Abstract
Nowadays, cellular bioenergetics has become a central issue of investigation in cancer biology. Recently, the metabolic activity of the cancer cell has been shown to correlate with a proteomic index that informs of the relative mitochondrial activity of the cell. Within this new field of investigation, we report herein the production and characterization of high-affinity monoclonal antibodies against proteins of the “bioenergetic signature” of the cell. The use of recombinant proteins and antibodies against the mitochondrial β-F1-ATPase and Hsp60 proteins and the enzymes of the glycolytic pathway glyceraldehyde-3-phosphate dehydrogenase and pyruvate kinase M2 in quantitative assays provide, for the first time, the actual amount of these proteins in normal and tumor surgical specimens of breast, lung, and esophagus. The application of this methodology affords a straightforward proteomic signature that quantifies the variable energetic demand of human tissues. Furthermore, the results show an unanticipated finding: tumors from different tissues and/or histological types have the same proteomic signature of energetic metabolism. Therefore, the results indicate that cancer abolishes the tissue-specific differences in the bioenergetic phenotype of mitochondria. Overall, the results support that energetic metabolism represents an additional hallmark of the phenotype of the cancer cell and a promising target for the treatment of diverse neoplasias.
Introduction
Cancer is a complex genetic disease orchestrated by the acquisition of gain-of-function mutations in oncogenes and loss-of-function mutations in tumor suppressor genes. Even so, early in this century, the phenotype of the cancer cell was summarized in six main traits [1]. Almost at the same time, a rebirth of the interest in the energetic metabolism of cancer spurred [2–4]. Specifically, mechanisms contributing to the “abnormal” aerobic glycolysis of the cancer cell are being characterized [2–4]. Within this frame, mitochondria have become central players of these studies [5], and consistently, it has been reported that the relative cellular expression of the bottleneck β-F1-ATPase (β-F1) protein of oxidative phosphorylation is significantly diminished in tumors when compared with its levels in normal tissues [6–9]. In certain types of carcinomas, the down-regulation of β-F1 is accompanied by an increased expression of some of the markers of the glycolytic pathway [6,8]. This proteomic feature of cancer, which is fulfilled by more than 95% of the carcinomas analyzed in large cohorts of different tumors [7,8], defines a “bioenergetic signature” of potential clinical value as an indicator of disease progression in colon [6,9], lung [7,10], and breast [8] cancer patients. Furthermore, the bioenergetic signature also affords a predictive marker of the cellular response to chemotherapy [9,11]. Indeed, the cell death response to chemotherapeutic agents highly correlated with both the activity of oxidative phosphorylation [12,13] and the expression of β-F1 [9,11,14]. This study represents an effort to favor the translation of the bioenergetic signature of the cell into the clinical setting by developing the tools that could quantitatively establish the pathological range limits of these markers in human neoplasias. Interestingly, it reveals for the first time and irrespective of the cancer type being considered that energy metabolism has a common protein signature that provides a generic marker of the cancer cell that might be exploited in the combat of the disease.
Materials and Methods
Patient Specimens and Protein Extraction
Frozen tissue sections obtained from surgical specimens of untreated cancer patients with primary breast and lung adenocarcinomas and squamous esophageal and lung carcinomas were obtained from the Banco de Tejidos y Tumores, IDIBAPS (Instituto de Investigaciones Biomédicas Pi y Suñer), Hospital Clinic, Barcelona, Spain. The tissue sections of the tumor and normal tissue of each patient were analyzed previously by an expert pathologist and the protein extracted [8]. All tissue samples were anonymized and received in a coded form to protect patient confidentiality. The institutional review board approved the project. Twenty tissue sections (15 µm) were extracted in 300 µl of 50 mM Tris-HCl pH 8.0 containing 150 mM NaCl, 0.02% (w/v) sodium azide, 0.1% (w/v) SDS, 1% (v/v) Nonidet P40, 0.5% (w/v) sodium deoxycholate, 1 µg/ml leupeptin, 1 µg/ml antitripsin, 0.4 mM EDTA, 10 mM NaF, and 0.75 mM PMSF at 4°C for 30 minutes. After protein extraction, the samples were centrifuged (15,000g) at 4°C for 25 minutes. Protein concentration in the supernatants was determined with the Bradford reagent (Bio-Rad, Hercules, CA) using bovine serum albumin as standard.
Cloning Strategies and Protein Expression and Purification
The cDNAs encoding human β-F1 (NP_001677.2), Hsp60 (NP_002147.2), pyruvate kinase (PK; AAH35198), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; AAF99678) proteins were amplified by polymerase chain reaction using the MGC-5231, MGC-4084, MGC-15908, and MGC-9949 clones, respectively, derived from a choriocarcinoma (β-F1), uterine (Hsp60), and lung (PK and GAPDH) tumor cell lines of the ATCC (Manassas, VA) collection. The sequences of the forward (F) and reverse (R) primers used were as follows: β-F1 (F: 5′-CGGGAGCTCATGTTGGGGTTTGTG-3′; R: 5′-ATAGTTTAGCGGCCGCCGATGAATGCTCTTC-3′); Hsp60 (F: 5′-CGGGAGCTCATGCTTCGGTTACCC-3′; R: 5′-CACCACGTGAGAACATGCCACCTCC-3′); PK (F: 5′-CGGGAGCTCATGTCGAAGCCCCAT-3′; R: 5′-ATAGTTTAGCGGCCGCCGGCACAGGAACAAC-3′); and GAPDH (F: 5′-CGCGAGCTCATGAACGAAAACCTGTT-3′; R: 5′-CACCACGTGACTCCTTGGAGGCCAT-3′). The Sac I, Not I, and Pml I restriction sites are shown underlined. Amplicons were first cloned into pGEM-Teasy vector (Promega, Madison, WI) and after into pQE-Trisystem. The resulting plasmids, pQE-β-F1, pQE-Hsp60, pQE-PK, and pQE-GAPDH, which encode for the proteins with C-terminal 6xHis and streptavidin tags, were used to transform Escherichia coli M15/pREP4 cells. After induction of protein expression by adding isopropyl-β-thio galactopranoside (1 mM), the cells were resuspended in buffer A (100 mM NaH2PO4·H2O, 300 mM NaCl, pH 8.0 supplemented with lysozyme 1 mg/ml). The expressed proteins were purified using either Strep-Tactin or metal ion affinity chromatography Ni-NTA superflow resins (Qiagen, Hilden, Germany). The purity of the proteins was estimated by fractionation on SDS-PAGE.
Monoclonal Antibody Production and ELISAs
BALB/c mice were immunized by intraperitoneal injection with various dosages of the purified proteins (20 µg). Hybridomas were produced by fusing spleen cells with myeloma SP2 or NS-1 cells with polyethylene glycol in HAT-RPMI 1640 medium according to standard hybridoma techniques. Supernatants of the hybridomas were screened by indirect ELISA on polystyrene plates coated with the recombinant proteins (0–150 ng per well). Bound antibodies were detected using horseradish peroxidase-labeled goat antimouse antibodies (1:1000; DAKO, Carpinteria, CA). After the final washing, 100 µl of OPD solution (Sigma, St. Louis, MO) was added, and the color reaction was developed for 15 minutes and stopped by the addition of 18 M H2SO4. Optical density at 490 nm was determined in a FluoStar Optima (BMG Labtech, Offenburg, Germany) apparatus. The positive colonies were cloned by limiting dilution. Mouse monoclonal antibodies were purified with Montage antibody purification kit (Millipore, Billerica, MA) according to the supplier's instructions.
Western and Slot-Blot Procedures
Protein samples were subjected to SDS-PAGE followed by immunoblot analysis. A slot-blot apparatus (Schleicher & Schuell, Keene, NH) was used to print bands of uniform protein density. Protein samples of purified recombinant proteins (0–40 ng) or of human tissue extracts (0–25 µg) were applied in a final volume of 100 µl to the wells (Figure 1, D and E), and the excess of protein binding sites was blocked with a solution containing 3% bovine serum albumin in Tris-buffered saline (50 mM Tris-HCI, pH 7.4, 150 mM NaCl). The membranes were incubated with 0.4 µg/ml of primary antibody for 60 minutes. The monoclonal antibodies described in this work are clone 11/21-7 A8 for anti-β-F1, clone 17/9-15 G1 for anti-Hsp60, clone 273A-E5 for anti-GAPDH, and clone 14/5-21/24 for anti-PK. Secondary horseradish peroxidase-conjugated goat antimouse antibodies (1:3000) were used for detection by a chemiluminescence detection method (ECL; Amersham Bioscience, Buckinghamshire, UK). Membranes were exposed to x-ray films, and the light signals obtained in the films were quantified only when falling within the linear response range of the film as shown in the standard curves of the recombinant proteins and tissue extracts (Figure 1). Quantification of the immunoreactive bands (arbitrary units) was accomplished using a Kodak DC120 Zoom digital camera and the Kodak 1D Image Analysis Software for Windows (Kodak, Rochester, NY).
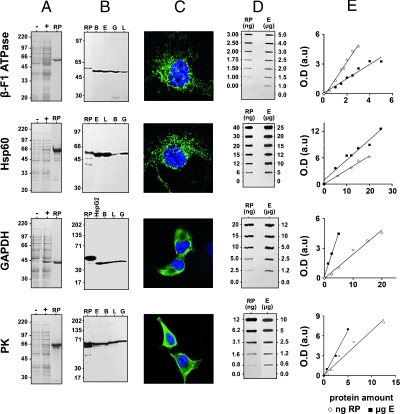
Figure 1.
Expression and purification of recombinant proteins and characterization of the monoclonal antibodies produced. (A) The M15 E. coli strain was used for the expression of recombinant proteins. Samples were collected before (-) and after 2 hours of 0.1 mM IPTG induction (+) and the cellular proteins analyzed by SDS-PAGE. The recombinant proteins were affinity-purified using the streptavidin tag (Hsp60) or the His tag (β-F1 ATPase, GAPDH, and PK) and the purity of the eluted protein estimated by SDS-PAGE (RP). (B) Western blot analysis showing the reactivity of the different antibodies produced. The antibodies (0.4 µg/ml) exclusively recognized the recombinant (RP; 1 ng of protein) as well as the native cellular protein in normal and/or tumor breast (B), esophagus (E), gastric (G), and lung (L) tissues or cell lines (HepG2; 10–30 µg of protein). (C) Immunofluorescence microscopy using the antibodies produced (0.4 µg/ml) revealed (green) the mitochondrial (β-F1 and Hsp60) or cytoplasmic (GAPDH and PK) localization of the cellular proteins in human liver (HepG2) and breast (Hs578T) cancer cells. Nuclear DNA (blue) was stained with ToPro3. Original magnification, x60. (D) Representative slot-blots with the antibodies produced for the quantification of the amount of the different biomarkers. A linear increase in the signal is observed as the amount of recombinant protein (RP in ng) or protein from cellular extracts (E in µg) is augmented. (E) Graphs of the experiments in panel D for both the recombinant protein (○, RP) and the cellular antigen (▪, CA) within the linear range of protein. The equations and correlation coefficients are as follows: β-F1 ATPase [RP: y = 1.7372x - 0.284/R2 = 0.9823; CA: y = 0.7228x + 0.0542/R2 = 0.9571], Hsp60 [RP: y = 0.3224x - 0.0161/R2 = 9879; CA: y = 0.4594x + 0.9/R2 = 0.9632], GAPDH [RP: y = 0.243x - 0.0404/R2 = 0.9804; CA: y = 0.8742x + 0.1839/R2 = 0.992], and PK [RP: y = 0.6673x + 0.179/R2 = 0.9664; CA: y = 1.3235x + 0.3378/R2 = 0.9803].
Immunofluorescence Microscopy
Human liver HepG2 and breast Hs578T cancer cells were fixed in freshly prepared 4% paraformaldehyde in PBS and permeabilized with a solution containing 0.1% Triton X-100 in PBS for 10 minutes. Coverslips were incubated for 1 hour with the indicated concentration of the antibodies produced and, after which, were incubated for 45 minutes with a 1:1000 dilution of goat antimouse immunoglobulin Gs conjugated to Alexa 594. Nuclei were stained with ToPro3 (Molecular Probes, Eugene, OR). Cellular fluorescence was analyzed by confocal microscopy using a Bio-Rad Radiance 2000 Zeiss Axiovert S100TV [15].
Statistical Analysis
Statistical analysis was performed using Student's t test for paired samples. One-way analysis of variance (ANOVA) was used to detect differences in the bioenergetic signature within the normal and tumor biopsies. Standard F test was used to assess significance. Statistical tests were two-sided at the 5% level of significance. The grouping of biopsies into different subsets with similar degree of similarity was carried out by unsupervised hierarchical clustering based on protein contents and derived indices of the metabolic biomarkers [8]. Raw data were normalized to either the mean value of the normal lung samples that accompany squamous lung carcinomas or the paired normal tissue and the log 2 of these values used to build up the data matrix. We used the Cluster Program from “Expression Profiler Clustering home page” (http://ep.ebi.ac.uk/EP/EPCLUST) using the Euclidean distances and average distance method. In this agglomerative clustering procedure, a series of partitions of the data into Pn, Pn - 1, …, P1 clusters is achieved. The Pn cluster consists of n single-biopsy clusters and the last P1 cluster consists of a single group containing all n biopsies. At a particular stage of the clustering procedure, the algorithm joins the clusters that are most similar. Similarity in this procedure is defined as the average of distances between all pairs of objects, where each pair is made up of one biopsy from each group.
Results
We have recently described [6,8,10] that markers of the bioenergetic signature of the cell are the catalytic β-F1 of the mitochondrial H+-ATP synthase, the structural mitochondrial protein Hsp60 (heat shock protein 60), and the enzymes of the glycolytic pathway GAPDH and the fetal/tumor M2-PK isoform of PK. To obtain the recombinant proteins to be used for immunization and as standards in quantitative assays, we cloned the human cDNA and expressed the full-length proteins carrying carboxy-terminal 6xHis and streptavidin tags (Figure 1A). The soluble (Hsp60) and insoluble (β-F1, GAPDH, and PK) recombinant proteins were purified on Strep-Tactin and Ni-NTA resins, respectively (Figure 1A), and then used for mice immunization. Mice were killed for the production of hybridomas only when a 104 dilution of the serum provided a strong positive response against the corresponding cellular protein on Western blots (data not shown). Supernatants from the growing hybridomas were screened by indirect ELISA and Western blot analysis. Only clones producing antibodies of high affinity and specificity in Western blots (Figure 1B) and in immunocytochemical techniques (Figure 1C) were preserved and used for the development of quantitative slot-blot assays (Figure 1D). Slot-blots had enough sensitivity to assess the tissue quantity of these proteins because they have a relatively high cellular representation. Moreover, the signals used for quantification purposes always fell within the linear response range of the film for both the recombinant protein assayed and the cellular antigens (Figure 1E), validating the assays for quantitative purposes.
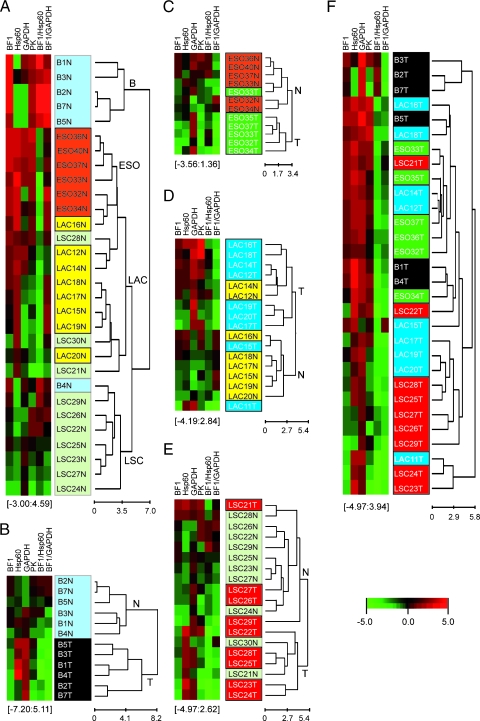
The results in Table 1 summarize the content in nanograms per microgram of cellular protein of β-F1, Hsp60, GAPDH, and PK in normal and tumor tissues of the lung, esophagus, and breast. The normalized nondimensional expression values of β-F1 per unit of structural mitochondrial protein (β-F1/Hsp60 ratio) and per unit of cellular protein (β-F1/GAPDH ratio) in each tissue are also presented (Table 1). These two proteomic indexes respectively inform of the mitochondrial competence and overall cellular activity of mitochondria [6,8,10]. The latter ratio, also defined as the bioenergetic signature of the cell [6], is a proteomic gauge of the activity of aerobic glycolysis in carcinomas [10]. As shown in Table 1, there is significant variability in some of these ratios in the normal tissues, especially in breast, suggesting the existence of heterogeneity in the bioenergetic phenotype of this tissue as a result of the physiological state of the patients. The content in normal tissues of the mitochondrial proteins (β-F1 and Hsp60) is variable (breast > esophagus > lung; Table 1), most likely reflecting the differential tissue dependence on the bioenergetic activity of mitochondria for provision of metabolic energy. Moreover, normal lung biopsies from adenocarcinomas and squamous carcinomas also revealed significant differences in the expression of Hsp60 and in the two ratios derived from β-F1 (Table 1), further suggesting a heterogeneous distribution/activity of mitochondria within the normal lung. Unsupervised hierarchical clustering of normal biopsies according to the expression pattern of proteins of the bioenergetic signature (Table 1) resulted in their distribution into two main groups (Figure 2A). One of them encompassed breast tissue only and displayed high mitochondrial activity as revealed by the two ratios used to normalize β-F1 expression (Table 1). The second group, including the rest of the biopsies (Figure 2A), showed a significantly lower content of β-F1 per cell (Table 1), suggesting the existence of subtle differences in the bioenergetic phenotype of these tissues. Within this large group, the analysis also illustrates the preferential clustering of esophageal biopsies in a group different from the normal lung (Figure 2A). Consistent with a heterogeneous activity of mitochondria within the normal lung, the normal lung tissue that accompanies adenocarcinomas or squamous carcinomas clustered into two different groups (Figure 2A). The significance of these differences in the tissue content of all the markers determined and derived ratios was confirmed by standard F test in one-way ANOVA model (P < .001).
Table 1.
Protein Content of Metabolic Biomarkers in Paired Normal and Tumor Samples of the Breast, Esophagus, and Lung.
| Tissue | Biopsy | β-F1 | Hsp60 | GAPDH | PK | β-F1/Hsp60 | β-F1/GAPDH |
| Lung | Normal (9) | 1.37 ± 0.16* | 0.19 ± 0.03* | 0.30 ± 0.07 | 0.45 ± 0.07* | 7.80 ± 0.84* | 5.95 ± 0.91* |
| Adenocarcinoma (9) | 1.43 ± 0.28 | 0.25 ± 0.05 | 0.94 ± 0.12† | 1.22 ± 0.41 | 5.63 ± 0.78 | 2.18 ± 0.86† | |
| Lung | Normal (9) | 0.90 ± 0.17* | 0.06 ± 0.02‡ | 0.27 ± 0.03 | 0.52 ± 0.13* | 26.20 ± 5.31*,‡ | 3.53 ± 0.61*,‡ |
| Squamous (9) | 0.77 ± 0.28 | 0.18 ± 0.04† | 0.51 ± 0.02† | 0.55 ± 0.19 | 6.01 ± 1.65† | 1.54 ± 0.49† | |
| Esophagus | Normal (6) | 3.36 ± 0.56* | 0.36 ± 0.04* | 0.54 ± 0.14 | 1.73 ± 0.39 | 9.57 ± 1.89 | 8.48 ± 1.87 |
| Squamous (6) | 1.85 ± 0.25† | 0.34 ± 0.08 | 1.09 ± 0.10† | 1.44 ± 0.65 | 6.94 ± 1.26 | 1.80 ± 0.27† | |
| Breast | Normal (6) | 6.10 ± 0.98 | 0.03 ± 0.01 | 0.31 ± 0.11 | 1.51 ± 0.25 | 264.33 ± 61.69 | 39.14 ± 13.95 |
| Adenocarcinoma (6) | 2.26 ± 0.45† | 0.34 ± 0.13† | 2.37 ± 0.39† | 0.98 ± 0.29 | 14.70 ± 4.75† | 1.11 ± 0.20† |
The protein content of β-F1, Hsp60, GAPDH, and PK is expressed in nanograms per microgram of total cellular protein. The ratios derived are dimensionless. The number of biopsies in each group is shown in parenthesis. The results shown are the mean ± SEM.
Significance of P ≤ .05 by Student's t test when compared with normal breast biopsies.
Significance of P ≤ .05 by Student's t test when compared with paired normal samples.
Significance of P ≤ .05 when comparing the two sets of normal lung biopsies.
Figure 2.
Graphical hierarchical clustering analysis of the bioenergetic signature as quantitatively determined by slot-blot procedures. Rows indicate type of sample; columns, proteins and derived ratios. Protein expression scores are shown normalized to the mean value of the normal lung samples that accompany squamous lung carcinomas (LSC-N) in panels A, E, and F and to the corresponding normal tissue in panels B, C and D, according to a color scale (below panel F): red indicates high; black, normal; and green, low. The dendogram (to the right of the matrix) represents overall similarities in expression profiles. The maximum and minimum values of the markers for each cluster are shown in brackets. (A) Clustering of normal breast (B-N, light blue), esophageal (ESO-N, orange), and normal lung samples from adenocarcinomas (LAC-N, yellow) and squamous carcinomas (LSC-N, light green). (B) Clustering of normal (B-N, light blue) and adenocarcinomas (B-T, black) of the breast. (C) Clustering of normal (ESO-N, orange) and squamous carcinomas (ESO-T, green) of the esophagus. (D) Clustering of normal (LAC-N, yellow) and adenocarcinomas of the lung (LAC-T, blue). (E) Clustering of normal (LSC-N, light green) and squamous carcinomas of the lung (LSC-T, red). (F) Clustering of breast, esophageal, and lung tumors.
Consistent with previous qualitative findings on the changes of these markers in tumors [6–8,10], we observed a significant decrease in the absolute amount of β-F1 in breast and esophageal carcinomas when compared with paired normal tissues (Table 1). In breast cancer, the sharp reduction in the bioenergetic signature resulted also from the concurrent and pronounced increase in the content of Hsp60 and GAPDH in the tumor (Table 1). In esophageal carcinomas, only the amount of GAPDH was significantly augmented (Table 1), contributing also to the reduction of the bioenergetic signature in this set of carcinomas (Table 1). The changes in the amount of metabolic markers allowed the correct classification of normal and tumor breast biopsies by their bioenergetic signature (Figure 2B). Likewise, the bioenergetic signature also allowed the correct classification of normal and tumor biopsies of the esophagus with high specificity (Figure 2C).
In adenocarcinomas and squamous carcinomas of the lung, we observed no significant differences in the absolute content of β-F1 when compared with paired normal tissues (Table 1). In squamous carcinomas of the lung, the absolute tumor content of Hsp60 and GAPDH were significantly augmented (Table 1), whereas in lung adenocarcinomas, the two glycolytic markers showed an increase, although only GAPDH revealed a significant increase when compared with normal lung (Table 1). However, the overall cellular activity of mitochondria (β-F1/GAPDH) in adenocarcinomas and squamous carcinomas of the lung showed a significant reduction when compared with paired normal lung (Table 1), consistent with previous reports in lung cancer [7,10]. As indicated for the case of breast and esophageal cancer (Figure 2), the changes in the amount of metabolic markers allowed the classification of normal and tumor lung biopsies by their bioenergetic signature (Figure 2, D and E, for adenocarcinomas and squamous carcinomas, respectively). It seems that the power of the classification of the biopsies by markers of energetic metabolism diminishes because the tissue of origin of the neoplasia is less dependent on the bioenergetic activity of mitochondria (breast > esophagus > lung; Figure 2). It is also possible that the tumor biopsies misclassified within the normal groups (Figure 2) could be derived from patients in early stages of their disease, i.e., when the bioenergetic phenotype of the cancer cell is still poorly affected [6–8].
Overall, it was remarkable to observe that tumors derived from different tissues and histological types had the same bioenergetic signature (β-F1/GAPDH ratio in Table 1; and compare the preferential red color under β-F1/GAPDH ratio in Figure 2A vs the green color under β-F1/GAPDH ratio in Figure 2F), as confirmed by F test in a one-way ANOVA model. Moreover, unsupervised clustering of the tumors did not reveal the preferential clustering of the biopsies within defined subgroups according to the tissue of origin or the histological subtype (Figure 2F), suggesting the existence of a common metabolic trait for these types of tumors.
Discussion
Previous studies have qualitatively shown that the expression of proteins of energetic metabolism are useful markers of the metabolic activity of cancer cells and tumors [10] and in predicting the prognosis of breast [8,16], colon [6,9], and lung [7,10] cancer patients. Moreover, the tumor expression of β-F1 has been shown to be an independent marker of survival in breast and lung cancer as assessed by multivariate Cox regression analysis [8,10]. Recently, these findings have been confirmed in a different large cohort of colon cancer patients [9]. Thus, the alteration of the bioenergetic signature strongly supports a relevant role for the mitochondrial impairment of the cancer cell in progression of the disease. In addition, these markers also predict the response to chemotherapy in various cancer cells [11,14] and in colorectal tumors [9]. Clinical translation of many of the protein biomarkers of cancer that have been discovered in recent years is scarce. A reason that limits translation is the lack of quantitative assays that could minimize the variability in assessing the expression of the biomarkers between different assays and/or laboratories. Herein, we describe the production of the recombinant proteins and the monoclonal antibodies against the markers of the bioenergetic signature as well as the implementation of these tools in simple quantitative assays based on slot-blot procedures to stimulate the translation of the bioenergetic signature into the clinics. Slot-blot procedures were chosen as initial methodology to assess the quantity of the biomarkers because they were easy to implement owing to the relatively high cellular abundance of these proteins and the high affinity and specificity of the antibodies produced. The availability of the recombinant proteins and of a set of well-validated monoclonal antibodies against these markers could now favor its further use in the development of a variety of other proteomic assays [17,18] to quantify the bioenergetic signature of human tissues.
It is well established that mitochondrial content is very different depending on the tissue type being considered [19]. This heterogeneity in the phenotype of energetic metabolism in normal human tissues is illustrated by the normalized expression of β-F1 relative GAPDH (Table 1 and Figure 2A). Moreover, mitochondria itself are structurally and molecularly different in the various mammalian cell types, supporting the existence of cell type-specific programs for controlling the biogenesis and activity of the organelle in each tissue [20]. In fact, it has been shown that whereas liver carcinogenesis involves a depletion of the cellular mitochondrial content [6], in kidney, breast, colon, gastric, esophageal, and lung carcinomas, there is a selective repression of the expression of β-F1 [6–8,21]. Both mechanisms limit the mitochondrial activity in the cancer cell, and consistently, we observe that cancer affects the mitochondrial phenotype of the cell in a tissue-specific way (Table 1). Prostate adenocarcinomas failed to reveal significant changes in the bioenergetic signature when compared with paired normal prostate tissue [21], which might suggest that the regulation of enzymatic activities by covalent and/or allosteric modification mediates the regulation of the metabolic flux in carcinogenesis in prostate and, perhaps, in some other tissues. Because the static protein signature of the β-F1/GAPDH ratio in the tumors is depicting the dynamic nature of glucose capture and utilization by aerobic glycolysis [10] and the bioenergetic signature is applicable in a wide range of neoplasias that include some of the most prevalent cancers (breast, lung, and colon), we strongly support its translation to the clinics.
We show that changes in the absolute amount of metabolic biomarkers provide a clear-cut classification of normal and tumor biopsies in a variety of tissues. Consistent with these findings, we have previously shown by Fischer discriminant analysis in large cohorts of breast [8] and lung [7] cancer patients that markers of the bioenergetic signature have classification sensitivity higher than 95%. More recently, we have shown that the bioenergetic signature (β-F1/GAPDH ratio) determined in tumors inversely correlates with the in vivo glucose uptake of lung carcinomas as assessed by positron emission tomography using 2-deoxy-2-[18F]fluoro-d-glucose as probe [10]. Furthermore, in the same study, we showed that the bioenergetic signature also correlates inversely with the rates of aerobic glycolysis [10] and that the β-F1/Hsp60 ratio informs of the rates of oligomycinsensitive respiration in colon cancer cells (Sanchez-Aragó and Cuezva, unpublished observation). Therefore, these results strongly support that changes in the expression of markers of the bioenergetic signature are indeed related with functional changes in the metabolic flux of glycolysis and of oxidative phosphorylation in cells and tumors. The frequent genetic alterations that occur in tumors and that are known to affect both mitochondrial and/or glycolytic metabolism are expected to superimpose to the changes on the expression of the markers of energetic metabolism that are required to sustain cellular proliferation.
Recent findings have established a relevant role for the M2 type of PK in oncogenesis [22,23] and in the promotion of the metabolic shift required for tumorigenesis [22]. Interestingly, the data presented herein with a specific antibody against the M2-PK isoform indicate that normal breast and esophageal tissues have, when compared with lung, a higher quantity of this oncofetal protein, providing the first indication that these human tissues are already outfitted with the less active isoform of PK [22]. This situation would suggest that breast, esophageal, and lung tissues are more prone to malignant transformation. Consistent with this suggestion, carcinogenesis in these tissues, albeit in lung adenocarcinomas, was not accompanied by changes in the content of the PK-M2, thus indicating that they are already equipped with enough complement of PK-M2 to sustain cellular proliferation.
Unexpectedly, we find that tumors from different tissues and/or histological subtypes have the same cellular content of these markers and, therefore, the same bioenergetic signature. It seems that cancer alters the expression of the markers of energetic metabolism of the cell in a tissue-specific manner (Table 1), consistent with the variable cellular response that oncogenes [3,24,25] and tumor suppressors [2,5,26] have on the phenotype of energetic metabolism. However, it is noteworthy that the bioenergetic signature is basically the same regardless of the tissue of origin and the histological type of the tumor (see β-F1/GAPDH ratio in Table 1 and Figure 2F). These findings could support that a common origin for tumors arises from an undifferentiated progenitor cell and that cancer cells undergo a process of dedifferentiation to acquire the traits of embryonic stem cells [27]. In this regard, we suggest that the bioenergetic signature of the tumors and, hence, the expression of markers of energetic metabolism partially respond to the installment of the reductive metabolic program (mainly glycolytic) that sustains cellular proliferation [15,28]. Conversely, the suppression of the tissue-specific differences in the bioenergetic signature of the tumors and its drastic reduction in certain tissues (Table 1) strongly support that containment of the mitochondrial bioenergetic activity in the cancer cell is an event required for tumor progression. Indeed, tumors with a low bioenergetic signature have a worse prognosis [6–8,10] and the activity of mitochondria has been shown to act as a tumor suppressor [12,14,29].
Owing to the convergence of breast, lung, and esophageal tumors on the same bioenergetic signature, it seems that energetic metabolism affords a common target for cancer therapy. In this regard, several groups and biotech companies are currently targeting the proteins of energetic metabolism as a promising approach to eradicate different types of tumors especially in combined therapy [30–33]. Overall, and because the bioenergetic signature provides a predictive marker of the response of tumors to chemotherapy [9], in agreement with the role of mitochondrial oxidative phosphorylation in the execution of cell death [11,13,14], we suggest that its translation to the clinics will benefit cancer patients.
Acknowledgments
The authors thank J. Satrústegui (Centro de Biología Molecular Severo Ochoa) for comments.
Footnotes
This work was supported by grants from Fina Biotech, S.L. and Ministerio de Educación y Ciencia (PET2005-0446; BFU2007-65253), Ministerio de Sanidad y Consumo (PI041255), the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) y RET-2039, del ISCIII, Madrid and Comunidad de Madrid (S-GEN-0269), Spain. Centro de Biología Molecular Severo Ochoa receives an institutional grant from Fundación Ramón Areces.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL, Artemov D, Bedi A, Bhujwalla Z, Chiles K, Feldser D, Laughner E, Ravi R, Simons J, Taghavi P, et al. “The metabolism of tumours”: 70 years later. Novartis Found Symp. 2001;240:251–260. [PubMed] [Google Scholar]
- 5.Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 2008;18:165–173. doi: 10.1016/j.tcb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaria G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 7.Cuezva JM, Chen G, Alonso AM, Isidoro A, Misek DE, Hanash SM, Beer DG. The bioenergetic signature of lung adenocarcinomas is a molecular marker of cancer diagnosis and prognosis. Carcinogenesis. 2004;25:1157–1163. doi: 10.1093/carcin/bgh113. [DOI] [PubMed] [Google Scholar]
- 8.Isidoro A, Casado E, Redondo A, Acebo P, Espinosa E, Alonso AM, Cejas P, Hardisson D, Fresno Vara JA, Belda-Iniesta C, et al. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis. 2005;26:2095–2104. doi: 10.1093/carcin/bgi188. [DOI] [PubMed] [Google Scholar]
- 9.Lin PC, Lin JK, Yang SH, Wang HS, Li AF, Chang SC. Expression of beta-F1-ATPase and mitochondrial transcription factor A and the change in mitochondrial DNA content in colorectal cancer: clinical data analysis and evidence from an in vitro study. Int J Colorectal Dis. 2008;23:1223–1232. doi: 10.1007/s00384-008-0539-4. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Rios F, Sanchez-Arago M, Garcia-Garcia E, Ortega AD, Berrendero JR, Pozo-Rodriguez F, Lopez-Encuentra A, Ballestin C, Cuezva JM. Loss of the mitochondrial bioenergetic capacity underlies the glucose avidity of carcinomas. Cancer Res. 2007;67:9013–9017. doi: 10.1158/0008-5472.CAN-07-1678. [DOI] [PubMed] [Google Scholar]
- 11.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65:3162–3170. doi: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]
- 12.Matsuyama S, Xu Q, Velours J, Reed JC. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol Cell. 1998;1:327–336. doi: 10.1016/s1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 13.Tomiyama A, Serizawa S, Tachibana K, Sakurada K, Samejima H, Kuchino Y, Kitanaka C. Critical role for mitochondrial oxidative phosphorylation in the activation of tumor suppressors Bax and Bak. J Natl Cancer Inst. 2006;98:1462–1473. doi: 10.1093/jnci/djj395. [DOI] [PubMed] [Google Scholar]
- 14.Santamaria G, Martinez-Diez M, Fabregat I, Cuezva JM. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Diez M, Santamaria G, Ortega AD, Cuezva JM. Biogenesis and dynamics of mitochondria during the cell cycle: significance of 3′UTRs. PLoS ONE. 2006;1:e107. [Google Scholar]
- 16.Ortega AD, Sala S, Espinosa E, Gonzalez-Baron M, Cuezva JM. HuR and the bioenergetic signature of breast cancer: a low tumor expression of the RNA-binding protein predicts a higher risk of disease recurrence. Carcinogenesis. 2008;29:2053–2061. doi: 10.1093/carcin/bgn185. [DOI] [PubMed] [Google Scholar]
- 17.Blow N. Antibodies: the generation game. Nature. 2007;447:741–744. doi: 10.1038/447741a. [DOI] [PubMed] [Google Scholar]
- 18.Haab BB. Applications of antibody array platforms. Curr Opin Biotechnol. 2006;17:415–421. doi: 10.1016/j.copbio.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 20.Cuezva JM, Sanchez-Arago M, Sala S, Blanco-Rivero A, Ortega AD. A message emerging from development: the repression of mitochondrial beta-F1-ATPase expression in cancer. J Bioenerg Biomembr. 2007;39:259–265. doi: 10.1007/s10863-007-9087-9. [DOI] [PubMed] [Google Scholar]
- 21.Isidoro A, Martinez M, Fernandez PL, Ortega AD, Santamaria G, Chamorro M, Reed JC, Cuezva JM. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem J. 2004;378:17–20. doi: 10.1042/BJ20031541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 23.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 24.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci USA. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govindarajan B, Sligh JE, Vincent BJ, Li M, Canter JA, Nickoloff BJ, Rodenburg RJ, Smeitink JA, Oberley L, Zhang Y, et al. Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J Clin Invest. 2007;117:719–729. doi: 10.1172/JCI30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261:702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 29.Schulz TJ, Thierbach R, Voigt A, Drewes G, Mietzner B, Steinberg P, Pfeiffer AF, Ristow M. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J Biol Chem. 2006;281:977–981. doi: 10.1074/jbc.M511064200. [DOI] [PubMed] [Google Scholar]
- 30.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Geschwind JF, Ko YH, Torbenson MS, Magee C, Pedersen PL. Novel therapy for liver cancer: direct intraarterial injection of a potent inhibitor of ATP production. Cancer Res. 2002;62:3909–3913. [PubMed] [Google Scholar]
- 32.Simons AL, Ahmad IM, Mattson DM, Dornfeld KJ, Spitz DR. 2-Deoxy-d-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells. Cancer Res. 2007;67:3364–3370. doi: 10.1158/0008-5472.CAN-06-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernlund E, Ihrlund LS, Khan O, Ates YO, Linder S, Panaretakis T, Shoshan MC. Potentiation of chemotherapeutic drugs by energy metabolism inhibitors 2-deoxyglucose and etomoxir. Int J Cancer. 2008;123:476–483. doi: 10.1002/ijc.23525. [DOI] [PubMed] [Google Scholar]