Summary
Kinesin-2 motors, which are involved in intraflagellar transport and cargo transport along cytoplasmic microtubules, differ from motors in the canonical Kinesin-1 family in having a heterodimeric rather than homodimeric structure and in possessing a three amino acid insertion in their neck linker domain. To determine how these structural features alter the chemomechanical coupling in Kinesin-2, we used single-molecule bead experiments to measure the processivity and velocity of mouse Kinesin-2 heterodimer (KIF3A/B) and the engineered homodimers KIF3A/A and KIF3B/B, and compared their behavior to Drosophila Kinesin-1 heavy chain (KHC). Single motor run lengths of Kinesin-2 were four-fold shorter than Kinesin-1. Extending the Kinesin-1 neck linker by three amino acids led to a similar reduction in processivity. Furthermore, Kinesin-2 processivity varied inversely with ATP concentration. Stochastic simulations of the Kinesin-1 and Kinesin-2 hydrolysis cycles suggest that “front head gating”, in which rearward tension prevents ATP binding to the front head when both heads are bound to the microtubule, is diminished in Kinesin-2. Because the mechanical tension that underlies front head gating must be transmitted through the neck linker domains, we propose that the diminished coordination in Kinesin-2 is a result of its longer and hence more compliant neck linker element.
Results and Discussion
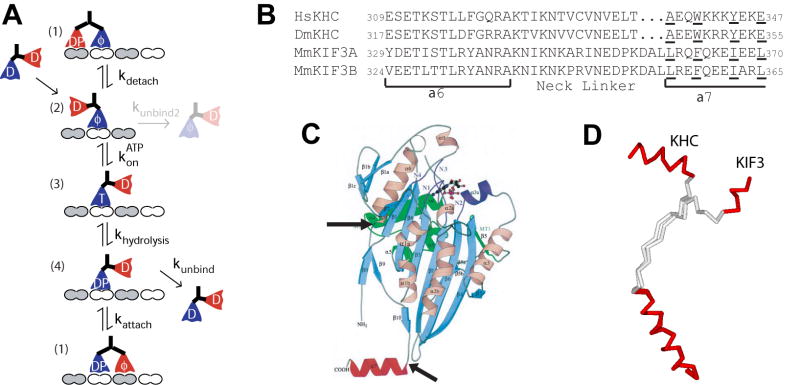
Kinesin processivity relies on maintaining the hydrolysis cycles of the two heads out of phase such that one head remains bound to the microtubule at all times. Figure 1A shows our working model for the Kinesin-1 hydrolysis cycle that accounts for a large body of kinesin mechanical and biochemical experiments [1]. Features of the hand-over-hand model that ensure processivity can be described by two nonexclusive mechanisms – front-head-gating and rear-head-gating* [2–5]. Both of these mechanisms involve mechanical tension between the two heads that is transmitted through the flexible neck linker of each head and their shared neck coiled-coil domain. In front-head gating, rearward strain on the leading head in State 1 prevents ATP from binding until the trailing head detaches (State 2). This mechanism prevents premature ATP hydrolysis and subsequent detachment of the leading head, and ensures that the trailing head is primed to advance to the next binding site when ATP binds to the leading head. In the rear-head gating model, the bound head in State 4 dissociates slowly in comparison to the overall cycle time, and binding of the second head (State 1) produces forward strain that leads to rapid detachment (State 2).
Figure 1.
A) Working model for the Kinesin-1 chemomechanical pathway. Nucleotide abbreviations are T = ATP, D = ADP, DP = ADP.Pi, φ = No nucleotide. Motors in solution have high affinity for ADP, and upon binding to the microtubule (State 2) release one ADP molecule [20]. The motor waits in this state until ATP binds to the front head, which results in docking of the neck linker and a displacement of the tethered head towards the next binding site (State 3) [21]. While the tethered head diffusively searches for the next binding site, the ATP on the bound head is hydrolyzed (State 4). Following hydrolysis (State 4) there are two possibilities. Most of the time (~99% for Kinesin-1), the tethered head will bind to the next biding site and release its ADP (State 1), and then the rear head will detach (State 2) completing an 8 nm step. Alternatively, the bound ADP.Pi head in State 4 will unbind from the microtubule, terminating the processive run. While detachment from State 4 is the predominant termination step, at limiting ATP concentrations the motor can occasionally detach from State 2. B) Neck linker sequences for Kinesin-1 and Kinesin-2 motors. The sequences of human conventional kinesin heavy chain (HsKHC), Drosophila melanogaster kinesin (DmKHC), and mouse KIF3A and KIF3B were aligned based on the known crystal structures of KHC (PDB: 3KIN, 2KIN and 1MKJ) and KIF3 (3B6U). The Kinesin-1 neck linker is defined as the 14 amino acids that span between the end of α6 and the first hydrophobic residue of the α7 coiled-coil (A323 to T336 in human numbering). The start of the α7 coiled-coil was taken from the rat Kinesin-1 dimer structure (3KIN) and the human KIF3B (3B6U) and Giardia KIF3A (GiKIN2a) monomer structures [22]. Hydrophobic a and d residues in the heptad repeat are underlined. C) Crystal structure of rat monomeric conventional kinesin from [23], showing the start and end of the 14-residue neck-linker in Kinesin-1. D) Comparison of the length and conformation of the KHC and KIF3B neck linkers. Figure was created by aligning the α6 helix of the 2KIN and 3B6U structures.
Because both front-head gating and rear-head-gating involve mechanical tension between the head domains, modifications that increase the mechanical compliance of the flexible neck linker are predicted to reduce motor processivity. Based on sequence alignments and comparisons of crystal structures, the neck linker domain in Kinesin-2 motors are three amino acids longer than the neck linker domain of Kinesin-1 (17 versus 14 amino acids) (Figure 1B). If this extension increases the compliance of the neck linker, then either front head gating, rear head gating, or both mechanisms may be diminished in Kinesin-2. This prediction was tested by measuring the processivity of Kinesin-1 and Kinesin-2 motors and interpreting the results using a stochastic model of the hydrolysis cycle.
Kinesin-2 is less processive than Kinesin-1
The velocities and run lengths of individual KIF3 and KHC motors attached to polystyrene beads were analyzed first at saturating ATP levels. Motor dilution profiles were generated to ensure that experiments were carried out in the single molecule regime (see Supplemental Data). The mean velocity of the KIF3A/B heterodimer was 436 ± 129 nm/s (mean ± SD, n = 90) and the velocity of KHC motors was 703 ± 136 nm/s (n = 58). The KIF3 homodimers, KIF3A/A (455 ± 115 nm/s, n = 101) and KIF3B/B (458 ± 106 nm/s, n = 102) had similar velocities to the wild type heterodimer. While the KIF3B/B velocity was consistent with previous measurements from gliding assays, the KIF3A/B and KIF3A/A were significantly higher than previously reported [6]. The velocity differences are due to mutations that were discovered in the motors used in the previous work (see Supplemental Data); all of the velocities presented here are from the corrected sequences.
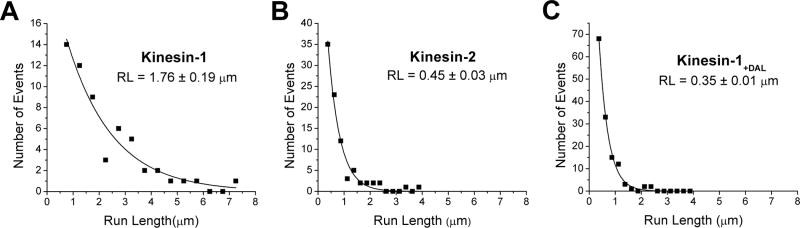
In contrast to velocity differences, which were within a factor of two, the run length of wild type Kinesin-2 was approximately four-fold shorter than Kinesin-1. The mean run length for KIF3A/B was 449 ± 30 nm (mean ± SEM, n = 88) compared to 1747 ± 199 nm (n = 57) for KHC (Figure 2). The reduced processivity of Kinesin-2 is consistent with the hypothesis that the longer neck-linker domain in Kinesin-2 reduces the degree of mechanical communication between the two heads and “uncouples” the hydrolysis cycles of the two heads. To test this hypothesis more directly, we extended the 14 amino acid neck linker of Kinesin-1 by inserting the last three amino acids of the Kinesin-2 neck linker (DAL) into Kinesin-1 at the neck-linker/neck-coil junction. The run length of this Kinesin-1+DAL construct (355 ± 14 nm) was five-fold shorter than wild-type (Figure 2), consistent with disruption of the mechanochemical coupling between the two heads. Despite the significant reduction in processivity, Kinesin-1+DAL moved at 552 ± 103 nm/s, only 20% slower than wild-type Kinesin-1. The behavior of Kinesin-1+DAL is qualitatively consistent with work from Hackney, who found that the biochemical processivity of Drosophila Kinesin-1 was reduced when the neck linker was artificially extended [7]. However, it contrasts with recent results from Yildez et al who extended the neck linker of Cys-lite human Kinesin-1 and found that the processivity fell by less than a factor of two for inserts as large as 29 amino acids [8]. Possible explanations for why the Yildiz results differ from ours include: 1) their use of a cys-lite modified kinesin, which has been shown to have altered motor kinetics [4], 2) the use of axonemes rather than taxol-stabilized microtubules, 3) the inclusion of two positively charged lysines in the inserts, and 4) the low ionic strength buffer (12 mM PIPES versus 80 mM PIPES used here), which will enhance electrostatic tethering of the motors to the axonemes.
Figure 2.
Run length distributions for single Kinesin-1, Kinesin-2 and Kinesin-1+DAL motors attached to beads. Run lengths were estimated by fitting the data to an exponential where the first bin (0–0.5 μm for Kinesin-1 and 0–0.25 μm for Kinesin-2 and Kinesin-1+DAL) was ignored due to uncertainties in detecting events below 250 nm.
The measured run lengths of KIF3A/A and KIF3B/B were in the range of KIF3A/B, but the KIF3A/A run length (410 ± 35 nm, n = 85) was moderately shorter than KIF3A/B, while the KIF3B/B run length (704 ± 81, n = 83) was somewhat longer (See Supplemental Data for further details). Hence, although the heterodimeric structure of KIF3A/B is highly conserved between diverse species, the ability of the motor to take multiple steps before detaching does not require the presence of two different motor domains.
KIF3A/A run length is ATP dependent
We next measured the ATP dependence of the motor run lengths and velocities from 1 μM to 1 mM ATP. We measured a weak positive dependence of Kinesin-1 processivity on ATP, with slightly longer run lengths observed at higher ATP concentrations (Figure 3A). However, differences between the run lengths at 1 μM and 1 mM ATP were not statistically significant (p = 0.07 from paired t-test), and these data are essentially consistent with Yajima et al who found the processivity of Kinesin-1 to be independent of ATP [9].
Figure 3.
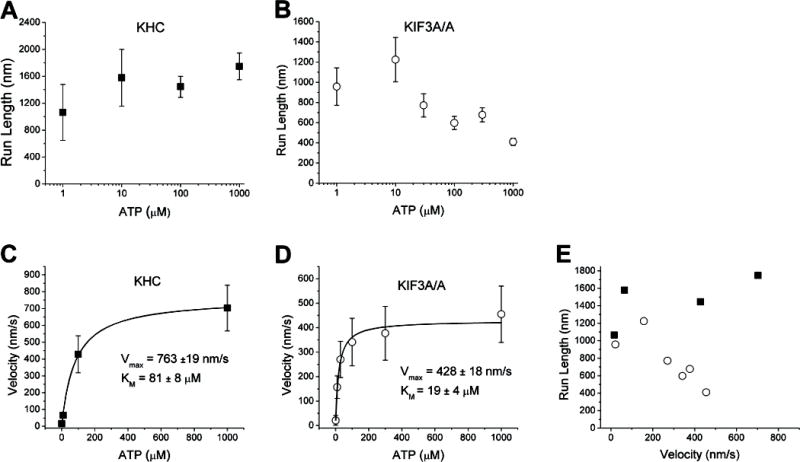
ATP dependence of motor velocity and run length for KHC and KIF3A/A. Run lengths (A and B) were obtained by fitting exponentials to the run length distribution at each ATP concentration, errors are SE of fits. Velocity data (C and D) are plotted as mean +/− SD for each concentration along with a fit to the Michaelis-Menten equation. The total number of events for KHC was 290 and for KIF3A/A was 467. E) Data are plotted as run length versus velocity to emphasize the different trends for KHC (solid squares) and KIF3A/A (open circles).
While low ATP concentrations had no effect on the relative activity of Kinesin-1, at ATP concentrations of 10 μM and below no binding events were observed for KIF3A/B in bead assays and no microtubule landing was observed in microtubule gliding assays (data not shown). Nucleotide-free Kinesin-1 is known to denature or misfold over time [10], and we interpret the KIF3A/B inactivation as an analogous process that has a greater nucleotide sensitivity. Interestingly, while KIF3B/B displayed similar inactivation at low ATP levels, KIF3A/A retained its function down to 1 μM ATP, suggesting that it is the KIF3B head that inactivates in low nucleotide environments. Therefore, to investigate the ATP dependence of motor velocity and processivity, the behavior of KHC and KIF3A/A were compared.
In contrast to KHC, KIF3A/A run lengths fell from roughly 1 micron at low ATP concentrations to ~400 nm at 1 mM ATP (Figure 3B). A paired t-test showed statistically significant differences when comparing the run length at 1 mM to run lengths at 1 μM and 10 μM ATP (p = 0.02 and p = 0.002, respectively). To exclude the possibility that multi-motor interactions occur preferentially at low ATP, the KIF3A/A motor/bead ratio was decreased by a factor of two at 1 μM ATP; no change in the run length was observed (run length 1260 ± 160 nm (mean ± SEM, n = 20)). To normalize for the different values and better show the run length differences, the data are plotted in Figure 3E as run length versus velocity, showing the clear processivity differences between KHC and KIF3A/A.
Kinetic model simulations
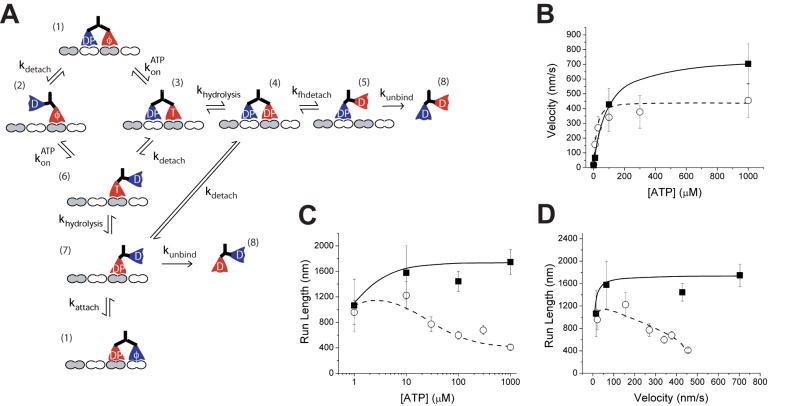
To better understand the chemomechanical cycle of Kinesin-2 and to interpret processivity and velocity differences between Kinesin-1 and Kinesin-2 motors, we developed a stochastic computational model of the kinesin chemomechanical cycle. Existing kinetic models that use sets of linked ordinary differential equations are valuable for interpreting enzyme kinetic data [11], but these models are suboptimal for modeling processivity and they are unable to model heterodimers with heads possessing different kinetics. Instead, we developed a Monte Carlo model that contains discrete states, corresponding to different nucleotide states in the kinesin hydrolysis cycle, and uses the Gillespie algorithm [12] to convert rate constants to expected transition times between states. To compare to the experimental data, many “runs” are modeled and the mean velocity and run length are calculated. Initial rate constants for the Kinesin-1 pathway (Figure 1) were taken from the literature, and selected rate constants were then systematically varied to match the experimentally observed velocity, run length and of KHC. As seen in Figure 4 the model is able to reproduce the ATP dependence of run length and velocity for KHC. Final model parameters and details of the simulations are described in Supplementary Data.
Figure 4.
A) Proposed model for the Kinesin-2 chemomechanical pathway. Due to the enhanced compliance between the heads, ATP binding to the leading head in State 1 is allowed, leading to a bifurcation in the pathway that dominates at saturating [ATP]. As in the Kinesin-1 model, slow detachment from State 2 waiting state is allowed, but for clarity is not shown. B) Experimental results (symbols) and model simulations (lines) of motor velocity versus ATP concentration for KHC (solid squares and solid line) and KIF3A/A (open circles and dashed line). Model parameters are given in Supplementary Data. C) Run length versus ATP concentration. D) Results at different ATP concentrations are replotted as run length versus velocity to emphasize the qualitatively different behavior of KHC and KIF3A/A.
We next investigated whether this model is sufficient to account for the KIF3A/A results. By adjusting specific rate constants, the reduced KIF3A/A run length and ATP dependence of velocity could easily be accounted for, but the model was unable to reproduce the ATP dependence of the run length. Inspection of the model in Figure 1 makes this is clear – limiting ATP slows the kinetic cycle by extending the wait in the nucleotide-free State 2, which either has no effect or increases the probability of detachment at low ATP. Hence, to account for the observed behavior of KIF3A/A, the kinetic model has to be expanded to include other mechanochemical states.
As discussed in the Introduction, Kinesin-1 processivity can be explained either by gating of the front head (tension-induced inhibition of ATP binding) or gating of the rear head (detachment triggered by attachment of the front head) [1–5]. While the shorter run lengths for Kinesin-2 could be accounted for in the model by diminishing the degree of rear-head gating (i.e. increasing kunbind and/or decreasing kattach), those modifications did not reproduce the observed ATP dependence of processivity (data not shown). As a second approach, the front-head gating mechanism was disrupted by allowing ATP binding by the leading head to precede detachment of the trailing head (Figure 4A). If ATP binds to the leading head and is hydrolyzed before the trailing head detaches, this puts both heads in a lower affinity state, which can lead to the motor detaching from the microtubule. This diminished front head gating has negligible effect at low ATP concentrations because the trailing head is able to detach before ATP binds (State 2), and the kinetic cycle collapses to the standard cycle shown in Figure 1. In contrast, at saturating ATP concentrations, ATP binds to the front head before the trailing head has time to detach (State 3), leading to possible premature termination of the cycle and shorter run lengths.
Initial rate constants for the KIF3A/A kinetic cycle were taken from the KHC simulations, and kon(ATP), k′on(ATP), khydrolysis and kunbind were iteratively adjusted to fit the measured KIF3A/A velocity, run length, and . The fits of the expanded model to the experimental KIF3A/A velocity and run data are shown in Figure 4. Hence, relaxing the requirement that ATP binding to the leading head precedes detachment of the trailing head introduces a second detachment pathway that can account for the measured ATP dependence of KIF3A/A processivity. Below we argue that the longer neck linker of Kinesin-2 is sufficient to explain this partial uncoupling of the Kinesin-2 mechanochemical cycle.
Mechanical Properties of the Neck Linker Domain
Disordered regions of proteins can be modeled as entropic springs, and the Worm-Like-Chain (WLC) model is commonly used to describe their force-extension characteristics [8, 13, 14]. Modeling the neck linker as a WLC, the force necessary to extend the polymer a distance x is:
where kBT is Boltzmann’s constant times the absolute temperature (4.1 pN-nm), lp is the persistence length of the polymer, estimated to be 0.5 nm [13–15], and L is the contour length of the polymer, which we take as 0.38 nm per amino acid [16]. If a) both heads of the motor are bound to a microtubule, b) the two core motor domains take on identical conformations such that the neck linkers originate from α6 at the same relative position, and c) the neck coil does not unfold, then each flexible neck linker must extend to 4.1 nm to span the 8.2 nm tubulin repeat distance. From the above WLC model, the resulting tension on a 14-residue neck linker would be 43 pN and the tension on a 17-residue neck linker would be 18 pN; both of these values are significantly greater than the ~6 pN stall force of Kinesin-1. Although these forces can be adjusted down somewhat by assuming different WLC model parameters or allowing elastic deformation of the heads or partial unfolding of the coiled–coil domain (see Supplementary Data), the important result here is that extending the neck linker is expected to significantly reduce the tension between the head domains when both heads are bound to a microtubule (State 1).
Because both front-head and rear-head gating rely on mechanical tension transmitted through the neck linker domain, the extended neck linker in Kinesin-2 is expected to diminish both of these mechanisms. Gating of the rear head will be reduced because the tension that accelerates rear head detachment will be diminished, while gating of the front head will be reduced because the tension that blocks ATP binding will be diminished. Because the predicted forces differ significantly (43 pN versus 18 pN), extending the neck linker could significantly alter the rate constants governing these transitions. For instance, if the characteristic distance associated with rear head detachment is 0.5 nm, then from the Bell Model , the load-induced detachment rate will be 20-fold slower in Kinesin-2 [17, 18]. Hence, it is reasonable to expect that the extended neck linker in Kinesin-2 motors will result in significant differences in mechanochemical coupling between the two motor domains compared to Kinesin-1.
Implications for in vivo motor behavior
What are the implications of this work for the function of Kinesin-2 motors in vivo? Whereas the role of Kinesin-1 is to transport cargo along axons that can be centimeters in length, Kinesin-2 primarily transports cargo along flagella and cilia, which are only tens of microns. However, both of these transport distances are much longer than the single-molecule run lengths, and in cells motor accumulations that enhance processivity as well as competition from cargo-associated dyneins that reverse the direction of transport will play important roles. This single-molecule characterization of Kinesin-2 serves as an important foundation for future cell biological experiments aimed to understand the roles of Kinesin-2 motors in cells.
Experimental Procedures
Protein Purification and Bead Assay
Full length His-tagged Drosophila melanogaster kinesin heavy chain and Kinesin-1+DAL were bacterially expressed and KIF3 motors were baculovirus-expressed and purified as previously described [2, 6]. Mutations discovered in the KIF3 plasmids used previously [6] were corrected using QuikChange® Multi Site-Directed Mutagenesis (Stratagene, Inc.) (See Supplemental Data).
For bead assays, motors were adsorbed to 560 nm diameter, casein-passivated carboxylated polystyrene beads as previously described [19]. A weak optical trap was used to facilitate motor-microtubule interactions. Motor:bead ratios were chosen such that ≤ 20% of the beads moved when brought in contact with microtubules. Bead positions were tracked manually using Meta-Vue. Run length data were fit using Origin 7.0. Details on the bead assays, optical trap, and motor dilution profiles are provided in Supplemental Data.
Model Simulations
Stochastic models were implemented in MATLAB. Transition times between states were determined using the Gillespie algorithm, , where k is the first order rate constant and rand# is a random number between 0 and 1 [12]. Runs were terminated when the motor detached, and the mean velocity and run length were calculated from 10,000 simulated runs for each condition. Further details are given in Supplementary Data.
Supplementary Material
Acknowledgments
The authors thank John Fricks for data analysis insights; Kihong Ahn, Chris Lengrich and Husam Katnani for their work on microscopy instrumentation; and Edgar Meyhöfer for helpful comments on the manuscript. This work was funded by NIH Grant GM076476 to W.O.H.
Abbreviations
- KHC
Kinesin Heavy Chain
Footnotes
For consistency with the literature we use the term “gating” but emphasize that this refers to gating of a given head and not gating by a given head.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hancock WO, Howard J. Kinesin: Processivity and chemomechanical coupling. In: Schliwa M, editor. Molecular Motors. Weinheim, Germany: Wiley-VCH; 2003. pp. 243–269. [Google Scholar]
- 2.Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc Natl Acad Sci U S A. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Hancock WO. The two motor domains of KIF3A/B coordinate for processive motility and move at different speeds. Biophys J. 2004;87:1795–1804. doi: 10.1529/biophysj.104.039842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackney DD, Stock MF, Moore J, Patterson RA. Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry. 2003;42:12011–12018. doi: 10.1021/bi0349118. [DOI] [PubMed] [Google Scholar]
- 8.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yajima J, Alonso MC, Cross RA, Toyoshima YY. Direct long-term observation of Kinesin processivity at low load. Curr Biol. 2002;12:301–306. doi: 10.1016/s0960-9822(01)00683-2. [DOI] [PubMed] [Google Scholar]
- 10.Huang TG, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J Biol Chem. 1994;269:16493–16501. [PubMed] [Google Scholar]
- 11.Gilbert SP, Johnson KA. Pre-steady-state kinetics of the microtubule-kinesin ATPase. Biochemistry. 1994;33:1951–1960. doi: 10.1021/bi00173a044. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- 13.Oberhauser AF, Hansma PK, Carrion-Vazquez M, Fernandez JM. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc Natl Acad Sci U S A. 2001;98:468–472. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc Natl Acad Sci U S A. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 16.Pauling L, Corey RB, Branson HR. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci U S A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 18.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. 1. Sunderland, MA: Sinauer Associates, Inc.; 2001. [Google Scholar]
- 19.Hancock WO, Howard J. Kinesin’s processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci U S A. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci U S A. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 22.Hoeng JC, Dawson SC, House SA, Sagolla MS, Pham JK, Mancuso JJ, Lowe J, Cande WZ. High Resolution Crystal Structure and in vivo Function of a Kinesin-2 Homolog in Giardia intestinalis. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-11-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sack S, Muller J, Marx A, Thormahlen M, Mandelkow EM, Brady ST, Mandelkow E. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry. 1997;36:16155–16165. doi: 10.1021/bi9722498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.