Abstract
Integrating signals from the extracellular matrix through the cell surface into the nucleus is an essential feature of metazoan life. To date, many signal transducers known as shuttle proteins have been identified to act as both a cytoskeletal and a signaling protein. Among them, the most prominent representatives are zyxin and lipoma preferred (translocation) partner (LPP). These proteins belong to the LIM domain protein family and are associated with cell migration, proliferation, and transcription. LPP was first identified in benign human lipomas and was subsequently found to be overexpressed in human malignancies such as lung carcinoma, soft tissue sarcoma, and leukemia. This review portrays LPP in the context of human neoplasia based on a study of the literature to define its important role as a novel protooncogene in carcinogenesis.
Genetic Background
Specialized cell adhesion sites not only play a role in the architectural organization and polarity of the cell but also are dynamic units directly involved in communicational processes. Adhesion receptors and their cytoskeletal partners can regulate nucleocytoplasmatic trafficking of signaling proteins and are thereby capable of influencing gene expression [1–4]. One such protein that may be involved in this process is the LIM-containing protein LPP (lipoma preferred (translocation) partner) [1].
Petit et al. [5] initially described LPP as the preferred translocation partner in a cytogenetic subgroup of lipomas that is characterized by translocations that mainly involve chromosome 12. Several chromosomes were identified as translocation partners of chromosome 12 with 3q27-q28 being preferentially involved. Moreover, it was shown that the high-mobility group (HMG) protein gene HMGA2 at 12q15 is consistently rearranged as a consequence of these translocations. Fusion transcript analysis of HMGA2 in the lipoma cell line Li-501/SV40 unmasked ectopic genetic sequences that originated from the chromosome segment 3q27-q28. These results subsequently yielded in the identification and characterization of the chromosome 3 translocation partner gene named lipoma preferred (translocation) partner (LPP) gene. Northern blot analysis detected a messenger RNA (mRNA) of more than 10 kb in a variety of human tissues [5,6]. The gene was found to span a genomic region of more than 400 kb. Detailed sequencing analysis of LPP revealed an open reading frame of 1836 nucleotides. The main promoter of the LPP gene is located in intron 2 leading to a full-length LPP protein of a highly modular organization [5]. An alternative promoter was found in murine intron 7 leading to a short form of LPP specifically expressed in testis [7]. Alternative LPP variants are also assumed to exist in human because high levels of two smaller LPP gene transcripts have also been detected specifically in the testis [5]. However, these smaller human LPP transcripts have not been characterized in all detail yet.
Functional Organization, Structure, and Binding Partners of LPP
LPP encodes an 80-kDa protein that was characterized as a novel member of group 3 proteins in the LIM family [5]. On the basis of the arrangement, position, and high sequence similarity of the LIM domains of LPP with those of zyxin, LPP was classified as a zyxin family member. This protein family consists of zyxin, ajuba, LIMD1, thyroid receptor-interacting protein 6 (TRIP6, also termed as zyxinrelated protein 1), WT1-interacting protein, migfilin, and LPP [8–14], all of which are strongly involved in cellular motility, proliferation, and tumorigenesis [12,13,15,16].
The LPP gene encodes a proline-rich protein containing a leucine zipper motif in its amino-terminal region and three LIM domains at the carboxy-terminal end (Figure 1) [5]. LIM domains are cysteineand histidine-rich domains that form two zinc fingers capable of mediating protein-protein interactions. Through binding to other partners, LIM proteins participate in diverse cellular processes [17,18].
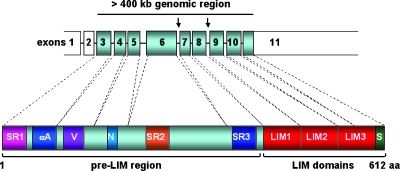
Figure 1.
Schematic illustration of the LPP gene (upper part) and LPP protein (lower part). LPP spans over a genomic region of >400 kb. The gene product consists of 612 amino acids. Several binding sites for other proteins are located within the N-terminal pre-LIM region: the first of two LPP/TRIP6 similar regions (SR1) followed by binding motifs for α-actinin (αA), VASP (V), a nuclear export signal (N), the second LPP/TRIP6 similar region (SR2), and a zyxin/LPP/TRIP6/LIMD1 similar region (SR3). The LIM domain region consists of three tandem LIM domains, each of which is equipped with a double zinc finger structure. The Scrib-binding motif (S) is located at the very C-terminal tail of LPP.
LPP mRNA is ubiquitously expressed in virtually all types of tissues with emphasis on organs of the reproductive tract [19,20]. In contrast to the ubiquitous LPP mRNA presence, the protein is selectively expressed in smooth muscle cells (SMCs), especially in the uterus, stomach, corpus cavernosum, portal vein, aorta, bladder, and ileum [21].
LPP not only is colocalized with vinculin at sites of cell adhesion but also translocalizes into the nucleus [1]. In various benign and malignant tumors, a mutant form of LPP is permanently present in the nucleus [22,23]. Considering the size of LPP, it is unlikely that the protein translocates into the nucleus by mass action and free diffusion. Thus, an active and selective import mechanism seems to be more likely [1,21,22,24].
All three LIM domains of LPP cooperate to provide robust targeting to focal adhesions, with the linker region between LIM domains 1 and 2 playing a pivotal role. Overexpression of the LIM domains results in the depletion of endogenous LPP and vinculin from focal adhesions [1]. Thus, the C-terminal LIM domains of LPP are required for targeting the protein to points of cell-cell and cell-matrix contacts. In contrast, the LIM domains are dispensable for nuclear LPP localization [1]. Beyond its structural function, the C-terminal LIM domains 2 and 3 exhibit transcriptional activity by enhancing transcription factors [25].
The proline-rich pre-LIM region of LPP harbors an intrinsic nuclear export signal [1,22,26] and contains binding sites for α-actinin, vasodilator-stimulated phosphoprotein (VASP), and LIM and SH3 domain protein 1 (LASP-1) [5,26,27]. LPP and its family member zyxin differ in their absolute and functional relevant number of their VASP binding repeats (FPPPPP repeats). Whereas LPP harbors two proline-rich motifs, of which only one binds VASP in vitro [22], zyxin features four functional active FPPPPP repeats [28–30].
VASP has been proposed to increase actin polymerization and force cell protrusion. By binding to the proline-rich repeats of LPP and zyxin, VASP is recruited to specific cellular locations, thus directing changes in actin dynamics [30]. The LIM region of zyxin and LPP acts as a negative autoregulatory domain that normally masks the VASP-recruiting function of the FPPPPP repeats, thereby preventing VASP incorporation into actin networks [31].
Similar to zyxin, LPP was found to bind to α-actinin in vitro and in vivo [26,32]. Studies using the three-hybrid system indicated that zyxin and LPP compete for the same binding site in the central rod of α-actinin containing spectrin-like repeats 2 and 3. On the LPP site, a conserved motif present at the N-terminus is involved in the interaction with α-actinin. Quantitative data obtained with the two- and three-hybrid systems suggested that LPP has a lower affinity for α-actinin than zyxin does. It is likely that this difference leads to slightly variant roles played by LPP and zyxin during the assembly and disassembly of focal adhesions [26].
Although both proteins localize at cell adhesions, zyxin is more prominently distributed along stress fibers. Moreover, there is a difference in the relative abundance of the two proteins. In fibroblasts, the level of zyxin is approximately five times higher than that of LPP, whereas epithelial cells show no significant difference in zyxin/LPP protein levels [22]. Despite the high sequence homology of zyxin and LPP, the observed differences in localization and affinity for the same binding partners might be due to the minor variants in their amino acid sequences.
Recently, it was demonstrated that, LPP-VASP binding is required for anchoring α-actinin at cell-cell contacts, whereas zyxin localization and function at cell-cell contacts is independent of the α-actinin binding site [32], leading to the assumption that zyxin is recruited to cell-cell contacts by other docking proteins, e.g., LASP-1 [33,34].
With its C-terminal tail, LPP interacts directly with the tumor-suppressor Scrib. Interestingly, although all zyxin family members share high sequence homology, only LPP and TRIP6 bind to Scrib [24]. LPP/Scrib will be discussed in more detail later.
Using the C-terminus of LPP as bait in a yeast two-hybrid system, another interaction partner of LPP and a key player in focal adhesion organization was identified — palladin. The palladin-interacting region of LPP was mapped to the first and second LIM domains, whereas the N-terminus of palladin interacts with LPP, both in vitro and in vivo. Like LPP, palladin is highly expressed in differentiated smooth muscle (SM) and localized at focal adhesions, at isolated lamellipodia, and at dense bodies [35]. Both LPP and palladin enhance cell migration and spreading [36], and their expression is markedly decreased in migration-defective focal adhesion kinase (FAK)-null cells [21,37].
Recently, LPP was identified as a substrate of the protein-tyrosine-phosphatase 1B, a negative regulator of multiple signaling pathways downstream of receptor tyrosine kinases and functionally linked to Ras signaling [38,39].
The modular organization of LPP and its multiple interaction sites for cytoskeletal proteins suggest an important role in focal adhesion architecture as a versatile scaffolding and adaptor protein.
An overview of the putative functions and binding partners of LPP is given in Tables 1 and 2, respectively.
Table 1.
Overview of the Putative LPP Functions in the Context of Intracellular LPP Localization.
| Location | Function | Reference(s) |
| Focal contacts/cell-cell contacts | Modulation of cytoskeleton and linker between membrane and cytoskeleton | [21,37] |
| Scaffolding and adaptor protein | [26] | |
| SMC migration | [21,37] | |
| Interaction partner for VASP, zyxin, palladin, Scrib, α-actinin, and LASP-1 | [24,26,27,30,32,35,51] | |
| Recruitment of VASP to cell-cell adhesions | [30,32] | |
| Nucleus | ndependent transcription factor | [1,22,25,51] |
| Transcriptional coactivator of transcription factors such as PEA3 and ER81 | [25,40] |
Table 2.
LPP Binding Partners and Their Putative Roles in Human Cancer.
| Protein | Expression | Localization | Putative Physiological Function | Putative Role in Human Cancer | Reference(s) |
| LASP-1 | Ubiquitous | cc, fa, nuc | scaffolding protein, zyxin-recruitment to fa and cc, NCST | Overexpression in breast, ovarian and liver cancer increases motility and proliferation and is associated with metastasis | [33,34,90–95] |
| VASP | Ubiquitous | cc and fa | Regulation of actin polymerization, involved in cellular migration | Overexpression in lung cancer is positively correlated with grading and staging | [96–100] |
| Overexpression in breast cancer cells enhances migration | |||||
| Palladin | Ubiquitous | cc, fa, Z-discs | Actin organization, involved in cell motility, embryonic development, scar formation in the skin, neuronal development | Palladin mutation causes familial pancreatic cancer | [101–105] |
| Overexpression is found in spontaneous pancreatic cancer | |||||
| Overexpression in breast cancer cells enhances migration | |||||
| Scrib | Ubiquitous | cc, fa, cy | Tumor suppressor; essential for cell shape, polarity, and directed cell migration in early embryonic development | Deregulation leads to breast cancer formation | [51,106–109] |
| Invasive cervical carcinomas show decrease in Scrib due to ubiquitin-mediated degradation by the high-risk papilloma virus E6 proteins | |||||
| Scrib is downregulated in colon cancer | |||||
| PEA3 | Epithelia | nuc | ETS transcription factor regulated by Ras and MAPK pathway | Transcription of prometastatic genes such as serine proteases, urokinase plasminogen activator, COX-2, and several MMPs in various cancers | [40–44,110] |
| ER81 | Mesenchyme | nuc | Related ETS transcription factor of the PEA3 group | Role in breast tumor metastasis as well as HER-2/neu-mediated mammary oncogenesis | [40,44–46] |
| α-actinin | Ubiquitous Z-discs, stress-fibers | cc, fa, F-actin | Scaffolding protein in fa and cc, recruitment of LPP and other signaling proteins involved in cellular motility, actin filament cross-linking, links cytoskeleton to transmembrane proteins | Overexpression of α-actinin isoforms is associated with worse prognosis of patients with astrocytomas, ovarian, breast, lung colorectal, and esophageal cancers | [32,111–115] |
All LPP binding partners are dysregulated in various cancer entities. Many of them contribute to tumor invasiveness and migration.
cc indicates cell-cell contact; cy, cytoplasm; fa, focal adhesion; NCST, nucleocytoplasmatic signal transduction; nuc, nucleus.
LPP and Its Effect on Gene Transcription
LPP has been shown to shuttle into the nucleus and to possess a Crm1-dependent nuclear export signal. The protein exhibits two domains harboring transcriptional activation capacity that coincides with the LIM domains and the proline-rich region of the LPP protein [1,22]. Within the nucleus, LPP is recruited to PEA3-dependent promoter regions and acts as a coregulatory protein enhancing the transactivational potential of PEA3 [40]. PEA3-binding sites have been identified in the regulatory regions of many genes associated with tumorigenesis as well as embryogenesis. A significant fraction of target genes encodes proteases required for degradation of the extracellular matrix (ECM), for example, serine proteases, urokinase plasminogen activator, COX-2, and several matrix metalloproteinases (MMPs) [41,42].
In a recent article, vascular endothelial growth factor was also identified as a potential PEA3 target gene [43]. Matrix metalloproteinases in turn contribute to metastatic dissemination of tumor cells by degrading the ECM, and their deregulated expression has been associated with the capability of tumor cells to metastasize [41,44,45].
LPP was shown to substantially increase the reporter gene activity of the target gene COX-2 in a dose-dependent manner when cotransfected with PEA3 and mitogen-activated kinase/ERK kinase [40]. However, the role of the mitogen-activated protein kinase (MAPK) pathway in controlling the activity of the PEA3-LPP complex is still under investigation.
LPP also interacts with the related ETS transcription factor ER81 by enhancing its transactivational potential [40]. Like PEA3, ER81 has been shown to play an important role in breast tumor metastasis as well as human epidermal growth factor receptor 2/neu-mediated mammary oncogenesis [46]. In contrast, less effect of LPP is seen on the third member of this subfamily, ets-related molecule, suggesting a specificity of action [40]. However, the knowledge about the effects of ER81 and its target genes in the context of LPP coactivation is fragmentary.
Recent work has shown that LPP can also act as an independent transcription factor with or without binding to other transcription factors. Moreover, HMGA2/LPP fusion proteins (see also LPP Fusion Genes section) retain the transactivational functions of the LPP LIM domains and thus function as genuine transcription factors [25,47,48].
LPP in Early Embryonic Development
LPP is localized at focal adhesions and cell-cell contacts and is involved in the regulation of SMC migration [49]. A known interaction partner of LPP in human is the tumor suppressor protein Scrib [11,24].
Knockdown of Scrib expression during zebrafish embryonic development results in defects of convergence and extension (C&E) movements, which appear during gastrulation and cause elongation of the anterior-posterior body axis. During vertebrate gastrulation and neurulation, C&E reflects the medial migration and intercalation of mesodermal and neuroectodermal cells. The mediolateral cell polarization mediated through C&E is controlled by the noncanonical wingless (Wnt) signaling pathway, which represents the vertebrate planar cell polarity pathway [50–52].
Interestingly, silencing of LPP in zebrafish embryos also results in impaired C&E movements, phenocopying noncanonical wingless-type MMTV integration site family member 11 (Wnt11) signaling mutants. The defective dorsal convergence movements are associated with a reduced ability of embryonic cells to migrate along straight paths, affecting both mesodermal and neuroectodermal structures. The embryos with decreased LPP expression showed a shortened body axis, a phenotype also observed for other genes involved in gastrulation [51,53,54]. Furthermore, the expression of LPP is significantly reduced in embryos with morpholino-mediated knockdown of Wnt11 and in embryos overexpressing Wnt11 or a dominantnegative form of Rho kinase 2 (ROK-2), a downstream effector of Wnt11. These data suggest that LPP expression is dependent on non-canonical Wnt signaling and that LPP acts downstream of Wnt11 and ROK-2 [51]. Likewise, an involvement of LPP in ROK-dependent signaling pathways was demonstrated in human iliac vein SMCs [37].
Surprisingly, Lpp knockout mice do not present defects of C&E during embryonic development. Moreover, Lpp-/- mice show no increased mortality compared with wild-type littermates and reach adulthood without displaying any obvious abnormalities [7]. This could be in part explained by the functional redundancy of zyxin family proteins because zyxin knockout mice also lack any macroscopic abnormalities [55]. However, Scrib and α-actinin mRNA levels were significantly reduced in Lpp-/- mice. In addition, Lpp-/- murine embryonic fibroblasts show reduced migration capacity and viability [7].
LPP in SM Physiology
Little is known about the physiological role of LPP and its operations to ensure normal protein signaling, but to date, LPP is best studied in SMCs [21,36,37,49,56].
Immunofluorescence microscopy demonstrated the almost selective expression of LPP in vascular and visceral SMC [21,37]. Consistently, single nucleotide polymorphisms in the LPP gene are associated with severe villus atrophy of the small intestine in celiac disease [57]. LPP has also been detected among a core of computationally predicted SM-specific genes from expressed sequence tag data [58] supporting the idea of LPP as a novel marker for SMC differentiation [37].
In other mature (noncultured) tissues, including heart and skeletal muscle, the protein is barely present, and its concentration correlates with the levels of the SM marker α-actin. LPP is present at 100-fold higher protein level in SM-rich tissues, including bladder, uterus, ileum, and aorta, than in non-SM organs, such as liver, heart, or brain. In healthy SM tissues, Gorenne et al. [21] reported detection of LPP in punctate foci at the cell surface where it colocalized with vinculin in peripheral membrane-dense bodies involved in actin filament attachment sites. Overexpression of LPP increased epidermal growth factor-stimulated migration of vascular SMCs, suggesting the participation of LPP in cell motility [21]. Conversely, LPP knockout led to reduced migration of murine embryonic fibroblasts [7].
trans-Retinoic acid treatment of A404 cells was shown to significantly increase LPP levels as well as SM α-actin, myosin heavy chain and smoothelin mRNA levels in a ROK-dependent manner. Treatment with the ROK inhibitor Y-27632 led to dissociated focal adhesions as well as reduced LPP staining at the cell periphery. Moreover, enhanced nuclear accumulation of LPP is observed after cell incubation with nuclear export inhibitor leptomycin B [37]. Similarly, treatment of human iliac vein SMCs (HIVS) with leptomycin B caused accumulation of LPP in the nucleus [21]. Therefore, LPP apparently has a potential for relocating to the nucleus through a shuttling mechanism that is sensitive to inhibition of ROK. Cytoplasmic-nuclear shuttling has also been demonstrated for zyxin [59] and TRIP6 [60], suggesting this as a common property for the group 3 LIM proteins.
Thus, it is intriguing to consider a possible signaling role for LPP in SMCs, possibly in the transcriptional feedback control of cytoskeletal proteins.
In adult pig hearts, intact coronary arteries exhibit strong immunostaining for LPP that colocalized with SM α-actin and smoothelin in the tunica media. In contrast, no expression of LPP was found in the adventitial layer. In injured vessels 28 days after stent implantation, neointimal cells migrating around the stent lesion were positive for LPP and SM α-actin but not for smoothelin [37]. Adventitial cells remained negative, whereas LPP expression could be detected in the walls of microvessels located in the adventitial layer [37]. Similar findings were reported for remodeling of injured rat aortic arteries [36]. These dynamic expression patterns in injured arteries suggest that LPP may act in concert with multiple players to facilitate cytoskeletal remodeling events needed to accomplish the transition from stationary to migrating SMCs in vivo [36,49,56].
Overexpression of myocardin, a well-known cofactor of the serum response factor (SRF) [61], significantly increased LPP mRNA expression in A404 cells [37]. Interestingly, inactivation of RhoA decreased myocardin mRNA expression in retinoic acid-treated A404 cells and HIVS. In addition, LPP silencing significantly decreased SMC migration. LPP expression was also markedly reduced in FAK-null cells known to show impaired migration [36]. Consistently, overexpression of LPP in cultured HIVS increased epidermal growth factor-stimulated cell migration by approximately 2.5-fold in a transwell chemotaxis assay. Moreover, expression of LPP in migration-defective FAK-null cells led to a significant increase in cell spreading on a fibronectin matrix. This is in agreement with data demonstrating an up-regulation of LPP in FAK-/- cells with restored FAK expression [36].
These results suggest that levels of LPP in SMCs are controlled by FAK signaling and raise questions about the role of FAK and LPP as determinants of SMC phenotype [21,37].
Recently, Gorenne et al. [37] reported the LPP gene being responsive to RhoA-mediated signaling pathways that activate myocardindependent transcription in SMC differentiation. In an extending work, Petit et al. [62] focused on the transcriptional regulation of the LPP gene by myocardin. The murine LPP gene contains three evolutionarily conserved CArG boxes. One box is part of an alternative promoter in intron 2. Petit et al. [62] showed that this promoter is directly regulated by SRF and myocardin, which thereby modulate transcription of LPP in SMC. Overexpression of myocardin resulted in an approximately three-fold increase of porcine LPP gene expression after 48 hours [37]. Thus, LPP was classified as a novel myocardin/SRF target gene [37,62]. In conclusion, LPP seems to be a SRF/myocardin- and RhoA/ROK-dependent SMC differentiation marker that plays a role in regulating SMC migration [21,37,62].
LPP in Tumorigenesis and Cancer
After the discovery of LPP in lipoma, many other human tumors have been identified, which take advantage of the physiological LPP properties to increase their malignant potential. To date, three different strategies have been identified, which allow tumors to usurp the physiological LPP signaling mechanism to enhance their runaway proliferation:
Formation of oncogenic LPP fusion genes
Overexpression of LPP leading to the disruption of the normal LPP signaling pathway
Abuse of the LPP shuttle mechanism for oncogenic signaling to the nucleus
A) LPP Fusion Genes
LPP is a frequent fusion partner for several oncogenes. The most prominent genes are members of the “high mobility group AT hook” HMGA protein family consisting of four proteins: HMGA1a, HMGA1b, HMGA1c, and HMGA2. All are nuclear factors characterized by three DNA-binding domains, called AT hooks, and an acidic carboxy-terminal tail. The proteins are architectural transcription factors that both positively and negatively regulate the transcription of a variety of genes [63–66], thereby influencing a considerable number of cellular processes including cell growth, proliferation, differentiation, and apoptosis. HMGAs do not directly activate transcription but rather regulate gene expression by changing DNA conformation [63].
Both HMGA1 and HMGA2 are barely detectable in normal adult tissue but are abundantly and ubiquitously expressed during embryonic development [63,65,66]. In malignant epithelial tumors as well as in leukemia, however, expression of HMGA1 is again strongly upregulated to embryonic levels, thus leading to ectopic expression of (fetal) target genes. HMGA2 overexpression has a causal role in inducing neoplasia and a malignant phenotype [5,63,67].
HMGA genes are often involved in chromosomal rearrangements. Such translocations are mostly detected in benign tumors of mesenchymal origin and are believed to be one of the most common chromosomal rearrangements in human neoplasia [63,68–70]. In most cases, HMGA2 alterations involve breaks within the third intron of the gene resulting in aberrant transcripts carrying exons 1 to 3, which encode the three DNA-binding domains, fused to ectopic sequences [64].
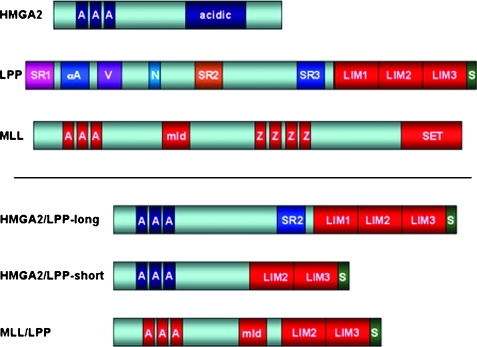
The LPP gene is the most frequent translocation partner of HMGA2 in a subgroup of lipomas. Moreover, LPP is also frequently rearranged in cases without cytogenetic detectable involvement of 3q27–q28. To date, two alternative HMGA2/LPP hybrid transcripts have been characterized. These two transcripts differ in the number of either two or three LIM domains in the predicted HMGA2/LPP fusion protein [71–73] (Figures 2 and 3, A and B). Interestingly, both forms are expressed only in the nucleus [22]. A truncated form of HMGA2 carrying the three DNA-binding domains of HMGA2 and the LIM domains of the LPP gene caused malignant transformation of NIH3T3 cells, whereas the wild-type HMGA2 does not exert any transforming activity [67]. These findings indicate that, specifically, the fusion of HMGA2 together with LPP achieves oncogenic potential [22,67].
Figure 2.
Schematic illustration of the HMGA2, LPP, and MLL proteins (upper panel) and the pathogenic fusion proteins found in human neoplasia to date (lower panel). AT hooks (A) are specialized DNA-binding motifs, acidic domain (acidic), α-actinin binding site (αA), VASP binding sites (V), nuclear export signal (N), LPP/TRIP6 similar region 1 (SR1), LPP/TRIP6 similar region 2 (SR2), zyxin/LPP/TRIP6/LIMD1 similar region (SR3), methyl-transferase-like domain (mld), PHD zinc fingers (Z), set domain (SET), and Scrib-binding motif (S).
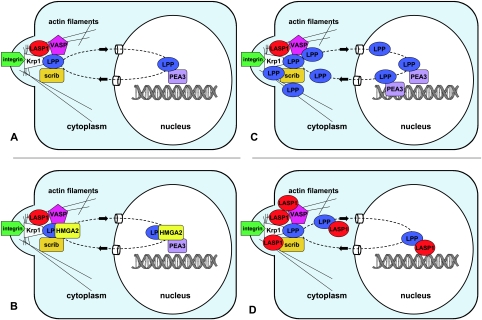
Figure 3.
Schematic illustration of the hypothetical, physiological (A), and pathological (C-D) LPP signaling in human neoplasia. (A) Physiological LPP signaling: LPP permanently shuttles from cytosol into the nucleus and back, thereby transducing signals from the cell surface to the transcription machinery and possibly vice versa. (B) Abnormal LPP signaling through formation of fusion proteins with altered transcriptional signature. As an example, the most common fusion protein of the truncated HGMA2 with the LIM domains of LPP is shown. (C) LPP overexpression leads to enhanced and possibly changed transcription of LPP target genes. (D) Overexpression of LPP binding partners: overexpression of, for example, LASP-1 in tumor cells may lead to enhanced protein binding to LPP, increased nuclear transportation of the shuttle partners, and potentially pathophysiological gene regulation.
The HMGA2/LPP fusion protein activates transcription through the well-characterized PRDII element, which is a part of the interferon-β (IFN-β) enhancer and known to bind to HMGA2. It was also shown that HMGA2/LPP activates transcription through the HLA-B associated transcript 1 element of the rhodopsin promoter, an HMGA1-binding element. Finally, in a number of lipomas, HMGA2/LPP and HMGA2 are coexpressed with wild-type HMGA2 even augmenting the transactivational functions of ectopic HMGA2/LPP. These results support the concept that the transactivational functions of the novel HMGA2/LPP transcription factor contribute to lipomagenesis [25,64,74].
Recent work showed that transforming growth factor β (TGF-β)-induced expression of HMGA2 was associated with the development of epithelial-mesenchymal-transition (EMT) [75]. Epithelial-mesenchymaltransition occurs during embryogenesis, carcinoma invasiveness, and metastatic dissemination and can be regulated by TGF-β signaling through intracellular Smad transducers. The specific molecular mechanisms that control the initiation of EMTare, to date, widely unknown. Transcriptomic analysis, however, revealed that the Smad pathway induces the HMGA2 gene during EMT. Endogenous HMGA2 mediates EMT by TGF-β, whereas ectopic HMGA2 causes irreversible EMT characterized by severe E-cadherin suppression. HMGA2 controls the expression of four known regulators of EMT, the zinc-finger proteins Snail and Slug, the basic helix-loop-helix protein Twist, and inhibitor of differentiation 2 [75]. Thus, ectopic overexpression of a hybrid HMGA2/LPP gene possibly provides a mechanistic explanation for a disruption of the physiological HMGA2 pathway resulting in tumor development.
The existence of identical fusion genes in different types of tumors is a very common strategy of these cells to promote proliferation. Consistently, HMGA2/LPP fusion transcripts have been reported in lipoma, pulmonary chondroid hamartoma (PHC), and soft tissue chondroma [47,48,71–73,76–78]. The high frequency of t(3q27–28;12q14–15) in lipomas and PCHs renders the HMGA2/LPP fusion gene the most common fusion gene in human tumors [77,79]. All tumors characterized by this HMGA2/LPP gene product share the same structure, that is, a protein composed of the AT hooks of HMGA2 and the LIM domains of LPP. Its common occurrence in PCHs indicates the absence of a larger deletion of the LPP locus accompanying the translocation [79,80], as described in a lipoma [73,74]. Moreover, in primary central nervous system lymphomas (PCNSLs, diffuse large B-cell lymphomas confined to the brain), LPP was fused to the BCL6 gene [81]. BCL6 is a protooncogene encoding a nuclear transcriptional repressor, with pivotal roles in germinal center formation and regulation of lymphocyte function, differentiation, and survival. BCL6 suppresses p53 in germinal center B cells and its constitutive expression can protect B-cell lines from apoptosis induced by DNA damage [82]. Little is known about chromosomal aberrations underlying PCNSLs. However, fluorescence in situ hybridization analysis of 41 PCNSLs revealed that 14 tumors (34%) carried a breakpoint in the BCL6 locus. All breakpoints were located within the BCL6 major translocation cluster. In some cases, a deletion in 3q leads to the loss of an 837-kb fragment extending from the first intron of BCL6 to the third intron of the LPP gene. This deletion may bring the BCL6 gene under the control of regulatory elements of the LPP gene. DNA sequencing analysis of the junctional sequences provided evidence that aberrant class switch recombination or somatic hypermutation may be involved in the generation of BCL6 translocations [81]. In addition, in a more recent study, a high LPP expression was associated with better treatment response of patients with B-cell lymphomas. This improved clinical outcome was possibly because of LPP-induced expression of genes responsible for the increased differentiation of the leukemic B cells [83].
Another gene, which was found to be rearranged with LPP, is MLL, the mixed lineage leukemia gene. MLL/LPP fusion transcripts were detected in a patient with secondary leukemia (AML-M5, FAB classification) after treatment with DNA topoisomerase II inhibitors. Fluorescence in situ hybridization and Southern blot analyses identified a rearrangement in the MLL gene because of a novel t(3;11) (q28;q23) chromosomal translocation in AML cells 3 years after chemotherapy for a follicular lymphoma. Through inverse polymerase chain reaction, the LPP gene on 3q28 was identified as the MLL fusion partner. The predicted MLL/LPP fusion protein included the AT hook motifs and methyltransferase domain of MLL joined to the two last LIM domains of LPP. The reciprocal LPP/MLL transcript, predicted to include the proline-rich and leucine zipper motifs, and the first LIM domain of LPP were also detected by reverse transcription-polymerase chain reaction. The new tumor-specific fusion proteins MLL/LPP and LPP/MLL contain many features present in other MLL rearrangements [84] (Figure 2) and, as such, could be used as promising novel drug targets owing to their unique presence in such tumors and absence in normal tissue. Interestingly, all these tumor-specific fusion proteins, composed of AT hooks from other proteins, are mainly localized within the nucleus [22] and possibly contribute to altered LPP transcriptional targeting [63,64] (Figure 3B).
B) Overexpression of LPP Disrupts Normal LPP Signaling
LPP is highly upregulated by gains on 3q in squamous cell lung carcinomas turning LPP into a potential candidate for pathogenesis and diagnosis of lung cancer [85]. Amplification of the 3q region accompanied by a 20-fold overexpression of LPP was detected in primary sarcomas and sarcoma cell lines pointing to an essential role of LPP overexpression in tumorigenesis not only of epithelia but also of tissues of mesenchymal origin [86].
LPP is a coregulatory binding partner for PEA3 [40]. PEA3 is a member of an ETS domain transcription factor subfamily and regulated by a number of signaling cascades including the MAPK pathways and Ras. PEA3 activates gene expression and is thought to play an important role in promoting tumor metastasis [44,45]. LPP forms a complex with PEA3 and is found to be associated with PEA3-regulated promoters. By manipulating LPP levels, it is possible to upregulate the transactivation capacity of PEA3 in a dose-dependent manner [40]. Therefore, overexpression of LPP leads to an up-regulation of PEA3 target genes. In a similar fashion, LPP can functionally interact with the related PEA3 family member ER81. Thus, beyond its own function as an independent transcription factor, LPP has an additional nuclear function as a transcriptional coactivator [40] (Table 1).
Among the PEA3 target genes are several MMPs [40]. Matrix metalloproteinases in turn contribute to metastatic dissemination of tumor cells by degrading the ECM, and their deregulated expression has been associated with the capability of tumor cells to metastasize. In fact, PEA3 group genes are often overexpressed in different types of cancer that also overexpress MMPs and that display a disseminating phenotype [41,44,45].
Hence, it is tempting to speculate that overexpression of LPP could cause an up-regulation of PEA3 targets such as MMPs resulting in an increased invasion and metastasis potential (Figure 3C). In support of this hypothesis, nonmetastatic breast cancer cells become metastatic when PEA3 is ectopically overexpressed [44,45]. LPP might be the link communicating changes in cytoskeleton or ECM contacts associated with invasion and metastasis into PEA3-mediated changes in gene expression profiles.
In comparison to LPP, zyxin overexpression has also been associated with increased invasiveness and migrational abilities of human hepatocellular carcinoma cells [87]. However, zyxin gene transfer into an Ewing sarcoma model resulted in the inhibition of tumor growth accompanied by reconstitution of zyxin-rich focal adhesions, reorganization of the actin cytoskeleton, and decreased cell motility [88]. Thus, overexpression does not represent per se a consistent mechanism for tumorigenesis but seems to depend on the cellular context.
C) LPP, an Abused Shuttle Protein
As LPP continually shuttles between the cell periphery and the nucleus, it represents a potential novel link between cell surface events and changes in gene expression [1]. LASP-1, another focal adhesion protein, that is found to be overexpressed in human breast and ovarian cancer, is known to interact with zyxin and LPP [27,33,34,89]. Although LASP-1 has no classic nuclear import signal of its own, it translocates into the nucleus. Therefore, it was speculated that LASP-1 might bind to shuttle proteins such as zyxin and LPP to use their transportation potential into the nucleus [2]. Hypothetically, overexpression of LPP binding partners such as LASP-1 in human neoplasia could interfere with normal LPP signaling, resulting in an altered transcriptional signature of LPP that in turn could contribute to tumorigenesis (Figure 3D). A summary of the putative roles of LPP binding partners in human cancer is depicted in Table 2.
Conclusions and Future Prospects
In synopsis, it seems to be a common mechanism for tumors showing a high rate of genetic instability to fuse protooncogenes such as HMGA2, MLL, and BCL6 with the LPP gene. These translocations lead to the formation of novel fusion gene products that are highly specific for tumor cells, turning these fusion proteins into interesting candidates for the rational design of specific antitumor medication. Furthermore, overexpression of LPP itself or its interacting binding partners seems to be another mechanism for tumorigenesis through interference with normal LPP signaling.
To date, there is an increasing line of evidence that LPP plays an important role in the formation of a variety of human cancers. The physiological role of LPP, however, remains largely unknown. Thus, more work has to be undertaken to reveal the physiological function of LPP and to delineate the functional differences between normal and altered LPP signaling.
Acknowledgments
The authors thank Barbara Lechner and Veit Buchholz for their helpful commentary and critical reading of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Authors' contributions: T.G.P.G. and S.M.P. drafted and wrote the paper and designed the tables and the figures. S.M.P. provided genetic guidance. E.B. corrected and finalized the manuscript. All authors read and approved the final manuscript.
References
- 1.Petit MM, Meulemans SM, Van de Ven WJ. The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J Biol Chem. 2003;278:2157–2168. doi: 10.1074/jbc.M206106200. [DOI] [PubMed] [Google Scholar]
- 2.Grunewald TG, Butt E. The LIM and SH3 domain protein family: structural proteins or signal transducers or both? Mol Cancer. 2008;7:31. doi: 10.1186/1476-4598-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benmerah A, Scott M, Poupon V, Marullo S. Nuclear functions for plasma membrane-associated proteins? Traffic. 2003;4:503–511. doi: 10.1034/j.1600-0854.2003.00102.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Gilmore TD. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim Biophys Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 5.Petit MM, Mols R, Schoenmakers EF, Mandahl N, Van de Ven WJ. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics. 1996;36:118–129. doi: 10.1006/geno.1996.0432. [DOI] [PubMed] [Google Scholar]
- 6.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 7.Vervenne HB, Crombez KR, Delvaux EL, Janssens V, Van de Ven WJ, Petit MM. Targeted disruption of the mouse lipoma preferred partner gene. Biochem Biophys Res Commun. 2008;379:368–373. doi: 10.1016/j.bbrc.2008.12.074. [DOI] [PubMed] [Google Scholar]
- 8.Srichai MB, Konieczkowski M, Padiyar A, Konieczkowski DJ, Mukherjee A, Hayden PS, Kamat S, El-Meanawy MA, Khan S, Mundel P, et al. A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J Biol Chem. 2004;279:14398–14408. doi: 10.1074/jbc.M314155200. [DOI] [PubMed] [Google Scholar]
- 9.Takafuta T, Saeki M, Fujimoto TT, Fujimura K, Shapiro SS. A new member of the LIM protein family binds to filamin B and localizes at stress fibers. J Biol Chem. 2003;278:12175–12181. doi: 10.1074/jbc.M209339200. [DOI] [PubMed] [Google Scholar]
- 10.Tu Y, Wu S, Shi X, Chen K, Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 11.Petit MM, Crombez KR, Vervenne HB, Weyns N, Van de Ven WJ. The tumor suppressor Scrib selectively interacts with specific members of the zyxin family of proteins. FEBS Lett. 2005;579:5061–5068. doi: 10.1016/j.febslet.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol Biol Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huggins CJ, Andrulis IL. Cell cycle regulated phosphorylation of LIMD1 in cell lines and expression in human breast cancers. Cancer Lett. 2008;267:55–66. doi: 10.1016/j.canlet.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Huggins CJ, Gill M, Andrulis IL. Identification of rare variants in the hLIMD1 gene in breast cancer. Cancer Genet Cytogenet. 2007;178:36–41. doi: 10.1016/j.cancergencyto.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Yi J, Beckerle MC. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics. 1998;49:314–316. doi: 10.1006/geno.1998.5248. [DOI] [PubMed] [Google Scholar]
- 16.Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 18.Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 19.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorenne I, Nakamoto RK, Phelps CP, Beckerle MC, Somlyo AV, Somlyo AP. LPP, a LIM protein highly expressed in smooth muscle. Am J Physiol Cell Physiol. 2003;285:C674–C685. doi: 10.1152/ajpcell.00608.2002. [DOI] [PubMed] [Google Scholar]
- 22.Petit MM, Fradelizi J, Golsteyn RM, Ayoubi TA, Menichi B, Louvard D, Van de Ven WJ, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–129. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 24.Petit MM, Meulemans SM, Alen P, Ayoubi TA, Jansen E, Van de Ven WJ. The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 2005;6:1. doi: 10.1186/1471-2121-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crombez KR, Vanoirbeek EM, Van de Ven WJ, Petit MM. Transactivation functions of the tumor-specific HMGA2/LPP fusion protein are augmented by wild-type HMGA2. Mol Cancer Res. 2005;3:63–70. doi: 10.1158/1541-7786.MCR-04-0181. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Zhuang L, Reinhard M, Trueb B. The lipoma preferred partner LPP interacts with alpha-actinin. J Cell Sci. 2003;116:1359–1366. doi: 10.1242/jcs.00309. [DOI] [PubMed] [Google Scholar]
- 27.Keicher C, Gambaryan S, Schulze E, Marcus K, Meyer HE, Butt E. Phosphorylation of mouse LASP-1 on threonine 156 by cAMP- and cGMP-dependent protein kinase. Biochem Biophys Res Commun. 2004;324:308–316. doi: 10.1016/j.bbrc.2004.08.235. [DOI] [PubMed] [Google Scholar]
- 28.Beckerle MC. Spatial control of actin filament assembly: lessons from Listeria. Cell. 1998;95:741–748. doi: 10.1016/s0092-8674(00)81697-9. [DOI] [PubMed] [Google Scholar]
- 29.Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 30.Hansen MD, Beckerle MC. Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J Biol Chem. 2006;281:16178–16188. doi: 10.1074/jbc.M512771200. [DOI] [PubMed] [Google Scholar]
- 31.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 32.Hansen MD, Beckerle MC. alpha-Actinin links LPP, but not zyxin, to cadherin-based junctions. Biochem Biophys Res Commun. 2008;371:144–148. doi: 10.1016/j.bbrc.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunewald TG, Kammerer U, Schulze E, Schindler D, Honig A, Zimmer M, Butt E. Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp Cell Res. 2006;312:974–982. doi: 10.1016/j.yexcr.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Grunewald TG, Kammerer U, Winkler C, Schindler D, Sickmann A, Honig A, Butt E. Overexpression of LASP-1 mediates migration and proliferation of human ovarian cancer cells and influences zyxin localisation. Br J Cancer. 2007;96:296–305. doi: 10.1038/sj.bjc.6603545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goicoechea S, Arneman D, Disanza A, Garcia-Mata R, Scita G, Otey CA. Palladin binds to Eps8 and enhances the formation of dorsal ruffles and podosomes in vascular smooth muscle cells. J Cell Sci. 2006;119:3316–3324. doi: 10.1242/jcs.03076. [DOI] [PubMed] [Google Scholar]
- 36.Jin L, Kern MJ, Otey CA, Wamhoff BR, Somlyo AV. Angiotensin II, focal adhesion kinase, and PRX1 enhance smooth muscle expression of lipoma preferred partner and its newly identified binding partner palladin to promote cell migration. Circ Res. 2007;100:817–825. doi: 10.1161/01.RES.0000261351.54147.de. [DOI] [PubMed] [Google Scholar]
- 37.Gorenne I, Jin L, Yoshida T, Sanders JM, Sarembock IJ, Owens GK, Somlyo AP, Somlyo AV. LPP expression during in vitro smooth muscle differentiation and stent-induced vascular injury. Circ Res. 2006;98:378–385. doi: 10.1161/01.RES.0000202802.34727.fd. [DOI] [PubMed] [Google Scholar]
- 38.Mertins P, Eberl HC, Renkawitz J, Olsen JV, Tremblay ML, Mann M, Ullrich A, Daub H. Investigation of protein-tyrosine phosphatase 1B function by quantitative proteomics. Mol Cell Proteomics. 2008;7:1763–1777. doi: 10.1074/mcp.M800196-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dube N, Cheng A, Tremblay ML. The role of protein tyrosine phosphatase 1B in Ras signaling. Proc Natl Acad Sci USA. 2004;101:1834–1839. doi: 10.1073/pnas.0304242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo B, Sallis RE, Greenall A, Petit MM, Jansen E, Young L, Van de Ven WJ, Sharrocks AD. The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol Cell Biol. 2006;26:4529–4538. doi: 10.1128/MCB.01667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higashino F, Yoshida K, Noumi T, Seiki M, Fujinaga K. Ets-related protein E1A-F can activate three different matrix metalloproteinase gene promoters. Oncogene. 1995;10:1461–1463. [PubMed] [Google Scholar]
- 42.Nerlov C, Rorth P, Blasi F, Johnsen M. Essential AP-1 and PEA3 binding elements in the human urokinase enhancer display cell type-specific activity. Oncogene. 1991;6:1583–1592. [PubMed] [Google Scholar]
- 43.Hua D, Chen B, Bai M, Yu H, Wu X, Jin W. PEA3 activates VEGF transcription in T47D and SKBR3 breast cancer cells. Acta Biochim Biophys Sin (Shanghai) 2009;41:63–68. doi: 10.1093/abbs/gmn007. [DOI] [PubMed] [Google Scholar]
- 44.de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 45.de Launoit Y, Chotteau-Lelievre A, Beaudoin C, Coutte L, Netzer S, Brenner C, Huvent I, Baert JL. The PEA3 group of ETS-related transcription factors. Role in breast cancer metastasis. Adv Exp Med Biol. 2000;480:107–116. doi: 10.1007/0-306-46832-8_13. [DOI] [PubMed] [Google Scholar]
- 46.Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr Biol. 2001;11:1739–1748. doi: 10.1016/s0960-9822(01)00536-x. [DOI] [PubMed] [Google Scholar]
- 47.Nilsson M, Mertens F, Hoglund M, Mandahl N, Panagopoulos I. Truncation and fusion of HMGA2 in lipomas with rearrangements of 5q32→33 and 12q14→q15. Cytogenet Genome Res. 2006;112:60–66. doi: 10.1159/000087514. [DOI] [PubMed] [Google Scholar]
- 48.Nilsson M, Panagopoulos I, Mertens F, Mandahl N. Fusion of the HMGA2 and NFIB genes in lipoma. Virchows Arch. 2005;447:855–858. doi: 10.1007/s00428-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 49.Majesky MW. Organizing motility: LIM domains, LPP, and smooth muscle migration. Circ Res. 2006;98:306–308. doi: 10.1161/01.RES.0000208059.16734.35. [DOI] [PubMed] [Google Scholar]
- 50.Barrow JR. Wnt/PCP signaling: a veritable polar star in establishing patterns of polarity in embryonic tissues. Semin Cell Dev Biol. 2006;17:185–193. doi: 10.1016/j.semcdb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Vervenne HB, Crombez KR, Lambaerts K, Carvalho L, Koppen M, Heisenberg CP, Van de Ven WJ, Petit MM. Lpp is involved in Wnt/PCP signaling and acts together with Scrib to mediate convergence and extension movements during zebrafish gastrulation. Dev Biol. 2008;320:267–277. doi: 10.1016/j.ydbio.2008.05.529. [DOI] [PubMed] [Google Scholar]
- 52.Dreesen O, Brivanlou AH. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- 53.Carreira-Barbosa F, Kajita M, Morel V, Wada H, Okamoto H, Martinez Arias A, Fujita Y, Wilson SW, Tada M. Flamingo regulates epiboly and convergence/extension movements through cell cohesive and signalling functions during zebrafish gastrulation. Development. 2009;136:383–392. doi: 10.1242/dev.026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tada M, Smith JC. Xwnt11 is a target of Xenopus Brachyury: regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development. 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- 55.Hoffman LM, Nix DA, Benson B, Boot-Hanford R, Gustafsson E, Jamora C, Menzies AS, Goh KL, Jensen CC, Gertler FB, et al. Targeted disruption of the murine zyxin gene. Mol Cell Biol. 2003;23:70–79. doi: 10.1128/MCB.23.1.70-79.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin L, Hastings NE, Blackman BR, Somlyo AV. Mechanical properties of the extracellular matrix alter expression of smooth muscle protein LPP and its partner palladin; relationship to early atherosclerosis and vascular injury. J Muscle Res Cell Motil. 2009 doi: 10.007/s10974-009-9173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romanos J, Barisani D, Trynka G, Zhernakova A, Bardella MT, Wijmenga C. Six new celiac disease loci replicated in an Italian population confirm association to celiac disease. J Med Genet. 2008;46:60–63. doi: 10.1136/jmg.2008.061457. [DOI] [PubMed] [Google Scholar]
- 58.Nelander S, Mostad P, Lindahl P. Prediction of cell type-specific gene modules: identification and initial characterization of a core set of smooth muscle-specific genes. Genome Res. 2003;13:1838–1854. doi: 10.1101/gr.1197303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Gilmore TD. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim Biophys Acta. 2001;1538:260–272. doi: 10.1016/s0167-4889(01)00077-5. [DOI] [PubMed] [Google Scholar]
- 61.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 62.Petit MM, Lindskog H, Larsson E, Wasteson P, Athley E, Breuer S, Angstenberger M, Hertfelder D, Mattsson E, Nordheim A, et al. Smooth muscle expression of lipoma preferred partner is mediated by an alternative intronic promoter that is regulated by serum response factor/myocardin. Circ Res. 2008;103:61–69. doi: 10.1161/CIRCRESAHA.108.177436. [DOI] [PubMed] [Google Scholar]
- 63.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions. Int J Oncol. 2008;32:289–305. [Review] [PubMed] [Google Scholar]
- 64.Ashar HR, Fejzo MS, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell. 1995;82:57–65. doi: 10.1016/0092-8674(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 65.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 66.Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A, Manfioletti G, Giancotti V. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 2004;574:1–8. doi: 10.1016/j.febslet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 67.Fedele M, Berlingieri MT, Scala S, Chiariotti L, Viglietto G, Rippel V, Bullerdiek J, Santoro M, Fusco A. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene. 1998;17:413–418. doi: 10.1038/sj.onc.1201952. [DOI] [PubMed] [Google Scholar]
- 68.Fedele M, Battista S, Manfioletti G, Croce CM, Giancotti V, Fusco A. Role of the high mobility group A proteins in human lipomas. Carcinogenesis. 2001;22:1583–1591. doi: 10.1093/carcin/22.10.1583. [DOI] [PubMed] [Google Scholar]
- 69.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 70.Young AR, Narita M. Oncogenic HMGA2: short or small? Genes Dev. 2007;21:1005–1009. doi: 10.1101/gad.1554707. [DOI] [PubMed] [Google Scholar]
- 71.Lemke I, Rogalla P, Bullerdiek J. Large deletion of part of the HMGIC locus accompanying a t(3;12)(q27 approximately q28;q14 approximately q15) in a lipoma. Cancer Genet Cytogenet. 2001;129:161–164. doi: 10.1016/s0165-4608(01)00441-1. [DOI] [PubMed] [Google Scholar]
- 72.Lemke I, Rogalla P, Bullerdiek J. A novel LPP fusion gene indicates the crucial role of truncated LPP proteins in lipomas and pulmonary chondroid hamartomas. Cytogenet Cell Genet. 2001;95:153–156. doi: 10.1159/000059338. [DOI] [PubMed] [Google Scholar]
- 73.Petit MM, Swarts S, Bridge JA, Van de Ven WJ. Expression of reciprocal fusion transcripts of the HMGIC and LPP genes in parosteal lipoma. Cancer Genet Cytogenet. 1998;106:18–23. doi: 10.1016/s0165-4608(98)00038-7. [DOI] [PubMed] [Google Scholar]
- 74.Ida CM, Wang X, Erickson-Johnson MR, Wenger DE, Blute ML, Nascimento AG, Oliveira AM. Primary retroperitoneal lipoma: a soft tissue pathology heresy? report of a case with classic histologic, cytogenetics, and molecular genetic features. Am J Surg Pathol. 2008;32:951–954. doi: 10.1097/pas.0b013e318160cfbf. [DOI] [PubMed] [Google Scholar]
- 75.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kubo T, Matsui Y, Goto T, Yukata K, Yasui N. Overexpression of HMGA2-LPP fusion transcripts promotes expression of the alpha 2 type XI collagen gene. Biochem Biophys Res Commun. 2006;340:476–481. doi: 10.1016/j.bbrc.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 77.Rogalla P, Kazmierczak B, Meyer-Bolte K, Tran KH, Bullerdiek J. The t(3;12)(q27;q14–q15) with underlying HMGIC-LPP fusion is not determining an adipocytic phenotype. Genes Chromosomes Cancer. 1998;22:100–104. doi: 10.1002/(sici)1098-2264(199806)22:2<100::aid-gcc3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 78.Lemke I, Rogalla P, Grundmann F, Kunze WP, Haupt R, Bullerdiek J. Expression of the HMGA2-LPP fusion transcript in only 1 of 61 karyotypically normal pulmonary chondroid hamartomas. Cancer Genet Cytogenet. 2002;138:160–164. doi: 10.1016/s0165-4608(02)00595-2. [DOI] [PubMed] [Google Scholar]
- 79.Rogalla P, Lemke I, Kazmierczak B, Bullerdiek J. An identical HMGIC-LPP fusion transcript is consistently expressed in pulmonary chondroid hamartomas with t(3;12)(q27-28;q14–15) Genes Chromosomes Cancer. 2000;29:363–366. [PubMed] [Google Scholar]
- 80.von Ahsen I, Rogalla P, Bullerdiek J. Expression patterns of the LPP-HMGA2 fusion transcript in pulmonary chondroid hamartomas with t(3;12) (q27 approximately 28;q14 approximately 15) Cancer Genet Cytogenet. 2005;163:68–70. doi: 10.1016/j.cancergencyto.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 81.Schwindt H, Akasaka T, Zuhlke-Jenisch R, Hans V, Schaller C, Klapper W, Dyer MJ, Siebert R, Deckert M. Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J Neuropathol Exp Neurol. 2006;65:776–782. doi: 10.1097/01.jnen.0000229988.48042.ae. [DOI] [PubMed] [Google Scholar]
- 82.Jardin F, Gaulard P, Buchonnet G, Contentin N, Lepretre S, Lenain P, Stamatoullas A, Picquenot JM, Duval C, Parmentier F, et al. Follicular lymphoma without t(14;18) and with BCL-6 rearrangement: a lymphoma subtype with distinct pathological, molecular and clinical characteristics. Leukemia. 2002;16:2309–2317. doi: 10.1038/sj.leu.2402707. [DOI] [PubMed] [Google Scholar]
- 83.Jais JP, Haioun C, Molina TJ, Rickman DS, de Reynies A, Berger F, Gisselbrecht C, Briere J, Reyes F, Gaulard P, et al. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia. 2008;22:1917–1924. doi: 10.1038/leu.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daheron L, Veinstein A, Brizard F, Drabkin H, Lacotte L, Guilhot F, Larsen CJ, Brizard A, Roche J. Human LPP gene is fused to MLL in a secondary acute leukemia with a t(3;11) (q28;q23) Genes Chromosomes Cancer. 2001;31:382–389. doi: 10.1002/gcc.1157. [DOI] [PubMed] [Google Scholar]
- 85.Choi YW, Choi JS, Zheng LT, Lim YJ, Yoon HK, Kim YH, Wang YP, Lim Y. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the lung. Lung Cancer. 2007;55:43–51. doi: 10.1016/j.lungcan.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 86.Hussenet T, Mallem N, Redon R, Jost B, Aurias A, du Manoir S. Overlapping 3q28 amplifications in the COMA cell line and undifferentiated primary sarcoma. Cancer Genet Cytogenet. 2006;169:102–113. doi: 10.1016/j.cancergencyto.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 87.Sy SM, Lai PB, Pang E, Wong NL, To KF, Johnson PJ, Wong N. Novel identification of zyxin upregulations in the motile phenotype of hepatocellular carcinoma. Mod Pathol. 2006;19:1108–1116. doi: 10.1038/modpathol.3800626. [DOI] [PubMed] [Google Scholar]
- 88.Amsellem V, Kryszke MH, Hervy M, Subra F, Athman R, Leh H, Brachet-Ducos C, Auclair C. The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp Cell Res. 2005;304:443–456. doi: 10.1016/j.yexcr.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 89.Grunewald TG, Kammerer U, Kapp M, Eck M, Dietl J, Butt E, Honig A. Nuclear localization and cytosolic overexpression of LASP-1 correlates with tumor size and nodal-positivity of human breast carcinoma. BMC Cancer. 2007;7:198. doi: 10.1186/1471-2407-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salvi A, Bongarzone I, Micciche F, Arici B, Barlati S, De Petro G. Proteomic identification of LASP-1 down-regulation after RNAi urokinase silencing in human hepatocellular carcinoma cells. Neoplasia. 2009;11:207–219. doi: 10.1593/neo.81076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang B, Feng P, Xiao Z, Ren EC. LIM and SH3 protein 1 (Lasp1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J Hepatol. 2008;50(3):528–537. doi: 10.1016/j.jhep.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 92.Tomasetto C, Moog-Lutz C, Regnier CH, Schreiber V, Basset P, Rio MC. Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. FEBS Lett. 1995;373:245–249. doi: 10.1016/0014-5793(95)01040-l. [DOI] [PubMed] [Google Scholar]
- 93.Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11–q21.3 region of chromosome 17. Genomics. 1995;28:367–376. doi: 10.1006/geno.1995.1163. [DOI] [PubMed] [Google Scholar]
- 94.Grunewald TG, Kammerer U, Kapp M, Eck M, Dietl J, Butt E, Honig A. Nuclear localization and cytosolic overexpression of LASP-1 correlates with tumor size and nodal-positivity of human breast carcinoma. BMC Cancer. 2007;7:198. doi: 10.1186/1471-2407-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Turner DP, Findlay VJ, Kirven AD, Moussa O, Watson DK. Global gene expression analysis identifies PDEF transcriptional networks regulating cell migration during cancer progression. Mol Biol Cell. 2008;19:3745–3757. doi: 10.1091/mbc.E08-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Galler AB, Garcia Arguinzonis MI, Baumgartner W, Kuhn M, Smolenski A, Simm A, Reinhard M. VASP-dependent regulation of actin cytoskeleton rigidity, cell adhesion, and detachment. Histochem Cell Biol. 2006;125:457–474. doi: 10.1007/s00418-005-0091-z. [DOI] [PubMed] [Google Scholar]
- 97.Krause M, Bear JE, Loureiro JJ, Gertler FB. The Ena/VASP enigma. J Cell Sci. 2002;115:4721–4726. doi: 10.1242/jcs.00218. [DOI] [PubMed] [Google Scholar]
- 98.Dertsiz L, Ozbilim G, Kayisli Y, Gokhan GA, Demircan A, Kayisli UA. Differential expression of VASP in normal lung tissue and lung adenocarcinomas. Thorax. 2005;60:576–581. doi: 10.1136/thx.2004.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y, Han G, Fan B, Zhou Y, Zhou X, Wei L, Zhang J. Green tea (-)-epigallocatechin-3-gallate down-regulates VASP expression and inhibits breast cancer cell migration and invasion by attenuating Rac1 activity. Eur J Pharmacol. 2009;606:172–179. doi: 10.1016/j.ejphar.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 100.Han G, Fan B, Zhang Y, Zhou X, Wang Y, Dong H, Wei Y, Sun S, Hu M, Zhang J, et al. Positive regulation of migration and invasion by vasodilator-stimulated phosphoprotein via Rac1 pathway in human breast cancer cells. Oncol Rep. 2008;20:929–939. [PubMed] [Google Scholar]
- 101.Parast MM, Otey CA. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J Cell Biol. 2000;150:643–656. doi: 10.1083/jcb.150.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goicoechea SM, Bednarski B, Garcia-Mata R, Prentice-Dunn H, Kim HJ, Otey CA. Palladin contributes to invasive motility in human breast cancer cells. Oncogene. 2009;28:587–598. doi: 10.1038/onc.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pogue-Geile KL, Chen R, Bronner MP, Crnogorac-Jurcevic T, Moyes KW, Dowen S, Otey CA, Crispin DA, George RD, Whitcomb DC, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goicoechea SM, Arneman D, Otey CA. The role of palladin in actin organization and cell motility. Eur J Cell Biol. 2008;87:517–525. doi: 10.1016/j.ejcb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Otey CA, Rachlin A, Moza M, Arneman D, Carpen O. The palladin/myotilin/myopalladin family of actin-associated scaffolds. Int Rev Cytol. 2005;246:31–58. doi: 10.1016/S0074-7696(05)46002-7. [DOI] [PubMed] [Google Scholar]
- 106.Nakagawa S, Huibregtse JM. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakagawa S, Yano T, Nakagawa K, Takizawa S, Suzuki Y, Yasugi T, Huibregtse JM, Taketani Y. Analysis of the expression and localisation of a LAP protein, human scribble, in the normal and neoplastic epithelium of uterine cervix. Br J Cancer. 2004;90:194–199. doi: 10.1038/sj.bjc.6601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dow LE, Kauffman JS, Caddy J, Zarbalis K, Peterson AS, Jane SM, Russell SM, Humbert PO. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 2007;26:2272–2282. doi: 10.1038/sj.onc.1210016. [DOI] [PubMed] [Google Scholar]
- 109.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nerlov C, De Cesare D, Pergola F, Caracciolo A, Blasi F, Johnsen M, Verde P. A regulatory element that mediates co-operation between a PEA3-AP-1 element and an AP-1 site is required for phorbol ester induction of urokinase enhancer activity in HepG2 hepatoma cells. EMBO J. 1992;11:4573–4582. doi: 10.1002/j.1460-2075.1992.tb05559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Belot N, Rorive S, Doyen I, Lefranc F, Bruyneel E, Dedecker R, Micik S, Brotchi J, Decaestecker C, Salmon I, et al. Molecular characterization of cell substratum attachments in human glial tumors relates to prognostic features. Glia. 2001;36:375–390. doi: 10.1002/glia.1124. [DOI] [PubMed] [Google Scholar]
- 112.Sjoblom B, Salmazo A, Djinovic-Carugo K. alpha-Actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F, Ino Y, Ono M, Hirohashi S. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology. 2005;128:51–62. doi: 10.1053/j.gastro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 114.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fu L, Qin YR, Xie D, Chow HY, Ngai SM, Kwong DL, Li Y, Guan XY. Identification of alpha-actinin 4 and 67 kDa laminin receptor as stage-specific markers in esophageal cancer via proteomic approaches. Cancer. 2007;110:2672–2681. doi: 10.1002/cncr.23110. [DOI] [PubMed] [Google Scholar]