Abstract
The activity of tyrosine hydroxylase is regulated by reversible phosphorylation of serine residues in an N-terminal regulatory domain and catecholamine inhibition at the active site. Catecholamines such as dopamine bind very tightly to the resting enzyme; phosphorylation of Ser40 decreases the affinity for catecholamines by three orders of magnitude. The effects of dopamine binding and phosphorylation of Ser40 on the kinetics of deuterium incorporation into peptide bonds were examined by mass spectrometry. When dopamine is bound, three peptic peptides show significantly slower deuterium incorporation, 35-41 and 42-71 in the regulatory domain and 295-299 in the catalytic domain. In the phosphorylated enzyme, peptide 295-299 shows more rapid incorporation of deuterium, while 35-41 and 42-71 can not be detected. These results are consistent with tyrosine hydroxylase existing in two different conformations. In the closed conformation, the regulatory domain lies across the active site loop containing residues 295-298; this is stabilized when dopamine is bound in the active site. In the open conformation, the regulatory domain has moved out of the active site, allowing substrates access; this conformation is favored by phosphorylation of Ser40.
Tyrosine hydroxylase (TyrH) catalyzes the first and rate-limiting step of catecholamine biosynthesis, the conversion of tyrosine into dihydroxyphenylalanine, utilizing a tetrahydropterin as the source of electrons. The enzyme belongs to the small family of aromatic amino acid hydroxylases, which also includes phenylalanine hydroxylase (PheH) and tryptophan hydroxylase (TrpH) (1). All three enzymes play critical physiological roles; PheH is responsible for catabolism of excess phenylalanine in the diet, while TrpH is the first and rate-limiting enzyme in serotonin biosynthesis. The mammalian forms of these enzymes are homotetramers (2-4) in which each monomer contains a regulatory domain of 100-150 amino acids at the N-terminus and a larger catalytic domain of around 350 amino acids at the C-terminus (5-9). The homologous (5) catalytic domains contain all of the residues required for catalysis and for substrate specificity (10), while the regulatory domains exhibit low levels of sequence identity (5, 11). Structures have been determined for the catalytic domains of all three enzymes (12-14), but the only regulatory domain with an available structure is that of PheH (15). While critical residues are missing from that structure, it does show that the N-terminus of the regulatory domain extends across the active site cavity in the catalytic domain.
In light of the similar reactions they catalyze and the structural similarities of the catalytic domains, all three enzymes are generally accepted to have the same enzymatic mechanism (16). In contrast, their regulatory mechanisms are divergent (1). TyrH is regulated by phosphorylation of serine residues in the regulatory domain (17, 18). There are four residues in the enzyme which are phosphorylated in vivo. Using the numbering for the rat enzyme or the human isoform 1, these are Ser/Thr 8, Ser 19, Ser31, and Ser 40 (19-21). The phosphorylation states of the C-terminal three residues change in response to external modulators of catecholamine biosynthesis, while that of Ser8 appears to depend on other properties of the cell (20-25). The reported effects of phosphorylation on the catalytic activity of TyrH in the absence of other ligands are quite small (26-29); indeed, a mutant protein lacking the regulatory domain is fully active (8), establishing that phosphorylation does not activate the enzyme directly. Rather, phosphorylation appears to control the enzyme activity by altering the interaction with other ligands. Phosphorylation of Ser19 is reported to increase the affinity of the enzyme for 14-3-3 proteins (30-32); the physiological role of this interaction is not settled, although it may be to protect the phosphorylated enzyme from phosphatases (33). Phosphorylation of either Ser19 or Ser31 is also reported to increase the rate of phosphorylation of Ser40 by protein kinase A (PKA) (33-35) and to increase the stability of the enzyme (33, 34, 36, 37). The effects of phosphorylation of Ser40 are better understood. TyrH contains a non-heme iron atom in the active site cleft that must be ferrous for activity (38). The ferrous enzyme is readily oxidized to the ferric form (39, 40), and catecholamines bind to the ferric enzyme with dissociation constants of ∼1 nM (41, 42). Phosphorylation of Ser40 activates TyrH by increasing the rate constant for dissociation of catecholamines ∼500 fold (41, 43, 44). This allows tetrahydrobiopterin to reduce the iron, reactivating the enzyme (40).
The structural basis for the dramatic effect of phosphorylation of Ser40 on catecholamine binding is not known. The structure of the catalytic domain of PheH with dopamine bound shows the expected bidentate interaction of the catechol oxygens with the active site iron, while the amino moiety, required for an effect of phosphorylation on catechol binding to TyrH (42), extends out of the active site (46); a similar mode of binding is likely for TyrH. The isolated catalytic domain of TyrH has a dissociation constant for dopamine similar to that of phosphorylated TyrH2 demonstrating that the regulatory domain is required for tight binding of dopamine in the active site. Dopamine binding protects residues 33-50 in the regulatory domain of TyrH from trypsin cleavage, and Ser40 phosphorylation makes this region more susceptible (45), suggesting that phosphorylation and dopamine binding result in conformational changes that have opposite effects on the accessibility of this region to solvent. TyrH phosphorylated at Ser40 has a shorter retention time on gel filtration chromatography compared to TyrH alone, and dopamine-bound TyrH has a slightly longer retention time (34), further demonstrating that these modes of regulation are associated with opposing changes in the protein structure. These results have led to a model in which dopamine bound in the active site of TyrH interacts with residues in the regulatory domain; this interaction increases the affinity of the protein for the catecholamine and holds the N-terminal region over the active site, preventing substrate access (42). We describe here the use of hydrogen/deuterium exchange monitored by mass spectrometry to identify regions in TyrH affected by dopamine binding and phosphorylation at Ser40.
EXPERIMENTAL PROCEDURES
Materials
Dopamine and ATP were from Sigma-Aldrich Chemical Co. (Milwaukee, WI). Porcine stomach pepsin A was from Worthington Biochemical Co. (Lakewood, NJ). Deuterium oxide (D2O, 99% D) was from Cambridge Isotope Laboratories (Andover, MA). HPLC grade water was from Mallinckrodt Chemical Inc. (St, Louis, MO), and HPLC grade acetonitrile was from VWR-International (Darmstadt, Germany). Formic acid was from Michrom Bioresources Inc. (Auburn, CA). All other chemicals were of the highest purity commercially available.
Protein purification
The catalytic domain of PKA from beef heart was purified according to Flockhart et al. (47). Rat TyrH was purified as previously described (8, 48). The preparation of TyrH stoichiometrically phosphorylated at Ser 40 was performed as previously described (41) with minor modifications. Approximately 10 ml of TyrH (20 μM) in 50 mM Hepes, 200 mM KCl, and 10% glycerol (pH 7.3) was incubated with 50 μM ATP, 6 mM MgSO4, 2 μg/ml PKA at 4 °C for 1 h. An additional aliquot of ATP was then added to give a concentration of 100 μM, followed by another 1 h incubation. The degree of phosphorylation was monitored using a MonoQ column as previously described (41). The phosphorylated TyrH was purified using a Q-Sepharose column. To ensure that the enzyme contained a stoichiometric amount of iron, an equimolar amount of Fe(NH4)2(SO4)2 was added to purified TyrH or phosphorylated TyrH, followed by Q-Sepharose chromatography. The purified enzyme was stored in 10% glycerol, 200 mM KCl and 200 mM Hepes (pH 7.3) at a concentration of ∼ 0.3 mM at −80 °C.
To obtain the TyrH-dopamine complex, dopamine was added at a final concentration of 0.5 mM to 0.3 mM TyrH in 10% glycerol, 200 mM KCl, 200 mM Hepes (pH 7.3) at 4 °C. The binding of dopamine to TyrH was monitored by following the absorbance increase at 690 nm (41); the reaction was complete within 30 min.
Hydrogen/deuterium exchange and mass spectrometry
The exchange reaction was initiated by diluting 25 μl of 0.3 mM enzyme with 500 μl of D2O buffer (200 mM Hepes, pD 7.7) at 25 °C. Over the time course of the experiment, 20 μl aliquots were taken and quenched with 20 μl of 300 mM H3PO4 in H2O at 4 °C to lower the pH to 2.4. The quenched sample was immediately frozen with liquid nitrogen and stored at −80 °C for less than 24 h. Samples were thawed quickly and pepsin (15 mg/ml) was added to yield ratios of pepsin to TyrH (w/w) of 0.5, 1 or 2. The first 3 min of the pepsin digestion were performed on ice and the last 2 min in the 20 μL HPLC injection loop, which was submerged in ice. The resulting peptides were then injected onto a Vydac C18 column (2.1 mm × 150 mm). Most of the HPLC system was kept at 4 °C, including the injection loop, the tubing, the C18 column and the solvents. After a 3 min desalting with 98% solvent A (0.3% formic acid in H2O, pH 2.4) and 2% solvent B (0.3% formic acid in acetonitrile, pH* 2.4) at a flow rate of 0.3 ml/min, a gradient of 10-60 % solvent B over 9 min was applied at a flow rate of 0.2 ml/min. The majority of the peptides eluted at 15% to 50% acetonitrile within 6 min. The outflow from the HPLC column was injected directly into a Thermo Finnigan LCQ DECA XP ion-trap mass spectrometer. Singly, doubly and triply charged peptides, with m/z values of 400 to 2000, were analyzed.
To identify peptides, an independent tandem mass spectrometry (MS/MS) experiment was performed using the same digestion and elution conditions, except that H2O instead of D2O was used in the dilution step. Peptide assignment was performed using the program TurboSEQUEST (Thermo Finnigan, version 3.1).
To obtain fully-deuterated peptides, 10 μl of 0.3 mM TyrH was diluted with 200 μl of 200 mM Hepes buffer (pH 7.3) in H2O and 210 μl of 300 mM H3PO4 in H2O at 4 °C. Pepsin was added to give pepsin to protein ratios (w/w) of 1 or 2. After 5 min on ice, three 50 μl aliquots were taken out and loaded onto three 500 μl Bio-Spin 6 columns to isolate the peptic peptides. To remove the pepsin, the columns were centrifuged at 1,000 g for 30 s; the flow-through was discarded. To collect the peptides, each column was washed with 300 μl D2O and centrifuged at 1,000 g for 60 s. The washing was repeated twice, and all the flow-through was combined and lyophilized. The resulting peptides were resuspended in 20 μl D2O and heated to 90 °C for 90 min. After cooling, 5 μl of 500 mM phosphate buffer in D2O (pD 2.8) was added. Twenty μl of the peptides were loaded onto the HPLC, and peptide separation and analysis were performed as described above. Only 21 peptides were found that produced good spectra, suggesting that there was peptide loss due to the extra steps required to obtain fully deuterated peptides. The back-exchange level of the 21 well-resolved peptides was ∼ 35%. As a result, a value of 35% back exchange was used for the rest of the peptides.
Six independent experiments were performed for each condition. Due to the availability of the peptides in different pepsin to protein ratios (w/w), most peptides could be analyzed using data from four different experiments; all peptides were analyzed in at least two independent experiments with the same pepsin to protein ratio (w/w). The results were well reproduced in independent experiments. The results for individual peptides indicated in figures, including standard deviations, were calculated from experiments with the same pepsin to protein ratio.
Data analysis
The .raw MS files were processed using the Thermo Finnigan Xcalibur software. The spectra of individual peptides were transferred to Excel, and centroid data were identified using the program HX-Express (49); the program MagTran (50) was also used to analyze the peptides showing altered exchange and gave the same results as HX-Express. The deuterium contents of individual peptides were calculated using eq 1, where Mt is the measured mass of the peptide at time t, MH is the measured mass of the nondeuterated peptide, and MD is the measured mass of the completely deuterated peptide corrected for back-exchange. The time courses for deuterium incorporation of each peptide were fitted to eq 2, where N is the number of exchangeable amide hydrogens over the time course of the experiment, and A and B are the numbers of amide hydrogens with exchange rate constants k1 and k2, respectively.
| (1) |
| (2) |
RESULTS
Identification of peptides
Electrospray mass spectrometry of peptic peptides was used to analyze the extent of deuterium incorporation from solvent into peptide bonds in TyrH under different conditions. In preliminary experiments, the peptides generated by pepsin cleavage of TyrH were separated using a C18 reverse-phase HPLC column and identified by ms/ms analysis, varying the ratio of pepsin to TyrH from 0.5 to 2 (mass/mass). Slightly different mixtures of peptides were generated with the different ratios of pepsin to TyrH. Combining the peptides generated at ratios of pepsin to TyrH of 0.5, 1, and 2 allowed reproducible detection of peptides covering ∼70% of the protein (Figure 1). Most of the residues which were not found in peptic peptides by mass spectrometry are in three regions of the protein: residues 1-27, 143-174, and 191-209. The remainder of the protein is well-covered.
FIGURE 1.
Peptic peptides (arrows) of TyrH used in the hydrogen deuterium exchange mass spectrometry analyses.
Amide H/D exchange dynamics of the native enzyme
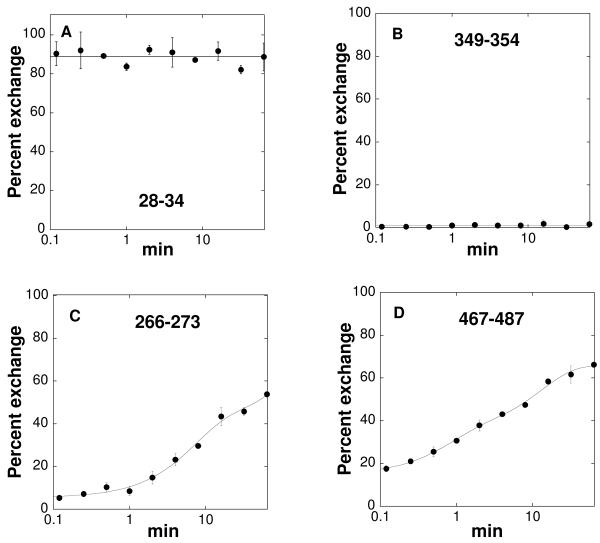
As a probe for the dynamics of the peptide backbone of TyrH, concentrated (0.3 mM) TyrH was mixed with 20 volumes of buffered D2O. Aliquots were removed at intervals from 7 s to 1 h, and the exchange reaction was quenched by mixing the sample with ice-cold 0.3 M H3PO4 to lower the pH to 2.4. The peptides generated by treatment for 5 min with pepsin were separated using a reverse-phase C18 column with a relatively short gradient, allowing the peptides to be injected into the mass spectrometer less than 10 min after the pepsin cleavage was complete. Figure 2 illustrates the three general exchange patterns that were observed for TyrH peptides. The time courses of incorporation of deuterium from solvent into all of the TyrH peptides which could be reproducibly analyzed under all conditions are given in Figure S1. The most N-terminal peptides, illustrated in Figure 2A by residues 28-34, exhibit complete exchange after only 7 s, the earliest time point used here. This suggests that the very N-terminus of the protein is highly dynamic. A much larger group of peptides shows no exchange even after 1 h incubation, consistent with their peptide bonds forming very stable hydrogen bonds. This behavior is illustrated in Figure 2B by peptide 349-354. Finally, the remainder of the protein shows time-dependent exchange, consistent with EX2 behavior due to transient formation of exchange-competent conformations (51). This behavior is illustrated in Figures 2C and 2D by peptides 266-273 and 477-487. The exchange kinetics of this group of peptides were typically well fit by eq 2, which separates the exchangeable hydrogens into a rapidly exchanging group (complete within 7 s) and two groups that exchange more slowly.
FIGURE 2.
Representative time courses of deuterium incorporation into TyrH peptides. The curves were obtained by fitting the data to eq 2.
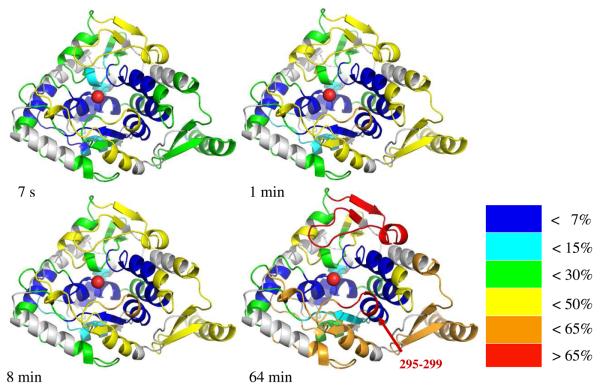
Figure 3 illustrates the kinetics of exchange of different peptides in the context of the structure of the catalytic domain; as noted above, no structure is available for the regulatory domain of TyrH. There is a central core around the active site which shows little exchange over the 1 h time course of the experiment: residues 226-259, 300-354, 364-377, and 387-393. The two histidines and one glutamate which act as iron ligands are all included in these peptides. The three peptides covering residues 28-71 of the regulatory domain exhibit complete exchange in less than 7 s, illustrating that this portion of the protein is highly mobile. This region contains two of the phosphorylation sites, Ser 31 and Ser 40. The remainder of the protein shows EX2 behavior with a range of rate constants. However, all the peptides in the tetramerization domain, residues 455-498, show very similar kinetics of exchange, consistent with a concerted motion associated with transient dissociation of the individual subunits into dimers or monomers.
FIGURE 3.
The extent of deuterium incorporation into peptides in the catalytic domain of TyrH at various times. Residues are color-labeled according to the deuterium content in the respective peptides. The figure was constructed using PDB file 1TOH. The active site is indicated as a sphere.
A similar analysis was carried with TyrH lacking the regulatory domain. Peptides covering almost 80% of this protein could be detected in the mass spectrometer. While there were some differences in the identities of the individual peptides from those seen in the intact protein, the overall patterns of exchange were very similar (results not shown).
Structural changes upon dopamine binding and phosphorylation
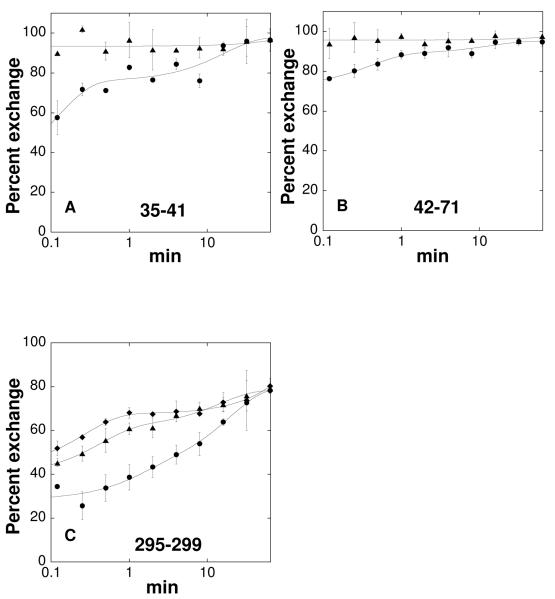
The deuterium exchange dynamics of TyrH with dopamine bound or phosphorylated at Ser40 were examined to gain insight into the structural changes accompanying regulation. The peptides surrounding Ser40, 35-41 and 42-71, were not detected in the phosphorylated protein. The reasons for this were not pursued, but phosphorylated peptides are typically quite difficult to detect by mass spectrometry, and phosphorylation of Ser40 makes this region of the protein more susceptible to proteolysis (45). With those exceptions, all of the peptides which could reproducibly be detected in the native enzyme could also be detected in the dopamine-bound and phosphorylated forms. Three peptides (35-41, 42-71, 295-299) show dramatic decreases in the rates of deuterium incorporation when dopamine is bound (Figure 4). The first two fragments are on the regulatory domain and show complete exchange within 7 s in the absence of dopamine. Peptide 295-299 is part of a loop near the active site opening in the catalytic domain (Figure 3). The altered exchange behavior of peptide 295-299 was confirmed by the overlapping peptide 295-301. The effect of phosphorylation on the exchange kinetics of peptide 295-299 is the reverse of the effect of dopamine, in that peptide 295-299 shows more rapid deuterium incorporation upon Ser40 phosphorylation. Two other peptides (133-142, 314-330) also show less deuterium incorporation upon dopamine binding (Figure S1), but the differences are small and may not be significant.
FIGURE 4.
Time course of deuterium incorporation into peptides 35-41 (A), 42-72 (B), and 295-299 (C) for TyrH alone (triangles), with bound dopamine (circles) or phosphorylated at Ser40 (diamonds).
DISCUSSION
TyrH was identified as a substrate for PKA over three decades ago (52, 53), and subsequent studies showed that Ser40 was the site of phosphorylation (19). The interplay between inhibition of the enzyme by catecholamines and activation by phosphorylation of Ser40 was established more recently (17, 18). In the present model for the effects of Ser40 phosphorylation on catecholamine inhibition of TyrH, the region of the regulatory domain near Ser40 interacts with the catecholamine chelated to the active site iron, preventing its dissociation (42, 45). Phosphorylation disrupts the interaction with the regulatory domain, decreasing the affinity of the enzyme for catecholamines and allowing the inhibitor to dissociate from the active site. Despite significant kinetic data regarding the interplay between phosphorylation and catecholamine inhibition, there is a lack of structural information regarding these regulatory events. The present results provide significant insight into the structural changes associated with regulation of TyrH.
Dopamine binding slows the H/D exchange of residues 35-71 in the regulatory domain and 295-299 in the catalytic domain. In contrast, phosphorylation of Ser40 results in faster exchange of residues 295-299. EX2 exchange kinetics such as those exhibited by these peptides are generally attributed to an equilibrium between a closed form of a protein which does not undergo exchange of amide protons with solvent during the course of the experiment and an open form which undergoes rapid exchange (51). Consequently, an increased rate of exchange reflects a shift in the equilibrium between the open and closed forms. Dopamine binding has previously been reported to protect Arg38, Arg39, and Arg49 from trypsin (45) and decrease the rate of phosphorylation of Ser40 by PKA (18). All these results can be explained by TyrH exhibiting an equilibrium between open and closed forms, with dopamine binding shifting the equilibrium toward the closed form. In the closed conformation, the peptide bonds of residues in the sequence 35-71 are protected from solvent and thus do not undergo H/D exchange and are not accessible to PKA or trypsin. Until now there has been a lack of evidence that this closed conformation involves an interaction between the regulatory and catalytic domains of TyrH. The observation that dopamine binding inhibits the exchange with solvent of peptides bonds on peptide 295-299 in the catalytic domain is consistent with a direct interaction between this peptide and residues in the regulatory domain.
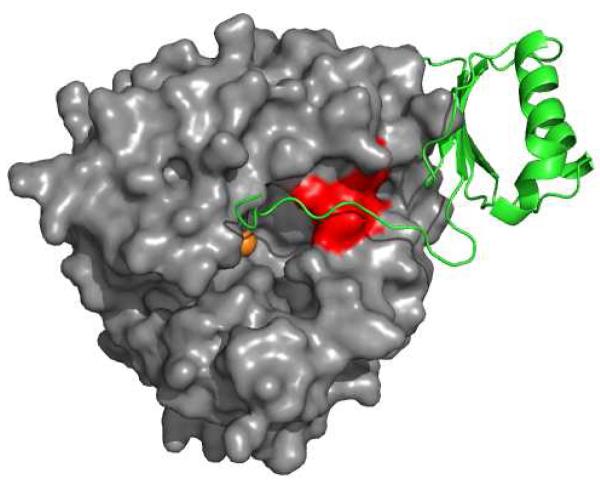
Peptide 295-299 is at the opening of the active site in TyrH (Figure 3). Residues 295-297 are part of a loop at the entrance of the active site, while residues 298 and 299 are at the beginning of a helix which extends into the protein. This is consistent with the extent of exchange seen in peptide 295-299 (Figure 4), in that about three amide bonds can exchange with solvent during the experiment. The complete lack of exchange of peptide bonds in peptide 300-305 (Figure S1) is also consistent with the expected stability of backbone hydrogen bonds in a helix. The most straightforward explanation for the decreased exchange of peptide 295-299 in the presence of dopamine is that the regulatory domain interacts directly with residues in this segment of the protein in the closed conformation. While the regulatory domain of TyrH is lacking from the available structures, the interaction between the regulatory and catalytic domains of the related protein PheH can provide a model for the interactions in TyrH. Figure 5 shows the structure of PheH, with the catalytic domain rendered in surface mode and the regulatory domain in cartoon fashion for clarity. This illustrates that the N-terminus of the regulatory domain of PheH extends across the active site cleft. The catalytic domains of TyrH and PheH are homologous, with essentially identical tertiary structures. Residues 249-253 of PheH align with residues 295-299 of TyrH. Figure 5 illustrates that the regulatory domain of PheH lies directly over the loop containing these residues in PheH. A similar arrangement in TyrH would explain the effect of dopamine on the H/D exchange kinetics of TyrH. Thus, the present data are consistent with a model in which the regulatory domain of TyrH lies across residues 295-299 of the catalytic domain in the closed form, thereby covering the active site, and with catecholamine binding stabilizing this conformation.
FIGURE 5.
Interactions between the regulatory and catalytic domains of PheH. The figure was generated using the PDB file 2PHM. The catalytic domain of PheH is shown as a surface rendering in gray, while the regulatory domain of PheH is shown as a green cartoon. Residues 249-253 are red. The active site iron is an orange sphere.
In the resting form of TyrH, residues 28-71 exchange fully with solvent within 7 s (Figures 4 and S1). The rate constants for exchange of the amides in unstructured peptides is ∼ 10 s−1 (54). Thus, the complete exchange of residues 28-71 within 7 s indicates a lack of significant secondary structure for this region in the absence of dopamine. The presence of the regulatory domain has little effect on the steady-state kinetic parameters of TyrH in the absence of dopamine (8, 10), suggesting that the regulatory domain does not hinder access to the active site cleft in the absence of catecholamines and that the equilibrium between the open and closed conformations lies toward the former in the resting enzyme. The effect of phosphorylation on the exchange kinetics of peptide 295-299 provides further support for a model in which the regulatory domain interacts with this region of the catalytic domain. Phosphorylation of Ser40 increases the exchange of peptide 295-299, consistent with phosphorylation shifting the equilibrium between the open and closed forms further toward the open form. Peptides 35-41 and 42-71 are not seen in the peptic digests of the phosphorylated enzyme. The inability to detect these two peptides in the phosphorylated enzyme can be attributed to phosphorylation of Ser40 hindering cleavage at the peptide bond between residues 41 and 42 and the difficulty of detecting phosphorylated peptides by mass spectrometry. A similar effect of phosphorylation on the sensitivity to trypsin of the peptide bond between residues 38 and 39 has been described previously (45).
The H/D exchange kinetics described here also provide insight into the dynamics of TyrH in the absence of ligands. There are four loops (177-193, 316-328, 421-429, and 290-297) at the entrance of the active site. Previous studies have established their importance for substrate specificity and catalytic activity (55-58). All four loops show a relatively high percentage of deuterium incorporation, suggesting that they are quite mobile. In contrast, the helices which surround the active site and make up the core of the protein show little exchange, consistent with the stability of the hydrogen bonds in alpha helices. The tetramerization domain of TyrH is composed of a long helix containing ∼ 26 amino acids and a two strand β-sheet (Figure 3). All the peptides in this region exhibit time-dependent incorporation of deuterium (Figure 2D) with similar kinetics (Figure S1). The similar patterns for both the helix and the β-sheet suggest that the deuterium incorporation into them is concerted; this can be explained by transient dissociation of the tetramer into dimers and/or monomers. To date, all descriptions of the quaternary structure of intact TyrH have found the enzyme to be a homotetramer (59), although truncation of the enzyme can generate dimers and monomers (60, 61). In contrast, PheH is reported to be a mixture of tetrameric and dimeric forms (62). The subunit interfaces in the two enzymes are similar, with the C-terminal helix playing a critical role in maintaining the tetrameric structure. The present results suggest that TyrH exhibits an equilibrium between tetramer and dimer similar to that of PheH, but that the equilibrium for the former lies far enough to the tetramer to be difficult to detect by methods less sensitive than H/D exchange.
In conclusion, we have found three peptides in TyrH showing substantially altered H/D exchange upon dopamine binding or phosphorylation. Two are in the regulatory domain surrounding the phosphorylation site Ser40, and the other is in one of four loops at the active site entrance. These data support a model in which dopamine and phosphorylation have opposite effects on the equilibrium between an open form of the enzyme and a closed form in which the N-terminus of the regulatory domain interacts with residues 295-298 in the catalytic domain.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Dr. Laura S. Busenlehner of the Univ. of Alabama for helping us develop the H/D exchange experiment and Dr. Zhang Zhongqi of Amgen Inc. for kindly providing the programs MagTran and HXPep. We thank Dr. Natalie Ahn of the Univ. of Colorado for helpful discussions. We also thank Sabrina Schmidtke of the Protein Chemistry Laboratory at Texas A&M University for her technical assistance with the mass spectrometry.
This work was supported in part by NIH grant R01 GM47291.
Footnotes
Abbreviations used: TyrH, rat tyrosine hydroxylase; PheH, phenylalanine hydroxylase; TrpH, tryptophan hydroxylase; PKA, protein kinase A.
Wang, S., and Fitzpatrick, P.F., unpublished observations.
Supporting Information Available: The effects of dopamine and phosphorylation on the kinetics of deuterium incorporation into all of the peptic peptides for TyrH. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Fitzpatrick PF. The tetrahydropterin-dependent amino acid hydroxylases. Ann. Rev. Biochem. 1999;68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- 2.Iwaki M, Phillips RS, Kaufman S. Proteolytic modification of the amino-terminal and carboxyl-terminal regions of rat hepatic phenylalanine hydroxylase. J.Biol.Chem. 1986;261:2051–2056. [PubMed] [Google Scholar]
- 3.Okuno S, Fujisawa H. Purification and some properties of tyrosine 3-monooxygenase from rat adrenal. Eur.J.Biochem. 1982;122:49–55. doi: 10.1111/j.1432-1033.1982.tb05846.x. [DOI] [PubMed] [Google Scholar]
- 4.Nakata H, Fujisawa H. Tryptophan 5-monooxygenase from mouse mastocytoma P815 a simple purification and general properties. Eur.J.Biochem. 1982;124:595–601. doi: 10.1111/j.1432-1033.1982.tb06636.x. [DOI] [PubMed] [Google Scholar]
- 5.Grenett HE, Ledley FD, Reed LL, Woo SLC. Full-length cDNA for rabbit tryptophan hydroxylase: Functional domains and evolution of aromatic amino acid hydroxylases. Proc. Natl. Acad. Sci. USA. 1987;84:5530–5534. doi: 10.1073/pnas.84.16.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daubner SC, Hillas PJ, Fitzpatrick PF. Expression and characterization of the catalytic domain of human phenylalanine hydroxylase. Arch. Biochem. Biophys. 1997;348:295–302. doi: 10.1006/abbi.1997.0435. [DOI] [PubMed] [Google Scholar]
- 7.Yang X-J, Kaufman S. High-level expression and deletion mutagenesis of human tryptophan hydroxylase. Proc. Natl. Acad. Sci. USA. 1994;91:6659–6663. doi: 10.1073/pnas.91.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daubner SC, Lohse DL, Fitzpatrick PF. Expression and characterization of catalytic and regulatory domains of rat tyrosine hydroxylase. Protein Expression Purif. 1993;2:1452–1460. doi: 10.1002/pro.5560020909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran GR, Daubner SC, Fitzpatrick PF. Expression and characterization of the catalytic core of tryptophan hydroxylase. J. Biol. Chem. 1998;273:12259–12266. doi: 10.1074/jbc.273.20.12259. [DOI] [PubMed] [Google Scholar]
- 10.Daubner SC, Hillas PJ, Fitzpatrick PF. Characterization of chimeric pterin dependent hydroxylases: Contributions of the regulatory domains of tyrosine and phenylalanine hydroxylase to substrate specificity. Biochemistry. 1997;36:11574–11582. doi: 10.1021/bi9711137. [DOI] [PubMed] [Google Scholar]
- 11.Abate C, Joh TH. Limited proteolysis of rat brain tyrosine hydroxylase defines an N-terminal region required for regulation of cofactor binding and directing substrate specificity. J. Mol. Neurosci. 1991;2:203–215. [PubMed] [Google Scholar]
- 12.Goodwill KE, Sabatier C, Marks C, Raag R, Fitzpatrick PF, Stevens RC. Crystal structure of tyrosine hydroxylase at 2.3 Å and its implications for inherited diseases. Nature Struct. Biol. 1997;4:578–585. doi: 10.1038/nsb0797-578. [DOI] [PubMed] [Google Scholar]
- 13.Erlandsen H, Fusetti F, Martinez A, Hough E, Flatmark T, Stevens RC. Crystal structure of the catalytic domain of human phenylalanine hydroxylase reveals the structural basis for phenylketonuria. Nature Struct.Biol. 1997;4:995–1000. doi: 10.1038/nsb1297-995. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Erlandsen H, Haavik J, Knappskog PM, Stevens RC. Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin. Biochemistry. 2002;41:12569–12574. doi: 10.1021/bi026561f. [DOI] [PubMed] [Google Scholar]
- 15.Kobe B, Jennings IG, House CM, Michell BJ, Goodwill KE, Santarsiero BD, Stevens RC, Cotton RGH, Kemp BE. Structural basis of intrasteric and allosteric controls of phenylalanine hydroxylase. Nature Struct.Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick PF. Mechanism of aromatic amino acid hydroxylation. Biochemistry. 2003;42:14083–14091. doi: 10.1021/bi035656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haavik J, Martinez A, Flatmark T. pH-dependent release of catecholamines from tyrosine hydroxylase and the effect of phosphorylation of Ser-40. FEBS Letts. 1990;262:363–365. doi: 10.1016/0014-5793(90)80230-g. [DOI] [PubMed] [Google Scholar]
- 18.Daubner SC, Lauriano C, Haycock JW, Fitzpatrick PF. Site-directed mutagenesis of Serine 40 of rat tyrosine hydroxylase. Effects of dopamine and cAMP-dependent phosphorylation on enzyme activity. J. Biol. Chem. 1992;267:12639–12646. [PubMed] [Google Scholar]
- 19.Campbell DG, Hardie DG, Vulliet PR. Identification of four phosphorylation sites in the N-terminal region of tyrosine hydroxylase. J. Biol. Chem. 1986;261:10489–10492. [PubMed] [Google Scholar]
- 20.Haycock JW, Bennett WF, George RJ, Waymire JC. Multiple site phosphorylation of tyrosine hydroxylase. Differential regulation in situ by 8-bromo-cAMP and acetylcholine. J.Biol.Chem. 1982;257:13699–13703. [PubMed] [Google Scholar]
- 21.Haycock JW. Phosphorylation of tyrosine hydroxylase in situ at serine 8, 19, 31, and 40. J.Biol.Chem. 1990;265:11682–11691. [PubMed] [Google Scholar]
- 22.Haycock JW, Meligeni JA, Bennett WF, Waymire JC. Phosphorylation and activation of tyrosine hydroxylase mediate the acetylcholine-induced increase in catecholamine biosynthesis in adrenal chromaffin cells. J.Biol.Chem. 1982;257:12641–12648. [PubMed] [Google Scholar]
- 23.George RJ, Haycock JW, Johnston JP, Craviso GL, Waymire JC. In vitro phosphorylation of bovine adrenal chromaffin cell tyrosine hydroxylase by endogenous protein kinases. J. Neurochem. 1989;52:274–284. doi: 10.1111/j.1471-4159.1989.tb10928.x. [DOI] [PubMed] [Google Scholar]
- 24.Haycock JW, Haycock DA. Tyrosine hydroxylase in rat brain dopaminergic nerve terminals. Multiple-site phosphorylation in vivo and in synaptosomes. J.Biol.Chem. 1991;266:5650–5657. [PubMed] [Google Scholar]
- 25.Haycock JW. Multiple signaling pathways in bovine chromaffin cells regulate tyrosine hydroxylase phosphorylation at Ser19, Ser31, and Ser40. Neurochem. Res. 1993;18:15–26. doi: 10.1007/BF00966919. [DOI] [PubMed] [Google Scholar]
- 26.Atkinson J, Richtand N, Schworer C, Kuczenski R, Soderling T. Phosphorylation of purified rat striatal tyrosine hydroxylase by Ca2+/calmodulin-dependent protein kinase II: Effect of an activator protein. J. Neurochem. 1987;49:1241–1249. doi: 10.1111/j.1471-4159.1987.tb10016.x. [DOI] [PubMed] [Google Scholar]
- 27.Le Bourdellès B, Horellou P, Le Caer J-P, Denèfle P, Latta M, Haavik J, Guibert B, Mayaux J-F, Mallet J. Phosphorylation of human recombinant tyrosine hydroxylase isoforms 1 and 2: An additional phosphorylated residue in isoform 2, generated through alternative splicing. J.Biol.Chem. 1991;266:17124–17130. [PubMed] [Google Scholar]
- 28.Almas B, Le Bourdelles B, Flatmark T, Mallet J, Haavik J. Regulation of recombinant human tyrosine hydroxylase isozymes by catecholamine binding and phosphorylation structure/activity studies and mechanistic implications. Eur. J. Biochem. 1992;209:249–255. doi: 10.1111/j.1432-1033.1992.tb17283.x. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland C, Alterio J, Campbell DG, Le Bourdelles B, Mallet J, Haavik J, Cohen P. Phosphorylation and activation of human tyrosine hydroxylase in vitro by mitogen-activated protein (MAP) kinase and MAP-kinase-activated kinases 1 and 2. Eur.J.Biochem. 1993;217:715–722. doi: 10.1111/j.1432-1033.1993.tb18297.x. [DOI] [PubMed] [Google Scholar]
- 30.Ichimura T, Isobe T, Okuyama T, Yamauchi T, Fujisawa H. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Letts. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. [DOI] [PubMed] [Google Scholar]
- 31.Kleppe R, Toska K, Haavik J. Interaction of phosphorylated tyrosine hydroxylase with 14-3-3 proteins: evidence for a phosphoserine 40-dependent association. J.Neurochem. 2001;77:1097–1107. doi: 10.1046/j.1471-4159.2001.00318.x. [DOI] [PubMed] [Google Scholar]
- 32.Obsilova V, Nedbalkova E, Silhan J, Boura E, Herman P, Vecer J, Sulc M, Teisinger J, Dyda F, Obsil T. The 14-3-3 protein affects the conformation of the regulatory domain of human tyrosine hydroxylase. Biochemistry. 2008;47:1768–1777. doi: 10.1021/bi7019468. [DOI] [PubMed] [Google Scholar]
- 33.Toska K, Kleppe R, Armstron CG, Morrice NA, Cohen P, Haavik J. Regulation of tyrosine hydroxylase by stress-activated protein kinases. J.Neurochem. 2002;83:775–783. doi: 10.1046/j.1471-4159.2002.01172.x. [DOI] [PubMed] [Google Scholar]
- 34.Bevilaqua LRM, Graham ME, Dunkley PR, von Nagy-Felsobuki EI, Dickson PW. Phosphorylation of Ser19 alters the conformation of tyrosine hydroxylase to increase the rate of phosphorylation of Ser40. J. Biol. Chem. 2001;276:40411–40416. doi: 10.1074/jbc.M105280200. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann IT, Bobrovskaya L, Gordon SL, Dunkley PR, Dickson PW. Differential regulation of the human tyrosine hydroxylase isoforms via hierarchical phosphorylation. J. Biol. Chem. 2006;281:17644–17651. doi: 10.1074/jbc.M512194200. [DOI] [PubMed] [Google Scholar]
- 36.Moy LY, Tsai L-H. Cyclin-dependent kinase 5 phosphorylates serine 31 of tyrosine hydroxylase and regulates its stability. J. Biol. Chem. 2004;279:54487–54493. doi: 10.1074/jbc.M406636200. [DOI] [PubMed] [Google Scholar]
- 37.Royo M, Fitzpatrick PF, Daubner SC. Mutation of regulatory serines of rat tyrosine hydroxylase to glutamate: effects on enzyme stability and activity. Arch. Biochem. Biophys. 2005;434:266–274. doi: 10.1016/j.abb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Fitzpatrick PF. The metal requirement of rat tyrosine hydroxylase. Biochem. Biophys. Res. Commun. 1989;161:211–215. doi: 10.1016/0006-291x(89)91582-9. [DOI] [PubMed] [Google Scholar]
- 39.Ramsey AJ, Hillas PJ, Fitzpatrick PF. Characterization of the active site iron in tyrosine hydroxylase: Redox states of the iron. J.Biol.Chem. 1996;271:24395–24400. doi: 10.1074/jbc.271.40.24395. [DOI] [PubMed] [Google Scholar]
- 40.Frantom PA, Seravalli J, Ragsdale SW, Fitzpatrick PF. Reduction and oxidation of the active site iron in tyrosine hydroxylase: kinetics and specificity. Biochemistry. 2006;45:2372–2379. doi: 10.1021/bi052283j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsey AJ, Fitzpatrick PF. Effects of phosphorylation of serine 40 of tyrosine hydroxylase on binding of catecholamines: Evidence for a novel regulatory mechanism. Biochemistry. 1998;37:8980–8986. doi: 10.1021/bi980582l. [DOI] [PubMed] [Google Scholar]
- 42.Ramsey AJ, Fitzpatrick PF. Effects of phosphorylation on binding of catecholamines to tyrosine hydroxylase: specificity and thermodynamics. Biochemistry. 2000;39:773–778. doi: 10.1021/bi991901r. [DOI] [PubMed] [Google Scholar]
- 43.Sura G, Daubner SC, Fitzpatrick PF. Effects of phosphorylation by protein kinase A on binding of catecholamines to the human tyrosine hydroxylase isoforms. J. Neurochem. 2004;90:970–978. doi: 10.1111/j.1471-4159.2004.02566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCulloch RI, Daubner SC, Fitzpatrick PF. Effects of substitution at serine 40 of tyrosine hydroxylase on catecholamine binding. Biochemistry. 2001;40:7273–7278. doi: 10.1021/bi010546d. [DOI] [PubMed] [Google Scholar]
- 45.McCulloch RI, Fitzpatrick PF. Limited proteolysis of tyrosine hydroxylase identifies residues 33-50 as conformationally sensitive to phosphorylation state and dopamine binding. Arch. Biochem. Biophys. 1999;367:143–145. doi: 10.1006/abbi.1999.1259. [DOI] [PubMed] [Google Scholar]
- 46.Erlandsen H, Flatmark T, Stevens RC, Hough E. Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 Å resolution. Biochemistry. 1998;37:15638–15646. doi: 10.1021/bi9815290. [DOI] [PubMed] [Google Scholar]
- 47.Flockhart DA, Corbin JD. Preparation of the catalytic subunit of cAMP-dependent protein kinase. In: Maranos PJ, Campbell IC, Cohen RM, editors. Brain Receptor Methodologies, Part A. Academic Press; New York: 1984. pp. 209–215. [Google Scholar]
- 48.Daubner SC, Fitzpatrick PF. Site-directed mutants of charged residues in the active site of tyrosine hydroxylase. Biochemistry. 1999;38:4448–4454. doi: 10.1021/bi983012u. [DOI] [PubMed] [Google Scholar]
- 49.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 51.Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch. Biochem. Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Lovenberg W, Bruckwick EA, Hanbauer I. ATP, cyclic AMP, and magnesium increase the affinity of rat striatal tyrosine hydroxylase for its cofactor. Proc. Natl. Acad. Sci. USA. 1975;72:2955–2958. doi: 10.1073/pnas.72.8.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldstein M, Bronaugh RL, Ebstein B, Roberge C. Stimulation of tyrosine hydroxylase activity by cyclic AMP in synaptosomes and in soluble striatal enzyme preparations. Brain Res. 1976;109:563–574. doi: 10.1016/0006-8993(76)90035-4. [DOI] [PubMed] [Google Scholar]
- 54.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins: Structure, Function & Genetics. 1996;17:87–92. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daubner SC, McGinnis JT, Gardner M, Kroboth SL, Morris AR, Fitzpatrick PF. A flexible loop in tyrosine hydroxylase controls coupling of amino acid hydroxylation to tetrahydropterin oxidation. J. Mol. Biol. 2006;359:299–307. doi: 10.1016/j.jmb.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sura GR, Lasagna M, Gawandi V, Reinhart GD, Fitzpatrick PF. Effects of ligands on the mobility of an active-site loop in tyrosine hydroxylase as monitored by fluorescence anisotropy. Biochemistry. 2006;45:9632–9638. doi: 10.1021/bi060754b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daubner SC, Melendez J, Fitzpatrick PF. Reversing the substrate specificities of phenylalanine and tyrosine hydroxylase: Aspartate 425 of tyrosine hydroxylase is essential for L-DOPA formation. Biochemistry. 2000;39:9652–9661. doi: 10.1021/bi000493k. [DOI] [PubMed] [Google Scholar]
- 58.Frantom PA, Fitzpatrick PF. Uncoupled forms of tyrosine hydroxylase unmask kinetic isotope effects on chemical steps. J. Am. Chem. Soc. 2003;125:16190–16191. doi: 10.1021/ja0383165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fitzpatrick PF, Chlumsky LJ, Daubner SC, O'Malley KL. Expression of rat tyrosine hydroxylase in insect tissue culture cells and purification and characterization of the cloned enzyme. J. Biol. Chem. 1990;265:2042–2047. [PubMed] [Google Scholar]
- 60.Lohse DL, Fitzpatrick PF. Identification of the intersubunit binding region in rat tyrosine hydroxylase. Biochem.Biophys.Res.Commun. 1993;197:1543–1548. doi: 10.1006/bbrc.1993.2653. [DOI] [PubMed] [Google Scholar]
- 61.Walker SJ, Liu X, Roskoski R, Vrana KE. Catalytic core of rat tyrosine hydroxylase: Terminal deletion analysis of bacterially expressed enzyme. Biochim.Biophys.Acta. 1994;1206:113–119. doi: 10.1016/0167-4838(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 62.Kappock TJ, Harkins PC, Friedenberg S, Caradonna JP. Spectroscopic and kinetic properties of unphosphorylated rat hepatic phenylalanine hydroxylase expressed in Escherichia coli. Comparison of resting and activated states. J.Biol.Chem. 1995;270:30532–30544. doi: 10.1074/jbc.270.51.30532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.