Abstract
The CTCF transcription factor is an 11 zinc fingers multifunctional protein that uses different zinc finger combinations to recognize and bind different sites within DNA. CTCF is thought to participate in various gene regulatory networks including transcription activation and repression, formation of independently functioning chromatin domains and regulation of imprinting. Sequencing of human and other genomes opened up a possibility to ascertain the genomic distribution of CTCF binding sites and to identify CTCF-dependent cis-regulatory elements, including insulators. In the review, we summarized recent data on genomic distribution of CTCF binding sites in the human and other genomes within a framework of the loop domain hypothesis of large-scale regulation of the genome activity. We also tried to formulate possible lines of studies on a variety of CTCF functions which probably depend on its ability to specifically bind DNA, interact with other proteins and form di- and multimers. These three fundamental properties allow CTCF to serve as a transcription factor, an insulator and a constitutive dispersed genome-wide demarcation tool able to recruit various factors that emerge in response to diverse external and internal signals, and thus to exert its signal-specific function(s).
Key Words: CTCF, transcription factor, insulator, loop domains, gene regulation.
INTRODUCTION
CTCF (also known as CCCTC-binding factor) [1] is a ubiquitously expressed vertebrate nuclear protein with numerous functions. CTCF was first detected as a protein specifically recognizing three regularly spaced repeats of the CCCTC sequence located ~200 bp upstream of the chicken c-myc gene transcription start site [2, 3] and binding to a chicken lysozyme silencer [4, 5]. CTCF contains three domains, one of which is a DNA-binding domain with 11 zinc fingers.
CTCF is evolutionally conserved "from Drosophila to humans" [6-8], and nearly 83-85% amino acid residues of the full-length protein are identical among human, rabbit, chicken, and frog. The identity rises up to 100 % in the zinc finger containing region [9].
The CTCF gene is expressed in multiple tissues. In the human genome it is located on chromosome 16q22.1 within the loss of heterozygosity region and is a suspected tumor suppressor gene in breast and prostate cancers [10, 11].
CTCF shows a dynamic distribution among cell compartments in a cell cycle-dependent manner. In interphase, CTCF is a nuclear protein mainly excluded from the nucleolus. During mitosis, especially in anaphase and metaphase, CTCF associates with the centrosome [12]. CTCF can be phosphorylated by protein kinase CK2 in vivo [13], as well as poly(ADP-ribosyl)ated (reviewed in [14]), and this latter modification is thought to regulate CTCF activity as a component of insulators [15]. In addition, CTCF is capable of activating self-modification of poly(ADP-ribose)-polymerase-1 [16]. Recently, sumoylation of CTCF was also reported [17].
Using different combinations of zinc fingers, CTCF binds diverse DNA cis-regulatory sequences [18] and participates in multiple cellular processes. A database of CTCF binding sites is constructed and now contains more than 30,000 characterized sites [19]. Binding of CTCF to a target DNA region can lead to either activation or repression of the transcription of the gene under its control [20].
Binding of CTCF to DNA can be methylation sensitive except when the binding site does not contain CpG dinucleotides. Moreover, CTCF binding can protect its sites from methylation (reviewed in [20]). This suggests the implication of CTCF in epigenetic regulation [21-23] and in X-inactivation choice and escape [23-26]. Binding of CTCF to DNA can be also regulated by nucleosome positioning [27] and vice versa [28].
CTCF AS A MULTIFUNCTIONAL REGULATOR
CTCF and Cancer-Associated Genes
The location of the human CTCF gene within the breast and prostate cancer loss of heterozygosity region on human chromosome 16q22.1 allowed Filippova et al. [10] to hypothesize that CTCF is a candidate tumor suppressor gene. The hypothesis was further supported by the finding of a tumor-specific rearrangement of CTCF exons [10] and several cancer-related mutations interfering with CTCF binding in a gene-specific manner [29]. However, a study of the CTCF mRNA level in breast carcinoma revealed no significant tumor-associated loss or decrease in expression, and therefore CTCF is unlikely to be a tumor suppressor gene targeted by the 16q22.1 loss in breast cancer [30]. Moreover, CTCF protein level was found to be elevated in breast cancer cell lines and tumors when compared with normal counterparts, also not suggesting that CTCF is a tumor suppressor gene [31]. However, the authors in [31] put forward the hypothesis that up-regulation of CTCF may be linked to the resistance of breast cancer cells to apoptosis. This suggestion was substantiated by the demonstration that overexpression of CTCF partially protects cells from apoptosis induced by the proapoptotic protein Bax. On the contrary, down-regulation of CTCF caused apoptotic cell death [31]. The experiments above thus indicate that CTCF behaves more like a regulator of other tumor-associated genes than a classical tumor suppressor. This is in accord with the observation that CTCF targets include various genes with regulatory functions, and among them the well known oncogene MYC [3], tumor suppressor gene RB1 [32] and other genes (see [20]).
CTCF and Development: Regulator of Master Regulators?
A complete knockout of the CTCF gene results in early embryonic lethality of mice indicating its essential function in early development [33, 34]. However, conditional deletion of the CTCF gene in fetal liver cells [34] and thymocytes [35] did not cause cell death but interfered with their growth and regulation. CTCF gene knockdown using antisense constructs shows inhibition of K562 cells differentiation [36] and apoptotic cell death in breast cancer cell lines [31]. Downregulation of CTCF expression by siRNA resulted in reduced expression of MHC class II genes [37]. The data above support the multifunctional nature of CTCF, and at the same time show that CTCF is not essential for cell growth in culture or for tumor cells proliferation.
CTCF regulates expression of the Pax6 gene coding for a highly conserved transcription factor of the paired box family, which is important in central nervous system development including development of eye [38]. A knockdown of CTCF in transgenic mice enhances the transcription of Pax6, whereas the overexpression of CTCF suppresses Pax6 transcription, possibly by the insulation of the Pax6 promoter from its enhancer [39]. Also, CTCF gene transcription was moderately (2-3 fold) induced in rabbit corneal epithelial cells by epidermal growth factor in a dose-dependent manner, and in human Rb cells by serum factors, this activation also resulted in suppression of Pax6 transcription [40, 41].
Depletion of maternal CTCF in mouse oocytes resulted in transcriptional misregulation of multiple genes, meiotic defects in the egg and mitotic defects in the embryo. The authors concluded that CTCF was an important maternal effect gene playing an essential role in early embryonic development [42].
The data above seem to be rather contradictory. However, they could be understood considering an essential difference between gene knockout leading to complete exclusion of CTCF from all cells of the organism and conditional gene knockout or gene transcript knockdown in a somatic tissue. In these latter cases a protein depot could exist for abundant and stable proteins, which is probably the case for CTCF. This depot might be sufficient to maintain several cell divisions, as with maternal CTCF in eggs [42]. However, there are no data allowing to estimate the CTCF content in various cells.
Collectively, these data indicate a critical role of CTCF in mammalian development. It can be argued that CTCF functions include (but are not limited to) regulation of other regulators, but the question how CTCF performs this regulation remains unsolved.
CTCF AND GENOME FUNCTIONING
CTCF and Chromatin Border Elements
Of all CTCF functions, the most thoroughly studied is undoubtedly formation of boundary elements in vertebrate genomes [43], which is implemented with the participation of transcriptional insulators. However, it would be incorrect to consider all CTCF-binding sequences as insulators.
There are two types of chromatin boundary elements identified so far - insulators and S/MARs. Insulators are DNA sequences that prevent activation of promoters by inappropriate enhancers and/or block the spread of condensed chromatin (for recent reviews see [44-47]). In some cases, however, these two activities can be linked to distinct parts of one and the same sequence [48, 49].
Insulators have been identified in various eukaryotic organisms, including vertebrates, Drosophila, sea urchin and yeast [50-54]. Some insulators can function when transferred in phylogenetically distant organisms, like sea urchin, plant and human [54, 55], and can interfere with heterologous enhancers [55, 56].
In contrast to a majority of known enhancers, the activity of different insulators can depend [52, 57, 58] or not depend [59-61] on their orientation relative to the cognate promoters. Moreover, if more than one insulator is located between enhancer and promoter, their combined enhancer-blocking activity can be much lower than that of a single insulator [62-64]. This neutralization effect can depend on the orientation of insulators relative to each other [65]. Consequently, insulators may not just subdivide a genome into domains, but rather form, in cooperation with genes, promoters, enhancers and other elements, a multilevel network regulating the transcriptional activity of the genome.
Although several reports indicated the existence of CTCF-independent insulators [66-68], CTCF participates in functioning of a great majority of these elements.
For example, CTCF plays an essential role in the activity of chicken [43], mouse [69, 70] and human [69, 71] beta-globins and chicken alpha-globin insulator elements [72], T-cell receptor alpha and delta insulators [68], an insulator of the Igf2/H19 locus [21, 22], and other insulators [73].
Another type of DNA elements considered to be capable of forming independently regulated chromatin domains are scaffold/matrix attachment regions or S/MARs (reviewed in [74]). S/MARs are operationally defined as the DNA sequences that in in vitro test preferentially bind nuclear matrix or scaffold [75]. S/MARs are hypothesized to be located at the base of chromatin loops and to anchor them to the nuclear matrix thus forming structurally and functionally independent chromatin domains (for review see [74, 76]). As both S/MARs and insulators can participate in the formation of chromatin domains, their interrelation has been investigated. It was reported that the same DNA fragments, at least in some cases, display both the S/MARs and insulator properties [72, 77-81]. However, it was also demonstrated that these activities can be separated [82] or are only observed for certain genetic constructs [83].
It was found recently that the CTCF protein can be associated with nuclear matrix [84]. The authors assumed that CTCF might demarcate nuclear matrix-dependent points of transition in chromatin, thereby forming topologically independent chromatin loops. Later, it was shown [85] that both CTCF and the chicken beta-globin HS4 insulator element can be incorporated in the nuclear matrix, and, moreover, HS4 incorporation depends on the presence of an intact CTCF-binding site. Recently, nuclear lamina associated domains (LADs) of the human genome were described [86], and CTCF was proposed to participate in the demarcation of these domains. All these data suggest a possible connection between the activities of S/MARs and insulators, but the problem of this relationship has not yet been solved and requires identification and comparative analysis of a greater number of these elements.
Genomic Distribution of CTCF Binding Sites
The publication of the human and other metazoan genome sequences opened a possibility for analysis of CTCF binding sites distribution within genomes. Using chromatin immunoprecipitation, more than 200 CTCF-binding DNA fragments were identified in the mouse genome. It was demonstrated that a considerable fraction of these fragments displayed insulator-like properties when assayed using an episome-based test [87]. Binding sites were found in intergenic DNA, in gene regulatory regions and in introns. The number of identified sites was, however, not sufficient to make a reliable conclusion about their distribution in the whole genome. An attempt to map the majority of CTCF binding sites in a 1 megabase human genome region was undertaken [88] using both in vitro and in vivo approaches. Ten binding sites were identified therein that allowed to estimate the total number of CTCF binding sites in the human genome at about 30,000. CTCF binding sites were mapped within gene introns and also within repeated elements, in particular Alu.
A whole genome chromatin immunoprecipitation followed by a detection with tiling microarrays (ChIP-on-chip) was used to identify CTCF binding sites in the human genome [89]. 13,804 CTCF binding sites were identified in the genome of IMR90 human fibroblasts. Their distribution was closely correlated with gene positions along the genome. 46% of the sites were found in intergenic sequences, 20% near transcription start sites, and 34% within genes, mostly in introns. It is noteworthy that more than 67% of CTCF binding sites occupied by proteins in IMR90 cells were also found to be occupied in human histiocytic lymphoma U937 cells, suggesting constitutive binding of CTCF to a majority of its sites [89]. Similar data was obtained by Barski et al. [90], who analyzed the whole genome distribution of CTCF binding sites using chromatin immunoprecipitation combined with the massively parallel sequencing (ChIP-seq) approach. More than 20,000 CTCF binding sites were identified within the genome of human CD4+ T-cells. About 40% of the sites were mapped within intergenic regions, 30% within genes and 30% near transcription start sites. A more detailed analysis performed later for three cell types (CD4+ T-cells, Jurkat and HeLa) [91] revealed that about 49-56% of sites bound to CTCF were located intergenically, 33% - intronically, and 3-4% - exonically. Most tissue specific characteristics were revealed for sites occupied by CTCF that were located near promoter regions. The occupancy was maximal in CD4+ T-cells (15%) and minimal in HeLa cells (7%). The authors also demonstrated that CTCF preferentially occupied boundaries of repressive chromatin regions enriched in histone H3K27me3 modification and that this occupancy was also cell type specific [91]. However, minor part of all CTCF-occupied sites were located at potential domain borders, in accord with the multifunctional nature of CTCF.
Chen et al. [92] used the ChIP-seq approach to identify and map CTCF and several other transcription factors binding sites across the whole mouse genome. The number of CTCF binding sites in the genome was estimated as 40,000, which somewhat exceeds the previous estimations (see above), and significantly (2-50 fold) exceeds the number of other transcription factors binding sites tested.
It was also demonstrated [90] that CTCF preferentially binds to regions containing specifically methylated histone H3 - CTCF binding regions were enriched in H3K4 (all methylation states) and H3K9me1, but not in H3K9me2 and H3K9me3. Also, an enrichment of CTCF binding sites with the H2A.Z histone variant was detected, in accord with earlier data showing preferable location of H2A.Z at chromatin boundaries in yeast [93].
A great majority of ubiquitous in many cell types DNase I hypersensitive sites are bound by CTCF [94] indicating that CTCF highly contributes to chromatin architecture and regulation.
CTCF Mediates Long-Distance Interactions of Regulatory Sites
Application of different variants of the chromosome conformation capture (3C) technique [95] allowed to identify CTCF-dependent chromatin loops (for recent review see [96, 97]). Within the mouse Igf2/H19 imprinted region, the loops are formed between the imprinting control region (ICR) and differentially methylated regions (DMR) [98]. This loop formation depends on CTCF binding with the maternal ICR, but its exact mechanism is unknown [99, 100]. Moreover, CTCF is necessary for the interaction of one ICR allele of the Igf2/H19 locus on chromosome 7 with an allele of the Wsb1/Nf1 locus on chromosome 11 [101].
CTCF may participate in the loop formation in several ways: (i) by forming dimers or oligomers able to interact with two or more different DNA regions; (ii) by interacting with other proteins capable of DNA or protein binding, and (iii) solely by CTCF-DNA interactions if a single CTCF molecule can bind at least two distant DNA regions. There is no experimental evidence for the latter case, and participation of CTCF in protein-protein interactions will be discussed below.
Interactions of CTCF with Other Proteins
Available in vitro and two-hybrid assay data indicate the formation of CTCF dimers or even multimers [102, 103], but the existence of CTCF di- and oligomers in vivo is still to be proved.
Affinity fractionation of nuclear extract on a column with immobilized CTCF produced a Y-box binding factor 1 (YB-1), although the role of this interaction is not quite clear [104].
Another example of CTCF-binding protein is Kaiso - a zinc-finger transcription factor of the POZ (pox virus and zinc-finger) family. Interactions between Kaiso and CTCF were documented using the two-hybrid system and co-immunoprecipitation [105]. Later, it was shown that the Kaiso factor recognizes unoccupied CTCF target sequences when CTCF binding is lost due to DNA methylation [32]. Recently, it was demonstrated that CTCF can associate with the transcription factor YY1 and transactivate Tsix (antisense of the Xist gene) thus playing a key role in X-inactivation [25]. This is the first example of a proved functionally significant interaction of CTCF with another protein. It was hypothesized that the CTCF-YY1 interaction may participate in regulation of other imprinting control regions [106].
According to in vitro and two-hybrid system data, CTCF also interacts with Suz12, a protein component of Polycomb repressive complex 2 (PRC2), which is responsible for methylation of histone H3 lysine 27 resulting in chromatin suppression. This mechanism is possibly responsible for the suppression of the maternal Igf2 promoters [107].
CTCF can interact with and activate automodification of poly(ADP-ribose)-polymerase-1 which, in turn, can affect activity of DNA (cytosine-5-)-methyltransferase 1 (DNMT1) and, consequently, chromatin structure [16, 108].
It was shown that the HeLa RNA polymerase II largest subunit (Pol II LS) co-immunoprecipitates with CTCF, and that CTCF and Pol II LS epitopes colocalize to the beta-globin insulator in vivo [109].
The in vitro immunoprecipitation and two-hybrid assay indicate that the SNF2-like chromodomain helicase protein CHD8 interacts through its carboxyl-terminal region with the zinc finger domain of CTCF. Using chromatin immunoprecipitation, CHD8 was also found at some CTCF binding sites in vivo. The authors suggested possible participation of CHD8 in CTCF-dependent insulation [110].
Using chromatin immunoprecipitation, CTCF was shown to colocalize [85] in vivo at insulator sites with nucleophosmin, a multifunctional acidic nucleolar protein that participates in regulation of cell growth and proliferation [111]. As nucleophosmin is a well known component of nuclear matrix [112], it was proposed that the interaction between this protein and CTCF leads to the appearance of CTCF in the cellular matrix fraction [85].
It is of interest to note that most of known CTCF-interacting partners are either transcriptional regulators (YB-1, Kaiso, YY1) or related to regulation of other cellular functions. However, it is worth noting that in most cases the data on CTCF-protein interactions were obtained by indirect or in vitro methods, like chromatin pull-down and two-hybrid analysis. Direct in vivo analysis of CTCF interactions is limited by the lack of adequate methods.
CTCF and Cohesin Complex
Cohesin is a highly conserved multi-protein complex whose main function is to hold together sister chromatids during S and G2 phases of the cell cycle to ensure proper chromosome segregation. The role of cohesin in gene regulation is also emerging (for reviews see [113, 114].
Recently, when studying the genome distribution of cohesin specific binding sites in the mouse and human chromatin, it was found that in 60-70% cases cohesin and CTCF are colocalized, and CTCF depletion disrupts positioning of cohesin. Importantly, the reverse is not true, and cohesin depletion does not significantly affect the chromatin distribution of CTCF. Depletion of cohesin somewhat inhibited activity of a chicken beta-globin insulator in transient transfection assays. The authors concluded that CTCF largely determined the localization of cohesins, and cohesin is involved in insulator function [115-117]. A putative interacting partner of CTCF is the Scc3/SA subunit of cohesin [118].
CTCF and Repeated Elements
CTCF binding sites were found to be located within repeating elements, such as the (GT)22(GA)15 microsatellite A9 in intron 2 of the HLA-DRB1*0401 gene [119], trinucleotide repeats, where CTCF binding modulated their instability [120], and Alu-elements [88, 121]. It was also shown that B2 repeated sequences were significantly overrepresented in CTCF binding regions of the mouse genome [122].
Interestingly, some Alu elements can function as insulators [121, 123]. Alu elements are capable of transposition, and their relocation within the genome together with CTCF binding sites may cause reorganization of the domain structure, as we proposed earlier for S/MARs-containing retroelements [74].
REGULATION OF THE CTCF GENE
Despite the obviously important regulatory function of CTCF in multiple cellular processes, very little is known about regulation of the CTCF gene itself. Transcription of the CTCF gene was moderately (2-3 fold) induced in rabbit corneal epithelial cells by epidermal growth factor in a dose-dependent manner, and in human Rb cells by serum, the activation resulted in Pax6 transcription suppression [40, 41]. Expression of CTCF in mice was reduced with age [124].
Treatment of human choriocarcinoma JAr cells with lithium resulted in a 2-3 fold increase in the CTCF mRNA content [125].
The CTCF promoter contains a CpG island, no TATA-box, a highly conserved YY1 transcription factor binding site and potential binding sites for GATA-1 and p53 [126, 127]. Within the chicken CTCF promoter several elements characteristic of cell cycle-regulated genes were found [126].
POSSIBLE MECHANISMS FOR CTCF VERSATILE REGULATORY ACTIVITY
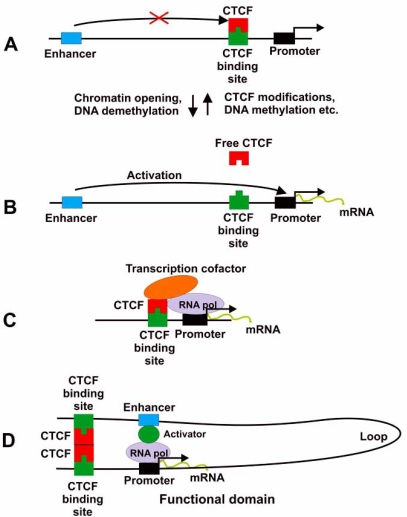
The regulatory effect of CTCF on gene transcription may involve several possible mechanisms some of which are illustrated in Fig. (1).
Fig. (1).
Possible mechanism of gene regulation with the participation of CTCF. See text for detail.
1). Conditional Transcriptional Insulation
In this mode CTCF regulates the access of an enhancer to the promoter region via either CTCF binding to a vacant binding site located between enhancer and promoter (Fig. 1A) or by release of its constitutively occupied binding site (Fig. 1B) due to modification (e.g. phosphorylation) of CTCF or modification (methylation) of its binding site. The former type is apparently used in the regulation of the H19-Igf-2 locus transcription, as well as in the Pax6 promoter regulation [40]. The latter regulation type is based on the ability of CTCF to be subject to phosphorylation [13], poly(ADP-ribosyl)ation [14, 15] and sumoylation [17].
2). Genome-wide Dispersed Anchor/Demarcation Function
CTCF may serve as an anchor protein whose binding to its binding sites can recruit other trans-acting regulatory elements to allow DNA bound functional (e.g. transcription) protein complex assembly (Fig. 1C). The abundance of CTCF and its numerous binding sites allow its participation in a multitude of functions involving the CTCF DNA binding domains and the capacity of interacting with other proteins. CTCF works as a constitutive demarcation tool of the genome/chromatin. Depending on particular genomic context and availability of various factors emerging in response to external or internal cellular signals, a particular CTCF could enable recruiting a particular set of proteins for executing a particular function. Therefore, the colocalization of CTCF with RNA-polymerase II as well as its capacity of binding various transcription factors might play a role in transcriptional regulation. This model is in agreement with constitutive binding of CTCF to a definite subset of its binding sites. Here, the induction of a transcription factor capable of interacting with promoter DNA-bound CTCF might activate the preinitiation complex.
3). Functional Domain Formation and Insulator Function
Finally, the capacity of CTCF to form di- or multimers and interact with other DNA binding proteins suggests its possible participation in DNA bending and directing enhancers to their cognate promoters as shown in Fig. (1D). In addition, two CTCF molecules in cooperation with other proteins can insulate the enhancer-promoter system from external regulatory interference.
Although all these possibilities seem quite reasonable, they certainly need additional experimental support.
CONCLUSIONS
CTCF is a striking example of a multifunctional regulator. It participates in activation/repression of gene activity, chromatin insulation, formation of chromatin loops, X-inactivation and escape, positioning of the cohesin complex etc. CTCF regulates a number of regulatory genes including oncogenes and tumor suppressors, as well as genes regulating development and differentiation. CTCF is subject to several important post-translational modifications. CTCF is not important for single cell proliferation, but it is very important for development of multicellular organisms. The number of CTCF genomic binding sites exceeds that for most other transcription factors.
At the same time, the mechanisms underlying the CTCF functions remain largely unknown. In particular, functioning of CTCF as a component of insulator complexes and its participation in formation of loop domains implies protein-protein interactions, but the number of the CTCF-interacting protein partners found is quite low. Therefore, search for proteins and/or protein complexes interacting with CTCF in vivo would contribute to better understanding its role in multiple cellular processes.
In this review, we tried to formulate possible lines of studies on the variety of CTCF functions probably based on three intrinsic properties: the ability of specific DNA binding, interacting with other proteins and forming dimers and multimers. These three fundamental properties allow CTCF to serve as a transcription factor, an insulator and as a constitutive dispersed genome-wide demarcation tool able to recruit various factors that emerge in response to diverse external and internal signals, and thus to exert its signal-specific function(s).
ACKNOWLEDGEMENTS
We are grateful to B.O. Glotov for critical reading of the manuscript. The work of authors was supported by the Scientific School program (project NSh 2395.2008.4), Program of the Russian Academy of Sciences on Molecular and Cellular Biology and by Russian Foundation for Basic Research (project 07-04-00709).
REFERENCES
- 1.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 2.Lobanenkov V V, Goodwin G H. CCCTC-binding protein: a new nuclear protein: factor which interaction with 5'-flanking sequence of chicken c-myc oncogene correlates with repression of the gene. Dokl. Akad. Nauk SSSR. 1989;309:741–745. [PubMed] [Google Scholar]
- 3.Lobanenkov V V, Nicolas R H, Adler V V, Paterson H, Klenova E M, Polotskaja A V, Goodwin G H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 4.Baniahmad A, Steiner C, Kohne A C, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–514. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- 5.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova G N, Lobanenkov V V, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray C E, Coates C J. Cloning and characterization of cDNAs encoding putative CTCFs in the mosquitoes, Aedes aegypti and Anopheles gambiae. BMC Mol. Biol. 2005;6:16. doi: 10.1186/1471-2199-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith S T, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke L J, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hore T A, Deakin J E, Marshall Graves J A. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 2008;4:e1000169. doi: 10.1371/journal.pgen.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke L J, Hollemann T, Pieler T, Renkawitz R. Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech. Dev. 2002;113:95–98. doi: 10.1016/s0925-4773(02)00005-9. [DOI] [PubMed] [Google Scholar]
- 10.Filippova G N, Lindblom A, Meincke L J, Klenova E M, Neiman P E, Collins S J, Doggett N A, Lobanenkov V V. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 11.Green A R, Krivinskas S, Young P, Rakha E A, Paish E C, Powe D G, Ellis I O. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res. Treat. 2009;113:59–66. doi: 10.1007/s10549-008-9905-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Burke L J, Rasko J E, Lobanenkov V, Renkawitz R. Dynamic association of the mammalian insulator protein CTCF with centrosomes and the midbody. Exp. Cell Res. 2004;294:86–93. doi: 10.1016/j.yexcr.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Klenova E M, Chernukhin I V, El-Kady A, Lee R E, Pugacheva E M, Loukinov D I, Goodwin G H, Delgado D, Filippova G N, Leon J, Morse H. C 3rd, Neiman P E, Lobanenkov V V. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol. Cell. Biol. 2001;21:2221–2234. doi: 10.1128/MCB.21.6.2221-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caiafa P, Zlatanova J. CCCTC-binding factor meets poly(ADP-ribose) polymerase-1. J. Cell. Physiol. 2009;219:265–270. doi: 10.1002/jcp.21691. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg A P, Lobanenkov V, Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 16.Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J. Biol. Chem. 2008;283:21873–21880. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacPherson M J, Beatty L G, Zhou W, Du M, Sadowski P D. The CTCF insulator protein is posttranslationally modified by SUMO. Mol. Cell. Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone P V. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 19.Bao L, Zhou M, Cui Y. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filippova G N. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 21.Bell A C, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 22.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 23.Chao W, Huynh K D, Spencer R J, Davidow L S, Lee J T. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 24.Filippova G N, Cheng M K, Moore J M, Truong J P, Hu Y J, Nguyen D K, Tsuchiya K D, Disteche C M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Donohoe M E, Zhang L F, Xu N, Shi Y, Lee J T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 26.Chadwick B P. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18:1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanduri M, Kanduri C, Mariano P, Vostrov A A, Quitschke W, Lobanenkov V, Ohlsson R. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell. Biol. 2002;22:3339–3344. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Sinha M, Peterson C L, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filippova G N, Qi C F, Ulmer J E, Moore J M, Ward M D, Hu Y J, Loukinov D I, Pugacheva E M, Klenova E M, Grundy P E, Feinberg A P, Cleton-Jansen A M, Moerland E W, Cornelisse C J, Suzuki H, Komiya A, Lindblom A, Dorion-Bonnet F, Neiman P E, Morse H. C 3rd, Collins S J, Lobanenkov V V. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 30.Rakha E A, Pinder S E, Paish C E, Ellis I O. Expression of the transcription factor CTCF in invasive breast cancer: a candidate gene located at 16q22.1. Br. J. Cancer. 2004;91:1591–1596. doi: 10.1038/sj.bjc.6602144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Docquier F, Farrar D, D'Arcy V, Chernukhin I, Robinson A F, Loukinov D, Vatolin S, Pack S, Mackay A, Harris R A, Dorricott H, O'Hare M J, Lobanenkov V, Klenova E. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 2005;65:5112–5122. doi: 10.1158/0008-5472.CAN-03-3498. [DOI] [PubMed] [Google Scholar]
- 32.De La Rosa-Velazquez I A, Rincon-Arano H, Benitez-Bribiesca L, Recillas-Targa F. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 2007;67:2577–2585. doi: 10.1158/0008-5472.CAN-06-2024. [DOI] [PubMed] [Google Scholar]
- 33.Fedoriw A M, Stein P, Svoboda P, Schultz R M, Bartolomei M S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 34.Splinter E, Heath H, Kooren J, Palstra R J, Klous P, Gros-veld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling K W, Gribnau J, Renkawitz R, Grosveld F, Hendriks R W, Galjart N. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torrano V, Chernukhin I, Docquier F, D'Arcy V, Leon J, Klenova E, Delgado M D. CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J. Biol. Chem. 2005;280:28152–28161. doi: 10.1074/jbc.M501481200. [DOI] [PubMed] [Google Scholar]
- 37.Majumder P, Gomez J A, Chadwick B P, Boss J M. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 39.Li T, Lu Z, Lu L. Regulation of eye development by transcription control of CCCTC binding factor (CTCF) J. Biol. Chem. 2004;279:27575–27583. doi: 10.1074/jbc.M313942200. [DOI] [PubMed] [Google Scholar]
- 40.Li T, Lu L. Epidermal growth factor-induced proliferation requires down-regulation of Pax6 in corneal epithelial cells. J. Biol. Chem. 2005;280:12988–12995. doi: 10.1074/jbc.M412458200. [DOI] [PubMed] [Google Scholar]
- 41.Li T, Lu Z, Lu L. Pax6 regulation in retinal cells by CCCTC binding factor. Invest. Ophthalmol. Vis. Sci. 2006;47:5218–5226. doi: 10.1167/iovs.06-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan L B, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham K E, Schultz R M, Bartolomei M S. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell A C, West A G, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 44.Wallace J A, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorman E R, Bushey A M, Corces V G. The role of insulator elements in large-scale chromatin structure in interphase. Semin. Cell. Dev. Biol. 2007;18:682–690. doi: 10.1016/j.semcdb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeda R K, Karch F. Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Maksimenko O G, Chetverina D A, Georgiev P G. Insulators of higher eukaryotes: properties, mechanisms of action, and role in transcriptional regulation. Russ. J. Genet. 2006;42:845–857. [PubMed] [Google Scholar]
- 48.Recillas-Targa F, Pikaart M J, Burgess-Beusse B, Bell A C, Litt M D, West A G, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh V, Srivastava M. Enhancer blocking activity of the insulator at H19-ICR is independent of chromatin barrier establishment. Mol. Cell. Biol. 2008;28:3767–3775. doi: 10.1128/MCB.00091-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung J H, Whiteley M, Felsenfeld G. A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 51.Zhong X P, Krangel M S. An enhancer-blocking element between alpha and delta gene segments within the human T cell receptor alpha/delta locus. Proc. Natl. Acad. Sci. USA. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bi X, Broach J R. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melfi R, Palla F, Di Simone P, Alessandro C, Cali L, Anello L, Spinelli G. Functional characterization of the enhancer blocking element of the sea urchin early histone gene cluster reveals insulator properties and three essential cis-acting sequences. J. Mol. Biol. 2000;304:753–763. doi: 10.1006/jmbi.2000.4273. [DOI] [PubMed] [Google Scholar]
- 54.Nagaya S, Yoshida K, Kato K, Akasaka K, Shinmyo A. An insulator element from the sea urchin Hemicentrotus pulcherrimus suppresses variation in transgene expression in cultured tobacco cells. Mol. Genet. Genomics. 2001;265:405–413. doi: 10.1007/s004380100448. [DOI] [PubMed] [Google Scholar]
- 55.Di Simone P, Di Leonardo A, Costanzo G, Melfi R, Spinelli G. The sea urchin sns insulator blocks CMV enhancer following integration in human cells. Biochem. Biophys. Res. Commun. 2001;284:987–992. doi: 10.1006/bbrc.2001.5082. [DOI] [PubMed] [Google Scholar]
- 56.Verona R I, Bartolomei M S. Role of H19 3' sequences in controlling H19 and Igf2 imprinting and expression. Genomics. 2004;84:59–68. doi: 10.1016/j.ygeno.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Akasaka K, Nishimura A, Takata K, Mitsunaga K, Mibuka F, Ueda H, Hirose S, Tsutsui K, Shimada H. Upstream element of the sea urchin arylsulfatase gene serves as an insulator. Cell. Mol. Biol. (Noisy-le-grand) 1999;45:555–565. [PubMed] [Google Scholar]
- 58.Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belozerov V E, Majumder P, Shen P, Cai H N. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yannaki E, Tubb J, Aker M, Stamatoyannopoulos G, Emery D W. Topological constraints governing the use of the chicken HS4 chromatin insulator in oncoretrovirus vectors. Mol. Ther. 2002;5:589–598. doi: 10.1006/mthe.2002.0582. [DOI] [PubMed] [Google Scholar]
- 61.Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- 62.Kuhn E J, Viering M M, Rhodes K M, Geyer P K. A test of insulator interactions in Drosophila. EMBO J. 2003;22:2463–2471. doi: 10.1093/emboj/cdg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maksimenko O, Golovnin A, Georgiev P. Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus. Mol. Cell. Biol. 2008;28:5469–5477. doi: 10.1128/MCB.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mongelard F, Corces V G. Two insulators are not better than one. Nat. Struct. Biol. 2001;8:192–194. doi: 10.1038/84905. [DOI] [PubMed] [Google Scholar]
- 65.Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomos-Klein J, Harrow F, Alarcon J, Ortiz B D. CTCF-independent, but not CTCF-dependent, elements significantly contribute to TCR-alpha locus control region activity. J. Immunol. 2007;179:1088–1095. doi: 10.4049/jimmunol.179.2.1088. [DOI] [PubMed] [Google Scholar]
- 67.Yao S, Osborne C S, Bharadwaj R R, Pasceri P, Sukonnik T, Pannell D, Recillas-Targa F, West A G, Ellis J. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res. 2003;31:5317–5323. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magdinier F, Yusufzai T M, Felsenfeld G. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor alpha and Dad1 genes. J. Biol. Chem. 2004;279:25381–25389. doi: 10.1074/jbc.M403121200. [DOI] [PubMed] [Google Scholar]
- 69.Farrell C M, West A G, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulger M, Schubeler D, Bender M A, Hamilton J, Farrell C M, Hardison R C, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanimoto K, Sugiura A, Omori A, Felsenfeld G, Engel J D, Fukamizu A. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol. Cell. Biol. 2003;23:8946–8952. doi: 10.1128/MCB.23.24.8946-8952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valadez-Graham V, Razin S V, Recillas-Targa F. CTCF-dependent enhancer blockers at the upstream region of the chicken alpha-globin gene domain. Nucleic Acids Res. 2004;32:1354–1362. doi: 10.1093/nar/gkh301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl. Acad. Sci. USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chernov I P, Akopov S B, Nikolaev L G. Structure and function of nuclear matrix associated regions (S/MARs) Russ. J. Bioorg. Chem. 2004;30:1–11. doi: 10.1023/b:rubi.0000015767.28683.69. [DOI] [PubMed] [Google Scholar]
- 75.Berezney R, Mortillaro M J, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- 76.Davie J R. The nuclear matrix and the regulation of chromatin organization and function. Int. Rev. Cytol. 1995;162A:191–250. doi: 10.1016/s0074-7696(08)61232-2. [DOI] [PubMed] [Google Scholar]
- 77.Phi-Van L, Stratling W H. Dissection of the ability of the chicken lysozyme gene 5' matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry. 1996;35:10735–10742. doi: 10.1021/bi9603783. [DOI] [PubMed] [Google Scholar]
- 78.Nabirochkin S, Ossokina M, Heidmann T. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 1998;273:2473–2479. doi: 10.1074/jbc.273.4.2473. [DOI] [PubMed] [Google Scholar]
- 79.Namciu S J, Blochlinger K B, Fournier R E. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol. 1998;18:2382–2391. doi: 10.1128/mcb.18.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sass A V, Ruda V M, Akopov S B, Snezhkov E V, Nikolaev L G, Sverdlov E D. Regulatory potential of S/MAR elements in transient expression. Russ. J. Bioorg. Chem. 2005;31:70–73. doi: 10.1007/s11171-005-0009-5. [DOI] [PubMed] [Google Scholar]
- 81.Majumder P, Gomez J A, Boss J M. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J. Biol. Chem. 2006;281:18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- 82.Scott K C, Taubman A D, Geyer P K. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 1999;153:787–798. doi: 10.1093/genetics/153.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Attal J, Cajero-Juarez M, Petitclerc D, Theron M C, Stinnakre M G, Bearzotti M, Kann G, Houdebine L M. The effect of matrix attached regions (MAR) and specialized chromatin structure (SCS) on the expression of gene constructs in cultured cells and in transgenic mice. Mol. Biol. Rep. 1995;22:37–46. doi: 10.1007/BF00996303. [DOI] [PubMed] [Google Scholar]
- 84.Dunn K L, Zhao H, Davie J R. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 2003;288:218–223. doi: 10.1016/s0014-4827(03)00185-x. [DOI] [PubMed] [Google Scholar]
- 85.Yusufzai T M, Felsenfeld G. The 5'-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guelen L, Pagie L, Brasset E, Meuleman W, Faza M B, Talhout W, Eussen B H, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 87.Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, Quitschke W, Chernukhin I, Klenova E, Lobanenkov V, Ohlsson R. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vetchinova A S, Akopov S B, Chernov I P, Nikolaev L G, Sverdlov E D. Two-dimensional electrophoretic mobility shift assay: identification and mapping of transcription factor CTCF target sequences within an FXYD5-COX7A1 region of human chromosome 19. Anal. Biochem. 2006;354:85–93. doi: 10.1016/j.ab.2006.03.052. [DOI] [PubMed] [Google Scholar]
- 89.Kim T H, Abdullaev Z K, Smith A D, Ching K A, Loukinov D I, Green R D, Zhang M Q, Lobanenkov V V, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barski A, Cuddapah S, Cui K, Roh T Y, Schones D E, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 91.Cuddapah S, Jothi R, Schones D E, Roh T Y, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega V B, Wong E, Orlov Y L, Zhang W, Jiang J, Loh Y H, Yeo H C, Yeo Z X, Narang V, Govindarajan K R, Leong B, Shahab A, Ruan Y, Bourque G, Sung W K, Clarke N D, Wei C L, Ng H H. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 93.Meneghini M D, Wu M, Madhani H D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 94.Xi H, Shulha H P, Lin J M, Vales T R, Fu Y, Bodine D M, McKay R D, Chenoweth J G, Tesar P J, Furey T S, Ren B, Weng Z, Crawford G E. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 96.Williams A, Flavell R A. The role of CTCF in regulating nuclear organization. J. Exp. Med. 2008;205:747–750. doi: 10.1084/jem.20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zlatanova J, Caiafa P. CCCTC-binding factor: to loop or to bridge. Cell. Mol. Life Sci. 2009 doi: 10.1007/s00018-009-8647-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 99.Kurukuti S, Tiwari V K, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoon Y S, Jeong S, Rong Q, Park K Y, Chung J H, Pfeifer K. Analysis of the H19ICR insulator. Mol. Cell. Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ling J Q, Li T, Hu J F, Vu T H, Chen H L, Qiu X W, Cherry A M, Hoffman A R. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 102.Yusufzai T M, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 103.Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chernukhin I V, Shamsuddin S, Robinson A F, Carne A F, Paul A, El-Kady A I, Lobanenkov V V, Klenova E M. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 2000;275:29915–29921. doi: 10.1074/jbc.M001538200. [DOI] [PubMed] [Google Scholar]
- 105.Defossez P A, Kelly K F, Filion G J, Perez-Torrado R, Magdinier F, Menoni H, Nordgaard C L, Daniel J M, Gilson E. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J. Biol. Chem. 2005;280:43017–43023. doi: 10.1074/jbc.M510802200. [DOI] [PubMed] [Google Scholar]
- 106.Kim J. Multiple YY1 and CTCF binding sites in imprinting control regions. Epigenetics. 2008;3:115–118. doi: 10.4161/epi.3.3.6176. [DOI] [PubMed] [Google Scholar]
- 107.Li T, Hu J F, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu T H, Hoffman A R. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reale A, Matteis G D, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- 109.Chernukhin I, Shamsuddin S, Kang S Y, Bergstrom R, Kwon Y W, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu S Y, Chiang C M, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 111.Okuwaki M. The structure and functions of NPM1/Nucleophsmin/ B23, a multifunctional nucleolar acidic protein. J. Biochem. 2008;143:441–448. doi: 10.1093/jb/mvm222. [DOI] [PubMed] [Google Scholar]
- 112.Mattern K A, Humbel B M, Muijsers A O, de Jong L, van Driel R. hnRNP proteins and B23 are the major proteins of the internal nuclear matrix of HeLa S3 cells. J. Cell. Biochem. 1996;62:275–289. doi: 10.1002/(sici)1097-4644(199608)62:2<275::aid-jcb15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 113.Onn I, Heidinger-Pauli J M, Guacci V, Unal E, Koshland D E. Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell. Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 114.Peters J M, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 115.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson H C, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb B S, Yokomori K, Dillon N, Aragon L, Fisher A G, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 116.Wendt K S, Yoshida K, Itoh T, Bando M, Koch B, Schir-ghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters J M. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 117.Stedman W, Kang H, Lin S, Kissil J L, Bartolomei M S, Lieberman P M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rubio E D, Reiss D J, Welcsh P L, Disteche C M, Filip-pova G N, Baliga N S, Aebersold R, Ranish J A, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arnold R, Maueler W, Bassili G, Lutz M, Burke L, Epplen T J, Renkawitz R. The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA-DRB1(*)0401 gene. Gene. 2000;253:209–214. doi: 10.1016/s0378-1119(00)00271-7. [DOI] [PubMed] [Google Scholar]
- 120.Libby R T, Hagerman K A, Pineda V V, Lau R, Cho D H, Baccam S L, Axford M M, Cleary J D, Moore J M, Sopher B L, Tapscott S J, Filippova G N, Pearson C E, La Spada A R. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ruiz-Narvaez E A, Campos H. Evolutionary rate heterogeneity of Alu repeats upstream of the APOA5 gene: do they regulate APOA5 expression? J. Hum. Genet. 2008;53:247–253. doi: 10.1007/s10038-008-0245-7. [DOI] [PubMed] [Google Scholar]
- 122.Bourque G, Leong B, Vega V B, Chen X, Lee Y L, Srini-vasan K G, Chew J L, Ruan Y, Wei C L, Ng H H, Liu E T. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Willoughby D A, Vilalta A, Oshima R G. An Alu element from the K18 gene confers position-independent expression in transgenic mice. J. Biol. Chem. 2000;275:759–768. doi: 10.1074/jbc.275.2.759. [DOI] [PubMed] [Google Scholar]
- 124.Fu V X, Dobosy J R, Desotelle J A, Almassi N, Ewald J A, Srinivasan R, Berres M, Svaren J, Weindruch R, Jarrard D F. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 2008;68:6797–6802. doi: 10.1158/0008-5472.CAN-08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roberts J, Scott A C, Howard M R, Breen G, Bubb V J, Klenova E, Quinn J P. Differential regulation of the serotonin transporter gene by lithium is mediated by transcription factors, CCCTC binding protein and Y-box binding protein 1, through the polymorphic intron 2 variable number tandem repeat. J. Neurosci. 2007;27:2793–2801. doi: 10.1523/JNEUROSCI.0892-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Klenova E M, Fagerlie S, Filippova G N, Kretzner L, Good-win G H, Loring G, Neiman P E, Lobanenkov V V. Characterization of the chicken CTCF genomic locus, and initial study of the cell cycle-regulated promoter of the gene. J. Biol. Chem. 1998;273:26571–26579. doi: 10.1074/jbc.273.41.26571. [DOI] [PubMed] [Google Scholar]
- 127.Pugacheva E M, Kwon Y W, Hukriede N A, Pack S, Flanagan P T, Ahn J C, Park J A, Choi K S, Kim K W, Loukinov D, Dawid I B, Lobanenkov V V. Cloning and characterization of zebrafish CTCF: Developmental expression patterns, regulation of the promoter region, and evolutionary aspects of gene organization. Gene. 2006;375:26–36. doi: 10.1016/j.gene.2006.01.036. [DOI] [PubMed] [Google Scholar]