Abstract
Biosynthesis of biosurfactant rhamnolipids by Pseudomonas aeruginosa depends on two hierarchical quorum sensing systems, LasRI and RhlRI, which synthesize and sense the signal molecules N-(3-oxododecanoyl)-L-homoserine lactone (3OC12-HSL) and N-butyryl-L-homoserine lactone (C4-HSL), respectively. The Pseudomonas Quinolone Signal (PQS) is a third cell-to-cell signal molecule connecting these two systems, and its precursor, 2-heptyl-4-quinolone (HHQ), also constitutes a signal. The chronology of the production of signal molecules and rhamnolipids was determined during growth in PPGAS medium. Hyperosmotic condition (0.5 M NaCl) moderately affected growth, and led to intra-cellular accumulation of compatible solutes. Production of signal molecules was delayed and their highest concentrations were 2.5 to 5 fold lower than in NaCl-free PPGAS, except for HHQ, the highest concentration of which was increased. The presence of NaCl prevented rhamnolipid synthesis. When the osmoprotectant glycine betaine was added to PPGAS/NaCl medium, it was imported by the cells without being metabolized. This did not improve growth, but reestablished the time-courses of HSL and HHQ accumulation and fully or partially restored the HSL and PQS levels. It also partially restored rhamnolipid production. Quantification of mRNAs encoding enzymes involved in HSL, PQS, and rhamnolipid biosyntheses confirmed the effect of hyperosmotic stress and glycine betaine at the gene expression level.
Keywords: Quorum sensing, Homoserine lactone, Pseudomonas Quinolone Signal, Rhamnolipid, Pseudomonas aeruginosa, Osmotic stress.
INTRODUCTION
Rhamnolipids are biosurfactants, ie amphipathic molecules reducing water surface tension and emulsifying oil, of glycolipidic nature. They are produced by the opportunistic pathogen bacterium Pseudomonas aeruginosa, which is in particular responsible for chronic lung infection of cystic fibrosis patients [1], and rhamnolipids are considered as virulence factors with multiple biological activities [2, 3]. Many other functions are attributed to rhamnolipids, including anti-microbial activities and biofilm maintenance, as reviewed by Soberón-Chávez et al. [2]. Rhamnolipid production is coordinated with that of other virulence factors [4]: the rhlAB operon and the rhlC gene encoding enzymes involved in rhamnolipid biosynthesis are regulated by the RhlRI quorum sensing (QS) system [5-7]. RhlI synthesizes the cell-to-cell communication molecule N-butyryl-L-homoserine lactone (C4-HSL), and RhlR activates transcription of target genes after C4-HSL binding. The RhlRI system itself depends on the LasRI QS system, which synthesizes and binds N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) [8-10]. These two hierarchical QS systems are embedded into a large regulatory network, which includes in particular a third communication molecule, 2-heptyl-3-hydroxy-4-quinolone more commonly known as the Pseudomonas quinolone signal (PQS) [8-12]. PQS synthesis occurs via a condensation of anthranilate and β-keto dodecanoate and requires the genes pqsABCD (forming an operon with the pqsE gene, which encodes a regulator involved in the QS network but not in PQS synthesis [13]) and pqsH [11, 12, 14]. The latter encodes the enzyme responsible for the last step of PQS synthesis: the modification of 2-heptyl-4-quinolone (HHQ) in PQS [11, 12, 14]. Since it is both released from, and taken up by bacteria, HHQ is itself suggested to act as an intercellular message molecule [15]. LasR and RhlR control PQS synthesis [16], whereas PQS positively regulates the RhlRI system [16-19]. In addition to this indirect action of PQS on rhamnolipid production via the RhlRI system, two other relationships were established between PQS and rhamnolipids: the two molecules share the common precursors β-keto-(do)decanoic acids [20], and rhamnolipids increase PQS solubility and bioactivity [21]. PQS, but not HHQ, was recently shown to both sensitize bacteria towards exogenous stresses and trigger an efficient stress response, thus leading to consider PQS as internal stress response molecule [22].
The fact that QS systems are part of a large regulatory network leads to the view that P. aeruginosa QS is not only dependent on cell density, but also on a wide variety of environmental signals, most of which remain to identify. We observed that hyperosmotic stress is one of these signals [23]. High salinity conditions are frequently encountered in P. aeruginosa habitats, such as soils, marshes and marine coasts. Furthermore, high NaCl concentrations are found in respiratory tract fluids from cystic fibrosis patients [24]. Osmoadaptation is thus critical to P. aeruginosa survival in the environment, and might play a role in its pathogenicity. Bacteria generally cope with hyperosmotic conditions by accumulating low molecular mass molecules that are compatible with cellular processes at high internal concentrations [25]. These molecules, termed compatible solutes or osmoprotectants, are either synthesized by the bacteria or imported from the environment. Osmotically-stressed P. aeruginosa PAO1 was shown to synthesize and accumulate glutamate, trehalose and N-acetylglutaminylglutamine amide (NAGGN) in the absence of exogenous osmoprotectant, but preferentially accumulated glycine betaine (GB) when the latter was added to the culture medium [26]. GB is the preferred compatible solute for most of the bacteria [25], and its uptake by P. aeruginosa PAO1 prevented the accumulation of the three endogenous osmoprotectants [26]. GB can also be used by P. aeruginosa PAO1 as carbon and nitrogen sources [27], and we showed that GB was metabolized in M63 minimal medium when the carbon source was glucose, whereas it was stably accumulated in succinate M63 [28]. Regarding HSL and rhamnolipid production, we reported that a hyperosmotic stress (0.5 M NaCl) applied to exponentially-growing P. aeruginosa PAO1 interrupted 3OC12-HSL production and prevented C4-HSL and rhamnolipid syntheses [23]. These defects were explained by reduced levels of rhlI, rhlAB, and rhlC mRNAs. The addition of GB partially restored rhlI expression and C4-HSL production, as well as rhlC expression, thereby reestablishing rhamnolipid synthesis. However, the rhamnolipid production level remained low, likely because GB had only a marginal positive effect on rhlAB expression [23]. The osmotic stress studies are generally performed in minimal media since components of rich media could bring osmoprotectants in an uncontrolled fashion. However, the use of a M63-based minimal medium (PLM63: M63 limited in phosphate in order to obtain rhamnolipid production) did not allow to fully appreciate the hyperosmotic stress effect on 3OC12-HSL production since P. aeruginosa PAO1 failed to grow if NaCl was added before the synthesis onset of this communication molecule. To obtain a more complete picture of the effects of hyperosmotic conditions on P. aeruginosa QS and on rhamnolipid production, we used here the rich PPGAS medium, which favors rhamnolipid production [29] and we extended our field of investigation to PQS since this signal molecule is part of the QS network and constitutes an internal stress response signal. We examined here the time-course production of rhamnolipids, of both HSLs, and of PQS and its precursor HHQ by P. aeruginosa PAO1 in PPGAS with or without hyperosmotic stress. The expression levels of genes encoding enzymes responsible for biosynthesis of these molecules were then compared in the two growth conditions.
MATERIALS AND METHODOLOGY
Bacterial Strain and Culture Conditions
P. aeruginosa PAO1 (obtained from M. Foglino, Marseille, France) was grown in PPGAS medium (NH4Cl 20 mM; KCl 20 mM; Tris-HCl 120 mM; MgSO4 1.6 mM; glucose 0.5 %; tryptone 1%, adjusted to pH 7.2 [29]) at 37°C with shaking, and growth was followed by measuring optical density at 600 nm (OD600). Hyperosmotic conditions were obtained by including 0.5 M NaCl into the medium before inoculation. GB was used at a final concentration of 1 mM. When indicated, C4-HSL (Sigma-Aldrich Co., St. Louis, USA) and PQS (P. Williams, University of Nottingham, UK) were respectively added at final concentrations of 10 µM and of 2 mM.
Extraction and Analysis of HSLs, PQS, HHQ, and Rhamnolipids
HSLs were extracted from culture supernatants as described by Bazire et al. [23], and quantified by liquid chromatography-mass spectrometry (LC-MS-MS) according to Morin et al. [30]. PQS and HHQ were extracted as the HSLs, and analyzed as described by Lépine et al. [31]. LC separation conditions were optimized as follows. The initial acetonitrile concentration in water was linearly increased from 20% to 59% in 10 min, kept stable for an additional 13 min, increased to 98% in 1 min, kept stable during 6 min, and returned to the initial value in 1 min and left to stabilize for another 9 min. Compounds were detected by an ion trap spectrometer (Bruker Daltonics, Germany). Operating conditions of the MS source were capillary voltage, 4.0 kV; nebuliser pressure, 12 psi; drying gas flow, 7 L/min at 300°C. The mass spectrometer was run in full scan MS. The trap parameters were set in ion charge control with a target of 30 000 and a maximum accumulation time of 400 ms at m/z ranging from 50 to 400. Quantification of HHQ and PQS was performed by using external standard. Rhamnolipids were identified and quantified from culture supernatants using LC-MS as previously reported [23, 32]. The proportion of each rhamnolipid was obtained from the corresponding m/z [M-H]- chromatograms by measure of peak areas.
RNA Quantification by Quantitative Reverse Transcription-PCR (qRT-PCR)
RNAs were extracted using RNAprotect bacteria reagent (Qiagen, Hilden, Germany) and the Master PureTm RNA purification kit (Epicentre® Biotechnologies, Madison, USA). RNAs were converted to cDNAs using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, USA). mRNAs were quantified by real-time PCR amplification of their cDNAs with the 7300 Real Time PCR System apparatus and SYBR Green PCR Master Mix (Applied Biosystems), using previously described procedures [33]. The primer sequences used for 16S rRNA, and rhlAB, rhlC and rhlI mRNAs were given in [23], whereas the sequences of the other primers are in Table 1. PCR reactions were performed in triplicate and the standard deviations were lower than 0.15 Ct. The relative quantification of the mRNAs of interest was obtained by the comparative Ct (2-ΔΔCT) method [34], using 16S rRNA as endogenous control. ΔCt values were calculated by subtracting the 16S Ct value from the Ct value of an mRNA of interest from the same sample. ΔΔCt values were then obtained by calculating the difference between: i) the ΔCt value of a given mRNA resulting from cells grown in the presence of NaCl with or without GB, and ii) the ΔCt value of the same mRNA from cells grown in NaCl-free PPGAS. Relative mRNA level values resulted from calculating 2–ΔΔCT values: values above and below 1 show a higher and a lower mRNA level in the presence of NaCl, respectively (e.g. a value of 2 indicates a 2 fold increased mRNA level in hyperosmotic condition; and a value of 0.5 reveals that the mRNA level was divided by 2 in hyperosmotic condition).
Table 1.
qRT-PCR Primers for lasI, pqsC, pqsE, and pqsH Genes
| Primers | 5'-3' Sequences |
|---|---|
| LasI1 | TTCGCCATCAACTCTGGACA |
| LasI2 | CGTACAGTCGGAAAAGCCCA |
| PqsC1 | AGCGACGCTACGTTCTATGAAGT |
| PqsC2 | TGAACAACGAGAAATAAAGCCG |
| PqsE1 | CTGCAGGTCATAGAGGCCCA |
| PqsE2 | GTAGAAAACCACGTGATCGTCG |
| PqsH1 | AGGCGAACGAGGGTATTCCT |
| PqsH2 | TCAGTGGGAATCGCCCTG |
GB Assimilation Assays
The fate of GB was studied with [methyl-14C]GB (2.07 GBq.mmol-1), which was prepared from [methyl-14C]choline chloride (2.07 GBq.mmol-1) as described by Bernard et al. [35]. Cells were grown in PPGAS medium without or with 0.5 M NaCl, and supplemented with 1 mM [14C]GB (3.3 MBq.mmol-1). After two to five generations, 1 ml of cell culture was harvested by centrifugation (10,000 x g for 10 min). After extraction in 80% ethanol, the radioactive components of the ethanol-soluble fraction were analysed by paper chromatography and/or electrophoresis [35]. The radioactive spots were visualised using a Packard Instant Imager. The radioactivity of the ethanol-insoluble fraction and of CO2 trapped on filter paper soaked with 6 M KOH was also determined.
Natural Abundance 13C-nuclear Magnetic Resonance Spectroscopy
Freshly harvested and washed cells were extracted in 80 % ethanol under stirring at room temperature for 30 min. After centrifugation, the supernatant (ethanol-soluble fraction) was evaporated to dryness at 40°C, the dry residue was dissolved into D2O and the 13C NMR spectrum was recorded as previously described [35].
RESULTS
Time-courses of Rhamnolipid, HSL, HHQ, and PQS Production in PPGAS Medium
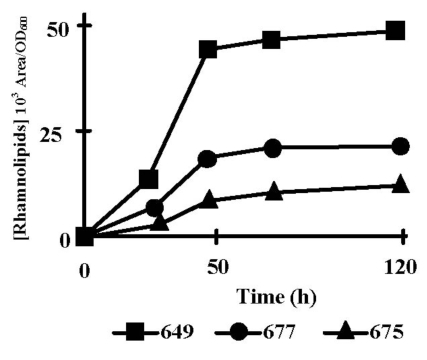
We first identified by LC-MS the rhamnolipid species which were produced by P. aeruginosa PAO1 grown at 37°C in PPGAS. Three mono-rhamnolipids and six dirhamnolipids were identified from supernatants of 48 h cultures with the following proportions: Rha-Rha-C10-C10, 63.67%; Rha-Rha-C10-C12, 19.39%; Rha-Rha-C10-C12:1, 9.33%. Rha-Rha-C10-C8, 2.39%; Rha-C10-C10, 2.31%; Rha-Rha-C12-C12, 1.16%; Rha-Rha-C12-C12:1, 0.94%; Rha-C10-C12, 0.28%. Rha-C10-C12:1, 0.27%. Like in PLM63 medium [23], the major species were, in decreasing order, the dirhamnolipids Rha-Rha-C10-C10, Rha-Rha-C10-C12, and Rha-Rha-C10-C12:1 corresponding to ions 649, 677, and 675, respectively. Their production occurred mainly during stationary phase, between 24 and 48 h of culture (Fig. 1). In the subsequent experiments, we only quantified the most abundant rhamnolipid (Rha-Rha-C10-C10) at 48 h of culture. Since transcription of rhamnolipid biosynthesis genes rhlAB and rhlC is dependent on the RhlRI QS system, which is itself controlled by the LasRI and PQS systems, we examined the time-course of extra-cellular accumulation of the corresponding signal molecules C4-HSL, 3OC12-HSL, as well as the quinolones PQS and HHQ. To take into account the bacterial population increase, the signal molecule concentrations were divided by the corresponding OD600 values (Fig. 2). The highest concentrations of these signal molecules are given in Table 2. 3OC12-HSL concentration per OD600 unit reached its peak during the exponential growth phase (Fig. 2B). 3OC12-HSL had almost disappeared from culture supernatants after 12 h of culture, at the entry into stationary phase. PQS and HHQ concentrations per OD600 unit reached their peaks at the end of exponential phase of growth (Fig. 2D and E). C4-HSL concentration per OD600 unit increased throughout the exponential phase and the beginning of the stationary phase, reaching its highest level after 20 h of culture (Fig. 2C). qRT-PCR assays showed that the amounts of rhlAB and rhlC mRNAs increased 6.5 and 24 fold, respectively, between 20 and 30 h of culture (not shown), ie subsequently to the C4-HSL peak and during the rhamnolipid production period.
Fig. (1).
Rhamnolipid production by P. aeruginosa PAO1. Time-course of production of the three major rhamnolipid species (103 Area/OD600): di-rhamnolipids Rha-Rha-C10-C10 (ion m/z 649, squares), Rha-Rha-C10-C12 (ion m/z 677, circles), and Rha-Rha-C10-C12:1 (ion m/z 675, triangles) in PPGAS medium. The results are representative of at least three independent experiments.
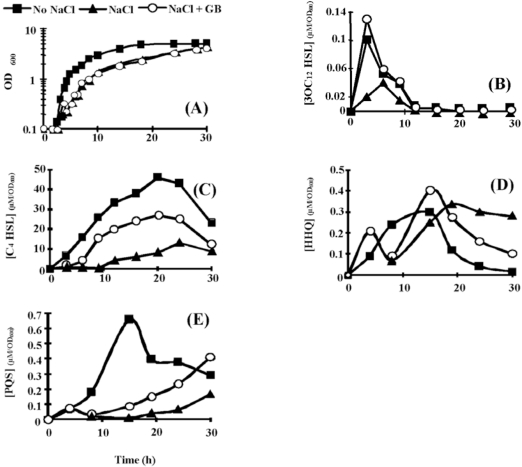
Fig. (2).
Effect of hyperosmotic condition on (A) P. aeruginosa PAO1 growth, and on the extra-cellular production of (B) 3OC12-HSL, (C) C4-HSL, (D) HHQ, and (E) PQS. Bacteria were grown in PPGAS medium (squares), in PPGAS containing 0.5 M NaCl (triangles), or in PPGAS containing 0.5 M NaCl and 1 mM glycine betaine (circles). The signal molecules concentrations (in µM) were divided by the OD600 values of the cultures at the sampling time. The points are averages of three experiments, and the standard deviations were lower than 10% of the mean values.
Table 2.
Highest Extra-Cellular Concentrations of HSLs and Quinolones in P. aeruginosa PAO1 Culture Supernatants
| Medium | 3OC12-HSL (nM)‡ | C4-HSL (nM)‡ | HHQ (nM)‡ | PQS (nM)‡ |
|---|---|---|---|---|
| PPGAS | 160 ± 10 | 2140 ± 230 | 893± 92 | 2054± 176 |
| PPGAS/NaCl* | 50 ± 4 | 410 ± 50 | 1525± 19 | 832± 67 |
| PPGAS/NaCl/GB† | 190 ± 12 | 860 ± 95 | 1002± 95 | 2162± 210 |
0.5 M NaCl was included in the PPGAS medium prior to bacterial inoculation.
0.5 M NaCl and 1 mM GB were included in the medium.
The values are averages of at least two experiments and the standard deviations are given.
Osmoadaptation of P. aeruginosa in PPGAS Medium
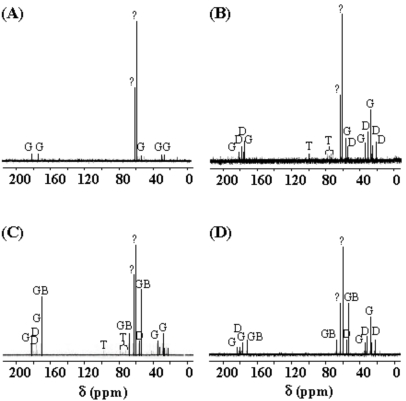
Hyperosmotic conditions were less detrimental in PPGAS medium than in the minimal PLM63 medium previously used: when 0.5 M NaCl was added before inoculating the bacteria, P. aeruginosa PAO1 failed to grow in PLM63, whereas its growth was only slightly affected in PPGAS (Fig. 2A). The addition of the osmoprotectant GB in PPGAS containing 0.5 M NaCl had no effect on bacterial growth (Fig. 2A). Growth was more strongly impaired by higher NaCl concentrations (0.7 and 0.9 M), but it was partially reestablished by the presence of GB (not shown). We chose to perform the subsequent experiments with 0.5 M NaCl, since this concentration only moderately affected growth and it allowed examining the GB effect on QS and rhamnolipid production independently from an action on growth. To understand how P. aeruginosa adapted to NaCl in PPGAS medium, we searched for the major osmolytes accumulated in the cells. In the absence of exogenously added GB, P. aeruginosa PAO1 accumulated glutamate, NAGGN, and trehalose (Fig. 3, compare B and A), as previously observed in minimal media [26, 28]. No GB accumulation was observed (Fig. 3B), showing that it was not provided by the PPGAS medium. By contrast, when GB was added to the culture medium, it was the main accumulated osmoprotectant (Fig. 3C). GB was still present in cells during stationary phase (Fig. 3D), indicating that it was not metabolized by P. aeruginosa in this medium. This was confirmed by following the fate of [methyl-14C]GB added to PPGAS (not shown).
Fig. (3).
Natural abundance 13C-nuclear magnetic resonance spectra of ethanolic extracts from P. aeruginosa PAO1 cells grown (A) in NaCl-free PPGAS medium, (B) in PPGAS containing 0.5 M NaCl, (C) in PPGAS/NaCl with 1 mM glycine betaine (GB) until exponential phase, or (D) in PPGAS/NaCl/GB until stationary phase. In (A) and (B) conditions, identical spectra were obtained from cells in exponential and stationary phase. Resonances due to glutamate (G), N-acetylglutaminylglutamine amide (D), trehalose (T), and GB are indicated. Unidentified peaks corresponding to none of the possible osmoprotectants are labelled “?”.
Hyperosmotic Condition Abolishes Rhamnolipid Production in the Absence of GB
In the presence of 0.5 M NaCl, we did not detect rhamnolipid in PPGAS culture supernatants of P. aeruginosa PAO1 (Table 3). We also failed to detect rhamnolipid from cell extracts whereas rhamnolipids were detected from extracts of PAO1 grown in NaCl-free PPGAS (not shown). This showed that the defect in rhamnolipid production in hyperosmotic condition was at the synthesis level rather than at the export level. After 20 h of culture with NaCl, the abundances of rhlAB (assayed using rhlA-targeting primers) and rhlC mRNAs were reduced 6.7 and 10 fold (Fig. 4), respectively, explaining at least in part the rhamnolipid biosynthesis defect. The addition of GB to PPGAS/NaCl fully restored the rhlAB mRNA level, and partially restored the rhlC mRNA amount, which was then only 1.5 fold lower than in PPGAS. GB also reestablished the rhamnolipid production, albeit to a 2.5 fold lower level than in NaCl-free PPGAS, as observed by assaying di-rhamnolipid Rha-Rha-C10-C10 (Table 3). Since RhlC is responsible for the formation of di-rhamnolipids from mono-rhamnolipids [7] and the expression of its gene was not fully restored by GB, we quantified the production of the most abundant mono-rhamnolipid, Rha-C10-C10, in PPGAS/NaCl/GB, but it remained lower than in PPGAS/NaCl (Table 3).
Table 3.
Highest Extra-Cellular Concentrations of Rhamnolipids in P. aeruginosa PAO1 Culture Supernatants
| Medium | Rha-Rha-C10-C10 (103 Area/OD600) ‡ | Rha-C10-C10 (103 Area/OD600) ‡ |
|---|---|---|
| PPGAS | 50 ± 7.8 | 1.83 ± 0.12 |
| PPGAS/NaCl* | 0 | 0 |
| PPGAS/NaCl/GB† | 20 ± 1.6 | 0.92 ± 0.1 |
0.5 M NaCl was included in the PPGAS medium prior to bacterial inoculation.
0.5 M NaCl and 1 mM GB were included in the medium.
The liquid chromatography-mass spectrometry peak surface areas were divided by the OD600 values of the cultures. The values are averages of at least two experiments and the standard deviations are given.
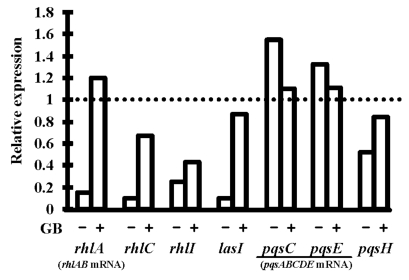
Fig. (4).
Relative expression levels of the genes lasI, rhlI, rhlC, pqsH and of the operons rhlAB and pqsABCDE in P. aeruginosa PAO1 submitted to 0.5 M osmotic shock without glycine betaine (GB) or in its presence. RNAs were extracted after 20 h of growth and the expression levels were determined by qRT-PCR, relatively to the expression in PPGAS medium without stress. Primers internal to rhlA, pqsC and pqsE were used to quantify rhlAB and pqsABCDE mRNAs. Relative values lower than 1 indicate reduction of gene expression upon stress (e.g. a value of 0.5 reveals that the mRNA level was divided by 2 in hyperosmotic condition). 16S rRNA was used as endogenous control to normalize the RNA input and reverse transcription efficiency. PCR reactions were performed in triplicate and the standard deviations were lower than 0.15 CT.
Hyperosmotic Condition Impairs HSL Accumulation
3OC12- and C4-HSL were still produced in the presence of NaCl, but their highest concentrations were 3.2 and 5.2 fold lower, respectively (Table 2). In addition, the 3OC12-HSL peak of concentration per OD600 unit was delayed by 2.5 h (from early to mid exponential phase), whereas C4-HSL was only detected from 12 h of culture and did not reach a peak at 20 h anymore (Fig. 2B and C). When GB was added to PPGAS/NaCl, the time-courses of 3OC12- and C4-HSL accumulation were similar to those seen in PPGAS (Fig. 2B and C). Since GB restored HSL time-course production without improving growth, the delays observed in PPGAS/NaCl compared to PPGAS cannot be attributed to the slower bacterial growth in hyperosmotic condition. GB furthermore reestablished the 3OC12-HSL production level, which was even slightly higher than in PPGAS (Table 2 and Fig. 2B). By contrast, the C4-HSL production level remained lower (2.5 fold) in PPGAS/NaCl/ GB than in PPGAS (Table 2 and Fig. 2C), indicating that hyperosmotic stress affects the C4-HSL production independently from that of 3OC12-HSL. The reductions in HSL levels in salt-containing media can be explained by the lasI and rhlI expression: their mRNA amounts were divided by 10 and 4.3 in PPGAS/NaCl and by 1.2 and 2.3 PPGAS/ NaCl/GB (Fig. 4), respectively, compared to PPGAS.
Hyperosmotic Condition Impairs Accumulation of PQS, but not of HHQ
In the presence of NaCl, PQS production was dramatically impaired during the end of growth and the entry into the stationary phase, and resumed only during the stationary phase (Fig. 2E). By contrast, the production of its precursor HHQ was markedly less affected by osmotic stress (Fig. 2D). Consistently, the osmotic condition led to a 2.5 fold decrease of the highest PQS concentration, whereas the highest HHQ concentration was 1.7 fold increased (Table 2). GB allowed P. aeruginosa to produce higher amounts of PQS in PPGAS/NaCl medium (Table 2), but this occurred mainly during stationary phase and GB therefore failed to restore the time-course of PQS production (Fig. 2E). We investigated the levels of pqsABCDE (using pqsC and pqsE primers) and pqsH mRNAs, the products of which are involved in PQS biosynthesis (pqsABCD and pqsH) and in regulation of a part of the QS network (pqsE). The presence of NaCl led to a decreased level of pqsH mRNA, which was 2 fold lower, but not of pqsABCDE mRNA (Fig. 4). Since PqsH is responsible for the conversion of HHQ to PQS, this result explains in part that hyperosmotic condition impairs the production of PQS, but not of HHQ.
DISCUSSION
We examined here the time-courses of HSL and quinolone accumulations in rich PPGAS medium by the wild type P. aeruginosa PAO1 strain. As expected from the hierarchy between the QS systems [8-10, 36] and as reported for HSLs in minimal M63 and PLM63 media [23], the peak of 3OC12-HSL per OD600 unit was first observed, followed by the peaks of HHQ and PQS, and finally of C4-HSL. These peaks were reached during the exponential growth phase for 3OC12-HSL, stationary phase for C4-HSL, and between these two phases for quinolones. Rhamnolipid production is dependent on both PQS and RhlRI QS system [5, 6, 36], which produces and senses C4-HSL. Consistently, we observed that rhamnolipids were mainly produced subsequently to the peak of C4-HSL per OD600 unit. In PPGAS medium, 0.5 M NaCl delayed and strongly reduced the accumulation of both HSLs and of PQS in the culture supernatants, whereas P. aeruginosa growth was only moderately affected. The osmotic stress effect on C4-HSL was however more dramatic in PLM63 [23] than in PPGAS, since this HSL was still produced in the latter medium containing 0.5 M NaCl. This can be correlated to the rhlI expression level, which was more strongly impaired by osmotic stress in PLM63 (14 fold reduced [23]) than in PPGAS (4.3 fold reduced). Furthermore, we investigated here gene expression levels of pqsABCD encoding HHQ synthesis enzymes, and pqsH encoding the enzyme responsible for the conversion of HHQ to PQS [11, 12, 14]. While pqsABCDE mRNA level was not decreased, the pqsH expression level was 2 fold lower in the presence of NaCl. This lower pqsH expression level might however not be sufficient to fully explain the lack of conversion of HHQ to PQS. In addition, it is possible that the activity of the PqsH enzyme might be more altered by osmotic stress than HHQ biosynthesis enzymes, which could be explained by the fact that PqsH is permanently exposed to the damaging effects of the external salt [25], due to it predicted periplasmic location [15]. As in PLM63 [23], NaCl abolished rhamnolipid production in PPGAS. This is at least in part due to the observed reduction of rhlAB and rhlC expressions (respectively 4 and 7 fold in PLM63/NaCl [23]; and 6.7 and 10 fold in PPGAS/NaCl compared to the corresponding NaCl-free media), which can in turn be explained by the lower C4-HSL and PQS accumulation. However, the addition of exogenous C4-HSL or PQS to the PPGAS/NaCl cultures, either at the beginning of the culture or at the end of the exponential growth phase, did not allow rhamnolipid production (data not shown), indicating that hyperosmotic stress acts also downstream of signal molecules production to prevent rhamnolipid synthesis, as previously suggested [23].
P. aeruginosa PAO1 adapted to 0.5 M NaCl in PPGAS by accumulating the K+ counter-ion glutamate, NAGGN, and trehalose, as previously observed in minimal media [26, 28]. By contrast, when GB was added to the culture medium, it was the main accumulated osmoprotectant although this did not improve growth. GB degradation by P. aeruginosa PAO1 was shown to depend on the carbon source: it occurred in M63 glucose medium, but not in M63 succinate [28]. Since glucose is the carbon source in PPGAS, we expected that GB would be metabolized by P. aeruginosa. This did not occur, suggesting that another component of this medium exerts a catabolite repression preventing GB degradation. Whereas it had no effect on growth, GB partially or fully reestablished all the observed defects: time-courses and levels of production of HSLs and PQS, mRNA amounts, and rhamnolipid biosynthesis. By contrast, GB had no effect on the rhamnolipid production level when PPGAS did not contain NaCl (not shown). Not all of GB effects were previously observed in PLM63/NaCl, since GB failed to restore the 3OC12-HSL production and the rhlAB mRNA amount in the latter medium [23].
In conclusion, this work shows that a hyperosmotic condition which has a moderate effect on P. aeruginosa growth nevertheless impairs the accumulation of cell-to-cell signal molecules of HSL type and PQS, but not of HHQ. Since QS-controlled phenotypes include production of multiple virulence factors, swarming and twitching motilities, and biofilm formation [4, 8, 10], hyperosmotic condition is likely to deeply affect the behavior of this bacterium, in agreement to a previous report showing that hyperosmotic condition inhibited biofilm development [37]. Rhamnolipid production is abolished by hyperosmotic condition, which cannot be only explained by the C4-HSL and PQS defects. GB, but not the other compatible solutes accumulated in the absence of GB, is able to restore at least partially these defects without improving growth, confirming previous results proposing GB as a chemical chaperonine [38].
ACKNOWLEDGEMENTS
This work was supported by the Region Bretagne, FEDER funds, and the Ministère de la Recherche et de la Technologie, France (RITMER grant and doctoral fellowships to AB and FD). We are grateful to M Foglino for the gift of P. aeruginosa PAO1.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soberón-Chávez G, Lépine F, Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–25. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 3.Zulianello L, Canard C, Köhler T, Caille D, Lacroix JS, Meda P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect Immun. 2006;74:3134–47. doi: 10.1128/IAI.01772-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 5.Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–8. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–67. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahim R, Ochsner UA, Olvera C, et al. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol. 2001;40:708–18. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- 8.Juhas M, Eberl L, Tümmler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol. 2005;7:459–71. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- 9.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2006;30:274–91. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 11.Diggle SP, Cornelis P, Williams P, Cámara M. 4-quinolone signaling in Pseudomonas aeruginosa: old molecules, new perspectives. Int J Med Microbiol. 2006;296:83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst. 2008;4:882–8. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 13.Farrow JM 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of the PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–51. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–80. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deziel E, Lepine F, Milot S, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101:1339–44. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wade DS, Calfee MW, Rocha ER, et al. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–80. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–8. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 19.Jensen V, Löns D, Zaoui C, et al. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and –independent pathways. J Bacteriol. 2006;188:8601–6. doi: 10.1128/JB.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bredenbruch F, Nimtz M, Wray V, Morr M, Müller R, Häussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol. 2005;187:3630–5. doi: 10.1128/JB.187.11.3630-3635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calfee MW, Shelton JG, McCubrey JA, Pesci EC. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect Immun. 2005;73:878–82. doi: 10.1128/IAI.73.2.878-882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Häussler S, Becker T. The Pseudomonas Quinolone Signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008;4:e1000166. doi: 10.1371/journal.ppat.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bazire A, Dheilly A, Diab F, et al. Osmotic stress and phosphate limitation alter production of cell-to-cell signal molecules and rhamnolipid biosurfactant by Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005;253:125–31. doi: 10.1016/j.femsle.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Berry A, DeVault JD, Chakrabarty AM. High osmolarity is a signal for enhanced algD transcription in mucoid and nonmucoid Pseudomonas aeruginosa strains. J Bacteriol. 1989;171:2312–7. doi: 10.1128/jb.171.5.2312-2317.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 26.D’Souza-Ault MR, Smith LT, Smith GM. Roles of N-acetyl-glutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl Environ Microbiol. 1993;59:473–8. doi: 10.1128/aem.59.2.473-478.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serra AL, Mariscotti JF, Barra JL, Lucchesi GI, Domenech CE, Lisa AT. Glycine betaine transmethylase mutant of Pseudomonas aeruginosa. J Bacteriol. 2002;184:4301–3. doi: 10.1128/JB.184.15.4301-4303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diab F, Bernard T, Bazire A, Haras D, Blanco C, Jebbar M. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology. 2006;152:1395–406. doi: 10.1099/mic.0.28652-0. [DOI] [PubMed] [Google Scholar]
- 29.Wild M, Caro AD, Hernández AL, Miller RM, Soberón-Chávez G. Selection and partial characterization of a Pseudomonas aeruginosa mono-rhamnolipid deficient mutant. FEMS Microbiol Lett. 1997;153:279–85. doi: 10.1111/j.1574-6968.1997.tb12586.x. [DOI] [PubMed] [Google Scholar]
- 30.Morin D, Grasland B, Vallée-Rehel K, Dufau C, Haras D. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J Chromatogr A. 2003;1002:79–92. doi: 10.1016/s0021-9673(03)00730-1. [DOI] [PubMed] [Google Scholar]
- 31.Lépine F, Milot S, Deziel E, He J, Rahme LG. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkyl-quinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom. 2004;15:862–9. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Déziel E, Lépine F, Milot S, Villemur R. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim Biophys Acta. 2000;1485:145–52. doi: 10.1016/s1388-1981(00)00039-1. [DOI] [PubMed] [Google Scholar]
- 33.Guyard-Nicodème M, Bazire A, Hémery G, et al. Outer membrane modifications of Pseudomonas fluorescens MF37 in response to hyperosmolarity. J Proteome Res. 2008;7:1218–25. doi: 10.1021/pr070539x. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Bernard T, Jebbar M, Rassouli Y, Himdi-Kabbab S, Hamelin J, Blanco C. Ectoine accumulation and osmotic regulation in Brevibacterium linens. J Gen Microbiol. 1993;139:129–38. [Google Scholar]
- 36.Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–23. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 37.Bazire A, Diab F, Jebbar M, Haras D. Influence of high salinity on biofilm formation and benzoate assimilation by Pseudomonas aeruginosa. J Ind Microbiol Biotechnol. 2007;34:5–8. doi: 10.1007/s10295-006-0087-2. [DOI] [PubMed] [Google Scholar]
- 38.Diamant S, Rosenthal D, Azem A, Eliahu N, Ben-Zvi AP, Goloubinoff P. Dicarboxylic amino acids and glycine-betaine regulate chaperone-mediated protein-disaggregation under stress. Mol Microbiol. 2003;49:401–10. doi: 10.1046/j.1365-2958.2003.03553.x. [DOI] [PubMed] [Google Scholar]