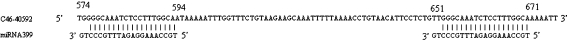Abstract
MicroRNAs (miRNAs) are a class of non-protein-coding small RNAs. Considering the conservation of many miRNA genes in different plant genomes, the identification of miRNAs from non-model organisms is both practicable and instrumental in addressing miRNA-guided gene regulation. Citrus is an important staple fruit tree, and publicly available expressed sequence tag (EST) database for citrus are increasing. However, until now, little has been known about miRNA in citrus. In this study, 27 known miRNAs from Arabidopsis were searched against citrus EST databases for miRNA precursors, of which 13 searched precursor sequences could form fold-back structures similar with those of Arabidopsis. The ubiquitous expression of those 13 citrus microRNAs and other 13 potential citrus miRNAs could be detected in citrus leaf, young shoot, flower, fruit and root by northern blotting, and some of them showed differential expression in different tissues. Based on the fact that miRNAs exhibit perfect or nearly perfect complementarity with their target sequences, a total of 41 potential targets were identified for 15 citrus miRNAs. The majority of the targets are transcription factors that play important roles in citrus development, including leaf, shoot, and root development. Additionally, some other target genes appear to play roles in diverse physiological processes. Four target genes have been experimentally verified by detection of the miRNA-mediated mRNA cleavage in Poncirus trifoliate. Overall, this study in the identification and characterization of miRNAs in citrus can initiate further study on citrus miRNA regulation mechanisms, and it can help us to know more about the important roles of miRNAs in citrus.
Keywords: Citrus, MicroRNAs, Northern blotting, 5′RACE
Introduction
MicroRNAs (miRNA) are endogenous tiny RNAs (about 22 nt in length) that can play important regulatory roles in animals and plants by targeting mRNAs for cleavage or translational repression. The miRNAs play a vital regulatory function in eukaryotic gene expression by binding specific sequences in target genes and suppressing their expression. Plant miRNAs were first identified in early 2002 (Llave et al. 2002a). Searching for these molecules could be accomplished by direct cloning and bioinformatics analysis, and hundreds of miRNAs have been identified in Arabidopsis thaliana (Rhoades and Bartel 2004; Sunkar and Zhu 2004), Oryza sativa (Sunkar et al. 2005), Populus trichocarpa (Lu et al. 2005), Malus domestica (Gleave et al. 2008), Zea mays (Mica et al. 2006; Zhang et al. 2008), Solanum lycopersicum (Pilcher et al. 2007; Moxon et al. 2008), Medicago truncatula (Szittya et al. 2008), and other plants (Zhang et al. 2005, 2006, 2007; Guo et al. 2007; Xie et al. 2007). Plant miRNAs bind to their target mRNAs and initiate mRNA degradation or mRNA translational repression with the help of an enzyme with ‘slicer activity’ that belongs to RISC (RNA-induced silencing complex) (Liu et al. 2004; Vaucheret et al. 2004; Baumberger and Baulcombe 2005). There have been some reports about the translational control of quite a few miRNAs (Palatnik et al. 2003; Chen 2004; Lu et al. 2005). In all known cases, most plant miRNAs bind to the protein-coding region of their target mRNAs with three or fewer mismatches and induce target mRNA degradation (Llave et al. 2002a; Rhoades et al. 2002) or repress mRNA translation (Chen 2004; Brodersen et al. 2008). The miRNAs are generated from long precursors suggesting that it is possible to find these precursors by searching expressed sequence tags (ESTs). The core principle is to look for the sequences containing conserved mature miRNAs and to check if these candidate sequences can fold into hairpins. Many known miRNAs are conserved in most plant species (Reinhart et al. 2002; Wang et al. 2004a, b; Sunkar and Jagadeeswaran 2008). There have been reports about the identification of miRNAs by mining the repository of available ESTs (Smalheiser 2003; Zhang et al. 2005, 2006, 2008). Different computational miRNA gene-finding strategies have been developed based on a comparative approach, and their core principle is to look for conserved sequences between different species that can fold into extended hairpins (Grad et al. 2003). In plants, miRNAs play important roles in regulating plant growth and development, including leaf growth (Palatnik et al. 2003), stem growth (Mallory et al. 2004), root growth (Subramanian et al. 2009), floral organ identity and reproductive development (Chen 2004; Millar and Gubler 2005), fleshy fruit development (Moxon et al. 2008), developmental transitions (Rhoades et al. 2002; Aukerman and Sakai 2003; Schmid et al. 2003), organ polarity (Juarez et al. 2004), auxin signaling (Rhoades and Bartel 2004), boundary formation/organ separation (Mallory et al. 2004), biotic and abiotic stress response (Phillips et al. 2007; Sunkar and Zhu 2007; Shukla et al. 2008; Jagadeeswaran et al. 2009), and species-specific metabolic pathways (Shukla et al. 2008). Despite their important roles of miRNAs in plant development, their expression profiles in non-model plants are still poorly studied and there have been few reports about miRNAs in fruit crops (Moxon et al. 2008). In the present study, we first research miRNA expression and characterization in different tissues of citrus for an understanding of the potential roles of the miRNAs studied.
Computational analysis based on sequence similarity has been proven to be a reliable and successful way to identify miRNA target genes, since the number of mismatches allowed between the small RNA and its target in plants is low. Identified target genes of plant miRNAs have shown that in most cases, the target genes were transcription factors that were mainly related to plant developmental processes and organ morphogenesis (Rhoades et al. 2002; Mallory et al. 2004; Lauter et al. 2005). Plant miRNAs generally interact with their targets through perfect or near-perfect complementarity (Reinhart et al. 2002; Subramanian et al. 2009). Based on these findings, we initiated the identification of the putative targets of the miRNAs studied in citrus.
Citrus aestivum L. fruit tree is usually propagated through asexual reproduction by grafting or budding the scion of a desired variety onto a suitable rootstock. The bud scion is usually taken from the fruiting tree, and the rootstock is a seedling from seed. We used adult fruiting trees and young rootstock seedlings as the plant materials in our study. The conservation of miRNAs in plant kingdom made it possible to carry out the miRNA-related study in different species of citrus. We studied the expression pattern of 27 Arabidopsis miRNAs in citrus, which had intriguing expression patterns in different tissues, indicating that they have important functions and might be involved in the regulation of gene expression. Moreover, four target genes were experimentally verified by detection of the miRNA-mediated mRNA cleavage sites in Poncirus trifoliate using RNA ligase-mediated 5′ rapid amplification of cDNA ends (RLM-RACE). Our achievement in the identification and characterization of miRNAs in citrus may initiate further study of citrus miRNA regulation mechanisms, and it can help us to learn more about the important roles of miRNAs in citrus.
Materials and methods
Plant materials
The samples were collected from a 5-year-old ‘Nules’ tree (Citrus reticulata) and 2-year-old trifoliate orange (Poncirus trifoliata (L.) Raf.) trees that were grown in Suzhou Evergreen Fruit Tree Research Institute, China and the University of California (UC) Lindcove Research and Extension Center (LREC), CA, USA. Leaves, young shoots, roots, flowers, and fruits (diameter 1 cm) were collected from ‘Nules’ trees, and roots, stems, and leaves from trifoliate orange trees which were important rootstock for the citrus scion cultivar. After collection, all the samples were immediately frozen in liquid nitrogen and stored at −80°C until used.
Prediction of fold-back structures and miRNA targets
Due to the phylogenetic conservation of the chosen miRNA sequences in plants, their predicted precursor secondary structures can be considered as an important validation for MIR genes (Ambros et al. 2003), which was an important parameter used to validate the stem-loops in citrus in this study. We used the set of conserved 27 miRNAs, of which miR158 and miR173 were reported as non-conserved, from Arabidopsis (Table 1; Rhoades and Bartel 2004; Griffiths-Jones et al. 2006) as a query set. We used citrus blast with an E-value cutoff of 1.0 for a similarity search against the citrus EST database, Version 1.20 of ‘HarvEST:Citrus’ which displays 89 libraries and 229,570 ESTs from Citrus and Poncirus and contains best BLASTX hits from UniProt (January 2007) the Arabidopsis genome (TAIR version 7; April 2007) and the poplar genome (JGI version 1.1). In the hairpin structures formed by miRNA precursors, all miRNAs were found in the stem region of the hairpins and had at least 75% sequence complementarity to their counterparts. All the predicted miRNAs were conserved with at least 90% sequence identity in citrus. G + C content in mature miRNAs ranged from 38 to 70%, and the loop lengths were between 20 and 75 nucleotides, which were same as the values reported elsewhere (Wang et al. 2004a, b). The blast hits of candidate miRNA genes and their candidate targets were performed at http://138.23.191.145/blast/index.html using the conserved miRNA sequences. Citrus miRNA fold-back secondary structures of the blast hits complementary to miRNAs (plus/plus) with 0–1 mismatches were predicted with the computer program MFOLD version 3 (Zuker 2003; http://www.bioinfo.rpi.edu/applications/mfold). The citrus mRNAs were predicted from the annotated contigs from the citrus EST database (Version 1.20 of HarvEST: Citrus, http://harvest.ucr.edu) with miRNA gene homology searching (Fig. 1; Tables 1, 2).
Table 1.
Putative miRNA target genes in citrus
| miRNA | miRNA(3′ → 5′)/mRNA(5′ → 3′) | Citrus putative target unigene no. (number of mismatches) | Target protein | Target function | Conserved gene in other plants (E-score) |
|---|---|---|---|---|---|
| ptr-miR156 (N) | 3′CACGAGUGAGAGAAGACAGU5′ | ||||
| GUGCUCUCUCUCUUCUGUCA | 12489 (1) | Squamosa promoter binding protein (SPB) | Transcription factor (TF) | At3g43270 (4e-07) | |
| GUGCUCUCUCUCUUCUGUCA | 11286 (1) | SPB | TF | At2g42200 (3e-033) | |
| GUGCUCUCUCUCUUCUGUCA | 9390 (1) | SPB | TF | At5g50670 (7e-066) | |
| GUGCUCUCUCUCUUCUGUCA | 8859 (1) | SPB | TF | At2g42200 (6e-064) | |
| AUGCUCUCUCUCUUCUGUCA | 41072 (2) | SPB | TF | At3g15270 (4e-013) | |
| AUGCUCUCUCUCUUCUGUCA | 39932 (2) | SPB | TF | At1g53160 (3e-038) | |
| AUGCUCUCUCUCUUCUGUCA | 18226 (2) | SPB | TF | At3g15270 (1e-034) | |
| miR157 (N) | 3′CACGAGAGAUAGAAGACAGUU5′ | ||||
| GUGCUCUCUCUCUUCUGUCAA | 12489 (1) | SPB | TF | At3g43270 (4e-07) | |
| GUGCUCUCUCUCUUCUGUCAA | 8859 (1) | SPB | TF | At2g42200 (6e-064) | |
| miR158 (N) | 3′ACGAAACAGAUGUAAACCCU5′ | No | |||
| miR159 (N) | 3′AUCUCGAGGGAAGUUAGGUUU5′ | No | |||
| cis-miR160 (N,R) | 3′ACCGUAUGUCCCUCGGUCCGU 5′ | ||||
| AGGCAUACAGGGAGCCAGGCA | 13966 (1) | Auxin response factor | TF | At2g28350 (1e-158) | |
| miR161 (N) | 3′GGGGCUACAUCAGUGAAAGUU5′ | No | |||
| miR162 (N) | 3′GACCUACGUCUCCAAAUAGCU 5′ | No | |||
| miR163 (N) | 3′UAGCUUCAAGGUUCAGGAGAAGUU5′ | No | |||
| cis-miR164 (N) | 3′ACGUGCACGGGACGAAGAGGU5′ | ||||
| CTTACGUGUCCUGCUUCUCCA | 26767 (4) | NAC domain protein | TF | At5g61430 (7e-065) | |
| CUCACG UGACCUGCU UCUCCG | 21315 (4) | NAC domain protein | TF | At5g61430 (6e-092) | |
| cis-miR165 (N) | 3′CCCCCUACUUCGGACCAGGCU5′ | ||||
| GACCCUAGUUGCCUGGUCCGG | 19387 (3) | HD-ZIP protein | TF | At5g60690 (1e-140) | |
| CCGGGAUGAAGCCUGGUCCGG | 5511 (3) | HD-ZIP protein | TF | At4g32880 (0.0) | |
| ptr-miR166 (N) | 3′CCCCUUACUUCGGACCAGGCU5′ | ||||
| CUGGGAUGAAGCCUG GUCCGG | 5509 (3) | HD-ZIP protein | TF | Populus trichocarpa (1e-62) | |
| ccl-miR167(N,R) | 3′AUCUAGUACGACCGUCGAAGU5′ | ||||
| UAGAUCAGGCUGGCAGCUUGU | 21883 (3) | Auxin response factor (ARF) | Transcription factor (TR) | At5g37020 (1e-045) | |
| UAGAUCAGGCUGGCAGCUUGU | 16450 (3) | ARF | TR | At5g37020 (1e-142) | |
| GAGAUCAGGCUGGCAGCUUGU | 8197 (4) | ARF | TR | Cucumis sativus (1e-81) | |
| GAGAUCAGGCUGGCAGCUUGU | 7960 (4) | ARF | TR | At1g30330 (2e-049) | |
| GAGAUCAGGCUG GCAGCUUGU | 7959 (4) | ARF | TR | At1g30330 (1e-121) | |
| ccl-miR168 (N) | 3′AAGGGCUGGACGUGGUUCGCU5′ | No | |||
| cis-miR169 (N) | 3′AGCCGUUCAGUAGGAACCGAC5′ | ||||
| UCGGCAAGUCAUUCUUGGCUC | 388432 (2) | Anthocyanidin synthase | Metabolism | ||
| miR170 (N) | 3′CUAUAACUGUGCCGAGUUAGU5′ | No | |||
| ccl-miR171 (N) | 3′CUAUAACCGCGCCGAGUUAGU5′ | No | |||
| cis-miR172 (N) | 3′UACGUCGUAGUAGUUCUAAGA5′ | ||||
| GUGCAGCAUCAUCAGGAUUCU | 10023 (2) | APETALA2-like protein (AP2) | DNA binding/transcription factor (DBF) | At2g28550 (1e-011) | |
| GUGCAGCAUCAUCAGGAUUCU | 10022 (2) | AP2 | DBF | At2g28550 (2e-010) | |
| CUGCAGCAUCAUCAGGAUUCC | 41024 (3) | AP2 | DBF | At4g36920 (7e-080) | |
| CUGCAGCAUCAUCAGGAUUCC | 27633 (3) | AP2 | DBF | At4g36920 (1e-121) | |
| CUGCAGCAUCAUCAGGAUUCC | 19697 (3) | AP2 | DBF | At4g36920 (2e-046) | |
| GUGCAGCAUCAUCAGGAUUCC | 10330 (3) | AP2 | DBF | At2g28550 (1e-114) | |
| GUGCAGCAUCAUCAGGAUUCC | 6618 (3) | AP2 | DBF | At2g28550 (1e-114) | |
| miR173 (N) | 3′CACUAAAGAGAGACGUUCGCUU5′ | No | |||
| ptr-miR319 (N) | 3′CCCUCGAGGGAAGUCAGGUU5′ | No | |||
| miR390 (N) | 3′CCGCGAUAGGGAGGACUCGAA5′ | ||||
| GGCGAUAUCUCUCCUGAGCUU | 27418 (2) | Unknown | |||
| miR391 (N) | 3′ACCGCGAUAGAGAGGACGCUU5′ | No | |||
| miR393(N,R) | 3′CUAGUUACGCUAGGGAAACCU5′ | ||||
| AGACAAUGCGAUCCCUUUGGA | 5599 (3) | Transport inhibitor response-like protein (TIR) | TF | At1g12820 (9e-072) | |
| AGACAAUGCGAUCCCUUUGGA | 5598 (3) | TIR | TF | At1g12820 (4e-064) | |
| AGACAAUGCGAUCCCUUUGGA | 5597 (3) | TIR | TF | At3g126810 (0.0) | |
| miR394(N,R) | 3′CCUCCACCUGUCUUACGGUU5′ | ||||
| GGAGGUUGACAGAAUGCCAA | 8269 (1) | F-box protein | TF | At1g27340 (4e-079) | |
| GGAGGUUGACAGAAUGCCAA | 8268 (1) | F-box protein | TF | At1g27340 (0.0) | |
| miR395(N) | 3′CUCAAGGGGGUUUGUGAAGUC5′ | No | |||
| ccl-miR396(N) | 3′GUCAAGUUCUUUCGACACCUU5′ | ||||
| AAGUUGAAGAAAGCUGUGGUG | 43399 (4) | Hypothetical protein | Medicago truncatula (5e-15) | ||
| miR397(N) | 3′GUAGUUGCGACGUGAGUUACU5′ | ||||
| UAUCAACGCUGCACUCAAUAA | 6145 (3) | IRX12 copper ion binding/oxidoreductase (IRX12CBO) | Metabolism | At3g09220 (0.0) | |
| CAUCAACGCUGCACUGAAUGA | 22908 (1) | IRX12CBO | Metabolism | At5g01190(1e-110) | |
| GAUCAACGCAGCACUGAAUGA | 23129 (2) | IRX12CBO | Metabolism | At5g60020 (1e-109) | |
| AAUCAAUGCUGCACUCAAUGA | 13453 (2) | IRX12CBO | Metabolism | At2g38080 (0.0) | |
| UGUCAAUGCUGCACUCAAUGA | 42850 (3) | IRX12CBO | Metabolism | At5g03260 (1e-120) | |
| UGUCAACGCUGCAGUCAAUGA | 15664 (3) | IRX12CBO | Metabolism | At2g38080 (0.0) | |
| cis-miR398 (N) | 3′UUCCCCACUGGACUCUUGUGU5′ | No | |||
| miR399 | 3′GUCCCGUUUAGAGGAAACCGU5′ | ||||
| UUGGGCAAAUCUCCUUUGGCA | 40592 (2) | Unknown | |||
| CAGGGCAACUCUCCUUUGGCA | 37301 (1) | Unknown |
N Northern blot of miRNA was done, R, 5′RACE of target mRNA was done
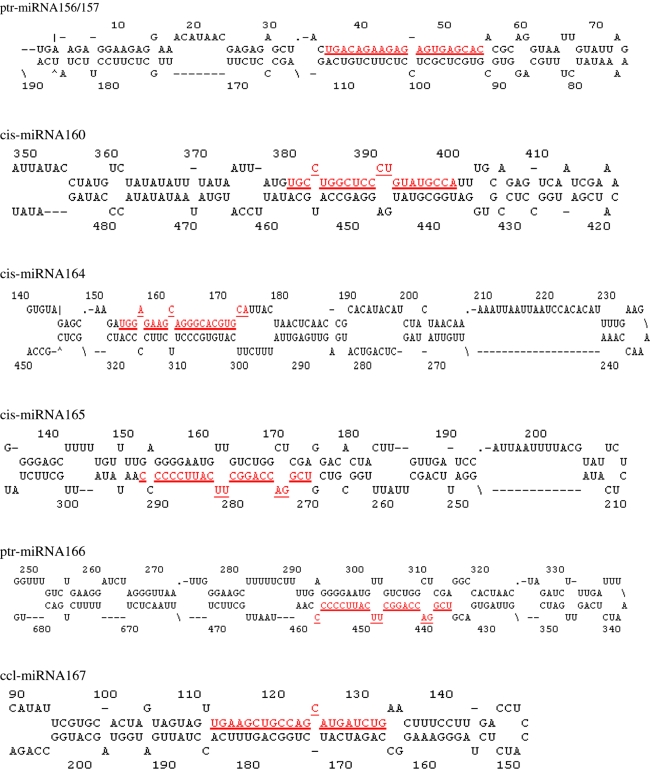
Fig. 1.
Predicted fold-back structures of identified citrus miRNAs. Mature miRNA sequences are shaded. miRNA precursors may be slightly longer than the sequences shown in this figure. Thirteen citrus miRNAs are from Poncirus trifoliate (ptr-miRNAs), Citrus clementine (ccl-miRNAs), and Citrus sinensis (cis-miRNAs), respectively
Table 2.
Expression patterns of miRNAs
| miRNA gene | miRNA sequence (5′→3′) | Length (nt) | Fold-back | Expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Citrusreticulata | Poncirus trifoliate | |||||||||||
| Leaf | Flower | Young shoot | Fruit | Mix | Leaf | Young shoot | Root | Mix | ||||
| ptr-miR156 | UGACAGAAGAGAGUGAGCAC | 20 | Y | se | he | he | se | he | se | se | se | he |
| miR157 | UUGACAGAAGAUAGAGAGCAC | 21 | Y | le | ne | ne | ne | ne | se | se | se | he |
| cis-miR160 | UGCCUGGCUCCCUGUAUGCCA | 21 | Y | me | le | le | le | le | he | he | me | me |
| cis-miR164 | UGGAGAAGCAGGGCACGUGCA | 21 | Y | me | ne | le | ne | le | se | he | he | he |
| cis-miR165 | UCGGACCAGGCUUCAUCCCCC | 21 | Y | he | me | me | he | he | se | se | se | he |
| ptr-miR166 | UCGGACCAGGCUUCAUUCCCC | 21 | Y | se | me | he | he | he | se | se | se | he |
| ccl-miR167 | UGAAGCUGCCAGCAUGAUCUA | 21 | Y | me | le | ne | ne | le | se | me | me | he |
| ccl-miR168 | UCGCUUGGUGCAGGUCGGGAA | 21 | Y | se | he | se | he | he | he | me | me | he |
| cis-miR169 | CAGCCAAGGAUGACUUGCCGA | 21 | Y | le | le | le | le | le | he | he | me | he |
| miR170 | UGAUUGAGCCGUGUCAAUAUC | 21 | Y | me | le | le | le | me | he | he | me | he |
| ccl-miR171 | UGAUUGAGCCGCGCCAAUAUC | 21 | Y | me | me | me | me | he | se | se | he | he |
| cis-miR172 | AGAAUCUUGAUGAUGCUGCAU | 21 | Y | se | me | se | me | he | me | me | me | he |
| ptr-miR319 | UUGGACUGAAGGGAGCUCCC | 20 | Y | he | me | he | he | he | se | se | se | he |
| ccl-miR396 | UUCCACAGCUUUCUUGAACUG | 21 | Y | le | ne | ne | ne | ne | le | ne | ne | ne |
| cis-miR398 | UGUGUUCUCAGGUCACCCCUU | 21 | Y | le | ne | ne | le | ne | le | le | le | le |
Predicted citrus miRNAs from Arabidopsis that showed blotting signals
Strength of expression: he high expression, le low expression, me moderate expression, ne no expression, se strong expression, N no, Y yes
Preparation of RNA and gel-blot analysis
Total RNA was isolated from different tissues using Trizol (Invitrogen, Life Technologies, Carlsbad, CA, USA), and low molecular weight RNA (LMW-RNA) was enriched with 4 M LiCl. Fifteen micrograms of each LMW-RNA sample was loaded per lane and resolved on a denaturing 15% polyacrylamide gel and electro-blotted onto Hybond N+ membranes (Amersham, Piscataway, NJ, USA) using a TransBlot-SD apparatus (Bio-Rad, Hercules, CA, USA). Membranes were UV cross-linked at 1,200 μJ × 100 in a Stratalinker 1800 Stratagene (Stratagene, La Jolla, CA, USA). Citrus miRNA oligonucleotide probes, including all the 27 miRNAs studied, were synthesized as sequences antisense to the mature miRNAs (Table 3) and were labeled at the 5′end with γ-32P-ATP using T4 polynucleotide kinase (Biolabs, Beverly, MA, USA). Membranes were pre-hybridized for 1 h and hybridized about 16 h using Perfect Hyb™ Plus buffer (Sigma, St Louis, MO, USA) at 38°C. Membranes were washed four times (two times with 1 × SSC and 0.1% SDS for 20 min and two times with 0.5 × SSC and 0.1% SDS for 50 min at 50°C). The washed membranes were air dried for a few minutes and then exposed to BIOMAX X-ray film using an intensifying screen for 48 h.
Table 3.
DNA oligonucleotides complementary to the predicted citrus microRNAs sequences
| Citrus miRNA probes | Sequence(5′ to 3′) | Size | Td |
|---|---|---|---|
| ptr-miR156 | GTGCTCACTCTCTTCTGTCA | 20 | 60 |
| miR157 | GTGCTCACTCTCTTCTGTCAA | 21 | 62 |
| miR159 | TAGAGCTCCCTTCAATCCAAA | 21 | 60 |
| cis-miR160 | TGGCATACAGGGAGCCAGGCA | 21 | 68 |
| miR161 | CCCCGATGTAGTCACTTTCAA | 21 | 66 |
| miR162 | CTGGATGCAGAGGTTTATCGA | 21 | 62 |
| miR163 | ATCGAAGTTCCAAGTCCTCTTCAA | 24 | 68 |
| cis-miR164 | TGCACGTGCCCTGCTTCTCCA | 20 | 64 |
| cis-miR165 | GGGGGATGAAGCCTGGTCCGA | 21 | 70 |
| ptr-miR166 | GGGGAATGAAGCCTGGTCCGA | 21 | 68 |
| ccl-miR167 | TCAGATCATGCTGGCAGCTTCA | 22 | 68 |
| ccl-miR168 | TTCCCGACCTGCACCAAGCGA | 21 | 68 |
| cis-miR169 | TCGGCAAGTCATTCTTGGCT | 20 | 60 |
| miR170 | GATATTGACACGGCTCAATCA | 21 | 60 |
| ccl-miR171 | GATATTGGCGCGGCTCAATCA | 21 | 64 |
| csi-miR172 | ATGCAGCATCATCAAGATTCT | 21 | 68 |
| miR173 | GTGATTTCTCTCTGCAAGCGAA | 22 | 64 |
| ptr-miR319 | GGGAGCTCCCTTCAGTCCAA | 20 | 68 |
| miR390 | GGCGCTATCCCTCCTGAGCTT | 21 | 68 |
| miR391 | TGGCGCTATCTCTCCTGCGAA | 21 | 66 |
| miR393 | GATCAATGCGATCCCTTTGGA | 21 | 62 |
| miR394 | GGAGGTGGACAGAATGCCAA | 20 | 62 |
| miR395 | GAGTTCCCCCAAACACTTCAG | 21 | 64 |
| ccl-miR396 | CAGTTCAAGAAAGCTGTGGAA | 21 | 60 |
| miR397 | CATCAACGCTGCACTCAATGA | 21 | 62 |
| cis-miR398 | AAGGGGTGACCTGAGAACACA | 21 | 64 |
Td dissociation temperature [Td (°C) = 4(G + C) + 2(A + T)]
Analysis of 5′RACE
For mapping the internal cleavage site in Unigene UC46-13966 (targeted by cis-miR160), UC46-16450 (targeted by ccl-miR167), UC46-5597 (targeted by miR393), and UC46-8268 (targeted by miR394) mRNA, RLM-RACE was performed using the GeneRacer Kit (Invitrogen). A modified procedure for RLM-RACE was carried out following the instruction of GeneRacer Kit (Invitrogen) as described previously (Llave et al. 2002b). Total RNA was extracted from leaf, stem, root, flower, and fruits of adult trifoliate orange using Trizol. Poly(A)+ mRNA was purified from the pooled of all tissues RNA using the PolyA kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The GeneRacer RNA Oligo adapter was directly ligated to mRNA (250 ng) without calf intestinal phosphatase and tobacco acid pyrophosphatase treatment. GeneRacer OligodT primer was then used to synthesize first strand cDNA in a reverse transcription reaction. This cDNA was subjected to an amplification procedure with the GeneRacer 5′Primer and the GeneRacer 3′Primer to generate a pool of non-gene-specific 5′RACE product. The conditions used for this amplification step were the same as those for gene-specific RACE recommended by the manufacturer, with the exception that an extension time of 3 min was used. Gene-specific 5′RACE reactions were performed with the GeneRacer 5′Nested Primer and gene-specific primers as follows: Pt-ARF10 (UC46-13966)-1094R (5′CTGCTCTGTGAGTATTGGTTGACCAAAGAGT3′), Pt-ARF8 (UC46-16450)-1376R (5′ATGACGGTCACTTACTCCCATGGGTCGT3′), Pt-AFB2 (UC46-5597)-2273R (5′TTAGAGAGTCCACACAAAATCTGGCGCAT3′), and Pt-F-box (UC46-8268)-1365R (5′TTAAGCTGTTGCCGTGAGGCATGGGT3′). In each case, a unique gene-specific DNA fragment was amplified. After amplification, 5′RACE products were gel-purified and cloned, and at least 15 independent clones were randomly chosen and sequenced.
Results
Identification of potential citrus miRNAs
BLASTN searches revealed that 13 sequences of miRNA156, miRNA160, miRNA164, cis-miRNA165, miRNA166, miRNA167, miRNA168, miRNA169, miRNA171, miRNA172, miRNA319, miRNA396, miRNA398 (Table 1) have at least one match in the citrus EST database. All blasted hits were non-coding sequences without annotation determined by BlastX searches against the NCBI database. All the blast hits were complementary to miRNAs (plus/plus) either with 0–1 mismatches or more than four mismatches, which could be due to the high-level conservation of miRNAs between Arabidopsis and Citrus. Based on the supposition that some miRNAs in both Citrus and Arabidopsis might differ by one base pair, we only considered and analyzed the first group to keep a higher threshold. We selected valid miRNA stem-loop structures based on the three general rules and five parameters reported by Bonnet et al. (2004). Thirteen of the 14 blast hits could fold into stem-loop structures (Fig. 1). The phenomenon that the blast hit of miRNA159 could not form this kind of structure might be due to the fact that the miRNA region was located at the 5′end of the sequence. The miRNA166 had different candidate precursors in Washington Navel orange and the hybrid of Citrus paradise × Poncirus trifoliate. Interestingly, their stem and complementary regions were the same, but the flanking sequences were different. From the stem-loop structures, only mature miRNA167 and miRNA169 in citrus were different at, respectively, the nucleotide at the 3′end (G) and 8th nucleotide from 5′ (A) compared to those in Arabidopsis (A and C). Overall, all the stem-loops are analogous to those of Arabidopsis.
In this study, such a frequency of candidate precursors matching sequences in citrus ESTs is reasonable, considering the limitation of the EST quantity, sequencing errors, conservation level between miRNAs of Arabidopsis and Citrus, and some other possibilities.
Detection and expression patterns of miRNAs
Preferential expression of a miRNA in specific tissues might provide clues about its physiological function. To assist with the determination of the citrus miRNA functions, we examined their expression in different organs (Fig. 2) by RNA gel blot analysis on LMW-RNA samples from various tissues of ‘Nules’ and trifoliate orange trees. These data can also be powerful evidence supporting the existence of these miRNAs in citrus. Overall, all the 25 conserved miRNAs and non-conserved miRNA173 were present in the RNA fractions with the expected size for the synthesized miRNAs, but the non-conserved miRNA158 could not be detected in any tissue analyzed, suggesting miRNA158 is probably not present in citrus similarly to other plant species. The expression of the 13 miRNAs with perfect predicted stem-loop structures could confirm them as the miRNAs in citrus, and they were designated as ptr-miRNA156, cis-miRNA160, cis-miRNA164, cis-miRNA165, ptr-miRNA166, ccl-miRNA167, ccl-miRNA168, cis-miRNA169, ccl-miRNA171, cis-miRNA172, ptr-miRNA319, ccl-miRNA396, cis-miRNA398, respectively, based on Poncirus trifoliate, Citrus clementine, and Citrus sinensis, three citrus species from which the miRNA precursor were sequenced (Fig. 1; Table 1), and their sequences were determined according to the corresponding stem-loops. As to other 13 Arabidopsis miRNAs expressed in citrus could be thought as potential ones in citrus and their expression information could suggest their existence in citrus. All the blotting results could also present us some information of their roles in citrus development by their preferential expression patterns.
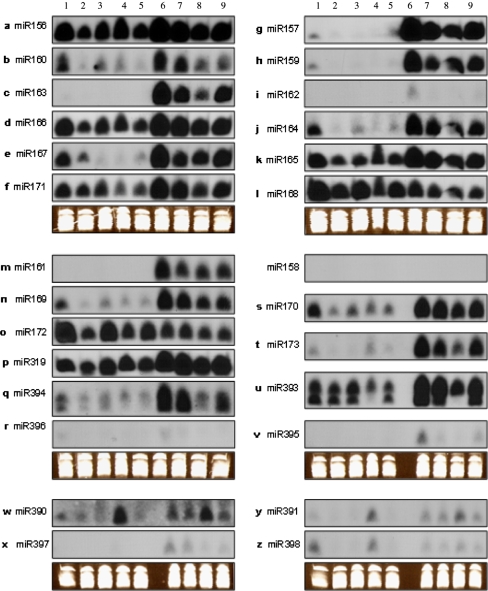
Fig. 2.
Expression patterns of miRNA predicted from citrus. RNA gel blots of total RNA isolated from different tissues were probed with labeled oligonucleotides. Lanes 1, 2, 3, 4, 5 are samples of leaf, flower, shoot, fruit, and the mix of these four organs of ‘Nules’ mandarin; 6, 7, 8, and 9 are leaf, shoot, root, and the mix of these tree organs of trifoliate orange seedling. The tRNA and 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls
The expression patterns of most miRNAs in citrus appear to be tissue or developmental-stage specific, and all the expression patterns of these miRNAs in citrus can be categorized into several situations. The expression patterns of ptr-miR156, cis-miR165, ptr-miR166, ccl-miR171, and ptr-miR319 are similar: strong expression in leaves, stems, and roots of trifoliate orange and high expression in leaves, flowers, young shoots, and fruits of ‘Nules’ (Fig. 2a, d, f, k p; Table 2). All these showed some minor organ specificity both in ‘Nules’ and trifoliate orange. However, the expression in trifoliate orange seemed stronger than in ‘Nules’. This might be species-specific or be preferentially expressed during a juvenile stage. Cis-miR160, cis-miR164, and cis-miR169 also displayed similar expression patterns with some difference of expression intensity in various organs: high expression in leaves, stems and roots of trifoliate orange and moderate expression in leaves of ‘Nules’, much weaker expression in other organs of ‘Nules’ (Fig. 2b, j, n; Table 2). The expression difference between ‘Nules’ and trifoliate orange seem to be species- and/ or growth stage-specific, which is also a very obvious phenomenon in the following group showing the similar expression. miR157, miR159, ccl-miR167, and miR173 seemed to be strongly expressed in leaves and moderately in stems and roots of trifoliate orange, with much weaker expression in all tissues of ‘Nules’ (Fig. 2e, g, h, t; Tables 2, 4). The miR390 was expressed in all tissues tested but at higher levels in the fruits of ‘Nules’ and young stems of trifoliate orange (Fig. 2w, Table 4). Ccl-miR168 and cis-miR172 expression were high in all tested tissues of ‘Nules’ and moderate in all organs of trifoliate orange (Fig. 2l, o; Table 2). Some of the miRNAs might display species-specific and/or developmental stage-specific expression patterns, and the best examples of it were observed for miR161 and miR163 (Fig. 2c, m; Table 4). miR395 and miR397 had preferential expression in leaves and stems of trifoliate orange (Fig. 2v, x; Table 4) in quite low intensity; miR162 had preferential expression only in trifoliate orange leaves (Fig. 2i; Table 4); miR396 had preferential expression in leaves of both trifoliate orange and ‘Nules’ (Fig. 2r; Table 2), even though the signals are very faint. The miR391 and cis-miR398 showed lower and ubiquitous expression in all tissues of trifoliate orange and lower expression in the leaves and fruits of ‘Nules’ (Fig. 2y, z; Tables 2, 4). Unexpectedly and interestingly, miR393 and miR394 had two bands that appeared to differ in length by one nucleotide in leaves, shoots, and flowers, with the larger band only in ‘Nules’ fruit and trifoliate orange roots (Fig. 2q, u; Table 4). This might imply that there were two copies that are one nucleotide different in length in citrus or that only the longer copy can be generated in fruit of ‘Nules’ and root of trifoliate orange.
Table 4.
Expression patterns of miRNAs without the fold-back predicted citrus miRNA from Arabidopsis
| miRNA gene | miRNA sequence (5′→3′) | Length (nt) | Fold-back | Expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Citrusreticulata | Poncirus trifoliate | |||||||||||
| Leaf | Flower | Young shoot | Fruit | Mix | Leaf | Young shoot | Root | Mix | ||||
| miR158 | UCCCAAAUGUAGACAAAGCA | 20 | N | ne | ne | ne | ne | ne | ne | ne | ne | ne |
| miR159 | UUUGGAUUGAAGGGAGCUCUA | 21 | N | le | ne | ne | ne | ne | se | he | se | he |
| miR161 | UUGAAAGUGACUACAUCGGGG | 21 | N | ne | ne | ne | ne | ne | he | me | me | me |
| miR162 | UCGAUAAACCUCUGCAUCCAG | 21 | N | ne | ne | ne | ne | ne | le | ne | ne | le |
| miR163 | UUGAAGAGGACUUGGAACUUCGAU | 24 | N | ne | ne | ne | ne | ne | se | he | me | he |
| miR173 | UUCGCUUGCAGAGAGAAAUCAC | 22 | N | le | ne | ne | le | me | he | me | le | he |
| miR390 | AAGCUCAGGAGGGAUAGCGCC | 21 | N | le | le | le | he | le | me | me | he | me |
| miR391 | UUCGCAGGAGAGAUAGCGCCA | 21 | N | le | ne | ne | le | le | le | le | le | me |
| miR393 | UCCAAAGGGAUCGCAUUGAUC | 21 | N | he | he | he | me | me | se | se | he | he |
| miR394 | UUGGCAUUCUGUCCACCUCC | 20 | N | me | le | le | le | me | se | se | le | he |
| miR395 | CUGAAGUGUUUGGGGGAACUC | 21 | N | ne | ne | ne | ne | ne | le | ne | ne | le |
| miR397 | UCAUUGAGUGCAGCGUUGAUG | 21 | N | ne | ne | ne | ne | ne | le | le | ne | le |
Strength of expression: he high expression, le low expression, me moderate expression, ne no expression, se strong expression, N no, Y yes
In summary, the RNA gel blot analysis confirmed the expression and sizes of 13 identified miRNAs in citrus and other 14 miRNAs except miR158 from Arabidopsis (Fig. 2). Some of them are expressed ubiquitously in all tissues with the characteristics of tissue-, species-, and/or growth-stage-specific reflected from different expression levels.
Prediction of the citrus miRNA target genes
Based on the homology between miRNAs and their target genes in plants, the citrus EST database was searched for homology to the selected miRNA sequences using a BLASTN algorithm employed for the discovery of citrus miRNA targets. Most antisense hits with three or fewer mismatches appeared to be potential miRNA targets, based on the high similarity with the corresponding orthologs in Arabidopsis (Rhoades et al. 2002). All searches of targets in citrus were performed using Arabidopsis as a reference organism for function conservation, and the putative candidate targets of the citrus miRNAs and the miRNAs might existed in citrus are listed in Table 1. We identified a total of 41 potential targets for 15 citrus miRNAs based on the fact that miRNAs perfectly or near-perfectly complement their target sequences. These 41 potential miRNA targets belong to a number of gene families that have different biological functions. The number of predicted targets per miRNA varied much, from one to seven. Only five (cis-miR160, ptr-miR166, ccl-miR168, miR390, and miR395) have one predicted target. All targets share high similarities, with a low E-value (Table 1), to their orthologs in Arabidopsis. In all cases when a miRNA was complementary to more than one mRNA, most of the potential targets were members of the same gene family (Table 1). Similar to the essential roles of miRNAs in regulating a variety of biological processes in plant reported (Chen 2005), it appeared that our predicted targets played similar roles not only in development but also in quite diverse physiological processes (Table 1). A majority of these targets were transcription factors that control plant development and phase change from vegetative growth to reproductive growth. Several studies indicate that miRNA156/157 targets squamosa promoter binding protein (SBP)-like (SPL) genes (Gandikota et al. 2007; Moxon et al. 2008), a plant-specific family of transcription factors involved in early flower development and vegetative phase changes. In this study, we found that seven citrus Unigenes were near-perfect matches of ptr-miRNA156/miRNA157 (Table 1), among which only two of them, Unigenes UC46-12489 and UC46-8859 were both simultaneously targeted by ptr-miRNA156/ miRNA157 (Table 1). Other five were targeted only by ptr-miRNA156 and they might have multiple unknown functions or be of the same gene. Another experimentally confirmed miRNA-targeted transcription factor is APETALA2 (AP2), which controls floral development and phase transition in Arabidopsis (Chen 2004). In this research, seven predicted targets of miR172 in citrus were found to be members of the AP2 gene family (Table 1). In addition to SPL and AP2, we also found that many other transcription factors are potential targets of citrus miRNAs. Class-III homeodomain leucine zipper proteins, NAC domain proteins, F-box protein and Transport inhibitor response-like protein have perfect or near-perfect complementary sites with certain identified citrus miRNAs (Table 1). UC46-26767 and UC46-21315 are potential targets of citrus cis-miRNA164; UC46-19387 and UC46-5511 are those of cis-miRNA165; UC46-8268 and UC46-8269 may be targeted by miRNA394; UC46-21833, UC46-16450, UC46-8197, UC46-7960 and UC46-7959 are all targeted by ccl-miR167; and UC46-5599, UC46-5598 and UC46-5597 are those targeted by miR393 (Table 1).
Auxin response factors (ARF), a plant-specific family of DNA binding proteins, are involved in hormone signal transduction (Hagen and Guilfoyle 2002). Currently, a large number of ARFs have been identified in A. thaliana (Okushima et al. 2005), of these, at least five have complementary sites with miRNAs. In this study, we found that two homologs of Arabidopsis ARF (ARF 8 and ARF 10) have one perfect or near-perfect complementary site with cis-miRNA160 and ccl-miRNA167, respectively.
In addition to some transcription factors, we predicted target genes that directly control anthocyanidin synthesis and a copper ion synthesis pathway. We found that UC388432, targeted by cis-miRNA169, might encode anthocyanidin synthase, and six Unigenes (UC6145, UC22908, UC23129, UC13453, UC42850, and UC15664), targeted by miRNA 397, encoded copper ion oxidoreductase (Table 1).
Interestingly, citrus unigene C46-40592, the potential target of miR399, was found to have two same complementary sites. One target site corresponding to the positions 574 to 594 is complementary to the miRNA with two mismatches and the other target site corresponding to the positions 651 to 671 is complementary to the miRNA with two mismatches, too. The two target sites are in frame and separated by a region of 55 nucleotides (Fig. 3).
Fig. 3.
Citrus unigene C46-40592, the predicted target of miRNA 399, was found to have two complementary sites
As to the situation that we were unable to predict targets for other 13 citrus miRNAs (ccl-miR168, ptr-miR319, cis-miR398, miR158, miR159, miR161, miR162, miR163, miR170, miR171, miR173, miR391, and miR395) by applying these criteria, this could be mainly due to the limitation of the citrus ESTs available. Non-target predicted for miR158 in citrus could also support the opinion that this miRNA might be truly absent in citrus.
In summary, predicted citrus targets might play a vital role in a variety of growth and developmental processes. The 41 identified citrus targets were not only transcription factors that control plant development and phase transition, but they were also involved in anthocyanidin synthesis and a copper ion synthesis pathway. Some of the newly identified miRNA targets, such as UC388432, may be unique to citrus.
Identification of miRNA-guided cleavage of target mRNAs in citrus
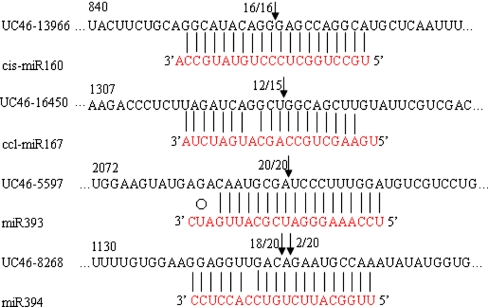
Most of the Arabidopsis miRNAs have been shown to guide cleavage of their target genes (Llave et al. 2002b; Jones-Rhoades and Bartel 2004; Sunkar et al. 2005). To verify the nature of the predicted miRNA targets and to study how the miRNAs in citrus regulate their target genes, a modified RLM-RACE experiment was set up, as described in the Sect. ’Materials and methods’. This is one of the most common and widely used methods in the literatures (Kasschau et al. 2003; Lauter et al. 2005) to support bioinformatics data. Target mRNA fragments resulting from miRNA-guided cleavage have a 5′ phosphate group, and cleavage usually occurs near the middle of the base-pairing interaction region between the miRNA and mRNA molecules. In this study, the RLM-RACE procedure was successfully used to map the cleavage sites in four predicted target genes in citrus. Given the clear tissue-specific pattern of expression of cis-miR160, ccl-miR167, miR393, and miR394 in all kinds of vegetative organs of trifoliate orange (Fig. 2b, e, u, q; Table 2), these analyses were performed on a few of their putative targets, using RNA extracted from leaves, stems, and roots of trifoliate orange, where cis-miR160, ccl-miR167, miR393 and miR394 were all abundantly expressed. UC46-13966, UC46-16450, UC46-5597, and UC46-8268 were confirmed as real targets of cis-miR160, ccl-miR167, miR393 and miR394, respectively, since all the 5′ends of the mRNA fragments were mapped to the nucleotide that pairs to the tenth nucleotide of each miRNA with higher frequencies than depicted for each pairing oligo (Fig. 4). All the four predicted targets were found to have specific cleavage sites corresponding to the miRNA complementary sequences (Fig. 4) and might be regulated by the four mRNAs in the style of small interfering RNAs (siRNAs; Elbashir et al. 2001) directing the cleavage of mRNA targets with extensive complementarity to the miRNAs (Kasschau et al. 2003). UC46-13966 and UC46-16450 are similar to Arabidopsis proteins coded by AUXIN RESPONSE FACTOR 10 (ARF10) and AUXIN RESPONSE FACTOR 8 (ARF8), a miRNA binding/transcription factor, respectively (Table 1). Similarly, UC46-5597 and UC46-8268 both coded for a protein highly homologous to a transport inhibitor response-like protein transcription factor and an F-box family protein transcription factor, respectively (Table 1).
Fig. 4.
Mapping of mRNA cleavage sites by RNA ligase-mediated 5′RACE. Each top strand (black) depicts a miRNA-complementary site in the target mRNA, and each bottom strand depicts the miRNA (red). Watson–Crick pairing (vertical dashes) and G:U wobble pairing (circles) are indicated. Arrows indicate the 5′ termini of mRNA fragments isolated from Citrus, as identified by cloned 5′RACE products, with the frequency of clones shown. Only cloned sequences that matched the correct gene and had 5′ ends within a 100-nt window centered on the miRNA complementary site are counted. Partial mRNA sequences from target genes were aligned with miRNAs. Numbers indicate the fraction of cloned PCR products terminating at different positions. UC46-13966 was similar to AT2G28350 (NP_180402) ARF10 (AUXIN RESPONSE FACTOR 10); UC46-16450 was similar to AT5G37020 (NP_198518) ARF8 (AUXIN RESPONSE FACTOR 8); UC46-16450 was similar to AT3G26810 (NP_566800) AFB2 (AUXIN SIGNALING F-BOX 2); UC46-8268 was similar to AT1G27340 (NP_564278) F-box family protein
Discussion
Although miRNAs have been extensively studied in the past several years, no systematic study has been performed on citrus, one of the most important fruits crops in the world. It was challenging to establish some degree of correlation between miRNAs from Citrus and Arabidopsis. Even though many miRNA genes might have evolved by duplication and/or translocation events or have been generated ex novo from target duplication (Allen et al. 2004), many plant mature miRNAs are conserved among plant species. Recently, Sunkar and Jagadeeswaran (2008) identified several miRNAs from citrus EST by bioinformatics analysis, but citrus miRNAs still remains largely unknown and the identified citrus miRNAs have not been confirmed by experiments. To elucidate the function and the regulation pathway of miRNAs in citrus, it is important to know the localization of the miRNA. Here, Northern blotting was employed to study the expression of the miRNAs in citrus. The RNA gel blot analysis confirmed the expression and sizes of 13 identified miRNAs and other 13 potential miRNAs in citrus except miR158 from Arabidopsis. The expression patterns were similar with those reported in Arabidopsis, such as the majority of them were expressed ubiquitously in all tissues, and some were expressed with the characteristics of tissue-, species-, and/or growth-stage-specific. The selection of putative miRNA gene transcripts with 0–1 mismatches to miRNAs of Arabidopsis was based on the hypothesis that some citrus miRNAs might be one nucleotide different from the same family in Arabidopsis. Our RNA blot analysis can validate the miRNA prediction in citrus, and their preferential expression can provide important clues about where these miRNAs function. Interestingly, miRNA173, a non-conserved one reported, was strongly expressed in Poncirus trifoliate. Most sequencing data for fruit crops are ESTs, so trying miRNA identification by searches against the EST database is a good start for more similar work to be carried out in other fruit crops. Our future work will be focused on the demonstration of the role of these citrus miRNAs in the control of citrus development.
Secondly, to assess and define a putative function for a miRNA in plant, a further step of target identification is necessary. Currently, the most efficient tool available for this is the bioinformatics approach facilitated by the high degree of homology between miRNA and its target sequences in plants (Rhoades et al. 2002). Analysis of several targets has now confirmed this prediction, making it feasible to identify plant miRNA targets (Llave et al. 2002a; Park et al. 2002; Reinhart et al. 2002). We first searched the candidate targets of the citrus miRNAs and the putative miRNAs by Blast, and confirmed them by their alignment with their orthologs in Arabidopsis. Our analysis reveals that most of the predicted targets in citrus have a conserved function with miRNA targets in Arabidopsis and these miRNA target sequences are also highly conserved among a wide variety of plant species as reported by Floyd and Bowman (2004). Consistent with previous reports, most of these targets in citrus were plant-specific transcription factors, such as AP2, NAC, SBP, and the ARF family. Only the putative targets of miR399 in citrus have distinct functions from Arabidopsis. Our computational identification of citrus miRNA targets is another strategy to predict the miRNAs, in which the identity level and the annotated function of the predicted targets with the orthologs of Arabidopsis can also be powerful reference to support the prediction of target genes. The numbers of putative targets of 15 citrus miRNAs ranged from 1 to 7, in which some targets of same miRNA might be of the same gene, and this needs to be verified by laboratory experiment. As to the fact that only 15 citrus miRNAs had sequences that exactly matched (in either orientation) one or more ESTs in the publicly accessible HarvEST (UC Riverside, USA), this might reflect a lack of coverage of ESTs in this database, or the fact that EST sequencing strategies favor long, stable, poly(A)+ transcription and processing pathways (Smalheiser 2003; Lee et al. 2002).
Thirdly, a growing number of plant miRNA targets predicted through bioinformatics have been experimentally confirmed. Even though miRNAs generally function as negative regulators of gene expression by mediating the cleavage of target mRNAs (Llave et al. 2002b) or by repressing their translation (Chen 2004), the cleavage of target mRNAs appears to be the predominant mode of gene regulation by plant miRNAs (Sunkar et al. 2005). Finding the cleavage site supposed to be located in the sequence complementary to miRNA in target gene has been one necessary work to verify the fact of the cleavage of target mRNAs. Of the methods used to observe miRNA-dependent cleavage of targets, RLM-RACE is the most useful one (Llave et al. 2002b). We tried performing the RACE on unigene to detect and clone the mRNA fragment corresponding precisely to the predicted product of miRNA processing. Just for a possible comparison between our results and those of others reported in rice and Arabidopsis, the targets of miR393 and miR394 together with cis-miR160 and ccl-miR167 were chosen to try this method of mapping the cleavage sites, and the results were the same as predicted in observing the location of cleavage sites. These results can also be thought as the criteria used to confirm the putative targets. Totally, we performed the 5′RACE assays on four predicted target genes: the representative targets of four conserved miRNAs (Unigene C46-50592, UC46-16450, UC46-5599, UC46-8269 targeted by miR160, miR167, miR393 and miR394, respectively). Unigenes C46-50592 and UC46-16450 are both ARFs that bind to auxin in response elements in promoters of early auxin response genes (Guilfoyle and Hagen 2001). Intriguingly, both cis-miR160 and ccl-miR167 regulate ARFs, but they have different complementary sites and unrelated sequences. In A. thaliana, miR167 was found to cause transcript degradation of ARF8, but not ARF6 (Ru et al. 2006), this was explained by the fact that the fewer base-pairs between ARF6 and miR167 may result in inefficient cleavage of ARF6 transcripts. All four predicted targets were found to have specific cleavage sites corresponding to the miRNA complementary sequences (Fig. 4), and the most common 5′ end of the mRNA fragments mapped to the nucleotides that pair with the 10th miRNA nucleotide from the 5′ ends. The validation was obtained by performing the modified 5′RACE protocol on mRNA extracted from pooled tissues of leaf, stem, and root, where it was previously demonstrated that cis-miR160, ccl-miR167, miR393, and miR394 are all abundant (Fig. 2b, e, u, q; Tables 2, 4). The miRNAs may directly target transcription factors that affect plant development and also specific genes that control metabolism. In our study, it appears that our predicted targets play roles not only in development, but also in diverse physiological processes.
Acknowledgments
This work was supported by the Science & Technology Key Project of China Ministry of Education (grant no. 109084) and the Program of NCET (grant no. NCET-08-0796).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AP2
APEATALA2
- ARF
Auxin response factor
- EST
Expressed sequence tag
- HD-ZIP
Homeodomain leucine zipper
- h
Hour(s)
- miRNA
MicroRNA
- RLM-RACE
RNA ligase-mediated 5′ rapid amplification of cDNA ends
- 5′RACE
5′Rapid amplification of cDNA ends
- RISC
RNA-induced silencing complex
- SBP
Squamosa promoter binding protein
- SPL
Squamosa promoter binding protein like
References
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290 [DOI] [PubMed]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshell M, Matzke M, Ruvkun G, Tusch T (2003) A uniform system for microRNA annotation. RNA 9:277–279 [DOI] [PMC free article] [PubMed]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741 [DOI] [PMC free article] [PubMed]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933 [DOI] [PMC free article] [PubMed]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y (2004) Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA 101:11511–11516 [DOI] [PMC free article] [PubMed]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190 [DOI] [PubMed]
- Chen X (2004) A miRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025 [DOI] [PMC free article] [PubMed]
- Chen X (2005) microRNA biogenesis and function in plants. FEBS Lett 579:5923–5931 [DOI] [PMC free article] [PubMed]
- Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev 16:1616–1626 [DOI] [PMC free article] [PubMed]
- Floyd SK, Bowman JL (2004) Gene regulation: ancient microRNA target sequences in plants. Nature (London) 435:441–445 [DOI] [PubMed]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49:683–693 [DOI] [PubMed]
- Gleave AP, Ampomah-Dwamena C, Berthold S, Dejnoprat S, Karunairetnam S, Nain B, Wang Y-Y, Crowhurst RN, MacDiarmid RM (2008) Identification and characterisation of primary microRNAs from apple (Malus domestica cv. Royal Gala) expressed sequence tags. Tree Genet Genomes 4:343–358 [DOI]
- Grad Y, Aach J, Hayes GD, Reinhart BJ, Church GM, Ruvkun G, Kim J (2003) Computational and experimental identification of C. elegans microRNAs. Mol Cell 11:1253–1263 [DOI] [PubMed]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144 [DOI] [PMC free article] [PubMed]
- Guilfoyle TJ, Hagen G (2001) Auxin response factors. J Plant Growth Regul 20:281–291 [DOI]
- Guo Q, Xiang AL, Yang Q, Yang ZM (2007) Bioinformatic identification of microRNAs and their target genes from Solanum tuberosum expressed sequence tags. Chin Sci Bull 52:2380–2389 [DOI]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385 [DOI] [PubMed]
- Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229:1009–1014 [DOI] [PubMed]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428:84–88 [DOI] [PubMed]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4(2):205–217 [DOI] [PubMed]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP (2005) MicroRNA 172 down-regulate glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 102:9412–9417 [DOI] [PMC free article] [PubMed]
- Lee Y, Jeon K, Lee JT, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21:4663–4670 [DOI] [PMC free article] [PubMed]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Sone J, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute 2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441 [DOI] [PubMed]
- Llave C, Kasschau KD, Rector M, Carrington JC (2002a) Endogeneous and silencing-associated small RNAs in plants. Plant Cell 14:1605–1619 [DOI] [PMC free article] [PubMed]
- Llave C, Xie Z, Kasschau KD, Carrington JC (2002b) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056 [DOI] [PubMed]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203 [DOI] [PMC free article] [PubMed]
- Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative and floral organs. Curr Biol 14:1035–1046 [DOI] [PubMed]
- Mica E, Gianfranceschi L, Enrico Pè M (2006) Characterization of five microRNA families in maize. J Exp Bot 57:2601–2612 [DOI] [PubMed]
- Millar AA, Gubler F (2005) The ArabidopsisGAMYB-Like Genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17:705–721 [DOI] [PMC free article] [PubMed]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609 [DOI] [PMC free article] [PubMed]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A (2005) Functional genomic analysis of the auxin response factor gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463 [DOI] [PMC free article] [PubMed]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263 [DOI] [PubMed]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a dicer homolog, and HEN I, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495 [DOI] [PMC free article] [PubMed]
- Phillips JR, Dalmay T, Bartels D (2007) The role of small RNAs in abiotic stress. FEBS Lett 581:3592–3597 [DOI] [PubMed]
- Pilcher RL, Moxon S, Pakseresht N, Moulton V, Manning K, Seymour G, Dalmay T (2007) Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226:709–717 [DOI] [PubMed]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) Micro-RNAs in plants. Genes Dev 16:1616–1626 [DOI] [PMC free article] [PubMed]
- Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799 [DOI] [PubMed]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520 [DOI] [PubMed]
- Ru P, Xu L, Ma H, Huang H (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res 16:457–465 [DOI] [PubMed]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130:6001–6012 [DOI] [PubMed]
- Shukla LI, Chinnusamy V, Sunkar R (2008) The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta 1779:743–748 [DOI] [PubMed]
- Smalheiser NR (2003) EST analysis predicts the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol 4:403 [DOI] [PMC free article] [PubMed]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Oliver Y (2009) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9:160 [DOI] [PMC free article] [PubMed]
- Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37 [DOI] [PMC free article] [PubMed]
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019 [DOI] [PMC free article] [PubMed]
- Sunkar R, Zhu JK (2007) MicroRNAs and short-interfering RNAs in plants. J Integr Plant Biol 49:817–826 [DOI]
- Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNAs from rice. Plant Cell 17:1397–1411 [DOI] [PMC free article] [PubMed]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9:593 [DOI] [PMC free article] [PubMed]
- Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581:1464–1474 [DOI] [PubMed]
- Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18:1187–1197 [DOI] [PMC free article] [PubMed]
- Wang J-F, Zhou H, Chen Y-Q, Luo Q-J, Qu L-H (2004a) Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res 32:1688–1695 [DOI] [PMC free article] [PubMed]
- Wang X-J, Reyes JJL, Chua N-H, Gaasterland T (2004b) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5:R65 [DOI] [PMC free article] [PubMed]
- Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA (2005) Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15:336–360 [DOI] [PubMed]
- Zhang BH, Pan XP, Anderson TA (2006) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762 [DOI] [PubMed]
- Zhang BH, Wang QL, Wang KB, Pan XP, Liu F, Guo TL, Cobb GP, Anderson TA (2007) Identification of cotton microRNAs and their targets. Gene 397:26–37 [DOI] [PubMed]
- Zhang BH, Pan XP, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182 [DOI] [PubMed]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 [DOI] [PMC free article] [PubMed]