Abstract
The Escherichia coli genome encodes at least 29 putative signal peptides containing a twin arginine motif characteristic of proteins exported via the twin arginine translocation (Tat) pathway. Fusions of the putative Tat signal peptides plus six to eight amino acids of the mature proteins to three reporter proteins (short-lived green fluorescent protein, maltose-binding protein (MBP), and alkaline phosphatase) and also data from the cell localization of epitope-tagged full-length proteins were employed to determine the ability of the 29 signal peptides to direct export through the Tat pathway, through the general secretory pathway (Sec), or through both. 27/29 putative signal peptides could export one or more reporter proteins through Tat. Of these, 11 signal peptides displayed Tat specificity in that they could not direct the export of Sec-only reporter proteins. The rest (16/27) were promiscuous and were capable of directing export of the appropriate reporter either via Tat (green fluorescent protein, MBP) or via Sec (PhoA, MBP). Mutations that conferred a ≥ +1 charge to the N terminus of the mature protein abolished or drastically reduced routing through the Sec pathway without affecting the ability to export via the Tat pathway. These experiments demonstrate that the charge of the mature protein N terminus affects export promiscuity, independent of the effect of the folding state of the mature protein.
The twin arginine translocation (Tat)4 pathway serves to transport folded proteins across energy-transducing membranes in archaea, bacteria, and the chloroplasts of plants (1–4). In bacteria, polypeptides exported via the Tat pathway include redox enzymes, virulence factors, periplasmic ligand-binding proteins, and enzymes involved in cell envelope biogenesis (5). Only proteins that can attain a folded state in the cytoplasm are competent for export via Tat (2, 6). Translocation across the membrane is initiated by the interaction of the signal peptide with the Tat machinery, which in Escherichia coli consists of the membrane proteins TatABC (7, 8). Bacterial Tat signal peptides are longer than signal peptides that target proteins to the Sec or to the signal recognition particle pathways (9). The amino-terminal (n-) region contains the Tat consensus motif (S/T) RRXFLK, which includes the signature twin arginine dipeptide. The hydrophobic (h-) region of Tat signal peptides is generally less hydrophobic than the respective region of Sec (and even less so than signal recognition particle-specific) signal peptides (9). Finally, the carboxyl-terminal (c-) region often contains basic residues, in contrast to Sec signal peptides, which are almost never charged in this region. The mean charge of the c-region of Tat signal peptides is +0.5 versus +0.03 for Sec signal peptides (10).
Bioinformatic analyses of the presence of putative Tat signal peptides suggest that the utilization of the Tat pathway varies widely between different organisms of the same phylum (11–14). The genome of E. coli K-12 is predicted to encode between 22–34 Tat signal peptides (11, 13, 14). However, only 15 have been confirmed experimentally (3, 4, 15–25). In general, identifying Tat substrates has proven difficult. Proteomic studies based on comparison of secreted proteins in WT and tat deletion strains are complicated because many Tat proteins are expressed only under specialized conditions (26). In addition, data from the localization of reporter protein fusions to Tat signal peptides have to be interpreted with caution because the nature of the reporter protein can misdirect the fusion to the Sec pathway (21, 27, 28).
In this report we evaluated the export pathway preference of the putative Tat signal peptides of E. coli by examining the localization of fusions to three complementary reporter proteins. This analysis, and additional localization studies of the respective epitope-tagged full-length proteins, revealed that the E. coli genome encodes at least 27 Tat-targeting signal peptides. Surprisingly, the majority of these signal peptides can direct export via either Tat or Sec depending on the export competence of the reporter polypeptide. We show that increasing the positive charge in the N terminus of the mature protein prevents export via Sec, without affecting Tat export. Thus, it appears that in E. coli the Tat pathway is utilized for the trans-location of proteins containing positively charged N-terminal regions that serve as “stop transfer” signals (29, 30) for Sec export.
EXPERIMENTAL PROCEDURES
Bioinformatics
Putative Tat signal peptides were identified by conducting a BLAST search using the strings: SRRRFLK, SRRXFLX, TRRXFLX, SRRXXLK, SRRXXLA, TRRXXLK, TRRXXLA, SRRXXLT, SRRXXIK, SRRXXIA, SRRXFIX, SRRXFMK, SRRXFVK, SRRXFVA, SRRQFLK, RRXFLA, and RRXFLK within the first 50 residues. Sequences were then analyzed for signal peptide characteristics using SignalP 1.1 (www.cbs.dtu.dk/services/SignalP/) (31). DmsA and YaeI, which had been assigned incorrect start codons in the NCBI data base (www.ncbi.nlm.nih.gov/), were also included in the list, as was YcdO, which did not match our criteria but had been predicted to be a Tat substrate (14).
Bacterial Strains and Plasmids
The strains and plasmids used in this study are listed in supplemental Table S2. Signal peptides were amplified from E. coli XL1-Blue genomic DNA by PCR using the primers 1–56 shown in supplemental Table S3 and cloned into pKKGS (32), a derivative of plasmid pBAD33 (33). Although SignalP predicts the likely signal peptidase cleavage site, there is evidence that such predictions are not always accurate for Tat signal peptides (34), and therefore reverse primers were designed to include the sequence encoding the first six to eight aa following the predicted cleavage site and ensure the actual cleavage site would be present in the fusions. Details of the construction of the gene fusions can be found in the supplemental data. Full-length proteins were tagged with a C-terminal FLAG epitope tag (DYKDDDDK) (Sigma) by PCR amplification (supplemental Table S3) and cloning into the SacI and XbaI sites of pBAD33 (33).
Cell Fractionations
Cell fractionations were performed by the cold osmotic shock procedure as described earlier (2). The soluble cytoplasmic fraction was obtained by resuspending the cell pellet obtained after fractionation into 300 µl of phosphate-buffered saline, sonication for 30 s, and recentrifugation to precipitate and remove insoluble cell debris.
General Procedures
Unless otherwise noted, cells were grown at 37 °C on Luria Bertani medium with 50 µg/ml chloramphenicol, 25 µg/ml kanamycin, or 100 µg/ml ampicillin as appropriate. To test growth on maltose, E. coli HS3018 or its derivatives containing plasmids encoding maltose-binding protein (MBP) fusions were grown overnight, diluted, plated on M9 minimal medium containing 0.4% maltose, and incubated at 37 °C for 2–3 days.
PhoA assays and Western blotting were performed as described earlier (2). The following primary antibodies were used: monoclonal mouse anti-FLAG-M2 (Sigma), monoclonal rabbit anti-DsbA (Stressgen), monoclonal rabbit anti-GroEL (Sigma), polyclonal rabbit anti-PhoA (Rockland), and monoclonal mouse anti-MalE (Sigma).
For flow cytometric analysis, E. coli MC4100-P and B1LK0-P containing plasmids encoding GFP-SsrA fusions were grown overnight in Luria Bertani medium as described above, and 500 µl of overnight culture were used to inoculate 10 ml of fresh medium. After 1 h of shaking at 37 °C, gene expression was induced with arabinose to a final concentration of 0.01%, the cells were incubated for an additional 4 h, and 1-ml samples were harvested by centrifugation, diluted in phosphate-buffered saline containing 5 µg/ml propidium iodide, and analyzed with a BD Biosciences FACSort flow cytometer.
RESULTS
Bioinformatic Identification of Putative Tat Signal Peptides
Putative Tat signal peptides were identified by first using BLAST to search for E. coli open reading frames encoding proteins that contained a twin arginine dipeptide in the first 50 aa and, in addition, any two of the additional four aa that comprise the conserved Tat motif. Hits were then analyzed for signal peptide characteristics using SignalP (31). A total of 26 sequences meeting ≥25% of the SignalP 1.1 threshold values were found. Two more were included that have misannotated start codons in the data base but otherwise fit the above criteria (Table 1). Furthermore, YcdO was included because it is in the same operon as another putative Tat protein and was previously predicted to be a Tat substrate itself (14). The final set contained all 15 confirmed Tat substrates (AmiA, AmiC, CueO, DmsA, FdnG, FhuD, HyaA, HybO, MdoD, NapA, NrfC, SufI, TorA, TorZ, and YcdB) and is in good agreement with the prediction from program TATFIND (11). However, TATFIND does not predict AmiC, DmsA, YaeI, YcdO, and YfhG to be Tat substrates; in addition, it predicts plasmid-encoded and membrane proteins that were not considered in the present study. Another program used to identify Tat signal peptides, TatP, is also in good agreement with our list but the latter missed FhuD, YagT, YcdO, and YfhG and predicted b3000, which was not considered in this study (14). Although SignalP predicts signal peptidase cleavage sites, there is evidence that such predictions are not always accurate for Tat signal peptides (34). To eliminate ambiguities, all signal peptide fusions in this study were designed to include the first six to eight aa following the SignalP-predicted cleavage site (Table 1).
TABLE 1.
Characteristics, reporter protein fusion assay results, and assigned export pathway for each putative Tat signal peptide
| Signal peptide |
Sequencea | Hydro- phobicity, h-regionb |
Charge, c-region |
Charge, mature N terminus |
Charge, net (cleavage area) |
GFP-SsrA FACS mean fluorescencec |
PhoA activityd |
MBP plate growthe |
Export pathway |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | tatC | WT | TatC | ||||||||
| % | |||||||||||
| FdnG | MDVSRRQFFKICAGGMAGTTVAALGFAPKQALAQARNYKL | 2.25 | +1 | +2 | +3 | 2 | 3 | n.e. | + | − | Tat |
| FdoG | MQVSRRQFFKICAGGMAGTTAAALGFAPSVALAETRQYKL | 1.91 | 0 | +1 | +1 | 2 | 4 | n.e. | + | − | Tat |
| NapG | MSRSAKPQNGRRRFLRDVVRTAGGLAAVGVALGLQQQTARASGVRLR | 2.00 | +1 | +2 | +3 | 2 | 2 | n.e. | + | − | Tat |
| NrfC | MTWSRRQFLTGVGVLAAVSGTAGRVVAKTLNIN | 2.28 | +1 | +1 | +2 | 9 | 3 | n.e. | + | − | Tat |
| HyaA | MNNEETFYQAMRRQGVTRRSFLKYCSLAATSLGLGAGMAPKIAWALENKPR | 1.36 | +1 | +1 | +2 | 27 | 3 | 0 | + | − | Tat |
| YnfE | MSKNERMVGISRRTLVKSTAIGSLALAAGGFSLPFTLRNAAAAVQQAREK | 2.01 | +1 | +1 | +2 | 9 | 3 | 0 | + | − | Tat |
| WcaM | MPFKKLSRRTFLTASSALAFLHTPFARALPARQS | 1.82 | +1 | +1 | +2 | 13 | 3 | 1 | + | − | Tat |
| TorA | MNNNDLFQASRRRFLAQLGGLTVAGMLGPSLLTPRRATAAQAATDA | 1.42 | +2 | −1 | +1 | 90 | 2 | 2 | + | − | Tat |
| NapA | MKLSRRSFMKANAVAAAAAAAGLSVPGVARAVVGQ | 1.53 | +1 | 0 | +1 | 2 | 3 | 3 | + | − | Tat |
| YagT | MSNQGEYPEDNRVGKHEPHDLSLTRRDLIKVSAATAATAVVYPHSTLAASVPAA | 1.63 | 0 | 0 | 0 | 57 | 5 | 3 | + | − | Tat |
| YcbK | MDKFDANRRKLLALGGVALGAAILPTPAFATLSTPR | 2.40 | 0 | +1 | +1 | 82 | 4 | 3 | + | − | Tat |
| DmsA | MKTKIPDAVLAAEVSRRGLVKTTAIGGLAMASSALTLPFSRIAHAVDSAIP | 1.56 | +1 | −1 | 0 | 90 | 6 | 2 | + | + | Tat+Sec |
| YdhX | MSWIGWTVAATALGDNQMSFTRRKFVLGMGTVIFFTGSASSLLANTRQEK | 2.31 | 0 | +1 | +1 | 7 | 5 | 2 | + | + | Tat+Secf |
| YahJ | MKESNSRREFLSQSGKMVTAAALFGTSVPLAHAAVAGTL | 2.17 | 0 | 0 | 0 | 100 | 6 | 3 | + | + | Tat+Sec |
| YedY | MKKNQFLKESDVTAESVFFMKRRQVLKALGISATALSLPHAAHADLLSWF | 1.73 | 0 | −1 | −1 | 47 | 6 | 4 | + | + | Tat+Sec |
| CueO | MQRRDFLKYSVALGVASALPLWSRAVFAAERPTL | 1.79 | +1 | 0 | +1 | 21 | 4 | 5 | + | + | Tat+Sec |
| SufI | MSLSRRQFIQASGIALCAGAVPLKASAAGQQQP | 1.74 | +1 | 0 | +1 | 29 | 2 | 5 | + | + | Tat+Sec |
| YcdB | MQYKDENGVNEPSRRRLLKVIGALALAGSCPVAHAQKTQSA | 2.66 | 0 | +1 | +1 | 123 | 5 | 6 | + | + | Tat+Sec |
| TorZ | MIREEVMTLTRREFIKHSGIAAGALVVTSAAPLPAWAEEKGGK | 1.98 | 0 | 0 | 0 | 56 | 4 | 6 | + | + | Tat+Sec |
| HybA | MNRRNFIKAASCGALLTGALPSVSHAAAENRPP | 1.98 | 0 | 0 | 0 | 33 | 3 | 11 | + | + | Tat+Sec |
| YnfF | MMKIHTTEALMKAEISRRSLMKTSALGSLALASSAFTLPFSQMVRAAEAPVE | 1.55 | +1 | −2 | −1 | 49 | 5 | 15 | + | + | Tat+Sec |
| HybO | MTGDNTLIHSHGINRRDFMKLCAALAATMGLSSKAAAEMAESV | 2.47 | +1 | −2 | −1 | 28 | 3 | 17 | + | + | Tat+Sec |
| AmiC | MSGSNTAISRRRLLQGAGAMWLLSVSQVSLAAVSQVV | 1.40 | 0 | 0 | 0 | 36 | 9 | 18 | + | + | Tat+Sec |
| AmiA | MSTFKPLKTLTSRRQVLKAGLAALTLSGMSQAIAKDELLK | 2.10 | 0 | 0 | 0 | 22 | 2 | 79 | + | + | Tat+Sec |
| YfhG | MRHIFQRLLPRRLWLAGLPCLALLGCVQNHNKPAIDT | 2.16 | 0 | 0 | 0 | 21 | 5 | 93 | + | + | Tat+Sec |
| MdoD | MDRRRFIKGSMAMAAVCGTSGIASLFSQAAFAADSDIA | 2.42 | 0 | −2 | −2 | 15 | 5 | 165 | + | + | Tat+Sec |
| FhuD | MSGLPLISRRRLLTAMALSPLLWQMNTAHAAAIDPN | 1.70 | 0 | −1 | −1 | 6 | 2 | 256 | + | + | Tat+Secf |
| YaeI | MISRRRFLQATAATIATSSGFGYMHYCEPGWFELIRHRLAFFKDNAAPFKIL | 2.80 | 0 | +1 | +1 | 2 | 2 | 20 | + | + | Sec |
| YcdO | MTINFRRNALQLSVAALFSSAFMANAADVPQVK | 1.63 | 0 | 0 | 0 | 5 | 4 | 107 | + | + | Sec |
Shown in bold is the signal peptide c-region and in italics the predicted N-terminal region of the mature protein.
Average hydrophobicity using Kyte Doolittle values is calculated for the h-region, defined as described by Cristóbal et al. (9).
Signal peptide-GFP-SsrA fusions were expressed both in wild type and tatC strains, and whole cell fluorescence values were measured by flow cytometry (fluorescence-activated cell sorter). The average mean fluorescence values of at least three replicate experiments are shown.
Activity of whole cell lysates of a phoAR strain expressing each signal peptide-PhoA fusion as percentage of activity of PhoA with its own signal peptide. FACS, fluorescence-activated cell sorter; n.e., no expression detected.
Colonies present (+) or absent (−) after 2 days at 37 °C on maltose minimal medium plates in malE (WT) and malE tatC (ΔtatC) strains. For ssDmsA and ssSufI, colonies appeared after 3 days in the ΔtatC strain.
Tat portion of assignment based on epitope tagging for ssYdhX and on the results of Ize et al. (18) for ssFhuD; Sec portion of assignment based on reporter fusion results for both ssYdhX and ssFhuD.
Export Pathway Analysis Using Reporter Fusions
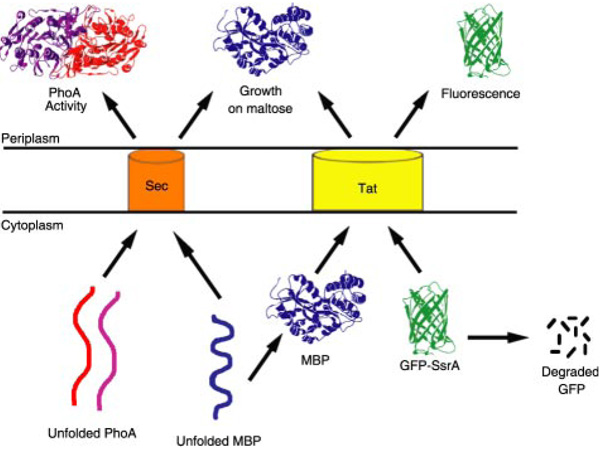
We sought to experimentally verify the export pathway preference of the putative 29 signal peptides above by systematic analysis using reporter fusions (Fig. 1). DeLisa et al. (32) have shown that a short-lived version of GFP containing an SsrA tag confers cell fluorescence only when it is exported to the periplasm by virtue of a Tat signal peptide. In the absence of export, GFP-SsrA is rapidly degraded by ClpXP and ClpAP in the cytoplasm, whereas targeting to the Sec apparatus also results in non-fluorescent cells. For these reasons, the fluorescence of cells expressing fusions between Tat signal peptides and GFP-SsrA is proportional to the efficiency of export into the periplasm (35).
FIGURE 1. Routing of PhoA, MBP, and GFP-SsrA via the Sec and Tat pathways.
PhoA and MBP are exported via Sec in an unfolded conformation, whereas MBP and GFP-SsrA are exported via Tat in a folded conformation. The SsrA tag targets GFP for degradation by ClpXP and ClpAP in the cytoplasm.
GFP-SsrA fusions to the set of 29 putative signal peptides were expressed under the control of the pBAD promoter. Because high level expression of SsrA-tagged proteins can saturate the ClpXP and ClpAP machinery, the fusions were expressed in E. coli MC4100 carrying a pcnB deletion that lowers the copy number of pBR322 and its derivatives (36). Cell fluorescence was analyzed by flow cytometry (supplemental Fig. S5), and the mean fluorescence values conferred by the 29 fusions are reported in Table 1. Signal peptides were classified as capable of mediating Tat export if they conferred a mean cell fluorescence ≥10 arbitrary units and the cell fluorescence decreased by more than 3-fold in an isogenic tatC strain. These criteria were derived from genetic studies in which we isolated TatC suppressor mutations that allow the export of twin lysine signal peptides.5 Nineteen signal peptides satisfied these criteria (Table 1). However, GFP-SsrA fusions to several experimentally confirmed Tat signal peptides (ssFdnG (signal sequence FdnG), ssFhuD, ssNapA, and ssNrfC), resulted in low cell fluorescence. Low whole cell fluorescence can result from poor expression and does not necessarily imply absence of Tat export. Indeed, when whole cell lysates were analyzed for GFP expression via Western blotting, no bands were detected for these four signal peptides or for other signal peptides with a mean fluorescence below 10 arbitrary units in the wild type strain (supplemental Fig. S6).
MBP is naturally a Sec substrate in E. coli but is also competent for Tat export (Fig. 1) (28). A low level of periplasmic MBP is sufficient to allow growth of malE cells on minimal medium with maltose as the sole carbon source. We reasoned that growth on maltose minimal medium might be a more sensitive reporter system for Tat export of fusions that are expressed poorly. However, Tat export of signal peptide-MBP fusions can only be inferred if a tatC mutation abolishes growth on maltose, thus ruling out the possibility that the fusion protein is instead exported via Sec. Eleven of the 29 fusions conferred growth on maltose only in WT cells (Table 1). These included ssFdnG, ssFdoG, ssNapA, ssNapG, ssNrfC, and ssYnfE, whose Tat export could not be assigned with GFP-SsrA fusions due to low fluorescence. To ensure that lack of growth in a tatC mutant strain was not due to lower protein expression levels, fusions to the six signal peptides above were further tested using maltose minimal medium plates containing inducer. Nonetheless, Tat-dependent growth was still observed only in WT cells and not in the tatC mutant strain. Western blotting revealed that expression levels are nearly equal in WT and tatC mutant cells (with the exception of ssTorA, for which expression was higher in the tatC mutant relative to the WT cells). Representative results are shown in Fig. 2A. The ssNrfC-MBP fusion could not be detected by Western blotting in either the WT or tatC strain, despite its ability to confer Tat-dependent growth on maltose plates with and without inducer. Collectively, these data indicate that the set of 11 signal peptides is capable of directing export via Tat but not via the Sec pathway.
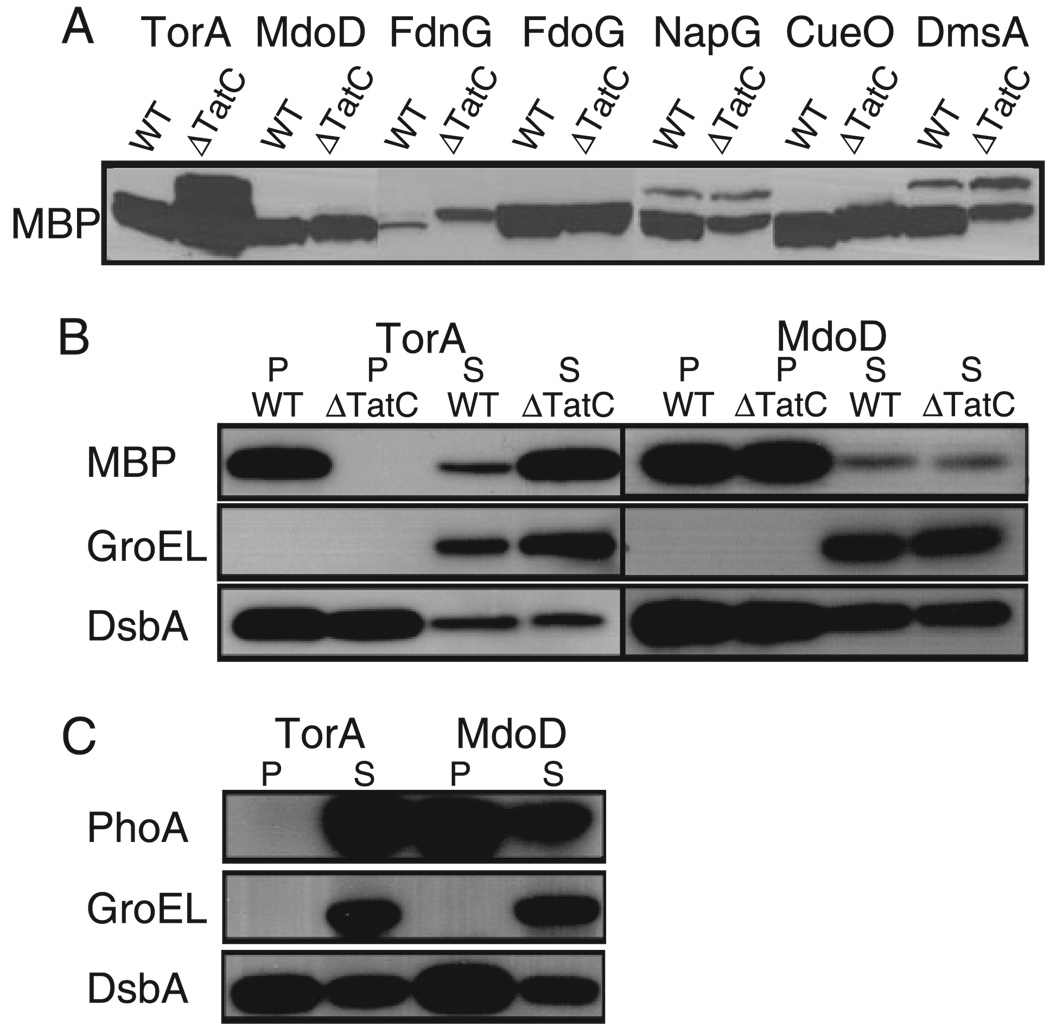
FIGURE 2. Subcellular localization of reporter protein fusions to ssTorA (Tat−specific) and ssMdoD (Tat+Sec) signal peptides.
A, Western blots of whole cell lysates from cells expressing selected signal peptide fusions to MBP. The protein in the lanes for ssNapG and ssDmsA fusions migrate as doublets, but the higher molecular mass band (~55 kDa) does not correspond to the molecular mass of the precursor (predicted to be 45.7 and 45.9 kDa, respectively). Western blots are shown of periplasmic and spheroplast fractions from cells expressing signal peptide fusions to MBP (B) and PhoA (C). P, periplasmic fraction; S, spheroplast fraction. PhoA fusions were expressed in E. coli strain DHB4, and MBP fusions were expressed in HS3018 and HS3018 tatC. GroEL and DsbA were used as fractionation markers by probing with anti-GroEL and anti-DsbA serums, respectively.
We tested whether the mal+ phenotype conferred by the set of 11 signal peptide fusions above was dependent on other Tat proteins. As expected, colony formation was abolished or severely retarded in a tatA strain but was unaffected in a tatE strain (data not shown). Interestingly, not all fusions were dependent on TatB for export. A tatB malE strain expressing ssHyaA-MBP or ssYcbK-MBP grew normally on minimal medium with maltose even though growth was abolished in the tatC strain. TatB-independent export of an ssHyaA fusion to a different reporter protein had also been reported earlier (2). These observations suggest that interactions with TatB may not be required for the initiation of export by some E. coli Tat signal peptides.
The remaining 18 MBP fusions conferred growth in both WT and tatC cells. Because Tat translocation is completely inactivated in a tatC strain, the export of MBP into the periplasm of these cells must have occurred through the Sec pathway. Export through Sec does not rule out the ability to target the Tat translocon as well, but these two possibilities cannot be distinguished from the phenotype of MBP fusions alone. However, 14 of these 18 signal peptides satisfied the criteria for Tat export of GFP-SsrA. Because they allow translocation of the MBP reporter in a tatC strain, these 14 signal peptides also possess the ability to direct export via Sec and therefore are assigned as Tat + Sec in Table 1. The ssFhuD-GFP-SsrA fusion gave 3-fold higher fluorescence in WT cells relative to the tatC strain but was lower compared with other fusions. Nonetheless, independent evidence has demonstrated that FhuD utilizes the Tat pathway (24), and therefore this signal peptide was also assigned as Tat + Sec.
Sec export was also evaluated quantitatively based on the alkaline phosphatase (PhoA) activity conferred by PhoA fusions. The two disulfide bonds that are essential for the folding of PhoA into its native conformation cannot form within the reducing cytoplasm of E. coli (37). Thus, PhoA is incompatible with Tat transport under normal cellular conditions. Unfolded PhoA is exported only via the Sec pathway and attains its active conformation within the periplasm (Fig. 1). Consequently, whole cell PhoA activity represents a measure of the efficiency of Sec export. The activity values for the fusions were normalized relative to the PhoA activity obtained when alkaline phosphatase was expressed with its own signal peptide (ssPhoA) from the same vector. Expression levels of each signal peptide-PhoA fusion were checked via Western blotting (supplemental Fig. S7). The ssFdnG, ssFdoG, ssNapG, and ssNrfC fusions could not be detected, but the other 25 proteins were expressed at comparable levels. All of the signal peptides that failed to confer growth on maltose when fused to MBP and expressed in a tatC mutant strain also gave background activity when fused to PhoA, further highlighting their specific targeting to the Tat pathway.
Eight signal peptides that conferred growth on maltose in a tatC strain when fused to MBP gave only background activity when fused to PhoA, underscoring the differences in sensitivity between the two reporter proteins. The other eight signal peptides that already had been assigned as Tat + Sec, namely ssHybA, ssYnfF, ssHybO, ssAmiC, ssAmiA, ssYfhG, ssMdoD, and ssFhuD, gave PhoA activities > 10% of those obtained when PhoA was exported with its own signal peptide. Notably, PhoA fusions to ssYfhG, ssMdoD, and ssFhuD gave 90, 160, and 260% activity relative to the control, suggesting that these signal peptides are as efficient as ssPhoA, a bona fide Sec signal peptide.
The localization of fusions that display Tat-specific or Tat + Sec targeting was also examined by cell fractionation and Western blotting; representative results are shown in Fig. 2, B and C. DsbA and GroEL were employed as periplasmic and cytoplasmic markers, respectively. For the TorA signal peptide, which our analysis showed to be Tat-specific, the PhoA fusion was localized exclusively in the spheroplast fraction whereas the MBP fusion was found mostly in the periplasm. In the tatC strain ssTorA-MBP accumulated exclusively in the cytoplasm. In contrast, with ssMdoD, a signal peptide that exhibits secretion pathway promiscuity, both the MBP and the PhoA fusions were localized mainly in the periplasm and localization was unaffected in a tatC strain.
Localization of Epitope-tagged Fusions
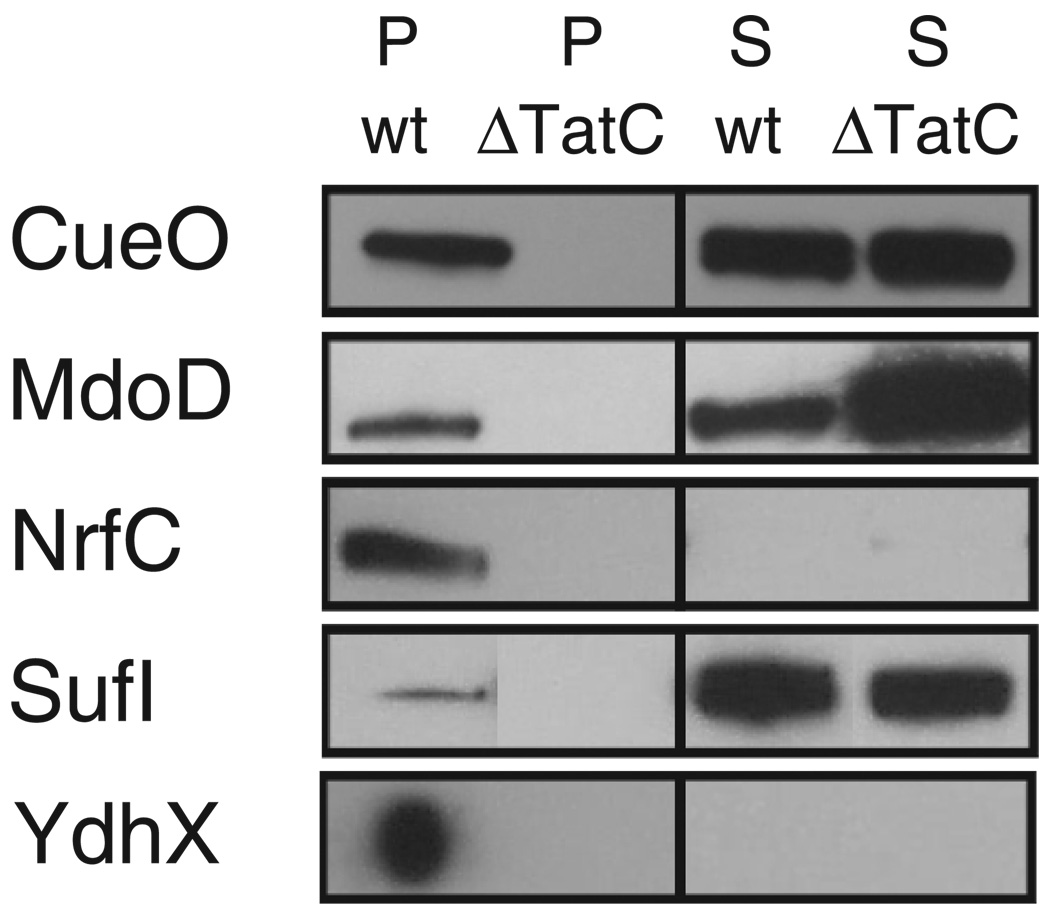
To test whether the secretion pathway assignments using reporter fusions conform to those of the authentic proteins, we constructed an additional set of Tat proteins C-terminal-tagged with the FLAG epitope. Even though the FLAG fusions were transcribed from the pBAD arabinose-inducible promoter on a low copy plasmid, for several proteins we failed to detect expression by Western blotting in cells grown in minimal or rich media, at various growth temperatures, with or without oxygen, and with different concentrations of inducer.6 Further complications arose from the propensity of some of the fusions to accumulate in an aggregated state and from the saturation of Tat export, a common phenomenon when Tat proteins are expressed from multicopy plasmids (2, 38). For all of the FLAG-tagged proteins that could be successfully expressed, the results were consistent with the reporter fusion analysis (see Fig. 3). Expression and export of NrfC-FLAG could be detected only in anaerobically grown cells (Fig. 3). In addition, whereas the ssYdhX-GFP fusion gave low fluorescence, the full-length protein was shown to be exported in a TatC-dependent fashion. Thus, YdhX must also be routed through the Tat pathway, bringing the total number of Tat-targeting signal peptides to 27.
FIGURE 3. Subcellular localization of FLAG-tagged full-length proteins.
Samples were normalized on the basis of the A600, resolved on SDS-12% polyacrylamide gels, and probed with anti-FLAG serum. All fractionations were confirmed with fractionation markers by probing with anti-GroEL and anti-DsbA serums (data not shown).
Determinants of Targeting Specificity
As discussed above, 11 of the 29 signal peptides tested in this study supported export only via the Tat pathway, eight signal peptides directed a small amount of Sec export (revealed by growth on maltose accompanied by low PhoA activity), and eight signal peptides appeared to be routed efficiently through both pathways. The eight signal peptides that displayed efficient Sec targeting had all the features of typical Tat signal peptides, i.e. a longer overall length, a twin arginine consensus motif, and a less hydrophobic h-region. Earlier, von Heijne and coworkers (9) had proposed that a positive charge in the c-region of the signal peptide serves as a “Sec-avoidance motif” similar to that found in substrates of the plant chloroplast ΔpH-dependent/cpTat pathway (39). However, on a genomic scale, the data shown in Table 1 indicate that the overall charge of the c-region does not correlate well with avoidance of Sec export. As explained above, the reporter fusions we constructed also contained the first six to eight predicted aa of the respective mature proteins. When the total charge of the predicted signal peptide c-region (shown in bold in Table 1) together with the first few predicted aa of the mature protein (shown in italics in Table 1) was taken into consideration, the correlation between charge and Sec avoidance improved significantly (Table 1). In particular, all fusions with a ≥+2 charge in this region showed exclusive Tat export. Fusions with a +1 charge in this area gave inefficient export via Sec as indicated by a low PhoA activity. Only signal peptide fusions that have a net neutral or negative charge in the entire region displayed efficient export of PhoA via Sec.
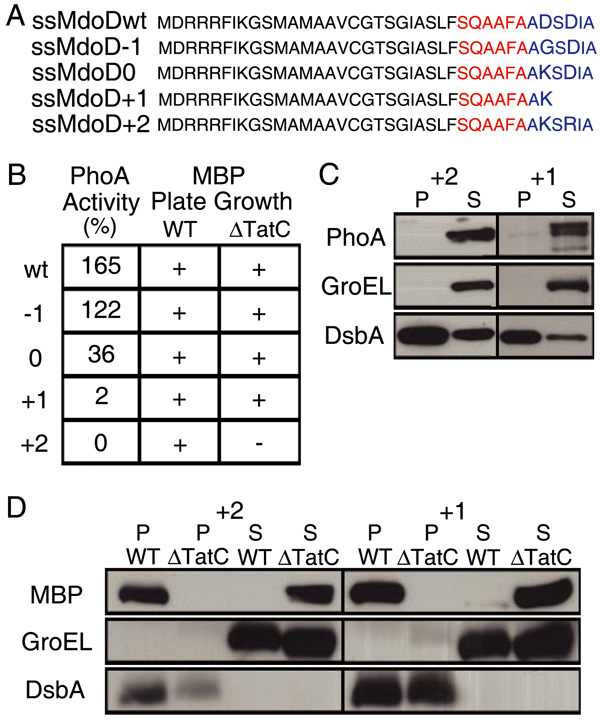
To further examine the effect of the N terminus of the mature protein on targeting specificity, we replaced the Asp residue at position +2 (relative to the predicted signal peptidase cleavage site) in the ssMdoD signal peptide with Gly or Lys, imparting a net −1 and neutral charge, respectively (Fig. 4A). Additionally, we replaced the Asp residue at +2 with Lys and truncated the remaining portion of the mature protein to confer a net +1 charge. Finally, we replaced the Asp residues at positions +2 and +4 with Lys and Arg, to impart a net +2 charge. The ssMdoD signal peptide was selected because it does not encode a redox enzyme and there is no evidence of specific chaperones involved in its export. The resulting signal peptides, ssMdoD−1, ssMdoD0, ssMdoD+1, and ssMdoD+2, were fused to PhoA and MBP, and the PhoA activities and growth on maltose were determined; the results are summarized in Fig. 4B. Additionally, for fusions to ssMdoD+1 and ssMdoD+2, the subcellular distribution of the respective proteins was determined by Western blotting (Fig. 4, C and D). As expected, an overall +2 charge completely abolished Sec targeting without affecting the efficiency of Tat translocation. A +1 charge allowed growth of the ssMdoD+1-MBP fusion on maltose in a tatC strain only after 3 days and gave very low PhoA activity. In contrast, the ssMdoD0 and ssMdoD−1 fusions gave progressively higher PhoA activities and growth on maltose in the tatC strain. These results support the notion that the charge of the first few aa of the mature protein together with the c-region of the signal peptide plays a role in avoiding routing to the Sec translocon.
FIGURE 4. A positive charge at the N terminus of the mature protein acts as a Sec-avoidance signal.
The charge of the first few amino acids of the mature MdoD was changed from −2 (ssMdoDwt) to −1, 0, +1, or +2 by site-specific mutagenesis (ssMdoD−1, ssMdoD0, ssMdoD+1, and ssMdoD+2). A, protein sequences of ssMdoDwt, ssMdoD−1, ssMdoD0, ssMdoD+1, and ssMdoD+2 up to the fusion site. The predicted c-region of the signal peptide is shown in red, and the predicted N terminus of the mature protein is shown in blue. B, PhoA activity and growth on maltose minimal medium. PhoA data are reported as percentage of activity of PhoA with its own signal peptide. Western blots are shown of periplasmic (P) and spheroplast (S) fractions from cells expressing fusions to PhoA (C) and MBP (D).
Interestingly, even though ssMdoD can target the Sec translocon efficiently, the FLAG-tagged full-length MdoD protein was localized in the periplasm only in strains with a functional Tat apparatus. Furthermore, attempts to export MdoD with the Sec signal peptide ssPhoA were unsuccessful (data not shown), providing additional evidence that the mature protein also prevents mislocalization to the Sec pathway under physiological conditions (19, 39).
DISCUSSION
In this work we employed reporter and epitope-tagged fusions to investigate the export routing of 29 putative Tat signal peptides identified by bioinformatics means. We analyzed the localization of a total of 116 fusions and found that at least 27 signal peptides can direct export via the Tat pathway. Thus, there are at least 27 Tat substrate proteins in E. coli in addition to “hitchhiker” substrates (DmsB, FdnH, HyaB, and HybC) (40) that are exported through Tat but do not contain a signal peptide. A Tat-targeting capability could not be confirmed or ruled out for two signal peptides, ssYcdO and ssYaeI.
Fusions to the Tat-specific reporter GFP-SsrA, the Sec-only reporter PhoA, and MBP, which is competent for export via both Sec and Tat, were used to obtain information on the ability of the various signal peptides to initiate export through the two translocons (Fig. 1). It should be noted that no single reporter protein is sufficient for this analysis because there are three experimental outcomes (i.e. Tat-only, Sec-only, or Tat + Sec). The maltose phenotype conferred by MBP fusions represents a very sensitive, but qualitative, way to detect export. In contrast, PhoA activity and GFP-SsrA fluorescence provide a quantitative measurement on the efficiency with which the signal peptides direct export to the Sec or Tat pathways, respectively. For five signal peptides, Tat export of the respective FLAG-tagged full-length proteins was confirmed by Western blotting of periplasmic fractions from wild type and tatC strains. However, most epitope-tagged full-length proteins showed poor expression under a variety of conditions or were prone to aggregation. Importantly, epitope-tagged full-length proteins do not provide information on the propensity of the signal peptide to target the Sec translocon, because the mature polypeptides can exhibit folding features that are not compatible with Sec export.
As mentioned above, earlier studies had led to the experimental verification of 15 E. coli Tat signal peptides. The GFP-SsrA and MBP fusions correctly predicted the Tat export of 14 of these signal peptides. ssFhuD is the only known Tat signal peptide for which Tat export could not be confirmed from reporter fusion analysis; therefore, the Tat portion of the assignment shown in Table 1 for that signal peptide had to be based on earlier experimental data (24). Our analysis provided definitive experimental evidence for 12 additional E. coli Tat substrates.
We addressed the targeting specificity of Tat signal peptides in a comprehensive manner. Approximately 60% of the signal peptides exhibited at least some level of promiscuous targeting to the Sec translocon, as manifested by the growth of MBP fusions on maltose in a tatC strain. Consistent with previous studies (2, 41), some of these signal peptides conferred a low level of PhoA activity. However, ssHybA, ssHybO, ssYnfF, ssAmiC, ssAmiA, ssYfhG, ssMdoD, and ssFhuD gave PhoA activities between 10–260% of the level obtained when PhoA is exported by its own signal peptide. Notably, ssFhuD, which gave the highest PhoA activity, has a less hydrophobic h-region typical of a Tat signal peptide yet evidently is efficient in targeting the Sec translocon and activating SecA.
Earlier it had been suggested that signal peptides containing an h-region sufficiently hydrophobic for Sec-mediated export require additional signals for Sec avoidance (9, 41). Clearly, such signals are not found in many Tat signal peptides and therefore other mechanisms, including the characteristics of the mature polypeptide and possibly interactions with specific chaperones, may be at play in order to prevent misrouting through Sec.
For 11 of the 27 signal peptides, we did not detect any Sec export of PhoA or MBP indicating that these signal peptides are Tat-specific. We note that for six of these signal peptides (ssFdnG, ssFdoG, ssNapA, ssNapG, ssNrfC, and ssYnfE), the assignment of Tat-only is based solely on their ability to confer growth on maltose in a TatC-dependent manner when fused to MBP. For these signal peptides, it was not possible to draw any conclusions based on their fluorescence levels in the wild type strain (means <10 arbitrary units) when fused to GFP-SsrA.
Although earlier data had suggested that the presence of a positive charge in the c-region serves as a Sec-avoidance signal (9, 39) the results shown in Table 1 indicate that additional factors must be important for Tat specificity. For example, ssFdoG fusions, which have an uncharged c-region, were routed only via Tat whereas ssHybO and ssYnfF fusions with positively charged termini showed appreciable Sec activity. Our data suggest that the charge of the first few aa of the mature protein also plays a role in Sec avoidance (Table 1). Indeed, site-specific mutagenesis of the first six aa of the MdoD mature protein, resulting in a change in the charge from −2 to −1, had no effect, whereas further increasing the charge to neutral and +1 severely reduced the Sec export of PhoA fusions. Finally, increasing the charge of this region to +2 completely abolished Sec export of both PhoA and MBP fusions (Fig. 4). Thus, alteration of the N terminus of the mature protein alone was sufficient to completely switch the export pathway.
It has long been known that a positive charge in the first few aa of the mature protein severely retards or even abolishes Sec export and that such a region serves as a stop transfer signal for Sec-dependent membrane proteins (29, 42–45). The critical window has been proposed to comprise the first five aa (42), and more recently up to sixteen aa (43). Only 4% of 191 Sec-exported proteins contain a net positive charge in the first sixteen aa of the mature protein (43). In contrast, 37% (10/27) of N-terminal regions of proteins with a Tat signal peptide display a charge ≥ + 1 within either the five aa or the sixteen aa windows. This observation raises the possibility that some proteins, particularly those that do not contain cofactors or form multimers, may have evolved to use the Tat pathway because they require positively charged N termini for function and thus are not compatible with Sec export.
We note, however, that one signal peptide, ssYagT, has no charge in the critical region yet exhibits Tat specificity, indicating that apart from charge an additional mechanism for specific Tat targeting must also exist. Signal peptide binding chaperones (46, 47) could potentially fulfill this role by sequestering the signal peptide from recognition by the Sec machinery.
Supplementary Material
Acknowledgments
We thank Drs. Philip Lee and C. Buddie Mullins for critical review of this manuscript, Joe Pizzini for help in assembling the figures, and Eva Strauch for many useful discussions.
Footnotes
This work was supported in part by grants from the Foundation for Research and National Institutes of Health (RO-01 GM069872).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S5–S7, Tables S2 and 3, and supplemental data.
Supported by a National Science Foundation graduate fellowship.
A Medical Research Council Senior Non-clinical Research Fellow.
The abbreviations used are: Tat, twin arginine translocation; Sec, general secretory pathway; ss, signal sequence; MBP, maltose-binding protein; GFP, green fluorescent protein; aa, amino acid; WT, wild type; c-region, carboxyl-terminal region; h-region, hydrophobic region.
E. Strauch and G. Georgiou, unpublished observations.
Y. Kawarasaki and G. Georgiou, data not shown.
REFERENCES
- 1.Berks BC. Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 2.DeLisa MP, Tullman D, Georgiou G. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6115–6120. doi: 10.1073/pnas.0937838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodrigue A, Chanal A, Beck K, Muller M, Wu LF. J. Biol. Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 4.Santini CL, Ize B, Chanal A, Muller M, Giordano G, Wu LF. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee PA, Tullman-Ercek D, Georgiou G. Ann. Rev. Microbiol. 2006;60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher AC, Kim W, DeLisa MP. Protein Sci. 2006 doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, Saibil HR, Berks BC. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. J. Biol. Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 9.Cristobal S, de Gier JW, Nielsen H, von Heijne G. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berks BC, Palmer T, Sargent F. Adv. Microb. Physiol. 2003;47:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 11.Dilks K, Rose RW, Hartmann E, Pohlschroder M. J. Bacteriol. 2003;185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose RW, Bruser T, Kissinger JC, Pohlschroder M. Mol. Microbiol. 2002;45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 13.Robinson C, Bolhuis A. Nat. Rev. Mol. Cell Biol. 2001;2:350–356. doi: 10.1038/35073038. [DOI] [PubMed] [Google Scholar]
- 14.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. BMC Bioinformatics. 2005;6:167–175. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 17.Bernhardt TG, de Boer PA. Mol. Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ize B, Stanley NR, Buchanan G, Palmer T. Mol. Microbiol. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 19.Stanley NR, Palmer T, Berks BC. J. Biol. Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 20.Gon S, Patte JC, Mejean V, Iobbi-Nivol C. J. Bacteriol. 2000;182:5779–5786. doi: 10.1128/jb.182.20.5779-5786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanley NR, Sargent F, Buchanan G, Shi J, Stewart V, Palmer T, Berks BC. Mol. Microbiol. 2002;43:1005–1021. doi: 10.1046/j.1365-2958.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 22.Lequette Y, Odberg-Ferragut C, Bohin JP, Lacroix JM. J. Bacteriol. 2004;186:3695–3702. doi: 10.1128/JB.186.12.3695-3702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambasivarao D, Turner RJ, Simala-Grant JL, Shaw G, Hu J, Weiner JH. J. Biol. Chem. 2000;275:22526–22531. doi: 10.1074/jbc.M909289199. [DOI] [PubMed] [Google Scholar]
- 24.Ize B, Porcelli I, Lucchini S, Hinton JC, Berks BC, Palmer T. J. Biol. Chem. 2004;279:47543–47554. doi: 10.1074/jbc.M406910200. [DOI] [PubMed] [Google Scholar]
- 25.Sturm A, Schierhorn A, Lindenstrauss U, Lilie H, Bruser T. J. Biol. Chem. 2006;281:13972–13978. doi: 10.1074/jbc.M511891200. [DOI] [PubMed] [Google Scholar]
- 26.Caldelari I, Mann S, Crooks C, Palmer T. Mol. Plant-Microbe Interact. 2006;19:200–212. doi: 10.1094/MPMI-19-0200. [DOI] [PubMed] [Google Scholar]
- 27.Blaudeck N, Sprenger GA, Freudl R, Wiegert T. J. Bacteriol. 2001;183:604–610. doi: 10.1128/JB.183.2.604-610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA. J. Bacteriol. 2003;185:2811–2819. doi: 10.1128/JB.185.9.2811-2819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamane K, Mizushima S. J. Biol. Chem. 1988;263:19690–19696. [PubMed] [Google Scholar]
- 30.Coleman J, Inukai M, Inouye M. Cell. 1985;43:351–360. doi: 10.1016/0092-8674(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Int. J. Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 32.DeLisa MP, Samuelson P, Palmer T, Georgiou G. J. Biol. Chem. 2002;277:29825–29831. doi: 10.1074/jbc.M201956200. [DOI] [PubMed] [Google Scholar]
- 33.Guzman LM, Belin D, Carson MJ, Beckwith J. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas G, Potter L, Cole JA. FEMS Microbiol. Lett. 1999;174:167–171. doi: 10.1111/j.1574-6968.1999.tb13564.x. [DOI] [PubMed] [Google Scholar]
- 35.DeLisa MP, Lee P, Palmer T, Georgiou G. J. Bacteriol. 2004;186:366–373. doi: 10.1128/JB.186.2.366-373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopilato J, Bortner S, Beckwith J. Mol. Gen. Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 37.Sone M, Kishigami S, Yoshihisa T, Ito K. J. Biol. Chem. 1997;272:6174–6178. doi: 10.1074/jbc.272.10.6174. [DOI] [PubMed] [Google Scholar]
- 38.Chanal A, Santini CL, Wu LF. J. Mol. Biol. 2003;327:563–570. doi: 10.1016/s0022-2836(03)00170-0. [DOI] [PubMed] [Google Scholar]
- 39.Bogsch E, Brink S, Robinson C. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berks BC, Palmer T, Sargent F. Curr. Opin. Microbiol. 2005;8:174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Kebir MO, Kendall DA. Biochemistry. 2002;41:5573–5580. doi: 10.1021/bi015798t. [DOI] [PubMed] [Google Scholar]
- 42.Li P, Beckwith J, Inouye H. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7685–7689. doi: 10.1073/pnas.85.20.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajava AV, Zolov SN, Kalinin AE, Nesmeyanova MA. J. Bacteriol. 2000;182:2163–2169. doi: 10.1128/jb.182.8.2163-2169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson H, von Heijne G. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9751–9754. doi: 10.1073/pnas.88.21.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geller B, Zhu HY, Cheng S, Kuhn A, Dalbey RE. J. Biol. Chem. 1993;268:9442–9447. [PubMed] [Google Scholar]
- 46.Jack RL, Buchanan G, Dubini A, Hatzixanthis K, Palmer T, Sargent F. EMBO J. 2004;23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oresnik IJ, Ladner CL, Turner RJ. Mol. Microbiol. 2001;40:323–331. doi: 10.1046/j.1365-2958.2001.02391.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.