SUMMARY
To obtain structural information on the early stages of V(D)J recombination, we isolated a complex of the core RAG1 and RAG2 proteins with DNA containing a pair of cleaved recombination signal sequences (RSS). Stoichiometric and molecular mass analysis established that this signal end complex (SEC) contains two protomers each of RAG1 and RAG2. Visualization of the SEC by negative staining electron microscopy revealed an anchor-shaped particle with approximate two-fold symmetry. Consistent with a parallel arrangement of DNA and protein subunits, the N-termini of RAG1 and RAG2 are positioned at opposing ends of the complex, and the DNA chains beyond the RSS nonamer emerge from the same face of the complex, near to the RAG1 N-termini. These first images of the V(D)J recombinase in its post-cleavage state provide a framework for modeling RAG domains and their interactions with DNA.
INTRODUCTION
V(D)J recombination is central to establishing a functional adaptive immune system. The large repertoire of immunoglobulins and T cell receptors is generated in many vertebrates by combinatorial rearrangement of an extensive array of variable (V), diversity (D), and joining (J) gene segments that are joined to encode the variable domains of the protein chains (Gellert, 2002).
The recombination signal sequences (RSS) that flank these gene segments are recognized, paired in a synaptic complex, and cleaved by collaboration of the lymphoid-specific proteins RAG1 and RAG2. Full-length mouse RAG1 contains 1040 aa and RAG2 527 aa. However, only the cores of RAG1 (384–1008 aa) and RAG2 (1–387 aa) are necessary for the basic recombination reaction (Cuomo and Oettinger, 1994; Sadofsky et al., 1994; Sadofsky et al., 1993; Silver et al., 1993) and for in vitro RSS binding and cleavage (McBlane et al, 1995). Most biochemical studies on the RAG proteins, including the work presented in this paper, have been done with these core proteins, because the full-length proteins are difficult to express and purify. The deleted portions of both proteins probably have regulatory functions (Jiang et al., 2005; Swanson et al., 2004). The RAG1 core contains the DDE triad that is found in many ‘cut and paste’ DNA transposases (Fugmann et al., 2000; Kim et al., 1999; Landree et al., 1999). The RAG2 core has been predicted to be a six-bladed beta propeller (Aravind and Koonin, 1999; Callebaut and Mornon, 1998). In the presence of Mg2+, the RAG1/2 complex catalyzes a double-strand break at the coding/signal junctions, leaving blunt cut signal ends (Schlissel et al., 1993) and DNA hairpins on the ends of the flanking coding sequences (McBlane et al., 1995; van Gent et al., 1997). DNA cleavage is a two-step reaction: hydrolysis of one DNA strand creates a nicked intermediate whose 3’ hydroxyl group goes on to attack the phosphodiester bond of the opposing DNA strand, resulting in a hairpin. Consistent with the two-metal-ion mechanism described for DDE phosphotransferases, Ca2+ supports binding of the RSS at the RAG1/2 active site but inhibits both cleavage steps (Hiom and Gellert, 1997; Nowotny and Yang, 2006).
There are two types of RSS, 12RSS and 23RSS, differing in the length of the spacer (12 or 23bp) between a conserved heptamer (consensus CACAGTG) and a nonamer, (preferably ACAAAAACC) (Figure S1). In all cases the heptamer sequence borders the coding V, D, or J segment. Efficient V(D)J recombination requires one RSS of each type; this is known as the 12/23 rule (Sakano et al., 1980). The preference is already evident when RAG1/2 binds to the RSSs (Hiom and Gellert, 1998), though there may be additional selectivity at the cleavage stage (West and Lieber, 1998).
In vivo, the two hairpinned coding ends are rapidly transferred to the Non-Homologous End Joining (NHEJ) DNA repair factors for joining (Lieber et al., 2006). Signal ends persist longer in vivo before being joined in a process requiring most of the same NHEJ factors. In vitro, the cleaved signal ends remain tightly bound to the RAG proteins in a particularly stable Signal-End Complex (SEC) that is resistant to high salt or heparin (Agrawal and Schatz, 1997; Jones and Gellert, 2001). The relationship between RAG1/2 and DDE transposases is also evident in the SEC since this complex is capable of transposing the signal ends into target DNA (Agrawal et al., 1998; Hiom et al., 1998; Jiang et al., 2004; Reddy et al., 2006).
The determination of the stoichiometry of RAG1 and RAG2 proteins bound to RSSs has previously been based largely on electrophoretic mobility shift assays. Results have varied, with two groups concluding that the pre-cleavage complex contains a RAG1 dimer and two RAG2 monomers, together with a 12RSS, a 23RSS and an uncertain amount of HMGB1 (Bailin et al., 1999; Swanson, 2002), while another study reported an excess of RAG1 over RAG2 (Mundy et al., 2002). The spatial conformation of the RAG1/2-DNA complex has also been obscure. In one study addressing this question by in-gel FRET measurements, distances between fluorescent probes placed at either end of 12RSS and 23RSS substrates were calculated (Ciubotaru et al., 2007). The results indicated that the DNA bends and the two RSSs cross over, but no orientation of the protein domains could be ascertained. RSS bending on binding to RAG1/2 has also been reported from circular permutation assays (Aidinis et al., 1999) and atomic force microscopy (Pavlicek et al., 2008). HMGB1, which is known to severely bend a DNA duplex, is required for efficient V(D)J cleavage in vitro and is postulated to stabilize a bent 23RSS spacer and facilitate 12/23 paired complex formation for DNA cleavage (Kim and Oettinger, 1998; van Gent et al., 1997; West and Lieber, 1998).
For quantitative biochemical characterization and to obtain three-dimensional structural information about RAG1/2 bound to RSS DNA, we generated and purified the stable post-cleavage SEC. We determined the precise stoichiometry of RAG1, RAG2, HMGB1, 12 and 23RSS in the SEC and analyzed the structure of the SEC by electron microscopy and atomic-force microscopy. By fusing the RAG1 and RAG2 cores with maltose binding protein (MBP), combined with antibody tagging, we have defined the arrangement of RAG1 and RAG2 in the complex, as well as the exit points of DNA. A model for the organization of the protein domains within the recombinase complex is proposed.
RESULTS
Purification of active Signal End Complex and removal of MBP fusions
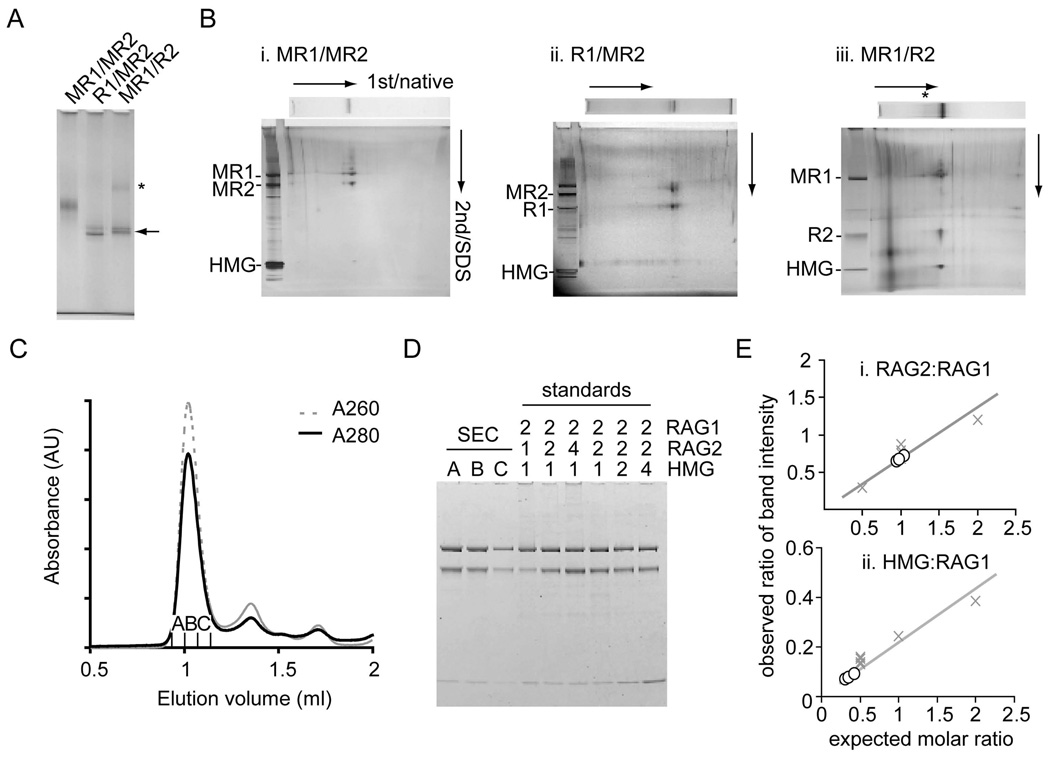
The increased stability of the RAG1/2-DNA complex after DNA cleavage was exploited to obtain sufficient quantities for characterization and structural analysis. To purify the post-cleavage complex, DNA with a 12RSS was bound to streptavidin-agarose via a biotin label on the coding flank. Catalytically active core RAG1 and RAG2 proteins, HMGB1 and 23RSS were then added step-wise, as described in Experimental Procedures, using Ca2+ to aid complex formation without DNA cleavage (Figure 1A). Unbound protein and DNA were removed with wash steps. After cleavage in Mg2+, the signal end complex (SEC) was released from the column matrix and collected. The eluate could be concentrated by ultrafiltration and contained both RAG proteins (Figure 1B). The concentrated eluate formed a major band on a native polyacrylamide gel (Figure 1C). In addition to the proteins, this band contained cleaved 12 and 23RSS in a 1:1 ratio (Figure 1D).
Figure 1. Purification of the Signal-End Complex (SEC).
(A) Schematic diagram of the SEC purification procedure: i. Biotinylated 12RSS was bound to streptavidin agarose. ii. The paired complex was then formed in-column by the addition of core RAG proteins (MR1/MR2), HMGB1, and 23RSS DNA. iii. Unbound protein and DNA were washed out. iv. Cleavage of the DNA was initiated by Mg2+ and the SEC was eluted.
(B) The purification procedure was monitored by Western blot using anti-MBP. The majority of the RAG proteins did not form a complex as seen in the flow through (FT). The column was washed until no protein was detected in the FT. Following cleavage, MR1 and MR2 were present in the eluted fraction, which could be concentrated 10-fold (Conc) by ultrafiltration.
(C) The SEC was visualized on a native 7% Tris-acetate polyacrylamide gel by Coomassie staining. The corresponding band was found to contain DNA when the SEC was prepared with radio-labeled RSSs. Free DNA 12*/23* migrated close to the dye front.
(D) A 15% TBE-Urea denaturing gel showed that the concentrated eluate (Conc) contained contaminating uncut RSS but the band excised from the native gel contained a 1:1 ratio of 12 and 23 signal-ends.
(E) The SEC retains transposition activity. Strand-transfer of radio-labeled signal ends into plasmid DNA was detected on an agarose gel as one-end (nicked plasmid, faint upper band) and two-end (linearized plasmid, predominant lower band) transposition products (tnp). The SEC eluate was more efficient at transposition than the control cleavage reaction prepared using the same DNA concentrations (1 nM) and same protein concentrations (2 nM) or 10-fold more protein (20 nM).
We tested radio-labeled SEC eluted from the streptavidin column for its ability to transpose the signal ends into plasmid DNA in vitro (Figure 1E). Transposition was more efficient with the purified SEC than starting from cleavage reactions with either the same or 10-fold greater RAG concentrations, indicating that the SEC was enriched for active RAG1/RAG2 species.
The N-terminal MBP tag on RAG1 or RAG2 protein could be removed in alternative constructs that contained a site for cleavage by the PreScission protease (Figure S2). Digestion following SEC purification resulted in a stable complex with faster migration (Figure 2A). Complexes that had MBP removed from either RAG1 or RAG2 had similar mobilities in the native gel, suggesting that the same number of MBP moieties had been removed, and therefore an equal number of RAG1 and RAG2 units were present in each SEC. When the lane from the native gel was excised and run in a second dimension on a denaturing gel, RAG1, RAG2 and HMGB1 were identified to be protein components of both digested and non-digested SEC bands (Figure 2B, i–iii). 2D electrophoresis also verified complete digestion of the alternative MBP fusions. Further purification of the SEC away from aggregates and contaminating protein and DNA was achieved using gel filtration (Figure 2C), resulting in a main peak containing RAG1, RAG2 and HMGB1 (Figure 2D).
Figure 2. Protein components and stoichiometry of SEC.
(A) MBP was removed from RAG1 (R1/MR2) or RAG2 (MR1/R2) of the purified SEC using PreScission protease. The mobilities of digested SEC were compared on a 3–8% Tris-acetate native gel (arrow). The SEC was less soluble when MBP was removed from RAG2 and a SEC dimer may be evident (*).
(B) Protein components of SEC and PreScission-digested SEC from a native gel (7% or 3–8%) were separated in the second dimension by SDS-PAGE. Silver staining confirmed the presence of RAG1, RAG2 and HMGB1 as components of each purified complex. Analysis by 2D gel electrophoresis confirmed complete removal of MBP from AR1 or LR2 fusions, referred to as R1 and R2.
(C) SEC made from MR1 and LR2 was purified by gel-filtration chromatography (Superdex 200) using binding buffer adjusted to 150 mM KCl as running buffer.
(D) Three fractions from the major peak were loaded onto an SDS-PAGE gel alongside standards of known molar stoichiometries (RAG1:RAG2:HMGB1). The gel was stained with Coomassie Blue and the intensity of the bands in each lane was measured with a fluorescence scanner.
(E). Relative ratios of band intensity of RAG2:RAG1 and HMGB1:RAG1 were calculated for each fraction (open circle) and compared to the stoichiometry standards (X). There are 2 RAG2s and ≤1 HMGB1 per RAG1 dimer.
Stoichiometry and molecular mass of the Signal-End Complex
Standards of known RAG1:RAG2:HMGB1 ratios were run to assess the stoichiometry of the protein components of the SEC (Figure 2D). Fluorescence of the Coomassie G-250 stained gel was used to quantify each protein, and the relative amounts of RAG2 and HMGB1 to RAG1 were compared to the standards (Figure 2D). In accordance with the digested complexes in the native gel, we found that there was a 1:1 ratio of RAG1 to RAG2. Slightly less than one HMGB1 was present per RAG1 dimer (~0.5/RAG1, Figure 2E). A small fraction of the SEC may lose HMGB1 during purification if it is not tightly bound. The empirical stoichiometry of the protein components of the SEC was estimated to be 2 RAG1: 2 RAG2: 1 HMGB1.
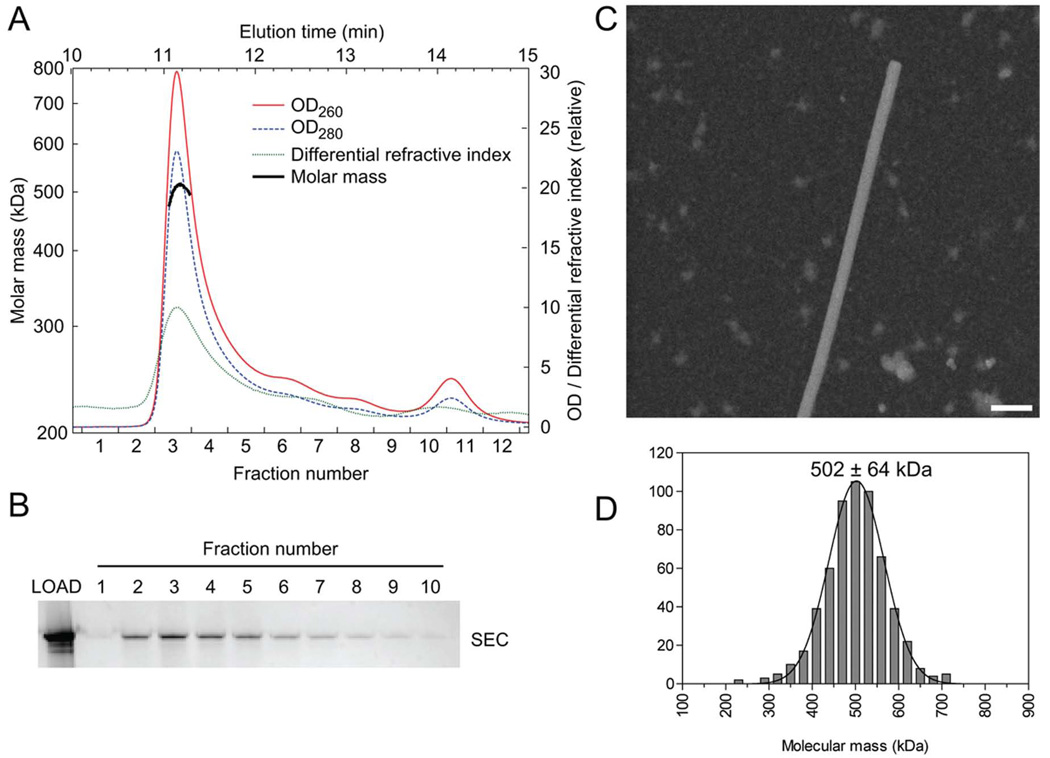
The molecular mass of the SEC was measured by multi-angle static light-scattering (MALS) analysis of the complex eluting from an in-line gel-filtration column (Figure 3A). The peak corresponding to the SEC, identified by native gel electrophoresis (Figure 3B), contained a species of average molecular mass 505 kDa (+/− 19 kDa) as averaged from 4 measurements. The measured value matches well with the calculated molecular mass (504 kDa) of an SEC comprising a 12 and 23RSS pair of DNAs (49 kDa), a single unit of HMGB1 (19 kDa), and two RAG1/2 (MR1/LR2) heterodimers (218 kDa). Given the equimolar ratio of RAG1 and RAG2, a SEC would be 722 kDa or 960 kDa if 3 or 4 RAG1/2 heterodimers were present, far outside the experimentally measured range. The A260/A280 ratio (1.27) of the SEC from gel filtration substantiated the presence of the two expected DNA fragments with the RAG1/2 heterotetramer (Table S1).
Figure 3. Molecular Mass of the Signal-End Complex.
(A) The molar mass of the SEC was measured using gel filtration coupled with light scattering. The measured mass of the SEC made from MR1 and LR2 was 505 kDa in this example. A range of 497 – 517 kDa was obtained from 4 experiments.
(B) Fractions from the gel filtration in (A) were analyzed by native gel electrophoresis, and silver-stained.
(C) A field of unstained SEC particles imaged in the scanning transmission electron microscope (STEM). TMV (rod-like structure) was included as an internal control. The molecular weight of the SEC was estimated by scaling to the known mass/length value of TMV.
(D) A histogram of the molecular weight of 580 SEC particles measured using STEM.
Further support for the molecular mass value was obtained by scanning transmission electron microscopy (STEM) of samples eluted from in-column complex formation (Figure 3C). The mass of the SEC as measured by STEM (calibrated against the mass per unit length of tobacco mosaic virus) presents a single peak centered at 502 ± 64 kDa (S.D.) (Figure 3D). The precision of the STEM data is insufficient to determine the amount of HMGB1 in each SEC but sufficient to corroborate that the SEC contains two RAG1–RAG2 heterodimers bound to the pair of RSS DNAs.
2D analysis of SEC by electron microscopy
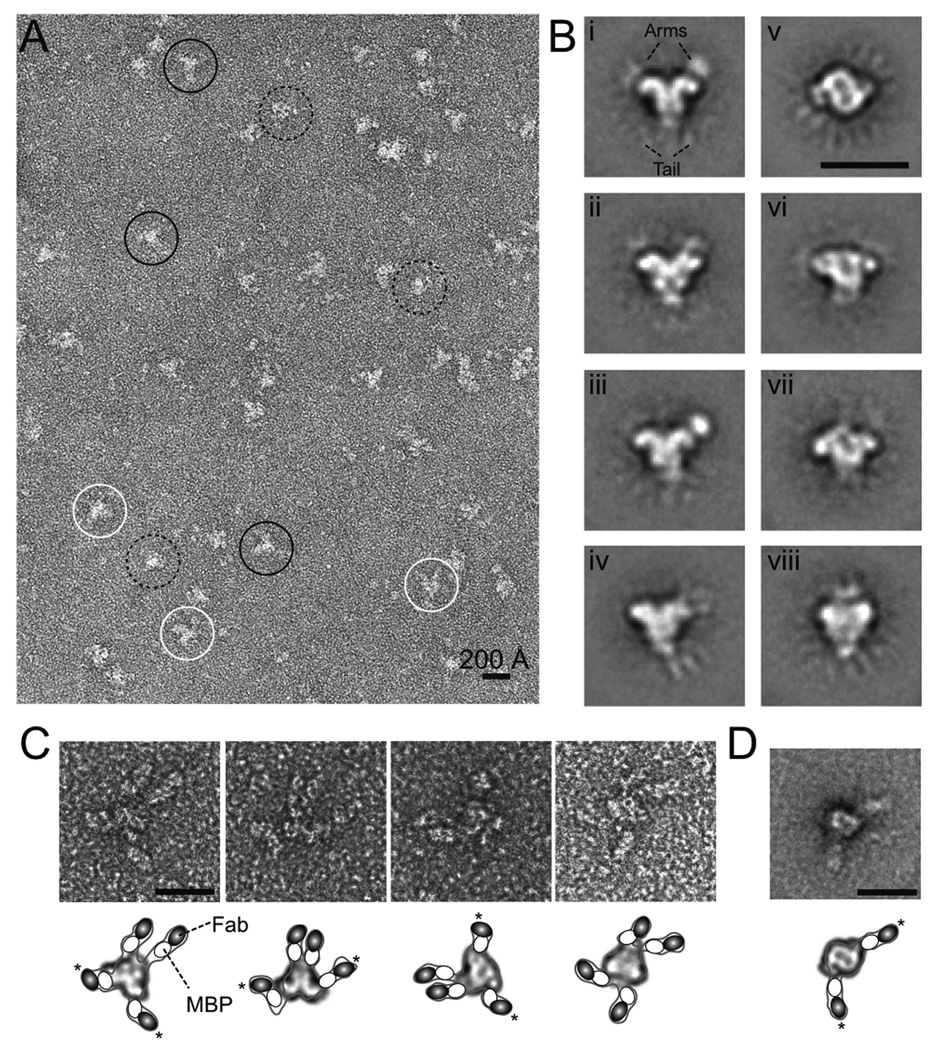
The purified SEC assembled from MBP-fused core RAG1 and RAG2 was adsorbed onto carbon-coated grids, negatively stained with uranyl acetate, and observed by transmission electron microscopy (EM). The micrographs showed an evenly spread population of particles of ~150 Å in the largest dimension (Figure 4A). Many of them exhibited a branched X or Y shape, while others appeared rounder. To enhance resolution, we selected 4200 SEC particles from an initially digitized set of > 10,000, and used reference-free alignment and classification methods to assign them to different internally consistent classes; the images in each class were then averaged. This procedure resulted in 8 predominant class-averages, each with distinctive features (Figure 4B). We experimented with larger numbers of classes but found this change in procedure resulted only in splitting of major classes into two or more closely related subclasses.
Figure 4. Electron microscopy and immunolabeling.
(A) Electron micrograph of SEC particles negatively stained with 1% uranyl acetate. Representative X-, Y-, and round-shaped particles are circled in white, black and dashed lines, respectively.
(B) Representative class-averaged images obtained from 4200 negatively stained SEC particles using image classification and averaging techniques. The numbers of particles in classes i to viii are 507, 519, 475, 681, 568, 513, 420 and 515.
(C) Gallery of images of SEC particles immunolabeled with Fab against MBP fused to RAG1 and RAG2.
(D) Class-average of 67 immunolabeled SEC particles, in which only RAG2 has an MBP fusion. Scale bars correspond to 200 Å in all panels. A schematic drawing of the Fab-bound particles is shown below the EM images in panels C and D. Inside the contoured particle, a possible 2D class-average is fitted, and MBP and Fab are depicted as white 60 × 45 Å ovals and black 70 × 45 Å ovals, respectively. The asterisks indicate the pair of arms projecting from the core particle in opposite directions.
The most prominent finding to emerge from the class-averages is detection of a two-fold rotational axis (C2) in the complex. This axis is either in the image plane in the projections of classes i–iv and viii, or perpendicular to the image plane (class v). This symmetry was also approximately observed in classes vi and vii, which likely represent projections between the two orthogonal views of SEC particles. Although the 11 bp difference between the 12 and 23RSS should in principle render the SEC particle asymmetric, such a difference is apparently too small to be discernible, given the limited resolution of the negatively stained EM data. Thus the SEC appears to be pseudo-symmetric.
In all 8 class-average images, a well-defined core structure is seen surrounded by several additional, mostly fainter, densities. These peripheral densities are labeled as “arm” and “tail” in Figure 4B (i) and appear as protuberances on opposite sides of the rounder core shape of class v. Further sub-classification did not result in better definition of the peripheral densities, indicating that their relative faintness might result in part from local mobility. This inference was supported by variance maps calculated from the particles in each class separately, or from combining classes i to iv, which confirmed that the arm and tail regions are more variable and thus more mobile than the core (Figure S3). Interestingly, classes i and iii exhibited a better-defined density on one of the “arms”, which did not obey the C2 symmetry (Figure 4B). While it is possible that this asymmetry may results from “locking on” to the density on one side during classification and smearing out the density on other side, we think it more likely that the distinction may arise from the ‘arm’ on one side being more fully immersed in the stain layer and consequently better contrasted than its counterpart on the other side (Figure S4). The dimensions of this feature were ~ 45 × 60 Å, roughly matching the expected 2D projection of the 40 kDa MBP. MBPs are linked to the N-termini of RAG1 and RAG2 by a 25 aa flexible tether and are expected to be mobile. The peripheral location of the MBPs is also consistent with the observation that MBP fusion does not alter the recombination activity of RAG1 and RAG2 (Kriatchko et al., 2006; Mundy et al., 2002).
Identification of MBP in the SEC by immuno-labeling
To better define the locations of MBPs in the SEC, we performed immuno-labeling with an anti-MBP Fab. Purified SEC particles were incubated with the Fab, negatively stained and visualized by EM. In agreement with the measured stoichiometry of two RAG1 and two RAG2 molecules in each SEC, four anti-MBP Fab extensions were observed for each SEC particle (Figure 4C). The extensions are near the “arms” and “tail” of the SEC, and their size approximately matches the combined dimensions of Fab (75 × 55 × 40 Å) and MBP (60 × 50 × 40 Å) (Figure 4C). Due to the flexibility of the MBPs’ linkers, alignment and class averaging of the Fab-bound SEC particles was not productive. However, the orientations of the Fabs are indicative of the configuration of the SEC. Two of the Fab-MBP extensions appear to protrude from adjacent points on the SEC particle and are nearly parallel, and the other two diverge from the opposite end of the SEC. Superimposed onto the unlabelled class-averages, it appears likely that the two parallel Fab extensions belong to the “tail” MBPs and the other two to the “arm” MBPs (Figure 4C). Assuming that the pseudo-C2 symmetry of SEC applies to RAG1/2 and MBP fusions, we surmised that one pair of MBPs is associated with RAG1 and the other pair to RAG2.
To distinguish the pairs of MBPs and their connections to RAG1 and RAG2, we used the AR1 and MR2 variations and removed the MBPs fused to RAG1 by protease digestion after assembly and purification of SEC particles (Figure S2). The remaining MBPs on RAG2 were labeled with the Fab and viewed by EM. The resulting images showed two extended densities for each SEC particle (Figure S5). Alignment and averaging of 67 similar particles showed that the core resembled the ‘round’ projection of SEC (class v) and the MBP-Fabs extended outwards on opposite sides (Figure 4D). Therefore MBPs fused to the RAG2s are most probably assigned as the “arms” in Figure 4B (i–iv), and the “tails” of the anchor-shaped SEC are most likely the MBPs linked to the N-termini of the RAG1 dimer.
Characterization of the SEC core
Disregarding the peripheral MBPs, the SEC particles in all 8 class-averages show well-defined features. When the projection is normal to the C2 axis, two distinct views of SEC are recorded: (1) an anchor shape with two thick tips and a shank bisected by the vertical C2 axis and further divided into three horizontal sections (Figure 4B, i–iv), and (2) a more compact triangular shape (Figure 4B, viii). Among these five projections, the length of the SEC along the C2 axis consistently measures 125 Å (Figure S6). The width of the shank measures ~80–90 Å. Its three horizontal sections appear most distinct in class ii, where the middle section is separated into two symmetric rounded densities. The widest dimension of the SEC is 150–160 Å between the tips of the anchor-shape projection (class i–iv), falling to 135 Å in the triangular projection. These two projections are probably related by a rotation around the C2 axis, with the triangle representing a side projection of the anchor-shaped SEC (Figure 5).
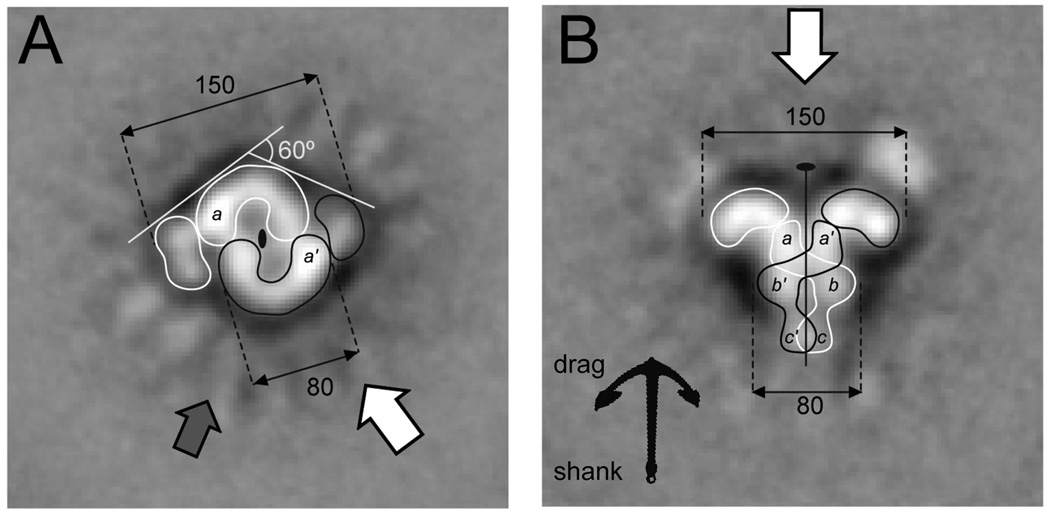
Figure 5. Orthogonal views of the SEC.
(A) The figure-eight and (B) the anchor-shaped class-average projections probably correspond to orthogonal views of the particle. This interpretation is supported by the relative disposition of the symmetry elements and correlated dimensions. The left image may result from the projection of the SEC in the direction indicated by the thick white arrow on the right panel, and vice versa.
(A) The figure-eight shape is depicted as two symmetry related C-shaped entities that encircle the darker regions in the center of the particle. One end of the C-entity is brighter than the other and is labeled with an a (a' in the other subunit). The grey arrow indicates the direction of the projection likely resulting in the triangular particle shown in class viii (Figure 4B). Thus the anchor- and triangle-shaped SEC are probably related by a ~ 60° rotation around the C2 axis.
(B) The C2 axis runs along the anchor-shaped projection dividing the particle in two nearly identical halves. This middle line shows an accumulation of stain, except for a region in the middle of the shank that we interpret as a crossover of the protein from one side of the shank to the other. Contours of two possible subunits related by the C2 symmetry are drawn. The densities on the tips of the anchor drag end that correspond to the side flaps of the figure-eight projection are contoured separately.
When the projection was along the C2 axis (Figure 4B, v and vii), the SEC had the appearance of an approximately rectangular figure-of-eight with two diagonally opposed side-flaps. Although there were two possibilities for the SEC being projected along the C2 axis (either end of the anchor towards the carbon grid), the mirror image of the projections v and vii was absent (Figure 4B), indicating a preferential orientation of SEC binding to the carbon grid. The central figure-of-eight is 110 Å long; its width, 80 Å (Figure S6), matches that of the shank in the orthogonal view, and the span of the side flaps (~150 Å) matches the width of the anchor. Accordingly, we suspect that the tips of the anchor drag correspond to the side-flaps of the figure-of-eight, and the shank of the anchor to the figure-of-eight itself.
The central figure-of-eight appear to be made of two C shaped entities around the C2 axis. Each C-shaped entity in the figure-of-eight projection encircles a dark disc of 25 Å in diameter, and the two discs are separated by ~10 Å. These accumulations of stain may indicate a depression, an internal cavity in the particle, or positively stained material such as DNA. Dark areas were also observed in the anchor-shape projections, either forming a line along the C2 axis in Figure 4B (i) or zigzags in Figure 4B (ii). Proteins are usually negatively stained, thus appearing bright in EM. Nucleic acids (DNA or RNA) are likely to be positively stained by uranyl-acetate, and thus may be invisible in these micrographs. In our hands, the 30 bp extensions of RSS DNAs in the SEC particles (thus ~100 Å additions to the core) remained “invisible” in the EM images.
Arrangement of DNA determined by atomic force microscopy
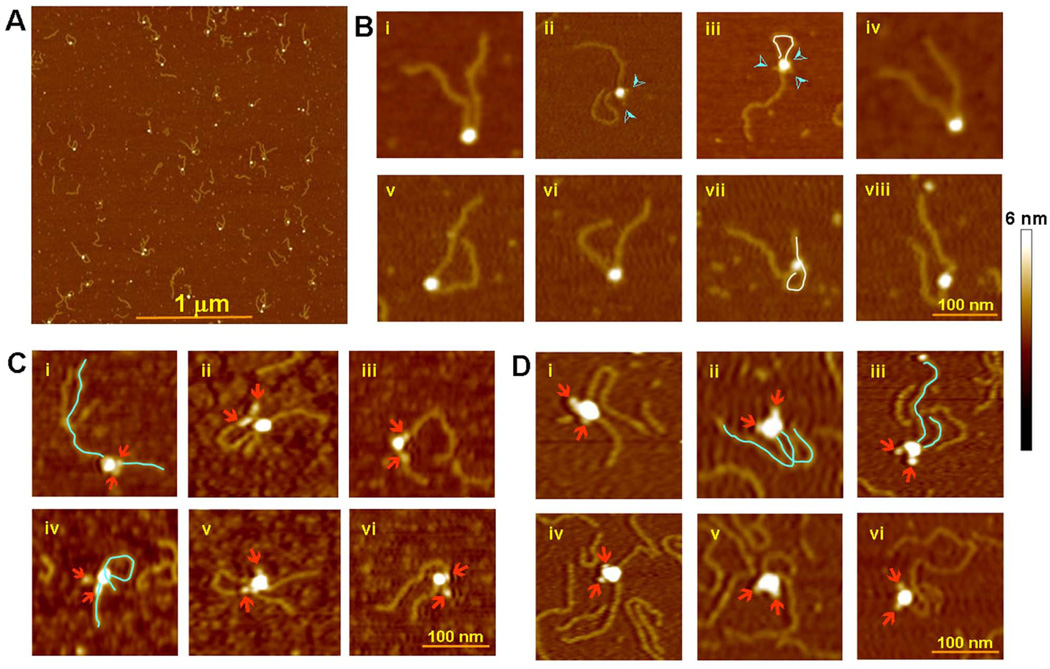
To locate the RSS DNAs, we subjected SEC particles to atomic force microscopy (AFM), a proven technique for visualizing DNA in complex with protein (Shlyakhtenko et al., 2003). To follow the DNA trajectory by AFM, the SEC was assembled using DNA substrates with 500 bp extensions beyond the RSS nonamers and purified as before. Figure 6A shows a population of SEC complexes detected by AFM. The SEC particles were of a uniform size, and the volume of the “headpiece” was consistent with the expected mass of 500 kDa (Figure S7). Depending on how the SEC particles and extended DNAs adsorbed onto the mica surface, a variety of projections was recorded. In a sample of 190 particles, however, it is clear that the two DNA chains predominantly (80% of cases) exit the SEC from adjacent points (Figure 6B, i,ii,iv–vii). More complicated configurations are seen in 10% of particles, where one of the DNA molecules loops and re-binds non-specifically to the protein complex, either at the free DNA end (Figure 6B, iii), or at an intermediate site (Figure 6B, vii). In another 10% the two DNA molecules seem to be exiting on opposite sides of the complex (Figure 6B, iii and viii). If DNAs truly exited from the SEC on opposite sides, they would never, or at least not often, appear to emerge near each other. We suspect that the apparent divergent trajectories of DNA molecules result when they emerge near each other on the SEC but bend and diverge upon attachment to the mica surface. The most populated view by AFM may correlate with the C2 axis in the mica plane and the less populated view to the C2 axis perpendicular to the plane.
Figure 6. Atomic Force Microscopy.
(A) Population of purified SEC (MR1/LR2) with 500 bp extensions to the nonamer end of the 12 and 23 RSSs.
(B) Representative SEC particles selected from (A). The majority of images support a parallel arrangement of the RSS DNAs in the SEC. The DNA chains in a few SECs appear to exit in anti-parallel direction (iii and viii). Occasionally, one of the DNA molecules loops around and binds non-specifically to the protein core (iii and vii), delineated in white. Protrusions often seen emanating from the protein core (indicated by cyan arrowheads) are most likely the MBP domains on RAG1 and RAG2 (ii, iii).
(C) MR1/R2 SEC with 250 bp 12RSS and 500 bp 23RSS DNA. The MBPs on RAG2 were removed and the MBPs on RAG1 were labeled with Fab fragments. The Fabs (indicated by arrowheads) are generally close to the point of DNA exit in the SEC. The DNA molecules are delineated for clarity in a couple of examples.
(D) R1/MR2 SEC with 250 bp 12RSS and 500 bp 23RSS DNA. The MBPs on RAG2 were labeled with Fab fragments. MBPs on RAG1 were pre-cleaved. Notice that the Fabs are on the opposite side from the DNA exits.
In order to identify where on the complex the DNAs exit in relation to RAG1 and RAG2, the MBPs on the SEC were selectively digested and the remaining MBPs were once again labeled with Fab (Figure 6C and D). These experiments also utilized 12 and 23RSS substrates of different lengths (250 bp and 500 bp). Fab-labeled MBPs were clearly identified as globular entities peripheral to the recombinase core. When only the RAG2 MBPs were labeled, the two DNAs most often exited together from the SEC on the opposite side to the Fabs (Figure 6D, i–vi). Consistent with this observation, when only the RAG1 MBPs are labeled, they are situated closer to the exiting DNAs (Figure 6C, i–vi). The MBPs attached to RAG1 appear to be more flexible and assume more varied positions relative to the SEC than the MBPs attached to RAG2.
DISCUSSION
Extensive purification of the SEC has allowed us to obtain detailed information about its composition and structure. Several lines of evidence show that the SEC contains two protomers each of RAG1 and RAG2, and independent measurements of its molecular mass by gel filtration, light scattering, and STEM all yield a value close to 500,000 Da, implying that the SEC is composed of a RAG1–RAG2 heterotetramer and two cleaved RSS DNAs (Figure 2–Figure 3). There is also one HMG1 protomer per SEC. Earlier studies had shown that HMGB1 stimulates cleavage of a 23RSS much more than a 12RSS, suggesting that HMGB1 is likely to be associated with the 23RSS in the SEC (Aidinis et al., 1999; van Gent et al., 1997).
Our results verifying a (RAG1)2–(RAG2)2 composition of the SEC are not strictly comparable to earlier data, which refer to the complex before DNA cleavage. However, when we examined pre-cleavage synaptic complexes by AFM (Figure S9), the majority of complexes containing the 12 and 23 RSSs were no larger than tetrameric in size (although more heterogeneous than the SEC particles). In addition, there was no separated subunit in the SEC sample eluted after in-column cleavage, and the SEC retains transposition activity, also making it unlikely that there is a change in subunit composition upon cleavage.
A recent paper (Shlyakhtenko et al., 2009) reports an equimolar RAG1–RAG2 hetero-octamer in a pre-cleavage synaptic complex, the size being estimated by AFM. However, this complex, is only detected in the absence of salt and after cross-linking of the proteins with glutaraldehyde, and may represent a less stable species. We note that in our hands, low salt concentrations in the sample buffer used for AFM increased aggregation and non-specific RAG1/2-DNA interactions.
We analyzed the SEC by negative staining EM. Classification of these data led us to identify three predominant projections, each with an apparent twofold axis. Two of these projections are perpendicular to this axis and respectively have an “anchor” shape and a “triangular” shape; the other is along the twofold axis and shows a “figure-of-eight” with two symmetrically disposed side-flaps (Figure 5). These three views of the same object are analyzed further below.
Because the core RAG1 and RAG2 constructs used for preparing the SEC were fusions with MBP, it was possible to tag the SEC with monovalent anti-MBP Fab fragments, and thus localize the MBP moieties. It appears that in the “anchor” projection, all four MBPs lie in approximately the same plane. Furthermore, by selectively removing MBP from RAG1 and again tagging with the anti-MBP Fab, we were able to distinguish the two RAG proteins in the SEC. These data indicate that RAG2 is at the “drag” end (the wide end), of the anchor and their MBPs form the “arms” and the MBP attached to RAG1 at the “shank” end and form the “tails” (Figure 4),
Further information obtained by AFM was used to determine the orientation of RSS DNAs in the SEC. Because the cleaved ends are presumably concealed within the complex, one can only detect the DNA extending beyond the nonamer of the RSS. In the AFM images, the pair of ends predominantly emerges from the same side of the SEC; indicating that the DNA fragments are arrayed in generally parallel rather than antiparallel (or oblique) orientation. This result dovetails with data showing that with a short spacing between two RSSs deletional recombination is more efficient than inversional joining (Lewis and Hesse, 1991; Sheehan and Lieber, 1993), presumably because deletion requires a shorter path and fewer turns of DNA than inversion. (Figure S8) (Kim and Oettinger, 1998). Additional AFM images of Fab-tagged complexes made with either MBP-RAG1 or MBP-RAG2 revealed that the exit points of the DNA lie close to the RAG1 end and opposite the RAG2 end.
Recently an antiparallel configuration of two nonamer DNAs crystallized with the “nonamer binding domain” (NBD) of RAG1 was reported (Yin et al., 2009). The contrast to the parallel configuration as shown in the EM and AFM analyses of SEC may be in part caused by the different context of these observations. The NBD fragment (aa 389–464) has no detectable binding to the nonamer in solution and displays few sequence-specific contacts in the crystal structure (Yin et al., 2009).
Model of the SEC
Having located the RAG1 and RAG2 N-termini and nonamer exits on the SEC particle, we were able to generate a fairly detailed model of the RAG1-RAG2-DNA complex (Figure 7). Like the bipartite RSS, the primary sequence of the core RAG1 can be considered in two parts: an N-terminal nonamer-binding domain (NBD) and a larger catalytic domain that interacts with the heptamer and coding sequence (Figure S2). The NBD is placed near the shank end based on the MBP mapping and the DNA extension beyond the nonamer (Figure 4C–D and Figure 6). Biochemical data showed that both RAG1 and RAG2 contact the RSS heptamer (Eastman et al., 1999); this would place the catalytic domain of RAG1 near the drag end of the anchor. In other words, the two units of the RAG1 dimer would extend from the shank end roughly side by side and then cross over (Figure 5B), each with a RAG2 protomer attached to form the drag end. Similarly, the two RSSs extend through the length of the “anchor”. To accommodate the extra 11 bp of the 23RSS, additional bending possibly facilitated by HMGB1 will be necessary.
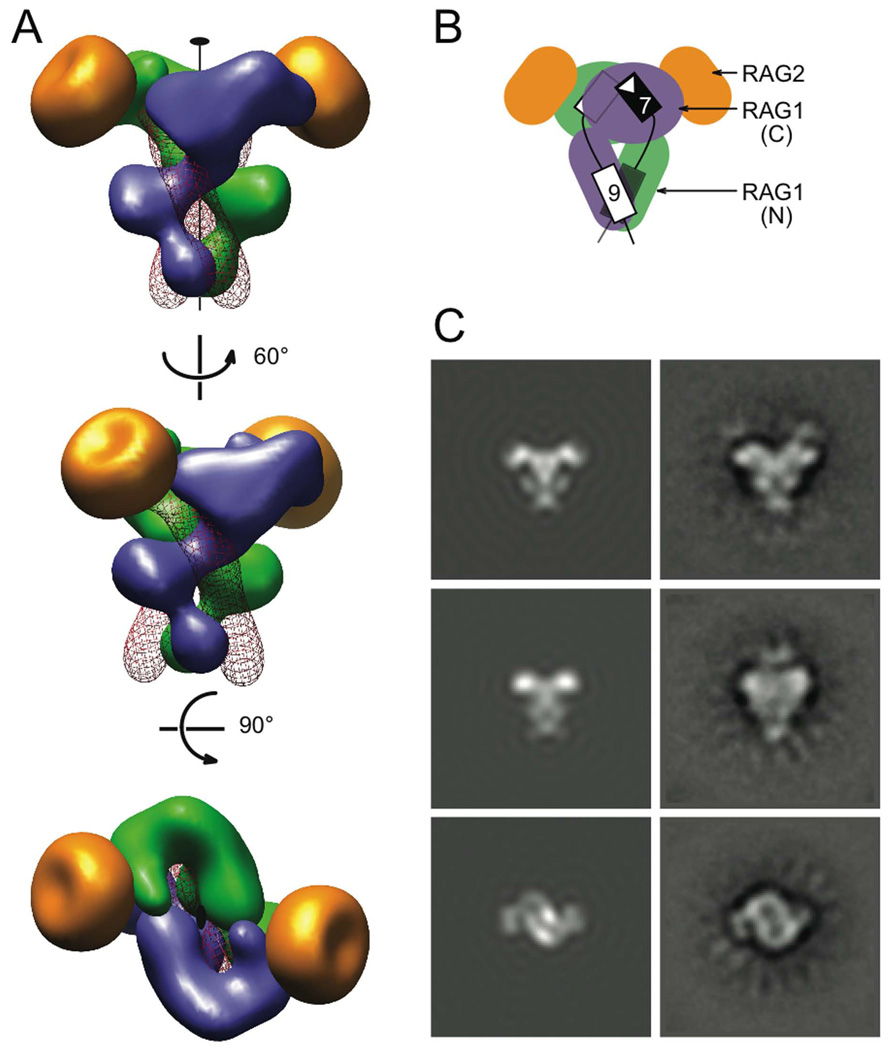
Figure 7. SEC model.
The model consists of two identical RSS DNAs, and core RAG1 and RAG2. (A) Three views of the SEC model. The two upper views are related by ~60 ° rotation around the C2 axis, and both are orthogonal to the bottom view. A RAG1 dimer (blue and green subunits), which was modeled after Tn5 transposase (PDB: 1MUR) and the HtH domains from the POU-DNA complex (PDB: 1GT0), forms the “shank” of the SEC. RAG2 molecules (gold) were modeled based on the gyrase A β-propeller domain (PDB: 1SUU). They form the tips of the “drag-end”. Two DNA molecules represented as red wires intertwine with the RAG1 dimer. The volume of each protein or DNA was calculated from the model coordinates with a filter of 25 Å resolution. (B) Cartoon depicting the location of the NBD (N) and catalytic (C) domains of RAG1, RAG2, and the DNA in the SEC. The heptamer and nonamer of the two RSSs (black and white) are shown as rectangles. (C) Comparison of 2D projections of the SEC model (left) and the EM class averages (right). The correlation coefficients for the three pairs are 0.79, 075 and 0.61 (details in SI).
To carry the modeling further, we made extensive use of sequence homologies of RAG1/2 with proteins of known structures. We used the Tn5 transposase post-DNA cleavage complex (PDB: 1MUR) as a model for the dimeric RAG1 catalytic domains associated with the heptamers. However, the DDE triad (D600, D708 and E962) essential for catalysis in RAG1 is split by a predicted C2H2 zinc finger and a helix-rich region, together including residues ~720–950. This inserted domain has been shown to be important for hairpin formation and heptamer/coding sequence contacts (Huye et al., 2002; Lu et al., 2006). The eukaryotic Hermes transposase also contain a DDE triad interrupted by a helical domain (Hickman et al., 2005). Superimposition of the Hermes transposase structure onto that of Tn5 (guided by the DDE motifs) does not alter the overall shape of the protein-DNA complex. The transposase dimer and two DNA ends (Davies et al., 2000) can be fit into the EM 2D projections as the “drag” end of SEC. The fitting is particularly pleasing in the “figure-eight” projection (Figure 5A), which gives evidence of “channels”, which were also seen in the EM structure of HIV integrase attributed to its catalytic domain (Michel et al., 2009). The Tn5-DNA complex has an antiparallel orientation of transposon ends at the two active sites. We envisage that a bend in 12 or 23bp spacer, possibly accompanied by further bending in the heptamer itself (Olson et al., 1998; Patel et al., 1987), allows the heptamers to adopt a similar head-to-head configuration in SEC.
Residues 390–460 of RAG1-NBD share sequence homology with the Helix-Turn-Helix (HTH) DNA-binding domain found in the HIN recombinase (Difilippantonio et al., 1996). A parallel arrangement of RSS DNAs associated with HTH domains is predicted for the Tc1/mariner family of transposases (Richardson et al., 2006). To model the NBD of RAG1 and its associated nonamer DNA, a pair of HTH domains and associated DNAs was adopted from the transcription factor POU-HMG1-DNA complex (PDB: 1GT0). Our interpretation is that the NBD (residue 390 – 590) may consist of more than one HTH domain. We adjusted the locations of the HTH domains along the DNA major groove and applied the C2 symmetry to the HTH-DNA complex to match the “b” and “c” densities (Figure 5B). The resulting placements were further adjusted to connect the nonamer to heptamer by 12 or 23 bp spacer DNA.
The essential core of RAG2 (residues 1–352) has been predicted to fold into a six-bladed beta propeller similar to those of the C-terminus of DNA gyrase A protein (Corbett et al., 2004). The GyrA beta propeller (60 Å in diameter and 35 Å deep) was used as a model, and fits reasonably well with the side flaps in the figure-eight projection and also with the tips of the anchor shape. The position of RAG2 would presumably allow it both to interact directly with RAG1 and to influence the heptamer binding to RAG1 (Aidinis et al., 2000; Eastman et al., 1999; Swanson and Desiderio, 1998, 1999).
After multiple rounds of building and rebuilding, we generated a SEC model whose dimensions and calculated two-dimensional projections approximately match the EM class-average images. Furthermore in this model, the DNA configuration is compatible with the trans mode of DNA binding by the RAG1 dimer (Swanson, 2001) where the nonamer binding domain and active site are provided by different RAG1 subunits.
In summary, by combining electron microscopy with information from mutational studies and homologous proteins, we have built a space-filling model of the SEC that is consistent with existing biochemical data, and helps to clarify the relation between the RAG proteins and transposases. Our results will serve as a platform for further experimental work on the structure and biological functions of the RAG1/2 complex and on its interplay with factors involved in DNA repair and in the regulation of V(D)J recombination.
EXPERIMENTAL PROCEDURES
Proteins and DNA
Core RAG1 (384–1008) and RAG2 (1–387) fused to MBP (MR1 and MR2) were made as described (McBlane et al., 1995). Alternative constructs of RAG1 (AR1) and RAG2 (LR2) contained a PreScission protease site between the MBP and core RAG sequence (Figure S2). MR2’ (with no c-myc tags) and LR2 coding regions were inserted into pLEXm vector (Aricescu et al., 2006) and transiently expressed in HEK293T cells (Swanson et al., 2004). Preparation of HMGB1 (1–163) and MBP Fab fragments is described in Supplemental Data. 23RSS and biotinylated-12RSS substrates were made from annealed oligonucleotides. DNA substrates for the AFM samples were prepared from pDVG54 plasmid (Supplemental Data, Figure S1).
Purification of SEC
500 pmol biotinylated 12RSS was bound to 0.2 ml streptavidin agarose (Novagen) in a 10 ml column. RAG-DNA complexes were assembled in the column in 10 ml binding buffer (25 mM MOPS-KOH pH 7, 50 mM potassium glutamate, 40 mM KCl, 4 mM CaCl2, 1% (v/v) glycerol, 4 mM dithiothreitol, 0.01% (w/v) dodecyl-D-maltoside, 0.4 µM AEBSF, 5 µM leupeptin, 0.5 µM pepstatin). 10 µg/ml HMGB1 then 200 µg/ml MR1/MR2 or AR1/MR2, or 100 µg/ml MR1/LR2, were added to the column at 37 °C for 30 minutes. 750 pmol 23RSS was included and incubated for another 30 minutes. The resin was washed with 20 ml of binding buffer. 5 mM MgCl2 in 0.5 ml binding buffer was added and incubated for 1 hour at 37 °C. Eluted SEC and a 0.5 ml wash were collected and concentrated in 100 mM potassium glutamate by ultrafiltration using a 100 kDa molecular weight cut-off membrane (Millipore). MBP was digested from AR1 or LR2 following SEC purification (minus protease inhibitors) with 20 µg/ml PreScission protease overnight at 4 °C. Purifications of radio-labeled SEC, analysis of SEC by gel electrophoresis, and transposition assays are detailed in Supplemental Data.
Molecular mass measurements
The methods are described in detail in Supplemental Data. Briefly, refractive index and multi-angle light scattering of samples eluted from a gel filtration column (TSK-G3000PWXL, Tosoh) were measured in-line using Optilab rEX and DAWN HELEOS instruments (Wyatt). Molar mass was obtained using Astra V software (Wyatt, 1993). STEM micrographs were recorded at the Brookhaven STEM facility (www.biology.bnl.gov/stem/stem.html). Intensities of SEC and TMV were measured using PCMass29 (Hainfeld et al., 1982), applying 115 Da/intensity unit. A scaling factor calculated from the measured (13.3 +/− 0.8 kDa/Å, Figure S10) and known (13.14 kDa/Å) TMV mass per length values was applied to correct the SEC mass.
AFM volume measurements were calculated from pixels values in ImageJ software (Figure S7).
Electron microscopy of negatively stained SEC
10–20 µg/ml of an SEC sample was applied to a glow-discharged carbon-coated copper grid, washed with five drops of deionized water and stained with two drops of 1% uranyl acetate. Micrographs of a final pixel size of 3.174 Å were produced, as described in Supplemental Data.
Single-particle image processing and 2D averaging
4,200 SEC images were selected from 87 micrographs for good particle distribution, staining, and optical quality (appropriate defocus, lack of drift, etc). SPIDER (Frank et al., 1996) was used to center and sort the particles by a reference-free alignment. Iterative cycles of correspondence analysis, hierarchical clustering by the Ward criteria, and multireference alignment produced 8 classes, each containing at least 420 particles. The resolution of the 2D averages, estimated by Fourier ring correlation at 0.5 cut-off, varied between 26 and 29 Å (Saxton and Baumeister, 1982). EMAN (Ludtke et al., 1999) and BSOFT (Heymann and Belnap, 2007) were used at multiple steps for image visualization and processing.
Atomic Force Microscopy
SEC preparation and AFM instrumentation is detailed in Supplemental Data. 5 µl of diluted sample was deposited on mica pretreated with 1-(3-amino-propyl)-silatrane (Shlyakhtenko et al., 2003).
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. J. S. Wall and M. M. Simon of Brookhaven National Laboratory for STEM data acquisition. S.R.-M. is indebted to Drs. N. Cheng, C. S. Smith, U. Baxa and D. Winkler for help with EM data acquisition, to Drs. G. Effantin, N. Mizuno and U. Baxa for guidance in image analysis, and to Drs. M. Nowotny and J. Y. Lee for insightful discussions. We thank C. Vander Kooi, P. Longo, J. McLellan and D. J. Leahy for help in establishing the mammalian expression system.
S.R.-M. was a recipient of a Human Frontiers Science Program fellowship. This work was supported by the intramural research programs of NIDDK, NIBIB and NIAMS of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi ME, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1–RAG2. Mol. Cell. Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aidinis V, Dias DC, Gomez CA, Bhattacharyya D, Spanopoulou E, Santagata S. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 2000;164:5826–5832. doi: 10.4049/jimmunol.164.11.5826. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Bailin T, Mo X, Sadofsky MJ. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell. Mol. Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciubotaru M, Kriatchko AN, Swanson PC, Bright FV, Schatz DG. Fluorescence resonance energy transfer analysis of recombination signal sequence configuration in the RAG1/2 synaptic complex. Mol. Cell. Biol. 2007;27:4745–4758. doi: 10.1128/MCB.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl. Acad. Sci. USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. (New York, N.Y. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- Eastman QM, Villey IJ, Schatz DG. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Hainfeld JF, Wall JS, Desmond EJ. A small computer system for micrograph analysis. Ultramicroscopy. 1982;8:263–270. doi: 10.1016/0304-3991(82)90242-x. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- Hiom K, Gellert M. A stable RAG1–RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol. Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Jiang H, Ross AE, Desiderio S. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J. Biol. Chem. 2004;279:8478–8486. doi: 10.1074/jbc.M311219200. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl. Acad. Sci. USA. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Oettinger MA. Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriatchko AN, Anderson DK, Swanson PC. Identification and characterization of a gain-of-function RAG-1 mutant. Mol. Cell. Biol. 2006;26:4712–4728. doi: 10.1128/MCB.02487-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Hesse JE. Cutting and closing without recombination in V(D)J joining. EMBO J. 1991;10:3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat. Struct. Mol. Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, Agapkina J, Ziganshin R, Gottikh M, Nazabal A, et al. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 2009;28:980–991. doi: 10.1038/emboj.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Shapiro L, Hare D. Nuclear magnetic resonance and distance geometry studies of DNA structures in solution. Annu. Rev. Biophys. Biophys. Chem. 1987;16:423–454. doi: 10.1146/annurev.bb.16.060187.002231. [DOI] [PubMed] [Google Scholar]
- Pavlicek JW, Lyubchenko YL, Chang Y. Quantitative analyses of RAG-RSS interactions and conformations revealed by atomic force microscopy. Biochem. 2008;47:11204–11211. doi: 10.1021/bi801426x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JM, Dawson A, O'Hagan N, Taylor P, Finnegan DJ, Walkinshaw MD. Mechanism of Mos1 transposition: insights from structural analysis. EMBO J. 2006;25:1324–1334. doi: 10.1038/sj.emboj.7601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5'-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- Sheehan KM, Lieber MR. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol. Cell. Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Gilmore J, Kriatchko AN, Kumar S, Swanson PC, Lyubchenko YL. Molecular mechanism underlying RAG1/RAG2 synaptic complex formation. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl. Acad. Sci. USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 2001;21:449–458. doi: 10.1128/MCB.21.2.449-458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC, Volkmer D, Wang L. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J. Biol. Chem. 2004;279:4034–4044. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RB, Lieber MR. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt P. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta. 1993;272:1–40. [Google Scholar]
- Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat. Struct. Mol. Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.