Summary
This review highlights recent progress in the use of embryonic stem cell (ESC) systems for studying and treating cardiovascular disease. Although ESCs represent an in vitro system, they can provide a rich source of progenitor cells, and this has been exploited recently to identify novel precursors and to investigate the lineage relationships among various cell types that comprise the developing heart, including cardiac muscle, endothelium, and smooth muscle. ESCs grown in aggregates (embryoid bodies) recapitulate normal developmental programs. Since they can be grown under defined culture conditions, they have been used to systematically identify specific genes and signaling pathways that promote cardiogenesis. A major goal is to optimize the production of cardiac progenitors and differentiated cell types, and to test their ability to promote healing in transplant assays, for example post-infarction. While many challenges remain, the development of iPS technology provides a means to generate cells for autologous transplant and for investigating patient-specific disease mechanisms. The development of new techniques to derive cardiac derivatives in vitro from ESC or iPS sources, coupled with novel tissue-engineering approaches and a better understanding of how explanted cells can survive and integrate in host tissue, should have a significant impact on the development of both cell-based and pharmacological therapies for cardiovascular disease.
Keywords: cardiogenesis, progenitors, cell therapies, iPS
Can heart development and disease be studied in vitro?
Organogenesis is a complex process that transforms relatively homogenous epithelial germ layers into functioning and highly integrated systems. From a developmental perspective, this can be viewed as a series of cellular transitions, including the commitment of progenitors to tissue-restricted fates, differentiation to express lineage-restricted genetic programs, morphogenesis to form appropriate tissue shapes, and system integration to incorporate the organ into the physiological state of the developing embryo. While the development of any organ system is remarkably complex, cardiogenesis requires extraordinary coordination of regulatory mechanisms in both time and space. In particular, both subtle and dramatic morphogenetic movements transform an initial primordial tube into a complex 3-dimensional organ consisting of septated chambers with distinct identities, a coronary vasculature and mature valves. Integration with both the venous and arterial systems must be timed perfectly with the developing hematopoietic system, and the slightest error can lead to early embryonic death. Even minor early morphogenetic defects surface at later stages to reveal debilitating septal or valvular defects. For this reason it seems intuitive that studying organogenesis requires an animal model, and indeed major progress in dissecting steps of heart development have derived from studies in a variety of animal models, including flies, zebrafish, frog, chick, and mouse [1–3].
Yet, there are issues that can be addressed effectively in vitro using cell culture systems, in particular those transitions related to early steps of stem and progenitor commitment, cardiac cell lineage differentiation, and cellular physiology. For this purpose, embryonic stem cell (ESC) systems provide unique advantages that have already contributed important insight into the derivation of cardiac cells, lineage relationships, and key cardiac developmental signaling pathways. The advent of induced pluripotent stem (iPS) cell technology has brought the promise of deriving patient-specific cardiac cells to the brink of reality, and this is likely to have a major impact on drug screening and for developing cellular therapies. Here the ESC system is reviewed with a focus on how it can be used to understand cardiac progenitor cell biology, and how this information can be exploited to understand normal cardiogenesis, relate this to cardiogenic defects, and enhance progress in the development of cellular, regenerative, and pharmacological therapies to treat cardiac disease. A separate but related issue that is not discussed here is the identity of putative cardiac-restricted stem cells that might be resident in the heart; this is a topic that has been reviewed elsewhere [4,5].
Embryonic Stem Cells
The primary advantage of the ESC model as an experimental system is that it represents a homogeneous cell population with the potential to generate any of the hundreds of distinct differentiated cell types [6]. This is true by definition, since for example mouse ESCs (mESCs), derived from the pre-gastrulation inner cell mass, are capable of deriving an entire animal. However, this has also been validated for numerous lineages in vitro, including cardiomyocytes [7]. From a developmental perspective, this permits 1) quantitative assessment of specification to a restricted progenitor fate, 2) fate-mapping experiments to establish lineage relationships, and 3) an experimental platform to direct lineage differentiation down one pathway or another, using for example forced expression of regulatory genes, defined signaling proteins, or small molecules. From a translational perspective, it allows the generation of potentially unlimited progenitors at various defined stages of developmental potency, for testing in cellular therapies to ameliorate cardiac disease by transplantation and engraftment.
A major challenge in adapting the insight gained from developmental studies to translational studies is fairly obvious. Cardiomyocytes are generated inefficiently from ESCs in vitro (typically 1–3% of differentiating cultures). After all, the ES derived cultures are multipotent and will spontaneously generate many other cell types. Therefore, the initial advantage of having a homogenous cell culture is rather quickly diminished by the generation of a heterogeneous mixture of lineages, many of which might be inhibitory to cardiogenesis or deleterious in cardiac transplant assays. A particularly insidious contaminant would be the ES cell itself, since it has the propensity to generate teratomas when transplanted in vivo [8]. Therefore, much effort in recent years has focused on optimizing the efficient generation of cardiac progenitors. Several experimental approaches are used to generate cardiomyocytes from ESCs, including directed differentiation in monolayer cultures using defined medium, culture in the presence of various stromal cell types, or by exploiting the ability of ESCs, when released from culture conditions that maintain ES identify (removal of LIF, in the case of mESCs) to generate aggregates or embryoid bodies (EBs). EB generation [4,5] somehow mimics the cellular interactions that occur during normal gastrulation and allows the specification of the primary germ layers, including mesoderm from which cardiomyocytes are derived. It has been nearly 25 years since Doetschman and colleagues described the appearance of myocardial tissue in a subset of EBs comprised also of cystic structures resembling visceral endoderm [9]. This ES-EB approach has been particularly fruitful in the mouse system for studying the “normal” development of cardiac progenitors and signaling pathways that can influence this pathway (Fig. 1).
Fig. 1. The ES-EB system provides a model to quantify the commitment of progenitors to specific fates.
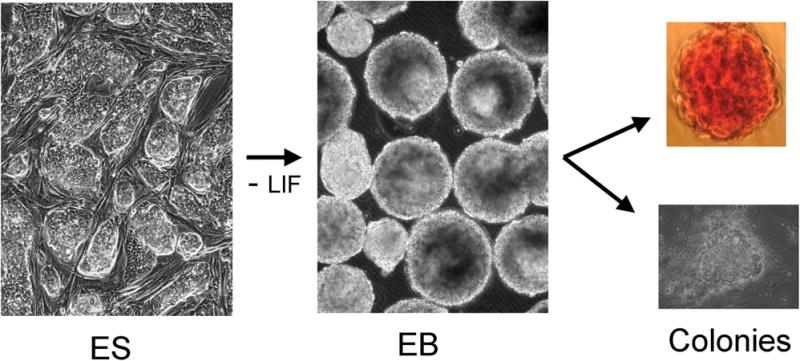
Embryonic stem (ES) cells are grown as colonies either in defined media that promotes maintenance of stem cell identify, or as shown here on feeder cells that support stemness. When the cells are harvested and allowed to reaggregate in the absence of stemness support (removal of feeders and LIF) they spontaneously form embryoid bodies (EB) in which progenitors commit to defined fates with a temporal progression similar to normal embryonic development. At any time during EB culture, the presence of committed progenitors can be quantified. Cells are harvested and plated onto soft agar in the presence of defined media and growth factors. As illustrated here, the EBs can derive hematopoietic colonies (top panel; erythroid colonies formed in the presence of erythropoietin) or cardiac muscle colonies (bottom panel shows a beating CM focus).
Lineage Relationships and Specification of Cardiomyocytes
The development of ESC-derived EBs in culture is synchronized, and importantly, largely recapitulates the temporal transitions that reflect specification to defined progenitor fates. This means that genetic cell surface markers will be coordinately expressed within the population of EBs, which facilitates the purification of subsets of progenitors that are at a defined developmental stage. This fact was exploited by Gordon Keller and colleagues for establishing the lineage relationships of progenitors that contribute to the hemato-vascular program, including characterization of bipotential hemangioblasts [10]. For example, an early committed hemato-vascular progenitor is highly enriched in the first cells that co-express the VEGF receptor FLK1 and the early mesoderm marker BRACHYURY (the latter monitored using as a surrogate marker expression of the GFP gene knocked into the Brachyury locus; Bry:GFP) [11]. Using the same approach, an early multipotent cardiovascular progenitor was identified within a subsequent second wave of FLK1+ cells that emerge within the Bry:GFP population [12]. This sorted population includes single cells that can be cultured to form colonies containing derivatives of three cardiovascular lineages: cardiomyocytes (CM), smooth muscle cells (SM), and endothelium (EN). These rare cells could also be identified in early mouse embryos, validating the in vitro model to identify what could be clinically relevant progenitors [13].
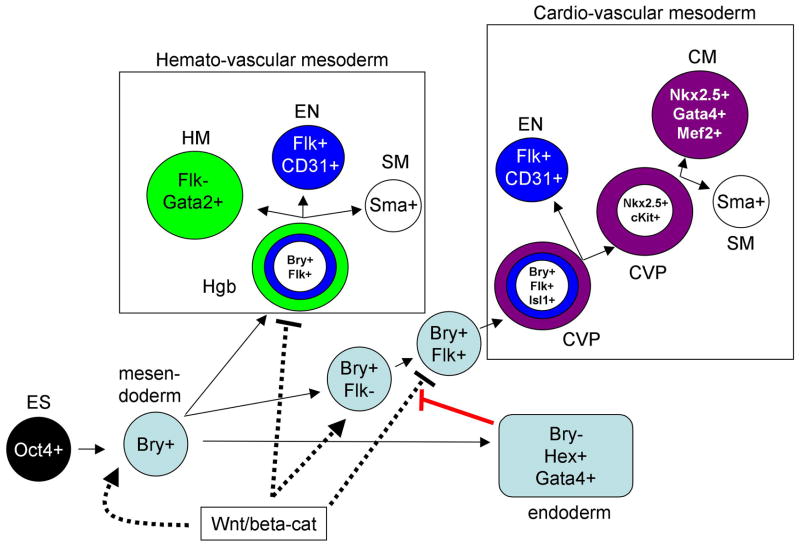
These data are consistent with two other reports that used the CM marker NKX2.5 to isolate multipotential progenitors. Cells that are positive for the second heart field marker ISLET1, once expanded on a feeder layer, include a subset of cells that are ISL1+/NKX2.5+/FLK1+ and can give rise to all three lineages [14,15], much like the FLK1+ BRY(late+) cells [12]. A seemingly more restricted, but still CM/SM bipotential progenitor was also isolated from EB-derived cells, using the Nkx2.5 promoter to express GFP as a surrogate early marker, and purifying the subset of NKX2.5+ cells that co-express the stem cell factor receptor, C-KIT [16]. A recent study also used Nkx2.5:GFP to isolate a CM/SM/EN tri-potential progenitor that is NKX2.5+/FLK1+/C-KIT+ [17]. A summary of how multipotent progenitors develop from Bry+ cells, based on colony assays in the ES system, is shown in Fig. 2. From a developmental perspective, it is very satisfying to find early cardiac progenitors that are multipotent, in order to understand the hierarchy of precursor relationships. But this may also be very important in a translational context, since an earlier, less restricted, progenitor might be more effective at integrating and healing damaged tissue (for example through coordinated development of necessary vasculature). The ESC system may be essential in a practical sense for the isolation of sufficient quantities of such early progenitors toward testing and developing cellular therapies.
Fig. 2.
The ES-EB system has made possible the identification of multipotent progenitors for both hemato-vascular and cardio-vascular derivatives. As indicated in the schematic, pluripotent Oct4+ ES cells commit to an early bipotential germ layer phenotype called mesendoderm that is brachyury (Bry) positive. The initial Flk1/Vegf receptor positive cells that emerge (Flk+) generate tri-potential hemangioblasts (Hgb) that are capable of generateing hematopoietic (HM), endothelial (EN), and smooth muscle (SM) cells, which are Gata2, CD31, or smooth muscle actin (Sma) positive, respectively. Subsequently, a second wave of Bry+/Flk+ cells gives rise to a distint cardio-vascular progenitor (CVP) that can generate EN or a more restricted Nkx2.5+ and cKit+ bipotential CVP. The latter is capable of generating CM or SM progenitors. Canonical Wnt signaling acting through beta-catenin (Wnt/beta-cat, dashed lines), functions throughout these transitions enhancing both early Hgb and CVP devlelopment, and in both cases subsequently repressing their generation. In the case of the CVP, endoderm that develops in association with the precardiac mesoderm expresses antagonists of this Wnt/beta-cat pathway and thereby induces cardiogenesis. The Hemato-vascular studies are primarily from G. Keller and colleagues, while the identification of CVPs is described in the text.
The transcriptional program that directs specification to a cardiomyocyte fate is likely to involve members of known regulatory families for cardiac-specific genes, including those encoding GATA, TBOX, NKX2, and MEF2 transcription factors, among others [18]. This concept is supported by loss-of-function studies in embryos [19,20]. However, a minimal cardiac-inducing gene set has not yet been defined, and this would seem to be a promising area of future research using the ESC system. Somewhat surprisingly, Oct-3/4 (POU5f1), one of the key genes responsible for maintaining ESC pluripotency, is sufficient when over-expressed to enhance significantly undifferentiated ESCs (maintained in LIF) toward mesoderm and then cardiac fate [21]. It is reasonable to think that several step-wise developmental transitions are required to take a pluripotent ESC to cardiac commitment, as indicated by the reporter-based progenitor studies discussed above. However, with the realization that differentiated cells can be reprogrammed back to pluripotency (iPS, discussed below), expression of the right combination of transcription factors (in the right ratio and at appropriate levels) might suffice for cardiac specification. In a practical sense, it may be more useful to exploit inducing factors or small molecules that can activate these programs without the need for introducing exogenous transgenes.
Identifying extrinsic regulators of cardiac specification
The key transcriptional regulatory genes for cardiac specification, for example Nkx2.5, provide excellent markers for the intrinsic program that drives cardiomyocyte fate. The ESC system therefore provides an outstanding platform for identification of extrinsic factors that promote cardiomyocyte fate from an uncommitted or multipotential progenitor. In developing EBs, there is a relatively low level of spontaneous differentiation that can be easily quantified as beating foci. This relatively inefficient program is actually an advantage for screening cells, factors, or small molecules that can stimulate the process, and significant progress has been made in recent years, mostly consistent with complementary genetic experiments evaluating normal cardiogenesis in animal models. One of the best characterized pathway is that regulated by WNT signaling [22]. Experiments in embryos have suggested that there are biphasic stage-specific functions for WNTs [23]. Two groups confirmed independently that WNT signaling has first positive and subsequently negative roles in cardiac specification in the ES-EB system. Early activation of WNT/BETA-CATENIN in EBs leads to enhanced cardiomyogenesis, whereas activation of the pathway at later stages of EB culture blocks cardiomyocyte differentiation [24,25]. Early, the pathway seems to expand cardiac-competent mesoderm, but at later stages this signal must be repressed in order for cardiac differentiation to proceed (see Fig. 2)
Other embryonic signaling pathways that are implicated in cardiomyocyte fate include those regulated by NOTCH, FGF and members of the TGF-β superfamily, including ACTIVIN and BMPs [26]. These signaling pathways play wide ranging roles in the generation and patterning of early germ layers and tissues, so that much of their function may be indirect through activation of yet unidentified specific cardiac inducers. Regardless, their utility in deriving efficient cardiac specification in vitro has been validated experimentally in the human ESC (hESC) system [27,28]. Following optimal induction with ACTIVIN-A, and subsequently with BMP4, cell populations containing up to 90% cardiomyocytes can be purified using a Percoll gradient centrifugation step [27]. At least some cardiac inducing factors appear to be normally generated by endoderm, as suggested also by previous experiments in animal and explant models [29,30]. Indeed, anterior endoderm secretes factors that can induce cardiac fate [31], and expression of the endoderm transcription factor SOX17 is required (indirectly) for the development of cardiac mesoderm in EBs, subsequent to initial mesoderm formation [32]. Studies from our own laboratory have shown that expression of GATA4 in developing EBs is sufficient to enhance cardiogenesis, although this is by a non-cell-autonomous mechanism (Fig. 3). GATA4 directs EBs to form definitive endoderm, and this is then able to induce other cells to a cardiac fate. The GATA4-expressing cells secrete inhibitors of the WNT pathway, including DKK1 and SFRP5, consistent with a role of WNT inhibition for cardiac specification. These results are consistent with the co-generation of beating cardiac cells with hepatocyte-like (endoderm-derived) cells when ESCs are induced to differentiate with FGF [33].
Fig. 3. The EB system can be used to define both cell intrinsic pathways, and to identify secreted cell extrinsic regulatory factors.
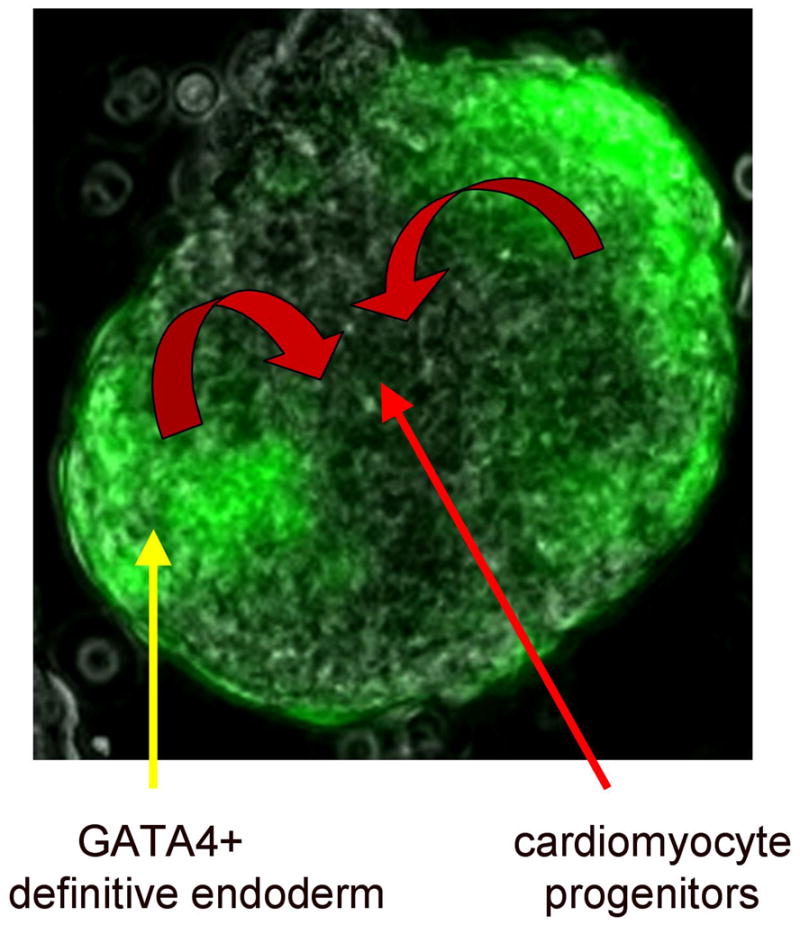
An ES line was created that allows GATA4 to be expressed conditionally by induction with doxycyclin. Shown is a “mixed” EB, consisting of GATA4-expressing ES-derived cells (that are also green because they co-express GFP) and cells from a parental line that does not contain the inducible GATA4 transgene. Colony assays demonstrated that the mixed EBs are enhanced at least 5-fold for the production of cardiomyocyte progenitors, but these are not derived from the GATA4-expressing cells. Instead, the GATA4+ cells differentiate as anterior definitive endoderm, and secrete cardiac-inducing factors (curved red arrows), including inhibitors of the WNT pathway. On their own, the ES cells induced to express GATA4 at the mesendoderm stage will generate Hex1+ endoderm at the expense of mesoderm. These cells also express high levels of the Wnt antagonist Dkk1. In the mixed cultures the cells that commit to express TroponinT and other cardiac markers do not express GFP (they were not induced) although they presumably do eventually express endogenous GATA4. Note (as shown in Fig. 2) GATA4 is a gene that marks both the cardiac-inducing endoderm, as well as the CM derivatives. (A. Holtzinger, G. Rosenfeld, and T. Evans, unpublished data).
Translating ESC Biology to Cellular Therapies
One major use for the ESC model is the generation of cardiomyocytes that can be tested for cellular therapies of heart damage. Initial experiments confirmed that ES-derived cardiomyocytes could survive as grafts when transplanted into the murine heart [34]. More recently, transplantation has typically been studied in an acute disease model following induced myocardial infarct (for example by ischemia-reperfusion injury caused by ligation of a coronary artery), followed by engraftment of ES-derived cells. In rodent models the effect can be measured functionally compared to controls by echocardiography and magnetic resonance imaging, and for integration and morphological resolution by histology. Several studies established that murine or human ESC-derived cardiomyocytes provide improved cardiac function following engraftment, documenting remuscularization, normalized contractility and conduction, and reduced susceptibility to arrhythmias (reviewed in [7,35,36]). A variety of stem and progenitor cell sources have been tested in patients for treating acute injury and chronic heart failure, based on the relative availability of defined progenitor populations, for example from bone marrow or skeletal muscle (reviewed in [37]). Clinical trials documented the safety of cell transplants and some physiological improvements to the damaged tissue. Yet, overall the clinical benefit using these cells might best be described as “mixed”. The results are likely to be from short-term paracrine-mediated relief of ischemia, rather than replacement of contractile cells [38]. Thus, it is unclear whether non-cardiac cell sources will be able to provide long-term clinically relevant regenerative therapies for a damaged heart. At least in animal models, ESC-derived cardiomyocytes can integrate and restore contractile function to infracted myocardium [39,40]. Using ESCs, potentially unlimited numbers of cardiomyocytes (or multipotent cardiac progenitors) could be generated in vitro and used to treat acute or chronically damaged hearts.
Human ES-derived Cardiomyocytes and iPS
As indicated above, protocols have been developed to generate in vitro cardiomyocytes from hESC lines [41]. A diversity of cardiac myocytes are generated in EB cultures [42], and commitment to cardiac fate can be achieved in defined media [43] and enhanced by co-culture with inducing stromal elements, such as the end2 endoderm-like cell line [44]. Similar to the lineage-tracing experiments, a useful approach has been to use a cardiac reporter (for example a cardiac-specific promoter regulating expression of GFP) to select for those cells that commit to cardiac fate [45–47]. However, this can also be achieved by modeling the normal developmental signals thought to direct cardiac fate [27,48] or using chemically defined media [28,49] so that highly enriched cardiomyocyte populations can be purified from density gradients. For unknown reasons, the human cells display features that are strikingly distinct from their murine ES-derived counterparts. For example, they proliferate very well in culture [50], dependent on PI-3K-AKT signaling pathway but not ERK [51]. This provides a fortuitous advantage for generating large numbers of cells for transplant and biochemical studies. At the single cell level, the cardiomyocytes progressively and reproducibly mature during the course of EB development [50]. While they display excitation-contraction coupling and generate calcium transients upon electrical stimulation, electro-physiological analysis suggests that the sarco/endoplasmic reticulum is active but immature [52–54]. This is perhaps not surprising, since the cells may represent the equivalent of early fetal cardiomyocytes, so it will be important to investigate the potential for further maturation in vitro. Nevertheless, several groups have shown that the cells survive when transplanted into rodent models and can improve function in the infracted rat heart [27,55,56], although further maturation or integration of the cells may be needed for long-term survival and successful therapy [57].
In 2006, Takahashi and Yamanaka demonstrated that it is possible to reprogram adult fibroblasts into so-called induced pluripotent stem-like (iPS) cells by over-expression of only 4 factors: Oct4, KLF4, c-Myc and Sox2 [58]. These iPS cells were shown to be very similar to ESCs based on gene expression and epigenetic patterns, and because they could form late stage chimeras upon injection into blastocysts, although they did not give rise to live mice. Less than a year later, this problem was solved by the same group and others through modifications of the selection procedure [59–61]. More recently, several reports demonstrated that reprogramming with four pluripotency factors is so efficient that it can be performed without any selection [62], and also using human cells [63–66]. These results are of considerable significance, since iPS cell lines might in principle be generated for individual patients, providing a source of custom stem cells for regenerative medicine. There are of course still many obstacles to be overcome before iPS achieves clinical significance. Minimally, risks of tumorigenesis caused by using insertional vectors and forced expression of proto-oncogenes will need to be solved. However, long before iPS will be ready for regenerative medicine, they already provide exceptional research tools. Both vascular components and cardiomyocytes can be generated from iPS lines [67,68].
In addition to cellular therapy, additional uses of ES-derived cardiomyocytes are for studying genetic requirements for cardiac development, and for evaluating disease progression and the response of cells to drugs and other small molecules. Therefore, ES cells deficient for a specific gene product can be tested for sufficiency to generate cardiomyocytes. This has been useful for showing that specific genes are (Sox17) or are not (Gata4) required for cardiac specification, but is somewhat limited in terms of dissecting disease mechanisms. However, the use of iPS technology promises to provide biochemical reagents that will be invaluable for understanding disease mechanism. Patient specific iPS can be used in the context of the ESC cardiac differentiation protocols to evaluate alterations in differentiation and to provide a rich source of materials for biochemical analysis. Furthermore, the iPS lines will provide a novel source for testing drugs and small molecules in a high throughput manner.
A major hurdle for translation to human therapies is that ESC derivatives represent a non-autologous transplant that would lead to tissue rejection in the absence of rigorous immune suppression. The experiments using iPS cells suggest that this limitation can eventually be overcome [69]. Unless the cardiac disease is caused by a specific germline mutation, a patient-derived iPS line can be generated (for example from biopsied fibroblasts), and used as a source to generate cardiac progenitors. In principle, this would provide unlimited numbers of cells for autologous transplant and regeneration of diseased tissue. While substantial development of iPS technology is needed before that can be achieved, already the cells provide a remarkable cellular source for screening small molecules and drug testing and discovery.
ESCs and pharmacogenetics
The ability to generate and culture in vitro mouse and human ESC-derived cardiomyocytes opens up new avenues of research for high throughput drug screening. The human cardiomyocytes can be maintained in defined media in the absence of fetal calf serum or feeder cells [43,49]; in addition to their proliferative capacity this should facilitate translational capabilities. Small molecule screens can be carried out [70] to identify inducers of cardiac development (or inhibitors of non-cardiac development), which should help to derive new protocols for induction under optimum and highly controlled conditions (see for example ref. [71]).
The availability of cultures of human cardiomyocytes derived from ESCs also provides tremendous potential for testing drugs that could modulate cardiac physiology. One good example relates to the inappropriate prolongation of the QT interval, which can trigger arrhythmias, or Torsade de Pointes (TdP). Patients suffering from the long QT syndrome (LQTS) face a serious and potentially deadly condition that reflects an abnormal cardiac ventricular repolarization interval [72]. LQTS is a major cause of arrhythmia and sudden cardiac death, for which genetic causes have been identified. So far at least 10 disease genes have been mapped; the majority of LQTS is caused by mutations in ion channels [73], e.g. KCNQ1 or KCNH2 (HERG). Cardiomyocytes derived from hESC respond to channel blockers as evaluated by single cell electrophysiology and microelectrode array mapping [74]. These cultures represent a vast improvement over artificial systems that employ non-cardiac cells forced to express ectopically specific channels. This should allow high throughput screens to identify new small molecule agonists and antagonists that could treat LQTS and other arrhythmic disorders. With the advent of iPS, drugs could be tested for efficacy in the context of patient specific genetic alterations.
However, hESC-derived cardiomyocyte cultures have pharmacogenetic implications far beyond drug discovery for known disorders. For reasons that are generally not well understood, TdP is often triggered by unpredicted drug interactions. Many of the drugs that cause TdP inhibit the IKr, which is the fast component of the delayed rectifier potassium current. It is a rather disturbing fact that current assays used to test new drugs for potential to trigger TdP are carried out in channel-expressing non-cardiac cells that are clearly not optimal in the context of the clinical manifestation of the arrhythmia. This has major implications for efforts of the pharmaceutical industry to gain approval for new drugs. In fact, the major reason that drugs fail to be approved, or approved drugs are withdrawn, is due to TdP. Based on current assays, inadequately tested drugs are dangerous, and inappropriate tests might invalidate potentially good drugs. Therefore, the availability of human cardiomyocytes derived from ES or patient-specific iPS lines should allow development of new in vitro assays to better predict drug-induced responses.
Challenges Ahead
Notwithstanding the exciting progress that has been made over the past several years with ESCs and iPS lines, it is perhaps not surprising that the research has raised more questions than have been answered. Clinical trials indicate that although stem or cardiac cells can survive as explants in damaged heart tissue, this provides little if any clinical benefit. Before the potential of cellular therapies is realized, a much better understanding is required of which cells should be transplanted, and how they should be supported in order to integrate and provide functional tissue for the long term. While new precursor cells are identified, they may need to be partially or fully committed toward defined lineages (in specific ratios) for maximal benefit. Studies are required to understand the relationship of first and second heart field progenitors, and the development of cells committed to atrial, ventricular, or valve tissue. Perhaps different combinations of differentially committed progenitors (CM, EN, SM, etc.) will provide the most benefit. It is also unclear how the equivalent of embryonic or fetal-staged cardiac cells will function in the context of an adult heart, and if the progenitors or derivatives can be appropriately “aged” to an adult phenotype prior to transplant.
While translating cellular therapies is understandably of high priority, it seems a safe bet that the ESC model systems will provide more immediate gains in our basic understanding of molecular mechanisms underpinning cardiac development and disease. The system is tailor-made for high-throughput screens to identify small molecules that promote cardiogenesis, or that alter lineage commitment or differentiation. Likewise, there is hope that iPS might be used for autologous transplants, although much more study is needed before knowing if iPS can form normal cardiac derivatives, and for their use in the absence of potential transforming agents. However, using iPS lines generated from patients with defined cardiac diseases should rather quickly generate lead compounds for treating patient-specific disorders, and for evaluating disease progression mechanisms from progenitors derived from these patients. The iPS lines provide unique biochemical sources for transcript profiling, proteomics, or epigenetic evaluation that was not feasible just a few years ago. Cardiac progenitors may be identified from additional ES-like sources [75], and based on our understanding of the key cardiac regulatory programs, it might be possible to reprogram somatic cells toward a defined cardio-vascular progenitor fate, without moving all the way back to an ES-like phenotype. These exciting possibilities, in addition to the major clinical need, should make the ESC system an attractive investigative cardiac model for years to come.
Acknowledgments
The author thanks Dr. Brian Zafonte and Dr. Audrey Holtzinger for their efforts to develop novel ESC models, and for providing figure panels for this manuscript. T.E. is supported by the National Institutes of Health (HL64282 and HL56182).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Chico TJ, et al. Modeling cardiovascular disease in the zebrafish. Trends Cardiovasc Med. 2008;18 (4):150–155. doi: 10.1016/j.tcm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 2.McFadden DG, Olson EN. Heart development: learning from mistakes. Curr Opin Genet Dev. 2002;12 (3):328–335. doi: 10.1016/s0959-437x(02)00306-4. [DOI] [PubMed] [Google Scholar]
- 3.Warkman AS, Krieg PA. Xenopus as a model system for vertebrate heart development. Semin Cell Dev Biol. 2007;18 (1):46–53. doi: 10.1016/j.semcdb.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barile L, et al. Endogenous cardiac stem cells. Prog Cardiovasc Dis. 2007;50 (1):31–48. doi: 10.1016/j.pcad.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Torella D, et al. Resident human cardiac stem cells: role in cardiac cellular homeostasis and potential for myocardial regeneration. Nat Clin Pract Cardiovasc Med. 2006;3(Suppl 1):S8–13. doi: 10.1038/ncpcardio0409. [DOI] [PubMed] [Google Scholar]
- 6.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19 (10):1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 7.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132 (4):661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Nussbaum J, et al. Transplantation of undifferentiated murine embryonic stem cells in the heart: teratoma formation and immune response. Faseb J. 2007;21 (7):1345–1357. doi: 10.1096/fj.06-6769com. [DOI] [PubMed] [Google Scholar]
- 9.Doetschman TC, et al. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 10.Lacaud G, et al. Tracking mesoderm formation and specification to the hemangioblast in vitro. Trends Cardiovasc Med. 2004;14 (8):314–317. doi: 10.1016/j.tcm.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Fehling HJ, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130 (17):4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 12.Kattman SJ, et al. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11 (5):723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Kattman SJ, et al. Specification of multipotential cardiovascular progenitor cells during embryonic stem cell differentiation and embryonic development. Trends Cardiovasc Med. 2007;17 (7):240–246. doi: 10.1016/j.tcm.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Laugwitz KL, et al. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135 (2):193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 15.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127 (6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 16.Wu SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127 (6):1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 17.Christoforou N, et al. Mouse ES cell-derived cardiac precursor cells are multipotent and facilitate identification of novel cardiac genes. J Clin Invest. 2008;118 (3):894–903. doi: 10.1172/JCI33942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313 (5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtzinger A, Evans T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev Biol. 2007;312 (2):613–622. doi: 10.1016/j.ydbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao R, et al. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317 (2):614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeineddine D, et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell. 2006;11 (4):535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Tzahor E. Wnt/beta-catenin signaling and cardiogenesis: timing does matter. Dev Cell. 2007;13 (1):10–13. doi: 10.1016/j.devcel.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Foley AC, et al. Embryonic heart induction. Ann N Y Acad Sci. 2006;1080:85–96. doi: 10.1196/annals.1380.008. [DOI] [PubMed] [Google Scholar]
- 24.Naito AT, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A. 2006;103 (52):19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueno S, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104 (23):9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;91 (6):457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- 27.Laflamme MA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25 (9):1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 28.Yao S, et al. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci U S A. 2006;103 (18):6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lough J, Sugi Y. Endoderm and heart development. Dev Dyn. 2000;217 (4):327–342. doi: 10.1002/(SICI)1097-0177(200004)217:4<327::AID-DVDY1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Schultheiss TM, et al. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121 (12):4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- 31.Rudy-Reil D, Lough J. Avian precardiac endoderm/mesoderm induces cardiac myocyte differentiation in murine embryonic stem cells. Circ Res. 2004;94 (12):e107–116. doi: 10.1161/01.RES.0000134852.12783.6e. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104 (10):3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal R, Khanna A. Role of hepatocyte-like cells in the differentiation of cardiomyocytes from mouse embryonic stem cells. Stem Cells Dev. 2005;14 (2):153–161. doi: 10.1089/scd.2005.14.153. [DOI] [PubMed] [Google Scholar]
- 34.Klug MG, et al. Genetically selected cardiomyocytes from differentiating embronic stem cells form stable intracardiac grafts. J Clin Invest. 1996;98 (1):216–224. doi: 10.1172/JCI118769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai J, et al. Transplantation of embryonic stem cell-derived cardiomyocytes improves cardiac function in infarcted rat hearts. Cytotherapy. 2007;9 (3):283–291. doi: 10.1080/14653240701247838. [DOI] [PubMed] [Google Scholar]
- 36.Leor J, et al. Human embryonic stem cell transplantation to repair the infarcted myocardium. Heart. 2007;93 (10):1278–1284. doi: 10.1136/hrt.2006.093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimmeler S, et al. Cell-based therapy of myocardial infarction. Arterioscler Thromb Vasc Biol. 2008;28 (2):208–216. doi: 10.1161/ATVBAHA.107.155317. [DOI] [PubMed] [Google Scholar]
- 38.Passier R, et al. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453 (7193):322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 39.Kolossov E, et al. Engraftment of engineered ES cell-derived cardiomyocytes but not BM cells restores contractile function to the infarcted myocardium. J Exp Med. 2006;203 (10):2315–2327. doi: 10.1084/jem.20061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roell W, et al. Engraftment of connexin 43-expressing cells prevents post-infarct arrhythmia. Nature. 2007;450 (7171):819–824. doi: 10.1038/nature06321. [DOI] [PubMed] [Google Scholar]
- 41.Xu C, et al. Growth and differentiation of human embryonic stem cells for cardiac cell replacement therapy. Curr Stem Cell Res Ther. 2006;1 (2):173–187. doi: 10.2174/157488806776956931. [DOI] [PubMed] [Google Scholar]
- 42.He JQ, et al. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93 (1):32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 43.Xu C, et al. Human embryonic stem cell-derived cardiomyocytes can be maintained in defined medium without serum. Stem Cells Dev. 2006;15 (6):931–941. doi: 10.1089/scd.2006.15.931. [DOI] [PubMed] [Google Scholar]
- 44.Mummery C, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107 (21):2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 45.Gallo P, et al. A lentiviral vector with a short troponin-I promoter for tracking cardiomyocyte differentiation of human embryonic stem cells. Gene Ther. 2008;15 (3):161–170. doi: 10.1038/sj.gt.3303017. [DOI] [PubMed] [Google Scholar]
- 46.Huber I, et al. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. Faseb J. 2007;21 (10):2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- 47.Xu XQ, et al. Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy. 2008;10 (4):376–389. doi: 10.1080/14653240802105307. [DOI] [PubMed] [Google Scholar]
- 48.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453 (7194):524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 49.Xu XQ, et al. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008 doi: 10.1111/j.1432-0436.2008.00284.x. [DOI] [PubMed] [Google Scholar]
- 50.Snir M, et al. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol. 2003;285 (6):H2355–2363. doi: 10.1152/ajpheart.00020.2003. [DOI] [PubMed] [Google Scholar]
- 51.McDevitt TC, et al. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J Mol Cell Cardiol. 2005;39 (6):865–873. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dolnikov K, et al. Functional properties of human embryonic stem cell-derived cardiomyocytes: intracellular Ca2+ handling and the role of sarcoplasmic reticulum in the contraction. Stem Cells. 2006;24 (2):236–245. doi: 10.1634/stemcells.2005-0036. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, et al. Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem Cells. 2007;25 (12):3038–3044. doi: 10.1634/stemcells.2007-0549. [DOI] [PubMed] [Google Scholar]
- 54.Sartiani L, et al. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25 (5):1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 55.Caspi O, et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol. 2007;50 (19):1884–1893. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 56.Dai W, et al. Survival and maturation of human embryonic stem cell-derived cardiomyocytes in rat hearts. J Mol Cell Cardiol. 2007;43 (4):504–516. doi: 10.1016/j.yjmcc.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Laake LW, et al. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circ Res. 2008;102 (9):1008–1010. doi: 10.1161/CIRCRESAHA.108.175505. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126 (4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 59.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448 (7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 60.Okita K, et al. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448 (7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 61.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1 (1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 62.Meissner A, et al. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25 (10):1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 63.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105 (8):2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451 (7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131 (5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 66.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318 (5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 67.Mauritz C, et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118 (5):507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- 68.Narazaki G, et al. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118 (5):498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- 69.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318 (5858):1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 70.Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132 (4):549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 71.Graichen R, et al. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76 (4):357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 72.Goldenberg I, Moss AJ. Long QT syndrome. J Am Coll Cardiol. 2008;51 (24):2291–2300. doi: 10.1016/j.jacc.2008.02.068. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz PJ. The congenital long QT syndromes from genotype to phenotype: clinical implications. J Intern Med. 2006;259 (1):39–47. doi: 10.1111/j.1365-2796.2005.01583.x. [DOI] [PubMed] [Google Scholar]
- 74.Caspi O, et al. In vitro Electrophysiological Drug Testing using Human Embryonic Stem Cell Derived Cardiomyocytes. Stem Cells Dev. 2008 doi: 10.1089/scd.2007.0280. [DOI] [PubMed] [Google Scholar]
- 75.Guan K, et al. Generation of functional cardiomyocytes from adult mouse spermatogonial stem cells. Circ Res. 2007;100 (11):1615–1625. doi: 10.1161/01.RES.0000269182.22798.d9. [DOI] [PubMed] [Google Scholar]