Abstract
Spatial regulation of C-function genes controlling reproductive organ identity in the centre of the flower can be achieved by adjusting the level of their expression within the genuine central expression domain in Antirrhinum and Petunia. Loss of this control in mutants is revealed by enhanced C-gene expression in the centre and by lateral expansion of the C-domain. In order to test whether the level of central C-gene expression and hence the principle of ‘regulation by tuning’ also applies to spatial regulation of the C-function gene AGAMOUS (AG) in Arabidopsis, we generated transgenic plants with enhanced central AG expression by using stem cell-specific CLAVATA3 (CLV3) regulatory sequences to drive transcription of the AG cDNA. The youngest terminal flowers on inflorescences of CLV3::AG plants displayed homeotic features in their outer whorls indicating ectopic AG expression. Dependence of the homeotic feature on the age of the plant is attributed to the known overall weakening of repressive mechanisms controlling AG. Monitoring AG with an AG-I::GUS reporter construct suggests ectopic AG expression in CLV3::AG flowers when AG in the inflorescence is still repressed, although in terminating inflorescence meristems, AG expression expands to all tissues. Supported by genetic tests, we conclude that upon enhanced central AG expression, the C-domain laterally expands necessitating tuning of the expression level of C-function genes in the wild type. The tuning mechanism in C-gene regulation in Arabidopsis is discussed as a late security switch that ensures wild-type C-domain control when other repressive mechanism starts to fade and fail.
Keywords: Arabidopsis, Boundary, C-function, Tuning
Introduction
In higher plants, reproductive development in the two inner floral whorls is governed by the C-function. Expansion of the C-domain towards the outer whorls conditions homeotic defects such as stamenoid petals and carpelloid sepals and is prevented by repressive mechanisms (Sridhar et al. 2006; Cartolano et al. 2007). In Arabidopsis, restriction of expression of the C-gene AGAMOUS (AG) to the inner whorls is achieved by controlling the balance between activation and repression, reinforced by region-specific activators and autoregulatory maintenance in the centre of the flower and by region-specific repressors in the outer whorls (Sridhar et al. 2006). Most of the proteins involved in this control as well as their cis-acting elements within C-function genes are conserved in different species including Antirrhinum (Navarro et al. 2004; Causier et al. 2009), suggesting similarities of C-domain control.
An additional way to spatially control the C-domain by fine-tuning the level of C-gene expression in the central expression domain has been detected in Antirrhinum and Petunia (Cartolano et al. 2007). Here, a ubiquitously expressed miR169-related microRNA fine-tunes C-gene transcription by controlling the expression of a positive regulator. In the absence of this control, C-gene expression increases in the centre, and likely due to C-product exchange between neighbouring cells (such as cell-to-cell trafficking or transmission to daughter cells by cell division) also ectopically expands towards the outer whorls. By these means, an outward extending gradient of C-gene product will form, where ‘recipient cells’ maintain C-gene expression by autoregulation, provided that a threshold for autoregulation is reached. The lateral extension of this gradient and hence the size of the C-domain will then depend on the level of C-gene expression in the central ‘donor cells’ where C-gene expression has been activated. Components of this miR-169-mediated circuit are conserved in Arabidopsis and likely in other species as well, but their function in the transcriptional control of AG and other C-genes is not established yet.
One limitation to the ‘tuning model’ is the lack of confirmation by independent experiments, for instance, observing C-domain expansion upon artificial enhancement of early C-activity in the centre of the flower. In order to address this question, we studied in Arabidopsis the effects of AG overexpression under the control of CLAVATA3 (CLV3) regulatory sequences whose function is confined to stem cells in the centre of vegetative and reproductive meristems (Brand et al. 2002).
Materials and methods
Transgenic lines and growth conditions
In order to obtain the CLV3::AG recombinant plasmid, the AG cDNA was amplified by RT-PCR using primers 5′-GTTACCTGCAGATGGCGTACCAATCGGAGCTAG-3′ and 5′-GCTAGGATCCTTACACTAACTGGAGAGCGGTTTGG-3′, and after restriction, the fragment was ligated into the PstI/BamHI sites of the pBU14 plasmid (Brand et al. 2002).
Arabidopsis thaliana L. Col-0 ecotype (stock Max-Planck Institut für Züchtungsforschung, Köln) was used for Agrobacterium-mediated transformation by vacuum infiltration. Several independent lines carrying one or more transgenes were obtained and selfed. Notably, only 50% of transgenic plants in the T2 and T3 progenies showed a modified phenotype. The copy number of the transgene had no influence on the phenotype.
In order to generate AG-I::GUS; CLV3::AG double transgenic lines AG-I::GUS plants (Ler background) carrying the KB9 construct with 3 kb of the AG intron sequence (Busch et al. 1999) were crossed with a CLV3::AG transgenic line. F1 plants were self-pollinated and the F2 progeny was screened by PCR for individuals containing both transgenes.
In order to generate the rbe; CLV3::AG double mutant, rbe-2 plants (line SALK_037010, Col-0 background) were crossed to a CLV3::AG plant. F2 individuals were selected by phenotype for the rbe mutant and screened by PCR for the presence of the transgene. Plants were grown in the greenhouse at 21–23°C under long day conditions (16 h light).
Real-time PCR
The copy number of transgenes was determined by quantitative PCR in the bulked T2 progeny of independent transformants. DNA was isolated by the CTAB method (Murray and Thompson 1980) and purified using the DNeasy Plant Mini kit (Qiagen Hilden, Germany). The transgenic cassette was amplified using primers spanning an exon sequence present in the genome as well as in the transgene using the IQ™5 Real-Time PCR Detection System (Biorad, München, Germany). For normalization, primers amplifying a region in the second intron of the AG gene was used. The number of transgenes was calculated with the Pfaffi method (Pfaffl et al. 2002) and corroborated by segregation analysis.
GUS staining
Samples were stained following the protocol described in Kosugi et al. (1990). Subsequently, the tissue was embedded in paraffin, 13-μm thick sections were prepared, deparaffinized and finally embedded in entellan. GUS signal was imaged by dark-field microscopy.
Microscopy
Scanning electron microscopy with a Zeiss EM10 microscope (Carl Zeiss, Oberkochen, Germany) was conducted by the CeMic service unit at the Max Planck Institut für Züchtungforschung.
Results and discussion
CLV3::AG plants display floral homeotic defects
Expression of AG outside its genuine central domain results in flowers, whose sepals become carpelloid and whose petals in the second whorl become stamenoid or are missing (Mizukami and Ma 1992). In order to observe whether enhancement of AG in the centre of the flower would convert wild-type flowers (Fig. 1a) to carpelloid flowers, we expressed AG in transgenic Arabidopsis plants under the control of CLV3 regulatory sequences. The pattern of transcription directed by the CLV3-construct is specific and confined to stem cells in all meristems as shown before in CLV3::GUS transgenic plants (Brand et al. 2002). Controlled by CLV3 regulatory sequences, AG will be expressed at stage 2 in the flower, slightly earlier than in the wild type, where AG onset is during stage 3 (Drews et al. 1991). Since stem cells divide slowly (Stahl and Simon 2005), we did not expect substantial broadening of the region of AG transcribing cells prior to genuine AG activation in the flower, even if the AG protein transmitted to daughter cells remained stable.
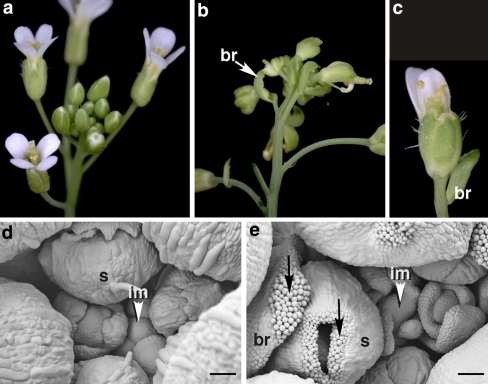
Fig. 1.
Terminal carpelloid flower phenotype conferred by the CLV3::AG transgene. The photographs show wild type (a, d) and transgenic (b, e) inflorescences, documented by SEM (d, e). A single flower with reduced number of petals is shown in c. Arrows in d point to stigmatic tissues in carpelloid bracts (br) and laterally fused carpelloid sepals (s). The inflorescence meristem (im) is indicated by arrowheads. Bar 50 μm
In the selfed progeny of a homozygous line carrying a single copy insert of the CLV3::AG transgene, plants developed flowers with carpelloid sepals and the number of petals was reduced or petals were absent (Fig. 1b, c). In addition, the carpelloid flowers were subtended by bracts decorated with stigmatic papillae (Fig. 1e) and the pedicels were often fused (Fig. 1b). The severity of the phenotype increased acropetally with the youngest flowers affected by more drastic homeotic changes revealed by laterally fused sepals tipped with stigmatic papillae (Fig. 1e). The old reproductive meristem became morphologically aberrant and developed bract-like leaves in place of flower primordia (Fig. 1e).
Floral defects became visible 10–15 days after opening of the first flower, indicating that the inflorescence meristem first produced a number of normal flowers and then switched to carpelloid flowers subtended by bracts. Carpelloid flowers were mainly visible on the main shoot and rarely on secondary shoots. The penetrance of the phenotype was incomplete in that only 50% of the transgenic progeny of selfed lines displayed the phenotype.
The observed phenotype shows remarkable similarity in terms of floral homeotic changes, carpelloid bracts, inflorescence anomalies, and incomplete penetrance of the phenotype to that of BELLRINGER (BLR) mutants carrying antimorphic alleles (Bao et al. 2004). blr mutants, like CLV3::AG transgenic plants develop the homeotic defects late during development, a feature termed terminal carpelloid flower (tcf). The tcf phenotype of blr is mediated by ectopic AG expression and it has been shown that BLR directly (negatively) regulates AG. In order to explain the tcf phenotype, Bao et al. (2004) suggest a time-dependent weakening of the transcriptional repression of AG, which is in line with the generally enhanced severity of phenotypes caused by mutation in negative regulators of AG during development. It appears, therefore, that phenotypic manifestation of the consequences of early enhanced AG expression in the centre of the CLV3::AG flower also depends on the late relaxation of AG repression.
Manifestation of the tcf phenotype is reinforced by slight reduction of AG repression in the second whorl
RABBIT EARS (RBE) is a second whorl-specific repressor of AG (Krizek et al. 2006), expressed early throughout second whorl primordia (Takeda et al. 2004). However, in situ detectable ectopic AG expression in the rbe mutant is restricted to the boundary between the second and the third whorl (Krizek et al. 2006). This suggests that derepression of AG in rbe flowers is more readily maintained in cells facing the central domain than in laterally more distant cells—complying with the C-product gradient extending from the centre to the periphery and its threshold-dependent autoregulatory component implied in the tuning model. If the tcf phenotype in CLV3::AG plants is due to lateral expansion of AG expression, then the chance of its manifestation and hence its penetrance should be enhanced in the rbe background due to additional slight and local weakening of AG repression. We tested this assumption by crossing the CLV3::AG transgene into the rbe-2 mutant background. rbe-2 mutants display a mild phenotype with very few or no petals formed (Takeda et al. 2004).
In a population of 139 CLV::AG plants segregating for rbe, 34 of the 35 rbe individuals displayed tcf showing the characteristic floral homeotic and inflorescence defects. In contrast, only 51 of the 104 CLV::AG plants carrying no or one rbe allele displayed these features. Enhancement of the phenotypic manifestation of the tcf phenotype of CLV::AG from 49% to 97% in the rbe background suggests that additional local derepression of AG in the second whorl facilitates lateral expansion of AG which is initiated primarily by enhanced central AG expression.
Notably, carpelloidy of sepals was not enhanced in rbe CLV3::AG flowers, although enhanced AG expression in the second whorl should promote lateral expansion to the first whorl according to the tuning model. One explanation could be that derepression of AG in the rbe mutant starts after initiation of sepals, as RBE expression commences at stage 3 of flower development (Takeda et al. 2004). Expression from the AP3 promoter also starts after the emergence of sepal primordia (Jack et al. 1992) and ectopic expression of AG in the second whorl of AP3::AG transgenic flowers had little effect on sepals whose weak carpelloidy has been attributed to a low AP3 promoter activity in sepals (Jack et al. 1997). It would then appear that physical separation from the second whorl impedes transmission of gene products from the second whorl area to the first.
Altered floral AG expression is independent of central AG expression in the inflorescence meristem
The defects observed in the CLV3::AG flower can be due to enhanced central AG expression and ‘spreading’ of the protein or transcript towards the periphery, as suggested by the tuning model, but they can also be the consequence of a primary effect of the transgene in the inflorescence meristem. For instance, assuming high stability of the AG protein or mRNA, it is possible that AG accumulates in cells giving rise to floral primordia, which then express AG in all cells from early on. In the former case, floral ectopic AG expression should be detectable in the absence of AG transcript in the inflorescence meristem while in the latter case, it should be always accompanied by AG expression in the inflorescence meristem.
In order to study AG expression in situ, we introduced the AG-I::GUS transgene (Busch et al. 1999) to the CLV3::AG background by crossing AG-I::GUS and CLV3::AG transgenic plants. The AG intron sequence contains regulatory elements that are necessary and sufficient for AG repression, activation and maintenance (Hong et al. 2003); thus, the GUS reporter monitors changes in the transcriptional regulation of AG in response to the enhanced or decreased function of AG-repressors and activators including AG itself.
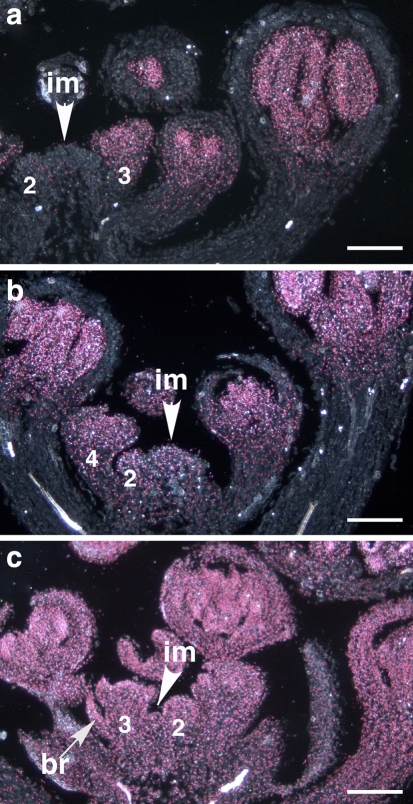
In order to observe the earliest events, inflorescences were harvested after opening of the first flowers, but before the tcf phenotype was visible. Samples with ectopic GUS signal were sorted after sectioning according to the criteria of the absence or presence of bracts. In samples where young flowers did not develop bracts, two patterns were observed. In some cases, GUS signal was detectable throughout the flower, but was not visible in the inflorescence meristem (Fig. 2a). This suggests that ectopic AG expression in the CLV3::AG flower is independent of AG expression in the inflorescence meristem. In other cases, weak GUS signal could also be detected in the inflorescence meristem (Fig. 2b). In the inflorescences marked by flowers subtended by bracts and misshapen inflorescence meristem, reporter expression was strong all over the tissues (Fig. 2c). The three patterns apparently reflect progression of ectopic AG expression during ageing of the inflorescence. During the earliest stages, expansion of the AG-expressing domain is only observable in flowers, likely because floral activation of endogenous AG expression can facilitate autoregulatory maintenance of expression. Domain expansion also occurs in the inflorescence meristem, but in the absence of endogenous activation, further age-dependent weakening of AG-repression is necessary to achieve enhanced AG expression outside the stem cell domain.
Fig. 2.
Ectopic AG expression in inflorescences of CLV3:AG transgenic plants. AG expression is followed by the GUS reporter using the AG-I::GUS transgene. a Ectopic AG expression in a stage 3 flower primordium (stages numbered after Smyth et al. 1990), but no or very low reporter expression in the inflorescence meristem (im). b AG expression in the inflorescence meristem is weak, but clear, and in c strong in all tissues. Notice aberrant morphology of the inflorescence meristem and the bract (br) subtending a stage 3 flower primordium, marking the onset of the tcf phenotype in the adult inflorescence. The typical CLV3-pattern in the centre of vegetative meristems (not shown) or in young inflorescence meristems and stage 2 floral meristems was not detectable (a), probably because expression of components other than AG needed for autoregulation was too low to govern reporter expression. Bar 100 μm
In conclusion, when AG is expressed in the stem cell domain of the transgenic plants, C-gene expression expands to neighbouring cells in both floral and inflorescence meristems. The mechanism of expansion cannot be addressed with these experiments, but it appears that the central cells are the source of AG protein/transcript and act as ‘donors’ of C-gene products to ‘recipient’ peripheral cells.
Mechanisms for C-domain control in Arabidopsis
The floral homeotic phenotype of CLV3::AG plants, its enhancement in the rbe mutant background as well as the pattern of ectopic AG expression suggest that the level of central C-gene expression can influence the extension of the C-domain even if the mechanism that accomplishes spreading of C-gene products remains elusive.
To which extent the control of central AG expression level by a tuning mechanism is meaningful in wild-type Arabidopsis is difficult to tell, however. Weakening of AG repression that accompanies aging of the inflorescence will change the activator/repressor balance in favour of activation both at the periphery and certainly also within the genuine activation domain in the centre of the flower. Enhanced central expression thus could further impair boundary control and the role of the miR169-related control of the C-gene expression level—as detected in Antirrhinum—could counteract this deleterious event. Thus, it seems that domain-specific activation of AG and the subsequent establishment of the specific activator/repressor ratio in the inner and outer whorls is the primary event in C-domain control in Arabidopsis which is reinforced by a self-tuning control system when repressive mechanisms start to decay.
Acknowledgments
We thank Rainer Franzen (CeMic, MPIZ Köln) for help with SEM, Rüdiger Simon (University Düsseldorf) for the pBU14 construct and Peter Huijser (MPIZ Köln) for help with photography. We are grateful to Brendan Davies (University Leeds) and Peter Huijser for critical reading of the manuscript.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AG
AGAMOUS
- CLV3
CLAVATA3
- SEM
Scanning electron microscopy
- tcf
Terminal carpelloid flower
- BLR
BELLRINGER
- RBE
RABBIT EARS
Contributor Information
Maria Cartolano, Email: maria.cartolano@plants.ox.ac.uk.
Nadia Efremova, Email: n.eferemova@phytowelt.com.
Markus Kuckenberg, Email: kuckenbe@mpiz-koeln.mpg.de.
Smita Raman, Email: S.Raman@uu.nl.
Zsuzsanna Schwarz-Sommer, Email: schwarzs@mpiz-koeln.mpg.de.
References
- Bao X, Franks RG, Levin JZ, Liu Z (2004) Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16:1478–1489 [DOI] [PMC free article] [PubMed]
- Brand U, Grunewald M, Hobe M, Simon R (2002) Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol 129:565–575 [DOI] [PMC free article] [PubMed]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285:585–587 [DOI] [PubMed]
- Cartolano M, Castillo R, Efremova N, Kuckenberg M, Zethof J, Gerats T, Schwarz-Sommer Z, Vandenbussche M (2007) A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet 39:901–905 [DOI] [PubMed]
- Causier B, Bradley D, Cook H, Davies B (2009) Conserved intragenic elements were critical for the evolution of the floral C-function. Plant J 58:41–52 [DOI] [PubMed]
- Drews GN, Bowman JL, Meyerowitz EM (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65:991–1002 [DOI] [PubMed]
- Hong RL, Hamaguchi L, Busch MA, Weigel D (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15:1296–1309 [DOI] [PMC free article] [PubMed]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68:683–697 [DOI] [PubMed]
- Jack T, Sieburth L, Meyerowitz E (1997) Targeted misexpression of agamous in whorl-2 of Arabidopsis flowers. Plant J 11:825–839 [DOI] [PubMed]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y (1990) An improved assay for beta-glucuronidase in transformed-cells-methanol almost completely suppresses a putative endogenous beta-glucuronidase activity. Plant Sci 70:133–140 [DOI]
- Krizek BA, Lewis MW, Fletcher JC (2006) RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J 45:369–383 [DOI] [PubMed]
- Mizukami Y, Ma H (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71:119–131 [DOI] [PubMed]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325 [DOI] [PMC free article] [PubMed]
- Navarro C, Efremova N, Golz JF, Rubiera R, Kuckenberg M, Castillo R, Tietz O, Saedler H, Schwarz-Sommer Z (2004) Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131:3649–3659 [DOI] [PubMed]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36 [DOI] [PMC free article] [PubMed]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2:755–767 [DOI] [PMC free article] [PubMed]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133:3159–3166 [DOI] [PubMed]
- Stahl Y, Simon R (2005) Plant stem cell niches. Int J Dev Biol 49:479–489 [DOI] [PubMed]
- Takeda S, Matsumoto N, Okada K (2004) RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131:425–434 [DOI] [PubMed]