Abstract
Background
Bacterial and fungal infections induce a potent immune response in Drosophila melanogaster, but it is unclear whether viral infections induce an antiviral immune response. Using microarrays, we examined the changes in gene expression in Drosophila that occur in response to infection with the sigma virus, a negative-stranded RNA virus (Rhabdoviridae) that occurs in wild populations of D. melanogaster.
Principal Findings
We detected many changes in gene expression in infected flies, but found no evidence for the activation of the Toll, IMD or Jak-STAT pathways, which control immune responses against bacteria and fungi. We identified a number of functional categories of genes, including serine proteases, ribosomal proteins and chorion proteins that were overrepresented among the differentially expressed genes. We also found that the sigma virus alters the expression of many more genes in males than in females.
Conclusions
These data suggest that either Drosophila do not mount an immune response against the sigma virus, or that the immune response is not controlled by known immune pathways. If the latter is true, the genes that we identified as differentially expressed after infection are promising candidates for controlling the host's response to the sigma virus.
Introduction
Viral infections in arthropods are widespread and are of considerable economic and medical importance. For example, viruses have had devastating economic consequences on honey-bee and shrimp populations [1], [2], and many viral pathogens in humans, crops and livestock are vectored by insects. Carefully chosen model systems could provide great insight into how arthropods combat viral infections. The principal model organism used to study invertebrate immune defenses is the fruit fly Drosophila melanogaster [3]. Although many studies have examined Drosophila's defenses against bacterial and fungal infections, relatively little is known about antiviral defenses. This is despite D. melanogaster being infected with a diverse range of viruses, including positive-stranded RNA viruses (several picornaviruses, including the Drosophila C virus), a negative-stranded RNA virus (sigma virus; Rhabdoviridae), and a double-stranded RNA virus (DFV; Reoviridae) [4].
The viruses that infect Drosophila have very different lifecycles and biology, which may have important implications for immune recognition and the immune response. The picornaviruses are released by lysing host cells, and the viral particles are non-enveloped [5]. In contrast, the Rhabdovirus sigma is released from host cells by budding, and the viral particles are enclosed in a lipid envelope with surface-exposed glycoproteins [4], [6]. Furthermore, the picornavirus DCV can cause severe pathology in infected flies, while the sigma virus is relatively benign [4]. Antiviral immune responses often recognize RNA viruses by the presence of dsRNA. Typically, positive-sense RNA viruses produce much more double stranded RNA than negative sense viruses, probably because the nucleocapsid protein of negatively sensed RNA viruses can prevent the two strands from annealing to produce dsRNA [7]. All of these factors may mean that the mechanisms by which flies can recognize viruses and protect themselves against infection may be differ between different viruses.
The only immune effector that has been found to target viruses in Drosophila is RNAi [8]–[10]. RNAi can distinguish self from non-self because RNA viruses often produce double-stranded RNA, while host cells typically do not. Some RNA viruses have a double-stranded genome, and even single stranded RNA viruses can produce double-stranded RNA during replication, gene expression or as a consequence of RNA secondary structure. RNAi processes this double-stranded RNA into short fragments, which are passed to an effector complex that recognizes and degrades viral RNA with the complimentary sequence. RNAi pathway genes are constitutively expressed, and are not known to be up-regulated by viral infection [11].
Other studies have examined the changes in gene expression that occur when flies are infected by viruses [11]–[13]. Microarray analyses of flies injected with DCV identified many up-regulated genes, raising the possibility that there is an induced immune response to viruses in addition to RNAi [11]. Detailed studies of one of the up-regulated genes identified in this study, vir-1, revealed that it was under the control of the Jak-STAT pathway, which is an important component of the antiviral response of vertebrates. Not only does DCV activate this pathway, but flies that are deficient for the Jak-kinase Hopscotch have a higher viral load and lower survival than wild-type flies [11]. It is currently unknown how the Jak-STAT pathway detects viral infection, or how it protects flies against DCV (knocking down vir-1 expression does not make flies more susceptible to DCV).
In addition to the Jak-STAT pathway, the Toll pathway, an immune signaling pathway activated by bacteria and fungi, has been implicated in antiviral immunity. A previous study has shown that the Drosophila X virus (DXV) activates the Toll pathway, and flies that are deficient for the Toll pathway transcription factor Dif are more susceptibility to DXV infection [13]. Furthermore, a gene called ref(2)P, which is required by the Toll immune response, has a naturally occurring polymorphism that reduces the rate at which sigma virus replicates within the fly [14], [15]. However, the role of the Toll pathway as a antiviral response remains uncertain because previous studies have shown that Toll pathway genes are not up-regulated by DCV infection [11], and not all Toll pathway mutants alter the flies' susceptibility to DXV [13].
In this study we have used microarrays to see which immune pathways, if any, are up-regulated when Drosophila is infected with the sigma virus—a naturally occurring pathogen that infects about 4% of D. melanogaster in the wild [16]. The sigma virus is transmitted only vertically, from parent to offspring through both eggs and sperm [4]. Typical for a vertically transmitted pathogen, the sigma virus is a fairly benign infection that causes a slight reduction in the survival and fecundity of infected flies [17]. There is however a great deal of genetic variation in the susceptibility of flies to sigma infection in natural populations [18], [19]. Several major effect resistance genes have been mapped [4], but only one of these has been identified (ref(2)P; see above). It is currently unclear whether these resistance genes are part of an antiviral immune response mounted by the flies or are host molecules that the virus exploits for its own replication and transmission. The only study which looked at the transcriptional response to sigma virus infection measured the expression of 15 immunity-related genes using quantitative real-time PCR (qPCR), and found that several antimicrobial peptides and the peptidoglycan recognition proteins (PGRP-SB1 and PGRP-SD) are up-regulated in infected flies [20]. However, it is unclear whether this transcriptional response has any effect on the replication or transmission of the virus.
Materials and Methods
Drosophila stocks and hybridizations
We compared patterns of gene expression in genetically identical flies that were either infected or uninfected with the sigma virus. The flies were an isogenic stock SM5/Pm;spapol that had been infected with the sigma virus isolate AP30 several generations before (see [19] for details). These flies have the susceptible allele of the ref(2)P gene which controls sigma virus replication. Four replicates of both the infected and uninfected flies were reared on Lewis medium [21] at a constant density of approximately 220 flies per bottle at 25°C on a 12 hour light-dark cycle for a minimum of three generations before the experiment. The flies were aged and allowed to mate for six days before being sexed on ice (they were not exposed to CO2), and RNA was extracted from pools of 180 males or 60 females using Trizol (Invitrogen, Carlsbad, CA, USA). We confirmed the infection status of a sample of flies from the same bottles using a standard CO2 test [19].
We performed 5 dye swap replicates (10 arrays) on the males and 4 dye swap replicates (8 arrays) on the females. Each dye-swap compared a different pair of RNA extractions from sigma-infected and -uninfected flies (i.e., biological replicates). A more detailed description of the hybridizations and statistical analysis is given by Hutter et al. [22], who used many of the same methods. We used a D. melanogaster microarray obtained from the Drosophila Genomics Resource Center (DGRC; Bloomington, IN, USA) known as DGRC-1. This consists of 13,921 exonic PCR amplicons (100–600 bp in length) representing 11,895 unique genes (∼88% of the genome).
The RNA was reverse transcribed and labeled with the SuperScript Plus Indirect cDNA Labeling System and Alexa Fluor 555 and 647 dyes (Invitrogen, Carlsbad, CA, USA). Hybridizations were performed following DGRC protocols and arrays were scanned using an aQuire microarray scanner (Genetix, New Milton, UK). All array data have been submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under series XXX.
Data analysis
To correct each spot on our arrays for local background effects, within-array variation and between-array variation, we normalized the signal intensity of the two dye channels using the three-step procedure described by [22] and implemented in CARMAweb [23]. In short, the relative expression level and probability of differential expression between infected and uninfected flies for each gene was estimated using the Markov Chain Monte Carlo algorithm implemented in BAGEL [24]. The results of the BAGEL runs are provided as Supporting Files S1 and S2. As some genes are represented by multiple probes, we defined a gene as differentially expressed if at least one of its probes displayed a significant difference. For each slide, only those spots displaying a signal greater than 95% of the negative control probes (182 probes from other species) in each dye channel were considered ‘expressed’, and ‘non-expressed’ data points were excluded from the analysis. To determine the experiment-wide false discovery rate (FDR), we repeated the BAGEL analysis on a randomized version of our final data-set [22]. To estimate the power of our experiment to detect expression differences between infected and uninfected flies, we calculated the GEL50 statistic [25].
To test if any gene ontology (GO) categories were over-represented in our list of differentially expressed genes, we used the web-based tool g:Profiler [26] that corrects for multiple testing while taking the hierarchical nature of GO terms into account. The analysis of gene ontology was based on the annotation of the D. melanogaster genome included in release v49 of ENSEMBL [27]. CG numbers were updated to match this version and these lists are provided separately in the Supporting File S3.
Quantitative real-time PCR
To confirm the results of the microarray analyses, we measured the expression of several genes using qPCR. These genes included PGRP-SC2 and Tudor-SN, whose expression differed between sigma-infected and -uninfected flies in our microarray experiment, and Attacin-A and Drosocin, which were found to be up-regulated in sigma-infected by Tsai et al. [20], but not in our experiments. As an endogenous control, we also measured the expression of RpL32 (Rp49).
To check the infection status of our flies and estimate their viral loads we also amplified viral genomic RNA by qPCR. The primers were designed to amplify a fragment spanning the sigma N and P genes to ensure that they amplified genomic RNA rather than mRNA. To allow our data on data to be compared to the results of Tsai et al. [20], we used Act88F as an endogenous control.
For the qPCR, 1.1 µg of total RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) and random hexamer primers. The resulting cDNA was used at 1∶10 dilution for qPCR using TaqMan probes and a 7500 Fast Real-Time PCR System (Applied Biosciences, Foster City, CA, USA). Pre-designed probe IDs were as follows: PGRP-SC2: Dm01818611_s1, Tudor-SN: Dm01834411_g1, Attacin-A: Dm02362218_s1, Drosocin: Dm01821449_s1, RpL32: Dm02151827_g1 and Act88F: Dm02362815_s1. Probes for quantifying viral loads were designed using the Custom TaqMan Assay Design Tool provided by Applied Biosciences. The region included in this assay corresponds to positions 3127 to 3239 of the AP30 isolate sequence (EMBL Accession AM689309) with the following primer and probe sequences: 5′-GCTCACAGTGAAGATCCATTACATG-3′ (forward), 5′- GCGGCTTCACAGAGAATTTGTC-3′ (reverse) and 5′- ACGAGATCTTAGTCAGCACCCT-3′ (probe). Seven replicate assays were performed for each of the 4 treatments: male, female, infected and uninfected and the threshold cycle value (Ct) was averaged across these replicates.
Results
Data quality and identification of differentially expressed genes
In total we performed 10 microarray hybridizations on males and 8 on females.
After removing probes that had no signal in either the sigma-infected or -uninfected flies, we were left with 4301 probes representing 3923 genes in females, and 5532 probes representing 4996 genes in males. The complete list of these genes is given in Supporting File S1. The higher number of genes detected as expressed in males may be explained by sex-related differences in gene expression, as we detected expression of a higher proportion of sex-biased genes in males than in females (see Discussion).
To assess the statistical power of the experiment, we calculated the GEL50 statistic, which is the fold-change in gene expression at which we have a 50% chance of detecting a difference (P<0.05) between the infected and uninfected flies. The GEL50 was 1.37 for the males and 1.52 for the females, which is similar to other microarray studies [22].
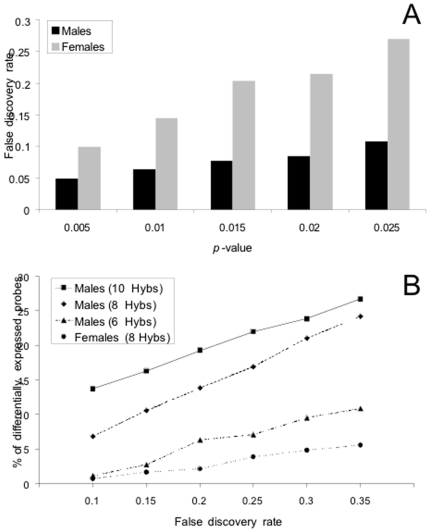
In order to define a significance threshold we calculated the FDRs for both the male and female datasets at different P-values (Figure 1a). We used a cut-off of P<0.02 for the male dataset, which corresponds to an FDR of 8.6%. Using this threshold we found that a total of 629 genes showed expression differences between sigma-infected and –uninfected flies (in infected flies 293 genes were up-regulated and 336 genes were down-regulated). Adjusting the P-value to produce a comparable FDR for the female dataset resulted in a very short list of only ∼30 differentially expressed genes. For further analysis we therefore decided to also use a cut-off of P<0.02 for females. This corresponds to an FDR of 21% (Figure 1a). At this threshold, we detected 134 differentially expressed genes (in infected flies 46 genes were up-regulated and 88 genes down-regulated). The excess of down-regulated genes in females was statistically significant (χ2-test, P = 0.0003).
Figure 1. Comparative statistical analysis of the male and female experiments.
(A) False discovery rates corresponding to several P-value cut-offs for both experiments. (B) Proportion of probes detected as differentially expressed at different false discovery rates. For the male experiment hybridizations which showed the best quality (defined as the number of spots with significant signal above background) were successively removed in order to determine the effect of replication on the detection of expression differences.
We found that the transcriptional response to sigma infection was very different between males and females. Of the 2862 genes detected as expressed in both males and females, only 41 showed a significant difference in expression in both sexes, and of these, 35 showed a consistent pattern of up- or down-regulation in both sexes (Table 1). Overall the magnitude of these differences was small; for both males and females, the maximum difference in expression was 2.5-fold (Figure 2) and the median difference was 1.28. Despite this, there seems to be a striking difference between males and females in the number of genes affected by infection. In order to investigate if this was due to differences between our replicates we removed the replicate hybridizations with the best quality signal from the male dataset and repeated the calculations. Removing the four best-quality hybridizations lowered the detection sensitivity to below that of the female dataset, and yet, males still showed consistently greater differences in expression (Figure 1b, see Discussion).
Table 1. Genes that are either up-regulated or down-regulated in both male and female flies infected with the sigma virus.
| Change in expression | Gene name |
| down | CG9350 |
| down | Ribosomal protein LP1 |
| down | CG6020 (NADH dehydrogenase) |
| down | CG2875 |
| down | CG1648 |
| down | Serine protease 6 |
| down | CG18594 |
| down | Defensin |
| down | CG12231 |
| down | CG8311 (dolichol kinase) |
| down | Ribosomal protein L13A |
| down | CG7675 |
| down | CG9572 |
| down | Antigen 5-related |
| down | tre oncogene-related protein |
| down | CG18179 (serine protease) |
| down | fau |
| down | CG4000 |
| down | CG1304 (serine protease) |
| down | Ard1 |
| down | CG9140 |
| down | yippee interacting protein 7 |
| down | CG7470 (glutamate 5-kinase) |
| down | PGRP-SC2 |
| down | CG10472 (serine protease) |
| down | Cytochrome P450-6a2 |
| down | CG17108 (acetyl-CoA carboxylase) |
| down | CG3088 (serine protease) |
| down | CG12736 (GTPase) |
| down | CG12057 |
| down | CG11314 |
| down | CG8343 (mannose binding) |
| up | rotund |
| up | Decondensation factor 31 |
| up | Actin 5C |
The molecular function of unnamed genes is given in brackets.
Figure 2. Volcano plots of the (A) male and (B) female experiments.
The X-axis defines the magnitude of expression difference between the infected and uninfected state, the Y-axis the corresponding P-value of the BAGEL analysis. Probes for which BAGEL assigned a P-value of 0 (i.e., P<0.0001), were set to P = 0.0001. Black dots represent probes up-regulated (log2 infected/uninfected>0) or down-regulated (log2 infected/uninfected<0) in the infected state at P<0.02.
Expression of known immunity genes
Previous studies have shown that a large number of genes are induced when flies are infected with bacteria or fungi, and that many of these genes are under the control of the Toll and IMD immune signaling pathways. To investigate whether these pathways might be activated in response to sigma virus infection, we have compared our results to previous studies that examined the fly's immune response to different pathogens. First, we compared data from a previous microarray study, which examined gene expression in flies infected with bacteria and fungi and identified some 400 up- or down-regulated genes [28], to our list of genes that showed a significant change in regulated in response to sigma-infection. From this comparison, it is clear that there is little overlap between the transcriptional response to bacteria and fungal infections and that of the sigma virus (Table 2). Of the genes that did overlap between the two studies, seven genes were up-regulated in both cases (Table 2). These include the antimicrobial peptide Metchnikowin, a translational regulator that is important in immune defense (Thor) and five other genes (CG13323, CG10912, CG16743, CG9928, CG15293). Seventeen genes were down-regulated in both studies including four Jonah proteases (Jonah 25Bii, Jonah 25Biii, Jonah 65Ai and Jonah 25Bi), two related calcium binding proteins that play an important role in many cellular processes (regucalcin and Smp-30) and 11 other genes (fit, CG18594, CG18179, CG12813, CG9090, CG4019, CG13947, CG7322, CG9672, CG9914 and CG3699).
Table 2. The effect of the sigma virus compared to bacterial and fungal infection on gene expression.
| Sigma virus infected flies | Bacteria and fungus infected males | |||
| Sex | Change expression | Up-regulateda | Down-regulateda | No change |
| Female | Up-regulatedb | 2 | 0 | 38 |
| Down-regulatedb | 4 | 7 | 67 | |
| No change | 77 | 69 | 3264 | |
| Male | Up-regulatedb | 5 | 5 | 230 |
| Down-regulatedb | 13 | 12 | 277 | |
| No change | 89 | 81 | 3743 | |
| Combined | Up-regulatedb | 7 | 5 | 266 |
| Down-regulatedb | 14 | 17 | 319 | |
| No change | 93 | 80 | 4447 | |
Only genes included in both datasets are shown. There is no significant association between the two datasets in any of the three comparisons (Fisher Exact tests on 2×2 contingency tables of genes showing a significant change in expression).
In the list of 400 ‘Drosophila Immune Related Genes’ identified by De Gregorio [28].
Significant at P<0.02.
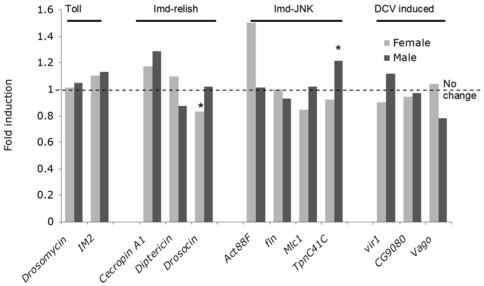
Next, we investigated whether sigma activates specific immune signaling pathways (Figure 3). Initially, we investigated how the expression of immunity genes that are known to be induced by either the Toll or IMD pathway changes in response to sigma infection. These genes were also selected to overlap with Figure 1 of Dostert et al. [11], who performed a similar analysis to this study, but investigated changes in gene expression in response to DCV-infection. The genes we selected fall into three groups—genes involved in the Toll pathway, the IMD pathway and the Jnk pathway. We looked at the regulation of IM2 and Drosomycin that are up-regulated by the Toll pathway [29], [30], and three antimicrobial peptides - Cecropin A1, Diptericin and Drosocin – that are up-regulated by the transcription factor Relish within the IMD pathway [29]. It should be noted that some of these antimicrobial peptides might also be under the control of the Toll pathway [31]. Finally, we looked at Act88F, fln, Mlc1 and TpnC41C, which are up-regulated by the Jnk pathway [29]. It is clear from Figure 3 that there is no evidence that any of these groups of genes have been up-regulated in response to sigma-infection. In a similar analysis, we compared our data to lists of genes under the control of the Toll and IMD-Relish pathways that were identified by De Gregorio et al. [28], and again there is no evidence that these pathways are activated (Table 3). We confirmed that there was no significant difference in the expression of Attacin A, Drosocin or Act88F by qPCR (Table 4).
Figure 3. The change in gene expression of genes controlled by known immune pathways in sigma virus infected flies relative to controls.
Genes showing a significant change (0.02<P<0.05) are labeled*.
Table 3. The effect of the sigma virus on the expression of genes controlled by the Toll and IMD pathways (identified by [31]).
| Sigma virus infected fliesa | Bacteria and fungus infected males | |||||||
| Up-regulated | Down-regulated | |||||||
| IMD | Toll | IMD+Toll | Neither | IMD | Toll | IMD+Toll | Neither | |
| Up-regulatedb | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Down-regulatedb | 0 | 0 | 3 | 1 | 2 | 2 | 1 | 3 |
| No change | 7 | 15 | 13 | 3 | 3 | 11 | 2 | 8 |
Male and female data combined, similar results were obtained using only male data.
Significant at P<0.02.
Table 4. Results of qPCR experiments.
| Gene | Male | Female | ||||
| Infecteda | Uninfecteda | Pb | Infecteda | Uninfecteda | Pb | |
| Attacin-A | 5.51 (1.50) | 5.45 (1.49) | 0.949 | 5.26 (1.73) | 4.52 (1.07) | 0.088 |
| Drosocin | 14.05 (1.60) | 13.65 (2.11) | 0.848 | 14.20 (1.96) | 13.05 (0.93) | 0.180 |
| PGRP-SC2 | 4.19 (0.45) | 4.08 (0.56) | 0.949 | 4.40 (0.33) | 3.79 (0.42) | 0.011 |
| Tudor-SN | 5.99 (0.78) | 6.18 (0.80) | 0.482 | 7.06 (1.03) | 7.61 (0.54) | 0.338 |
| Act88F | 7.82 (0.86) | 7.86 (0.83) | 0.610 | 10.46 (1.25) | 10.16 (1.04) | 0.522 |
Shown are the mean ΔCt values (standard deviation) relative to the control ribosomal protein gene, RpL32, for seven biological replicates of each gene/treatment.
Two-tailed P-value from Mann-Whitney test.
There has only been one published study on the transcriptional response of Drosophila to sigma virus infection, which used qPCR to measure the expression of a selection of immunity genes [20]. This study found that four antimicrobial peptides and two PGRPs were up-regulated in infected flies. In our study none of these genes were significantly up-regulated in infected flies, despite all six being detected as expressed in both males and females. For two of the genes (Drosocin and Attacin A), we confirmed this result using qPCR on both the male and female samples (Table 4). The difference between our results and those of Tsai et al. [20] is not due to our flies having lower viral titers, as when we measured the copy number of sigma virus genomic RNA relative to Act88F by qPCR, we found that our flies had higher viral titers than in this previous study (males: 3.62±1.20; females: 8.14±1.32; average ratio in Tsai et al.: 2.3±0.76).
Finally, we compared our data with a microarray analysis of DCV-infected flies (Dostert et al., 2005[32]; Jean-Luc Imler, personal communication). Few genes were up-regulated by both DCV and the sigma virus—of the 85 genes that were significantly up-regulated in response to DCV infection, only nine were up-regulated in response to the sigma virus, and seven were instead down-regulated in sigma-infected flies. There was a greater overlap in the genes that were down-regulated in both DCV and sigma-infected flies—of the 200 genes that were significantly down-regulated in response to DCV infection, 30 were down-regulated in response to the sigma virus, and 11 were up-regulated in sigma-infected flies. Overall, the association between the two studies is marginally non-significant (Fisher Exact Test on 2×2 contingency table of differentially expressed genes: P = 0.06).
There were however a few notable immune-related genes that were differentially expressed. The genes PGRP-SC2, which is important in dampening the immune response [33], was one of the few genes to be down-regulated in both our male and female datasets (females: ratio infected/uninfected = 0.67, P = 0.005; males: ratio = 0.81, P = 0.015). We replicated this result using qPCR (females: ratio = 0.66, P = 0.011; males: ratio = 0.93, P = 0.95). Similarly, Tudor-SN, which is involved in RNAi, was down regulated in females, but this was not repeatable using qPCR (Table 4).
Up- and down-regulated genes
To investigate which biological processes are affected by sigma virus infection, we identified gene ontology (GO) terms that were overrepresented among our up- and down-regulated genes (Table 5). A selection of the genes with these GO terms is listed in Table 6. Among the genes that were down-regulated in sigma-infected males, ribosomal proteins were overrepresented (29 of the 93 genes in one of the two ribosomal subunits). Virtually all of the other GO categories overrepresented among the genes down-regulated in infected males are related to mitochondria. In sigma-infected females, serine proteases were overrepresented among the down-regulated genes, including seven genes from the chymotrypsin superfamily, and six from the Jonah family (note that these categories overlap in the GO annotations). Six chymotrypsin genes were also down-regulated in infected males, including three of the same genes as were detected in infected females. Among the genes that were up-regulated in infected males, genes related to DNA binding and regulating transcription were overrepresented. While in infected females, chorion structural proteins are overrepresented, with three of the nine genes in the genome being significantly up-regulated. Furthermore, three additional chorion proteins (CP18, CP19 and CP36) showed up-regulation with P-values very close to our detection threshold (P = 0.023, P = 0.024 and P = 0.039 respectively).
Table 5. Overrepresented GO terms.
| P-value | Genes in Genome | Differentially expressed | Domaina | GO Termb |
| (a) Down-regulated in females (88 genes) | ||||
| 4.1E-10 | 184 | 13 | MF | serine-type endopeptidase activity |
| 1.1E-10 | 22 | 7 | MF | chymotrypsin activity |
| (b) Up-regulated in females (46 genes) | ||||
| 2.6E-06 | 9 | 3 | MF | structural constituent of chorion |
| (c) Down-regulated in males (336 genes) | ||||
| 6.5E-11 | 377 | 35 | BP | translation |
| 2.0E-11 | 773 | 54 | BP | cellular biosynthetic process |
| 5.9E-11 | 337 | 33 | BP | electron transport |
| 1.6E-06 | 130 | 15 | BP | monocarboxylic acid metabolic process |
| 1.3E-06 | 15 | 6 | BP | pyruvate metabolic process |
| 2.2E-15 | 74 | 20 | BP | oxidation reduction |
| 1.6E-10 | 941 | 59 | BP | biosynthetic process |
| 1.8E-06 | 489 | 32 | BP | phosphorylation |
| 1.6E-11 | 458 | 40 | BP | generation of precursor metabolites and energy |
| 2.2E-15 | 74 | 20 | BP | respiratory electron transport chain |
| 3.2E-14 | 155 | 26 | BP | oxidative phosphorylation |
| 9.2E-16 | 71 | 20 | BP | organelle ATP synthesis coupled electron transport |
| 5.6E-09 | 41 | 11 | BP | mitochondrial electron transport |
| 1.9E-12 | 333 | 35 | CC | ribonucleoprotein complex |
| 2.9E-17 | 165 | 30 | CC | ribosomal subunit |
| 3.7E-12 | 53 | 15 | CC | cytosolic large ribosomal subunit |
| 7.3E-13 | 40 | 14 | CC | cytosolic small ribosomal subunit |
| 8.3E-18 | 224 | 35 | CC | cytosol |
| 2.5E-14 | 128 | 24 | CC | lipid particle |
| 5.7E-17 | 491 | 50 | CC | mitochondrion |
| 2.1E-13 | 400 | 40 | CC | organelle membrane |
| 5.4E-16 | 370 | 42 | CC | mitochondrial part |
| 2.1E-14 | 223 | 31 | CC | mitochondrial envelope |
| 6.1E-14 | 187 | 28 | CC | mitochondrial inner membrane |
| 1.1E-15 | 81 | 21 | CC | mitochondrial respiratory chain |
| 2.8E-16 | 192 | 31 | MF | structural constituent of ribosome |
| 3.7E-14 | 705 | 56 | MF | oxidoreductase activity |
| 1.3E-08 | 235 | 24 | MF | electron carrier activity |
| 1.3E-08 | 44 | 11 | MF | NADH dehydrogenase activity |
| (d) Up-regulated in males (293 genes) | ||||
| 8.47E-06 | 239 | 18 | MF | sequence-specific DNA binding |
| 2.97E-07 | 395 | 27 | MF | transcription factor activity |
| 5.28E-08 | 938 | 48 | BP | regulation of gene expression |
| 7.48E-06 | 790 | 38 | BP | regulation of transcription, DNA-dependent |
| 8.84E-07 | 998 | 47 | BP | organ development |
| 1.56E-06 | 861 | 42 | BP | cell development |
| 1.19E-06 | 128 | 14 | CC | lipid particle |
BP: biological process; CC: cellular compartment; MF: molecular function.
Only GO categories overrepresented with P<10−6 are shown. GO categories that contain over 1000 genes in the genome and categories that were wholly or largely redundant with another category are not shown.
Table 6. Significantly up- and down-regulated genes with selected functions.
| Males | Females | |||||||
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | |||||
| Serine endopeptidases | ||||||||
| CG11037 | Serine protease 6 | - | Serine protease 6 | |||||
| Starving | yippee interacting protein 7 a | yippee interacting protein 7 a | ||||||
| TER94 | epsilonTrypsin | CG17571 | Jonah 25Bii a | CG10472a | ||||
| Tripeptidyl-peptidase II | Jonah 25Bi | CG18179 a | Jonah 25Biii | CG11911 a | ||||
| CG10472 a | CG3088 | Jonah 65Ai | CG1304 | |||||
| CG1304 | CG7542 a | Jonah 65Aiii | CG18179 a | |||||
| CG16996 a | CG9672 | Jonah 74E a | CG3088 | |||||
| CG16997 a | CG9673 | Jonah 99Ci | CG7829 | |||||
| CG8329 a | ||||||||
| Ribosomal proteins | ||||||||
| Ribosomal proteins: | string of pearls | - | Ribosomal proteins: | |||||
| L29 | stubarista | L10Ab | ||||||
| L3 | CG6764 | L13A | ||||||
| overgrown hematopoietic organs 23B | L18A | |||||||
| Ribosomal proteins: | LP1 | |||||||
| L12 | L30 | S10b | S19a | |||||
| L13 | L34b | S11 | S25 | |||||
| L13A | L7 | S14a | S5a | |||||
| L14 | L8 | S14b | S8 | |||||
| L19 | LP0 | S15Aa | S9 | |||||
| L23 | LP1 | S16 | ||||||
| L23A | LP2 | S18 | ||||||
| Transcription factors | ||||||||
| Hairy/E(spl)-related with YRPW motif | CG11876 | SoxNeuro | - | |||||
| optomotor-blind-related-gene-1 | Deformed | rotund | ||||||
| Zn finger homeodomain 1 | CG33097 | |||||||
| doublesex-Mab related 99B | bicaudal | |||||||
| Ecdysone-induced prot 74EF | ||||||||
| ftz transcription factor 1 | ||||||||
| luna | CG4136 | |||||||
| pannier | bunched | |||||||
| rotund | midline | |||||||
| homeobrain | bric a brac 1 | |||||||
| CG15455 | abdominal A | |||||||
| labial | caupolican | |||||||
| Mnf | POU domain prot 2 | |||||||
| squeeze | forkhead domain 3F | |||||||
| scribbler | ventral veins lacking | |||||||
| Chorion Proteins | ||||||||
| - | - | Chorion prot 38 | - | |||||
| Chorion prot 15 | ||||||||
| Chorion prot 16 | ||||||||
chymotrypsin family.
Discussion
Immune pathway activation
When Drosophila is infected by bacteria or fungi, the Toll and IMD pathways are activated, leading to the up-regulation of large numbers of genes. These genes include immune effectors such as antimicrobial peptides that are secreted into the hemolymph and defend the flies against the invading pathogens. It is currently unclear whether there is a comparable induced immune defense against viruses. We found that neither genes up-regulated by bacterial or fungal infection, nor the subset of these controlled by the Toll and IMD pathways, are induced in sigma-infected flies. As these pathways control the majority of the genes up-regulated by fungal and bacteria infections [31], any induced immune response to the sigma virus must be controlled by distinct regulatory mechanisms. And although it has been reported that DXV activates the Toll pathway [13] and sigma-virus activates the IMD pathway [20], our results are more similar to with microarray analyses of DCV that found no evidence for the Toll or IMD pathways being activated [11].
An additional pathway implicated in viral infection in Drosophila is the Jak-STAT pathway—genes within the Jak-STAT pathway have been shown to be up-regulated in response to DCV infection, and flies deficient in this pathway are more susceptible to DCV. However, we found that there is little overlap between the genes induced by DCV and the sigma virus, suggesting that the Jak-STAT pathway is not activated by sigma virus. Therefore, we conclude that it is unlikely that Drosophila mounts a general immune response to all viruses.
Why is there little overlap between the genes induced by sigma virus and DCV? One possibility is that DCV, unlike the sigma virus, causes cells lysis and it is this rupturing of cells, and the ensuing tissue damage, that induces an immune response [3]. This hypothesis is consistent with the observation that when flies are fed DCV that results in a relatively benign infection [4] and far fewer genes are induced compared to when flies are injected with the virus [11], [12]. Alternatively, the sigma virus may avoid recognition by the immune system for other reasons. As the sigma virus is only transmitted vertically, it must establish a persistent infection and therefore it must avoid being cleared by the immune response. Avoiding inducing an immune response may be essential for persistent vertically transmitted infections, as the vertically transmitted bacteria Spiroplasma poulsonii and Wolbachia do not induce an immune response in D. melanogaster either [34], [35]. Vertically transmitted infections will also be selected to minimize the harm that the cause to the host, as they rely on their host surviving and reproducing to be transmitted. This may also select for viruses that do not induce a costly transcriptional response in their host.
A previous study by Tsai et al. [20] found that four antimicrobial peptides and two PGRPs were strongly induced by the sigma virus. Despite our flies having a higher viral titer, none of these genes were induced in our study. Therefore, the apparent immune response observed by Tsai et al. is not always induce by sigma virus infection. The reasons why the results of the two studies differ are unclear. It is possible that only certain fly or viral genotypes induce an immune response, and it is known that fly lines differ greatly in their resistance to the sigma virus [19], at it is not known how the flies in the two studies differ in their resistance genes. Alternatively, the changes in gene expression observed by Tsai et al. may not be directly caused by viral infection (for example the sigma infected flies may be more prone to secondary bacterial infections).
Up- and down-regulated genes
What genes changed in expression in sigma infected flies? In males, the down-regulated genes were strongly enriched for proteins involved in translation, especially ribosomal proteins. Viruses rely on the host's translational machinery to produce proteins, and many viruses—including related virus vesicular stomatitis virus (Rhabdoviridae)—both inhibit host translation and cause viral mRNAs to be preferentially translated over host mRNAs [36]. Furthermore, depleting ribosomal proteins inhibits the replication of DCV in Drosophila cells [37]. The down-regulation of genes involved in translation by the sigma virus probably reflects this interaction, but the biological importance is unclear.
We found that serine proteases were down-regulated in infected flies. Serine proteases have a wide range of functions, including key roles in the regulation of the immune system. The Toll and IMD responses to bacteria and fungi induce and repress a number of serine proteases, some of which have important roles in immune regulation [31], [38]. In females there was an overrepresentation of serine proteases that were down-regulated. Of particular note was an excess of chymotrypsin superfamily serine protease, and several Jonah serine proteases. Several of these genes were also down-regulated in sigma-infected males, making these genes interesting candidates for controlling the fly's response to viruses.
We found that in infected females, genes encoding for chorion proteins were up-regulated. During oogenesis, the chromosomal copies of these genes are amplified up to 10 times, allowing high levels of gene expression [39]. The sigma virus is transmitted through the fly's eggs and it is possible that this relates to the up-regulation of these genes.
Sex differences in gene expression
Sigma virus infection appears to alter the expression of many more genes in males than in females. For example, with an FDR of 10% we detect over 10 times as many significant genes in males as in females (Figure 1b). Several factors could contribute to this difference. First, the two experiments differ slightly in their replication schemes (10 arrays for males versus 8 arrays for females) and statistical power (GEL50 = 1.37 for males and 1.52 for females). Thus, we have more power to detect expression differences in the male experiment. This alone, however, cannot explain the large discrepancy in the number of differentially expressed genes. If we exclude two (GEL50 = 1.43) or even four (GEL50 = 1.57) of the male replicates, we still detect a larger fraction of significant genes in the male experiment (Figure 1b). A second factor could be sex-biased gene expression, as the genes that we detected differ between the two experiments. Using the sex-bias classifications of Gnad and Parsch [40], we find that 59% of the male-biased genes on the array are detected in males, while only 16% are detected in females. In contrast, 43% of female-biased genes on the array are detected in females, while only 28% are detected in males. Sex-biased genes, however, are not over-represented among those whose expression was significantly altered by the sigma virus. Male-biased genes comprise 18% of the genes detected in males, but only 14% of the genes significantly affected by sigma infection. Similarly, female-biased genes comprise 18% of the genes detected in females, but only 11% of the genes significantly affected by the sigma infection.
It is also possible that the difference between males and females results from intersexual differences in the mechanism of viral transmission or host defense. Sigma virus is transmitted along with sperm and there appear to be specific barriers to it entering the male germline [4]. Furthermore, genes that cause variation in the transmission of the virus often affect transmission through sperm, but not eggs [19]. This may reflect a qualitative difference in the nature of the infection in males, or indicate that specific resistance responses are triggered in the male germline, leading to a greater transcriptional response to infection. Alternatively, males and females may invest differently in their immune defenses [41], and this may be reflected in sex differences in the transcriptional response to sigma infection.
A final possibility is that, in general, male gene expression is more sensitive to genetic and/or environmental perturbations than female gene expression. Some support for this comes from the observation that, among laboratory strains, greater gene expression variation is observed among males than among females [42]. However, more experimental comparisons of male and female transcriptional responses to a common treatment are necessary to determine the generality of this observation.
Supporting Information
The relative expression level, 95% confidence limits of expression levels in males and females, and probability of differential expression of each gene between infected and uninfected males
(1.28 MB XLS)
The relative expression level, 95% confidence limits of expression levels in males and females, and probability of differential expression of each gene between infected and uninfected females
(1.00 MB XLS)
The list of significantly differentially expressed genes (P<0.02) in which the CG numbers have been updated to match v49 of ENSEMBL
(2.20 MB XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a Wellcome Trust Grant (WT081279, http://www.wellcome.ac.uk/) and a Royal Society University Research Fellowship (NA http://royalsociety.org/) to FJ and by a Deutsche Forschungsgemeinschaft grant (PA 903/3, http://www.dfg.de/en/) to JP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, et al. A Metagenomic Survey of Microbes in Honey Bee Colony Collapse Disorder. 10.1126/science.1146498. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 2.Jesús Genaro Sánchez-Martínez, Gabriel Aguirre-Guzmán, Humberto Mejía-Ruíz. White Spot Syndrome Virus in cultured shrimp: A review. Aquaculture Research. 2007;38:1339–1354. [Google Scholar]
- 3.Lemaitre B, Hoffmann J. The Host Defense of Drosophila melanogaster. Annual Review of Immunology. 2007;25 doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 4.Brun G, Plus N. The viruses of Drosophila. In: Ashburner M, Wright TRF, editors. The genetics and biology of Drosophila. New York: Academic Press; 1980. pp. 625–702. [Google Scholar]
- 5.Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nature Immunology. 2004;5:81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ammar E-D, Tsai C-W, Whitfield AE, Redinbaugh MG, Hogenhout SA. Cellular and Molecular Aspects of Rhabdovirus Interactions with Insect and Plant Hosts*. doi:10.1146/annurev.ento.54.110807.090454. Annual Review of Entomology. 2009;54:447–468. doi: 10.1146/annurev.ento.54.110807.090454. [DOI] [PubMed] [Google Scholar]
- 7.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses 10.1128/JVI.80.10.5059-5064.2006. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nature Immunology. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 9.Wang XH, Aliyari R, Li WX, Li HW, Kim K, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nature Immunology. 2005;6:946. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 12.Roxstrom-Lindquist K, Terenius O, Faye I. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 2004;5:207–212. doi: 10.1038/sj.embor.7400073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. PNAS. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avila A, Silverman N, Diaz-Meco MT, Moscat J. The Drosophila Atypical Protein Kinase C-Ref(2)P Complex Constitutes a Conserved Module for Signaling in the Toll Pathway. Mol Cell Biol. 2002;22:8787–8795. doi: 10.1128/MCB.22.24.8787-8795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contamine D, Petitjean AM, Ashburner M. Genetic-Resistance to Viral-Infection - the Molecular-Cloning of a Drosophila Gene That Restricts Infection by the Rhabdovirus-Sigma. Genetics. 1989;123:525–533. doi: 10.1093/genetics/123.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter JA, Obbard DJ, Maside X, Jiggins FM. The recent spread of a vertically transmitted virus through populations of Drosophila melanogaster. Molecular Ecology. 2007;16:3947–3954. doi: 10.1111/j.1365-294X.2007.03460.x. [DOI] [PubMed] [Google Scholar]
- 17.Fleuriet A. Maintenance Of A Hereditary Virus - The Sigma-Virus In Populations Of Its Host, Drosophila-Melanogaster. Evolutionary Biology. 1988;23:1–30. [Google Scholar]
- 18.Bangham J, Knott SA, Kim KW, Young RS, Jiggins FM. Genetic variation affecting host-parasite interactions: major-effect quantitative trait loci affect the transmission of sigma virus in Drosophila melanogaster. Molecular Ecology. 2008;17:3800–3807. doi: 10.1111/j.1365-294X.2008.03873.x. [DOI] [PubMed] [Google Scholar]
- 19.Bangham J, Kim KW, Webster CL, Jiggins FM. Genetic variation affecting host-parasite interactions: Different genes affect different aspects of sigma virus replication and transmission in Drosophila melanogaster. Genetics. 2008;178:2191–2199. doi: 10.1534/genetics.107.085449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai CW, McGraw EA, Ammar E-D, Dietzgen RG, Hogenhout SA. Drosophila melanogaster Mounts a Unique Immune Response to the Rhabdovirus Sigma virus. Appl Environ Microbiol. 2008;74:3251–3256. doi: 10.1128/AEM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis EB. A new standard food medium. Dros Inf Serv. 1960;34:117–118. [Google Scholar]
- 22.Hutter S, Saminadin-Peter SS, Stephan W, Parsch J. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biology. 2008;9 doi: 10.1186/gb-2008-9-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Research. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Townsend J, Hartl D. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biology 3: 2002:research0071.0071–research0071.0016. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Townsend JP. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. Bmc Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g: Profiler - a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Research. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, et al. Ensembl 2008. Nucleic Acids Research. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boutros M, Agaisse H, Perrimon N. Sequential Activation of Signaling Pathways during Innate Immune Responses in Drosophila. Developmental Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 30.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann JA. The Dorsoventral Regulatory Gene Cassette spatzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 31.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyers F, Petitjean AM, Dru P, Gay P, Contamine D. Localization of domains within the Drosophila Ref(2)P protein involved in the intracellular control of sigma rhabdovirus multiplication. J Virol. 1995;69:4463–4470. doi: 10.1128/jvi.69.7.4463-4470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. Plos Pathogens. 2006;2:139–147. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurst GDD, Anbutsu H, Kutsukake M, Fukatsu T. Hidden from the host: Spiroplasma bacteria infecting Drosophila do not cause an immune response, but are suppressed by ectopic immune activation. Insect Molecular Biology. 2003;12:93–97. doi: 10.1046/j.1365-2583.2003.00380.x. [DOI] [PubMed] [Google Scholar]
- 35.Bourtzis K, Pettigrew MM, O'Neill SL. Wolbachia neither surpresses nor induces antimicrobial peptides. Insect Molecular Biology. 2000;9:635–639. doi: 10.1046/j.1365-2583.2000.00224.x. [DOI] [PubMed] [Google Scholar]
- 36.Whitlow ZW, Connor JH, Lyles DS. Preferential translation of vesicular stomatitis virus mRNAs is conferred by transcription from the viral genome. Journal of Virology. 2006;80:11733–11742. doi: 10.1128/JVI.00971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kambris Z, Brun S, Jang I-H, Nam H-J, Romeo Y, et al. Drosophila Immunity: A Large-Scale In Vivo RNAi Screen Identifies Five Serine Proteases Required for Toll Activation. Current Biology. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Spradling AC, Mahowald AP. Amplification of Genes for Chorion Proteins During Oogenesis in Drosophila-Melanogaster. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1980;77:1096–1100. doi: 10.1073/pnas.77.2.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- 41.McKean KA, Nunney L. Bateman's principle and immunity: Phenotypically plastic reproductive strategies predict changes in immunological sex differences. Evolution. 2005;59:1510–1517. [PubMed] [Google Scholar]
- 42.Baker DA, Meadows LA, Wang J, Dow JA, Russell S. Variable sexually dimorphic gene expression in laboratory strains of Drosophila melanogaster. Bmc Genomics. 2007;8 doi: 10.1186/1471-2164-8-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relative expression level, 95% confidence limits of expression levels in males and females, and probability of differential expression of each gene between infected and uninfected males
(1.28 MB XLS)
The relative expression level, 95% confidence limits of expression levels in males and females, and probability of differential expression of each gene between infected and uninfected females
(1.00 MB XLS)
The list of significantly differentially expressed genes (P<0.02) in which the CG numbers have been updated to match v49 of ENSEMBL
(2.20 MB XLS)