assay (Tables 1and 2).
Table 1.
Optimized Conditions of the khe, rmpA, and magA PCR Assays
| Final Concentration |
|||
|---|---|---|---|
| Reaction Components | khe | rmpA | magA |
| 10× PCR Buffer (Idaho Technologies) | 1× | 1× | 1× |
| MgCl2 | 4 mmol/L | 3 mmol/L | 4 mmol/L |
| Primers | 0.7 μmol/L | 0.6 μmol/L | 0.6 μmol/L |
| Probe | 0.2 μmol/L | 0.2 μmol/L | 0.1 μmol/L |
| Platinum Taq DNA Polymerase | 1 Unit | 1 Unit | 1 Unit |
| Total Rxn Volume | 20 μl | 20 μl | 20 μl |
| Melt | 95°C, 2 minutes | 95°C, 2 minutes | 95°C, 2 minutes |
| Denaturation | 95°C, 1 second | 95°C, 1 second | 95°C, 1 second |
| Annealing/Extension | 60°C, 20 seconds | 62°C, 20 seconds | 60°C, 20 seconds |
| Cycles | 45 | 45 | 45 |
| PCR Product Size | 77 base pairs | 106 base pairs | 121 base pairs |
Table 2.
Assay Performance Summary
| Target gene with accession number | Primer/probe sequences | Assay linearity | LOD |
|---|---|---|---|
| khe | F: 5′-GATGAAACGACCTGATTGCATTC-3′ | R2 = 0.98 | 50 fg |
| Hemolysin gene | R: 5′-CCGGGCTGTCGGGATAAG-3′ | Slope = −3.471 | (60/60) |
| (AF293352.1) | P: 5′-6FAM-CGCGAACTGGAAGGGCCCG-TAMRA-3′ | Efficacy = 1.94 | |
| PCR Product Size: 77 base pairs | Efficiency = 0.94 | ||
| Intercept = 42.56 | |||
| rmpA | F: 5′-AGAGTATTGGTTGACTGCAGGATTT-3′ | R2 = 0.98 | 50 fg |
| Regulator of mucoid phenotype gene | R: 5′-AAACATCAAGCCATATCCATTGG-3′ | Slope = −3.318 | (59/60) |
| (AB289644) | P: 5′-AGGAAAATGGAGAGGGTAC-NFQMGB-3′ | Efficacy = 2.00 | |
| PCR Product Size: 106 base pairs | Efficiency = 1.00 | ||
| Intercept = 42.69 | |||
| magA | F: 5′-CGAAAGTGAACGAATTGATGCT-3′ | R2 = 0.92 | 500 fg |
| Mucoviscosity associated gene | R: 5′-GTTTCTGCTGCAGATTCGAAGA-3′ | Slope = −3.055 | (58/60) |
| (AB117611) | P: 5′-CATCATGCAATAGCCACGT-NFQMGB-3′ | Efficacy = 2.12 | |
| PCR Product Size: 121 base pairs | Efficiency = 1.12 | ||
| Intercept = 43.69 |
Standard Curve Development
Genomic DNA at 50 pg, 10 pg, 5 pg, 1 pg, 500 fg, 100 fg, 50 fg, 10 fg, 5 fg, and 1 fg (per 5 μl) concentrations were prepared and run in triplicate with the optimized PCR conditions. All real-time PCR experiments were performed on the R.A.P.I.D. System and the LightCycler Data Analysis software version 3.5.3 was used to apply the standard curve to the results. The linearity was determined by calculating the efficacy and efficiency of the assays based on the standard curves.
Limit of Detection
Using the preliminary limit of detection (LOD) determined in the development of the standard curve, the true LOD was established by running a total of 60 samples that consisted of the following: two separate runs of 30 replicates each, performed on two different instruments. The lowest concentration that produced a positive signal in 97% of samples (58/60) was considered the assay LOD (Table 2).
Cross-Reactivity Testing
The khe, rmpA, and magA assays were tested against the USAMRIID general bacterial/eukaryotic DNA reference panel (Tables 3 and 4). This panel consisted of 139 organisms that included threat organisms; 10 K. pneumoniae strains; 10 non-pneumoniae Klebsiella species; nearest genetic neighbors to Klebsiella or threat organisms; organisms sharing an environmental niche with Klebsiella or threat organisms and thus likely to be found in environmental samples; organisms sharing a clinical niche with Klebsiella or a threat organism, particularly respiratory pathogens, opportunists, and typical respiratory flora; and organisms observed repeatedly in clinical and environmental samples. In all cases, 100 pg of genomic DNA was used to determine whether the assays cross-reacted with nucleic acids from other organisms.
Table 3.
List of the 119 DNAs that Encompass the General Cross-Reactivity Panel and the Eukaryote Panel
| Organism name | ATCC # | Organism name | ATCC # | Organism name | ATCC # |
|---|---|---|---|---|---|
| Acinetobacter baumanni | 19606 | Deinococcus radiodurans | 13939D | Pseudomonas aeruginosa | 47085D |
| Acinetobacter lwoffii | 17925D | Enterobacter aerogenes | NA | Pseudomonas putida | 47054D |
| Actinobacillus pleuropneumoniae | 27088D | Enterobacter aerogenes | 15038D | Ralstonia pickettii | 27511 |
| Actinomyces naeslundi | 12104D | Enterobacter agglomerans | 29904 | Rhizobium radiobacter | 33970D |
| Alcaligenes faecalis subsp. Faecalis | 8750D | Enterobacter cloacae | 13047D | Saccharomyces cerevisiae | 2601D |
| Alcaligenes xylosoxidans | 27061 | Enterobacter dissolvens | 23373D | Salmonella enterica subsp. enterica serovar Paratyphi A | 9150D |
| Bacillus anthracis Ames | NA | Enterococcus faecalis | 700802D | Salmonella enterica subsp. enterica serovar Typhimurium | 700720D |
| Bacillus cereus | 19637 | Escherichia coli | 25922 | Serratia marcescens | 13880 |
| Bacillus staerothermophilus | 7953 | Escherichia coli | 10798D | Serratia odorifera | 33077 |
| Bacillus subtilis var niger | NA | Escherichia coli | 700928D | Shewanella oneidensis | 700550D |
| Bacillus thuringiensis | 35646 | Feline DNA | NA | Shigella flexneri | 12022 |
| Bacteroides distasonis | 8503 | Francisella tularensis | NA | Shigella sonnei | 9290 |
| Bacteroides fragilis | 25285D | Francisella tularensis | NA | Staphylococcus aureus | 29247 |
| Bartonella henselae | 49882D | Francisella tularensis | NA | Staphylococcus epidermidis | 12228D |
| Bifidobacterium infantis | 15697D | Francisella tularensis LVS Lot 04 | NA | Stenotrophomonas maltophilia | 13637 |
| Bordetella bronchiseptica | 10580 | Haemophilus actinomycetemcomitans | 700685D | Streptococcus anginosum | 33397 |
| Bordetella parapertussis | BAA-587D | Haemophilus influenzae | 10211 | Streptococcus pneumoniae | 33400 |
| Bordetella pertussis | 9797D | Haemophilus influenzae | 51907D | Streptococcus pyogenes | 19615 |
| Borrelia burgdorferi | 35210D | Human DNA | NA | Streptococcus pyogenes | 12344D |
| Bovine DNA | NA | Legionella pneumophila | 33152D | Streptococcus sp (B) | 12386 |
| Brucella abortus | NA | Listeria monocytogenes | 15313 | Streptococcus sp (F2) | 12392 |
| Brucella canis | NA | Mannheimia haemolytica | BAA-410D | Ureaplasma urealyticum | 700970D |
| Brucella melitensis | NA | Moraxella catarrhalis | 25240 | Vaccinia virus | NA |
| Brucella ovis | NA | Moraxella lacunata | 17967D | Vibrio cholerae | 51394D |
| Budvicia aquatica | 35567 | Morganella morganii | 35200D | Vibrio parahaemolyticus | 17802D |
| Burkholderia cepacia | 25416 | Murine DNA | NA | Yersinia enterocolitica | NA |
| Burkholderia pseudomallei | NA | Mycobacterium gordonae | 35760D | Yersinia enterocolitica | 9610 |
| Burkholderia pseudomallei | Mycobacterium species | 19015D | Yersinia frederiksenii | 33641 | |
| Campylobacter jejuni | 33560D | Mycoplasma pneumoniae | 15531D | Yersinia kristensenii | 33638 |
| Candida albicans | 10231D | Neisseria lactamica | 23970 | Yersinia pestis (Antigua; Pgm+) | NA |
| Canine DNA | NA | Neisseria meningitidis | 53415D | Yersinia pestis (CO92;PW) | NA |
| Chryseobacterium meningosepticum | 33958D | Pantoea ananatis | 19321D | Yersinia pestis (Nairobi) | NA |
| Citrobacter freundii | 8090D | Pasteurella multocida | 43137 | Yersinia pestis (PBM19:Pgm+) | NA |
| Clostridium botulinum type A | 19397 | Porcine DNA | NA | Yersinia pestis (Pestoides B) | NA |
| Clostridium difficile | 9689D | Porphyromonas gingivalis | 33277D | Yersinia pestis Java 9 | NA |
| Clostridium perfringens | 13124 | Propionibacterium acnes | 25746D | Yersinia pestis Kim 5 wild type | NA |
| Comamonas terrigena | 8461 | Proteus mirabilis | 7002 | Yersinia pseudotuberculosis | 6904 |
| Comanonas acidovorans | 15668 | Proteus vulgaris | 49132 | Yersinia pseudotuberculosis | 6902 |
| Corynebacterium diphtheriae | 700971D | Providencia stuartii | 33672 | Yersinia ruckeri | 29908 |
| Coxiella burnetii | NA | Pseudomonas aeruginosa | 17933D |
All assays were analyzed with 100 pg of DNA from these panels to ensure specificity.
Table 4.
List of 20 Klebsiella DNAs Tested with khe, rmpA, and magA Assays to Ensure Specificity
| Organism | Strain/ATCC # | khe | rmpA | magA |
|---|---|---|---|---|
| Klebsiella pneumoniae subsp. pneumoniae | 700721D | + | neg | neg |
| Klebsiella oxytoca | 49131 | neg | neg | neg |
| Klebsiella pneumoniae subsp. pneumoniae | 13883 | + | neg | neg |
| Klebsiella ozaenae | 87A-02504 | + | neg | neg |
| Klebsiella ozaenae | 6406-10−70 | + | neg | neg |
| Klebsiella oxytoca | 96A-14214 | neg | neg | neg |
| Klebsiella rhinoscleromatis | 82A-0082A | + | neg | neg |
| Klebsiella oxytoca | 05X-00690 | neg | neg | neg |
| Klebsiella rhinoscleromatis | 3124-3-78 | + | neg | neg |
| Klebsiella oxytoca | 10787-1 | neg | neg | neg |
| Klebsiella ornithinolytica | 87A-03732 | + | neg | neg |
| Raeultella planticola | 87A-06657 | + | neg | neg |
| Klebsiella pneumoniae | 12705-1 | + | neg | neg |
| Klebsiella pneumoniae, magA- | 06X-03045 | + | neg | neg |
| Klebsiella pneumoniae, magA-,rmpA+ | AGM06189 | + | + | neg |
| Klebsiella pneumoniae, magA- | 92A-02214 | + | neg | neg |
| Klebsiella pneumoniae, rmpA- | 5805-1 | + | neg | neg |
| Klebsiella pneumoniae | V513 | + | + | neg |
| Klebsiella pneumoniae | 06X-03044 | + | neg | + |
| Klebsiella pneumoniae | 06X-03046 | + | neg | + |
In all cases 100 pg of DNA was used.
Sample Collection for Initial Screening of 45 African Green Monkeys
Research was conducted in compliance with the Animal Welfare Act and other federal statues and regulations relating to animals and experiments involving animals and adheres to principles stated in the guide for the Care and Use of Laboratory Animals, National Research Council, 1996. The facility where this research was conducted is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Sterile dacron-tipped swabs (Puritan, Guilford, ME), were used to collect dual samples from AGMs. The swabs were either oral swabs collected from live animals; or esophagus, cecum, ileum, stomach, bladder, colon, or lung swabs of animal tissue obtained at necropsy. The extracted DNA was tested with the khe assay and all khe+ samples were tested in triplicate with the rmpA and magA assays. A total of 99 samples were collected and tested from these 45 AGMs (Table 5).
Table 5.
Includes 42 AGM Samples Collected from 14 Animals with Positive khe Results
| Primate # | Source* | Culture results | IPC† (Inhibition) | Inhibition dilution | khe (CT) | rmpA | magA |
|---|---|---|---|---|---|---|---|
| 5772 | Oral | NEG | NEG | − | NEG | ND | ND |
| 5772 | Cecum | NEG | POS | 1:2 | POS (38.11) | NEG | NEG |
| 5772 | Esophagus | NEG | POS | 1:2 | POS (>41.00) | NEG | NEG |
| 5812 | Esophagus | NEG | NEG | − | NEG | ND | ND |
| 5812 | Cecum | POS | NEG | − | POS (32.62) | NEG | NEG |
| 5816 | Oral | NEG | NEG | − | POS (>41.00) | NEG | NEG |
| 5816 | Esophagus | NEG | NEG | − | POS (35.53) | NEG | NEG |
| 5816 | Ileum | NEG | NEG | − | NEG | ND | ND |
| 5816 | Stomach | NEG | NEG | − | POS (32.98) | NEG | NEG |
| 5816 | Bladder | NEG | NEG | − | NEG | ND | ND |
| 5816 | Cecum | NEG | NEG | − | NEG | ND | ND |
| 5816 | Colon | NEG | NEG | − | NEG | ND | ND |
| 5871 | Esophagus | NEG | NEG | − | POS (40.06) | NEG | NEG |
| 5871 | Cecum | NEG | NEG | − | NEG | ND | ND |
| 5907 | Oral | NEG | NEG | − | NEG | ND | ND |
| 5907 | Esophagus | NEG | NEG | − | NEG | ND | ND |
| 5907 | Cecum | NEG | NEG | − | POS (36.83) | NEG | NEG |
| 5907 | Bladder | NEG | NEG | − | NEG | ND | ND |
| 5954 | Cecum | POS | POS | 1:2 | POS (36.43) | NEG | NEG |
| 5954 | Esophagus | NEG | POS | 1:2 | NEG | ND | ND |
| 5994 | Oral | NEG | NEG | − | POS (39.09) | NEG | NEG |
| 6039 | Oral | NEG | NEG | − | POS (37.14) | NEG | NEG |
| 6068 | Oral | NEG | NEG | − | NEG | ND | ND |
| 6068 | Cecum | NEG | POS | 1:2 | NEG | ND | ND |
| 6068 | Esophagus | NEG | NEG | − | POS (34.93) | NEG | NEG |
| 6094 | Oral | NEG | NEG | − | NEG | ND | ND |
| 6094 | Cecum | NEG | NEG | − | POS (>41.00) | NEG | NEG |
| 6094 | Esophagus | NEG | NEG | − | NEG | ND | ND |
| 6183 | Oral | NEG | NEG | − | NEG | ND | ND |
| 6183 | Esophagus | POS | NEG | − | POS (40.87) | NEG | NEG |
| 6183 | Cecum | POS | NEG | − | POS (25.09) | NEG | NEG |
| 6193 | Oral | NEG | NEG | − | NEG | ND | ND |
| 6193 | Cecum | NEG | NEG | − | POS (34.31) | NEG | NEG |
| 6193 | Esophagus | POS | NEG | − | POS (37.82) | NEG | NEG |
| 6271 | Oral | NEG | NEG | − | NEG | ND | ND |
| 6271 | Cecum | NEG | POS | 1:2 | POS (39.25) | NEG | NEG |
| 6271 | Esophagus | NEG | POS | 1:2 | NEG | ND | ND |
| 7020 | Lung | NEG | NEG | − | NEG | ND | ND |
| 7020 | Esophagus | NEG | NEG | − | NEG | ND | ND |
| 7020 | Bladder | NEG | NEG | − | NEG | ND | ND |
| 7020 | Cecum | NEG | NEG | − | NEG | ND | ND |
| 7020 | Trachea | NEG | NEG | − | POS (38.17) | NEG | NEG |
All positive culture and PCR results are shown plus a small portion of negatives.
Oral: swab of live animal; Esophagus, cecum, ileum, stomach, bladder, colon, lung: swab of animal tissue following necropsy.
Internal positive control.
DNA Extraction
One swab from each dual sample was used for DNA extraction. The dacron tip was transferred to a 1.5 ml microcentrifuge tube containing 500 μl of PBS (Sigma-Aldrich, St. Louis, MO) with 0.3% Tween-20, and vortexed at maximum for 2 minutes. Swabs were transferred into 1.5 ml microcentrifuge tubes containing collection baskets (Costar, Corning, NY) and centrifuged at 8000 × g for 5 minutes. The swab eluate was combined with the remaining volume of PBS with 0.3% Tween-20 and centrifuged at 16,000 × g to concentrate the sample. Supernatants were removed and pellets were resuspended in 180 μl of Dulbecco's PBS. The Qiagen DNA Mini Kit (Valencia, CA) was then used according to manufacturer's instructions. Briefly, 200 μl of Buffer AL, and 20 μl of Proteinase K (17.8 mg/ml) were added to the samples and incubated at 55°C for 60 minutes. After incubation, 100% ethanol was added and the sample was mixed by vortex and loaded onto a QIAamp spin column by centrifugation. The columns were washed once each with buffers AW1 and AW2. After a drying centrifugation spin, the nucleic acid was eluted in 100 μl of AE buffer preheated to 70°C.
Microbiology Culture
The second swab was processed for microbiology culture by plating on 5% sheep blood agar, chocolate agar, and MacConkey agar. Suspect K. pneumoniae colonies were isolated and identified on the bioMerieux Vitek 2 (Durham, NC) with the GNI card. A DNA isolation boil-prep was also performed on the suspect colonies for testing with the khe assay with all khe+ samples then tested with the rmpA and magA assays in triplicate.
Real-Time PCR Testing
Each extracted monkey sample was tested for PCR inhibitors with an internal positive control assay.18,19 If inhibition was encountered, the eluate was diluted 1:2, 1:4, and 1:8 and retested with the internal positive control assay. The dilution that relieved the inhibition was used in all subsequent assays. All initial samples were tested with the khe assay with all khe+ samples then tested with the rmpA and magA assays in triplicate.
Additional NHP Study
After the initial screening of 45 AGMs, an additional 307 NHPs from the USAMRIID colony, consisting of Rhesus and cynomolgus macaques and AGMs who were not exhibiting any clinical signs of K. pneumoniae HMV disease were screened. During this study, a total of 1825 oral and rectal samples were collected and cultured on MacConkey agar plates. Suspect colonies were tested with the Vitek 2 GNI card and any K. pneumoniae isolates were examined with the string test to determine HMV phenotype. The string test was performed by touching the colony with a loop and pulling up. A colony was considered positive with the HMV phenotype when a string of ≥5 mm was observed.6,7 All string test-positive K. pneumoniae isolates were tested with the rmpA and magA real-time PCR assays (Table 5).
Results
PCR Assay Development and Optimization
Three real-time TaqMan and TaqMan-MGB PCR assays were developed and optimized. One was a K. pneumoniae-specific assay, and the other two were specific for genes known to be associated with HMV K. pneumoniae originally isolated in the monkey colony at USAMRIID.10 All primer combinations resulting in PCR products smaller than 160 bp were tested for amplification efficiency. The final primer and probe sequences, optimized assay conditions, LODs and assay linearity calculations are listed in Tables 1and 2.
Standard Curves
Linearity was established using purified genomic DNAs from representative strains containing each gene. Tenfold serial dilutions from 10 pg (1800 genome copies) to 1 fg (0.18 genome copies) along with a 50 fg (nine genome copies) standard of quantified genomic DNA were performed in triplicate to establish the linearity of each assay (Figure 1, A–C). Once established, 60 replicates were tested at the LOD to establish 97% sensitivity (≥ 58 positives). The khe and rmpA assays have LODs of approximately nine genome copies (50 fg), whereas the magA assay has an approximate LOD of 92 genome copies (500 fg), based on a 5.3-Mb K. pneumoniae genome with a GC ratio of 57.5%.
Figure 1.
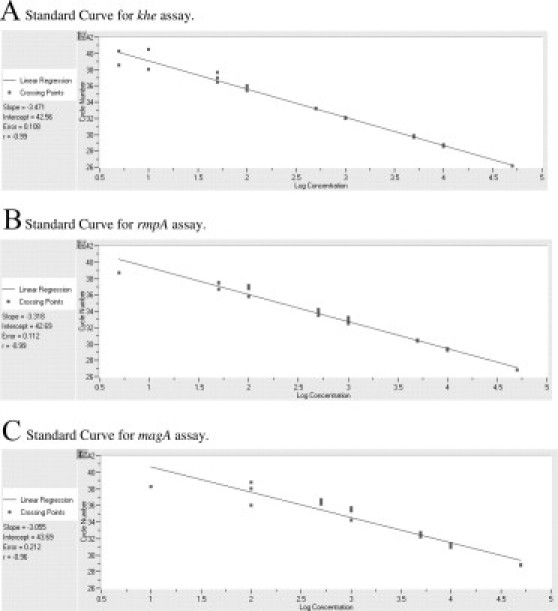
Standard curves for (A) khe, (B) rmpA, and (C) magA assays.
Cross-Reactivity Testing
The khe, rmpA, and magA assays were tested against the general bacterial/eukaryote USAMRIID DNA reference panel for specificity (Tables 3 and 4). The rmpA and magA assays showed no cross-reactivity with any of the panel DNAs and were specific for their targets. The khe assay detected some species of Klebsiella other than K. pneumoniae (Table 4).
Nonhuman Primate Testing
Initial 45 AGMs
An initial study of 45 AGMs involved 99 dual oral swabs processed by culture and DNA extraction. In this initial study, there were 19 khe+ samples from 14 different AGMs. The PCR was more sensitive than traditional culture. Of the 19 khe+ positive samples, only five were culture-positive. Of the five culture-positive samples, none were of the HMV phenotype. All of the khe+ samples were negative for rmpA and magA (Table 5).
Additional 307 NHPs
In a subsequent study of 307 Rhesus, cynomolgus macaques, and AGMs, a total of 1825 oral and rectal samples were collected and tested by culture and string test for HMV K. pneumoniae. This group resulted in 177 K. pneumoniae isolates, of which 64 were determined to be of the HMV phenotype by the string test. Real-time-PCR testing of the HMV K. pneumoniae resulted in 42 rmpA+ isolates and 15 magA+ isolates. Interestingly, there were an additional seven HMV K. pneumoniae isolates from four different NHPs that were rmpA− and magA− (Table 6).
Table 6.
Real-Time PCR Results from 307 NHP
| Number of primates tested | Source | Number of cultures | K. pneumoniae culture positive | String test positive (HMV phenotype) | rmpA Positive | magA Positive | HMV K. pneumoniae rmpA negative and magA negative |
|---|---|---|---|---|---|---|---|
| 307 | oral and rectal | 1825 | 177 | 64 | 42 | 15 | 7 |
Discussion
Invasive HMV K. pneumoniae is emerging as a significant threat to the health of NHPs. In this study, we developed real-time PCR assays for the detection of HMV K. pneumoniae on the R.A.P.I.D. and LightCycler using gene specific primers combined with either TaqMan or TaqMan-MGB probes and used these assays to screen NHPs for the presence of HMV K. pneumoniae. The khe assay targets the hemolysin gene on the chromosome of K. pneumoniae. This gene encodes a unique peptide of 20 kDa and is present in all strains of K. pneumoniae.11 Yin-Ching et al11 used Southern blot hybridization and determined that all of the strains of K. pneumoniae that were tested contained the hemolysin gene. We had similar results in that all of the strains of K. pneumoniae that we tested with the khe assay were positive. However, Yin-Ching et al tested only one strain of each of the following: K. oxytoca, K. planticola, K. terrigena, and K. ornithinolytica. They determined that all other strains of Klebsiella species do not have the khe gene. In our testing we determined that the khe assay is positive with two strains of K. ozaenae, two strains of K. rhinoscleromatis, one strain of K. ornithinolytica, and one strain of Raeultella planticola. Fortunately, the khe assay is simply a screening tool and can be used to detect the presence of Klebsiella. The rmpA and magA assays were found to be specific for the HMV K. pneumoniae.
The rmpA assay specifically targets the rmpA gene on a plasmid of K. pneumoniae. This gene encodes a unique peptide of 25 kDa and has been shown in human isolates to be highly associated with the hypermucoviscosity phenotype of K. pneumoniae.12 The magA assay specifically targets the magA gene on the chromosome of K. pneumoniae. This gene encodes a unique 43-kDa outer membrane protein that is significantly more prevalent in invasive human strains of K. pneumoniae.6
During the cross-reactivity testing in the development of the rmpA assay, positive results were seen with K. pneumoniae, ATCC 13883; however, this strain does not have the HMV phenotype. The sequence of the rmpA gene from the USAMRIID NHP isolate was compared with the sequence of the amplicon we obtained from K. pneumoniae ATCC 13883. Several mutations were seen in the sequence from K. pneumoniae ATCC 13883, thus we redesigned our probe as a shortened TaqMan-MGB probe to exactly match the USAMRIID NHP isolate. The MGB probe has several advantages. The non-fluorescent quencher is a much better quencher of reporter dyes than the TAMRA dye on traditional dual-labeled TaqMan probes. The MGB also increases the melting temperature of the oligonucleotide20,21 allowing the use of shorter probes. Consequently, the TaqMan-MGB probes can be designed to regions where GC content is low, which greatly increases the genetic regions available for assay development. In addition to the probe changes, the MgCl2 was lowered to 3 mmol/L and the annealing temperature was raised from 60°C to 62°C. The lower magnesium concentration and higher annealing temperature significantly increased the specificity of the assay, thereby eliminating the detection of the non-HMV K. pneumoniae ATCC 13883. In summary, all three assays were highly specific for their intended target with each possessing a very low LOD (9 to 92 genome copies).
The first occurrence of multisystemic abscesses in an AGM at USAMRIID in 2005 raised concern for K. pneumoniae infection in the USAMRIID NHP colony10 and all ongoing and planned infectious disease, vaccines, or therapeutics protocols involving NHPs. A semiannual screening of all NHPs for HMV K. pneumoniae in March 2007 and 2008 identified other positive, asymptomatic HMV K. pneumoniae NHPs. Samples (oral, rectal, and necropsy swabs) were then taken from all of 307 animals in the colony and cultured on MacConkey agar for HMV K. pneumoniae. MacConkey agar was chosen for its specificity for Gram-negative organisms. Colonies suspicious for K. pneumoniae were selected and identification was confirmed with the Vitek 2 GNI card.
An initial study of 45 AGMs involved 99 dual oral swabs. In addition to being processed with culture, the samples were extracted for DNA and tested for the presence of inhibitors with an internal positive control.18,19 This was done to eliminate the possibility of false negative real-time PCR results. If a sample was inhibitory, it was diluted in molecular biology grade water and retested. In this study, any inhibition was relieved with a 1:2 dilution of the sample DNA. A total of 99 samples were collected from the 45 AGMs. Nineteen samples from 14 different animals were positive for K. pneumoniae, but none of these isolates were positive for rmpA or magA. These results indicate that these 45 AGMs, though potentially exposed to the AGM that was subclinically infected with HMV K. pneumoniae, did not themselves become infected. The epidemiology and pathophysiology of the HMV strain(s) within the USAMRIID colony is currently being evaluated and will be reported elsewhere.
After this initial screening of 45 AGMs, an additional 307 NHPs, consisting of AGMs, Rhesus, and cynomolgus macaques who were not exhibiting any clinical signs of infection, were screened. Due to the large size of the second cohort and the request to take weekly samples from all 307 animals, the veterinarians decided to implement the screening in a more cost-effective manner. Swab samples were only cultured on MacConkey agar and K. pneumoniae identification was confirmed with the Vitek 2. All K. pneumoniae positive samples were then processed for further evaluation by real-time PCR.
Of 177 K. pneumoniae isolates, 64 exhibited the HMV phenotype as determined by a positive string test. Real-time PCR testing of the HMV K. pneumoniae resulted in 42 rmpA+ isolates and 15 magA+ isolates. Unexpectedly, seven HMV K. pneumoniae isolates from four different NHPs were rmpA− and magA− (four of the samples were from a single NHP). Other studies examining human clinical K. pneumoniae isolates have also identified HMV strains that were rmpA−/magA−.22 These strains typically possess the K1 or K2 capsular serotype and other genes including aerobactin, kfu, and allS.23 The involvement of a transcriptional regulator in serotype-specific extracapsular polysaccharide production, rmpA2, has also been reported to be important for the HMV phenotype of K. pneumoniae.24 Thus, it is likely that there are other regulator genes that play a role in the HMV phenotype.
In summary, it is currently unknown whether Rhesus and cynomolgus macaques infected with HMV K. pneumoniae will maintain a persistent subclinical infection or whether they will develop disease similar to that seen in AGMs. The ability to identify gene associations and correlate those findings with the presence or absence of clinical signs of disease greatly aids ongoing studies of the pathophysiology of HMV K. pneumoniae in NHPs. In addition, it is of the utmost importance to determine whether HMV K. pneumoniae in any way interferes with studies involving the pathophysiology of experimentally induced diseases (ie, Ebola, Marburg, Lassa, smallpox, monkeypox, etc) and the ongoing development of vaccines and therapeutics for these diseases.
Addendum
As this paper was being prepared for publication, a cynomolgus macaque (Macaca fascicularis) assigned to a research project appeared to have survived challenge to the test agent when the animal was unexpectedly found dead. Based on gross and histological lesions, HMV K. pneumoniae infection was suspected in conjunction with infection with the test agent. DNA was extracted from formalin-fixed tissues and PCR confirmed infection with rmpA+/magA− HMV K. pneumoniae. Continued surveillance at our Institute for HMV K. pneumoniae has identified several asymptomatic macaques, both Rhesus (Macaca mulatta), and cynomolgus; however, this is the first macaque with clinical disease leading to the death of the animal. Clearly, subclinical, chronic infections in AGMs and macaques have great potential to disrupt research protocols and pose risks to personnel.
Acknowledgements
We are grateful to Stephen Libby, Department of Laboratory Medicine, University of Washington, for providing us with the magA+ K. pneumoniae strains used as positive controls. Thanks to John Kondig and Len Wasieloski for assisting us with DNA sequencing. We also thank Katheryn Kenyon for reviewing the manuscript.
Footnotes
Supported by DTRA Project # 8.10030_07_RD_B.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von GA, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lederman ER, Crum NF. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol. 2005;100:322–331. doi: 10.1111/j.1572-0241.2005.40310.x. [DOI] [PubMed] [Google Scholar]
- 4.Chang SC, Fang CT, Hsueh PR, Chen YC, Luh KT. Klebsiella pneumoniae isolates causing liver abscess in Taiwan. Diagn Microbiol Infect Dis. 2000;37:279–284. doi: 10.1016/s0732-8893(00)00157-7. [DOI] [PubMed] [Google Scholar]
- 5.Braiteh F, Golden MP. Cryptogenic invasive Klebsiella pneumoniae liver abscess syndrome. International J Infect Dis. 2007;11:16–22. doi: 10.1016/j.ijid.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, Wang LR, Ko WC. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med. 2006;259:606–614. doi: 10.1111/j.1365-2796.2006.01641.x. [DOI] [PubMed] [Google Scholar]
- 8.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun. 1989;57:546–552. doi: 10.1128/iai.57.2.546-552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadasy KA, Domiati-Saad R, Tribble MA. Invasive Klebsiella pneumoniae syndrome in North America. Clin Infect Dis. 2007;45:e25–e28. doi: 10.1086/519424. [DOI] [PubMed] [Google Scholar]
- 10.Twenhafel NA, Whitehouse CA, Stevens EL, Hottel HE, Foster CD, Gamble S, Abbott S, Janda JM, Kreiselmeier N, Steele KE. Multisystemic abscesses in African green monkeys (Chlorocebus aethiops) with invasive Klebsiella pneumoniae–identification of the hypermucoviscosity phenotype. Vet Pathol. 2008;45:226–231. doi: 10.1354/vp.45-2-226. [DOI] [PubMed] [Google Scholar]
- 11.Yin-Ching C, Jer-Horng S, Ching-Nan L, Ming-Chung C. Cloning of a gene encoding a unique haemolysin from Klebsiella pneumoniae and its potential use as a species-specific gene probe. Microb Pathog. 2002;33:1–6. doi: 10.1006/mpat.2002.0499. [DOI] [PubMed] [Google Scholar]
- 12.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 13.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2006;193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 14.Yeh KM, Chang FY, Fung CP, Lin JC, Siu LK. magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumoniae serotype K1. J Med Microbiol. 2006;55:803–804. doi: 10.1099/jmm.0.46368-0. [DOI] [PubMed] [Google Scholar]
- 15.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol. 2008;190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Nassif X, Honore N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol. 1989;3:1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 18.Hartman LJ, Coyne SR, Norwood DA. Development of a novel internal positive control for Taqman based assays. Mol Cell Probes. 2005;19:51–59. doi: 10.1016/j.mcp.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Hartman LJ, Coyne SR, Norwood DA. Erratum: Development of a novel internal positive control for Taqman bassed assays [YMCPR 19(1):51–9] Mol Cell Probes. 2005;19:298. doi: 10.1016/j.mcp.2004.07.006. 298. [DOI] [PubMed] [Google Scholar]
- 20.Afonina I, Zivarts M, Kutyavin I, Lukhtanov E, Gamper H, Meyer RB. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 1997;25:2657–2660. doi: 10.1093/nar/25.13.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutyavin IV, Lukhtanov EA, Gamper HB, Meyer RB. Oligonucleotides with conjugated dihydropyrroloindole tripeptides: base composition and backbone effects on hybridization. Nucleic Acids Res. 1997;25:3718–3723. doi: 10.1093/nar/25.18.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42:1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 23.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62:1–6. doi: 10.1016/j.diagmicrobio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Wacharotayankun R, Arakawa Y, Ohta M, Tanaka K, Akashi T, Mori M, Kato N. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect Immun. 1993;61:3164–3174. doi: 10.1128/iai.61.8.3164-3174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


