Abstract
In the human adrenal cortex, the peptide hormone adrenocorticotropin (ACTH) directs cortisol and adrenal androgen biosynthesis by activating a cAMP/cAMP-dependent protein kinase (PKA) pathway. Carboxyl-terminal binding protein 1 (CtBP1) is a corepressor that regulates transcription of the CYP17 gene by periodically interacting with steroidogenic factor-1 in response to ACTH signaling. Given that CtBP1 function is regulated by NADH binding, we hypothesized that ACTH-stimulated changes in cellular pyridine nucleotide concentrations modulate the ability of CtBP1 to repress CYP17 transcription. Further, we postulated that PKA evokes changes in the phosphorylation status of CtBP1 that control the ability of the protein to bind to steroidogenic factor-1 and the coactivator GCN5 (general control nonderepressed 5) and repress CYP17 gene expression. We show that ACTH alters pyridine nucleotide redox state and identify amino acid residues in CtBP1 that are targeted by PKA and PAK6. Both ACTH/cAMP signaling and NADH/NAD+ ratio stimulate nuclear-cytoplasmic oscillation of both CtBP proteins. We provide evidence that PKA 1) induces metabolic changes in the adrenal cortex and 2) phosphorylates CtBP proteins, particularly CtBP1 at T144, resulting in CtBP protein partnering and ACTH-dependent CYP17 transcription.
CtBP12 was originally identified as a repressor of the adenoviral early antigen 1A oncogenic transcription activator (1) but has subsequently been shown to have 2-hydroxy acid dehydrogenase activity and a conserved lactate dehydrogenase structure (2, 3), although the endogenous substrates targeted by this activity are probably different (4). Perhaps via a preference for binding of reduced pyridine nucleotide (5, 6) or innate dehydrogenase activity (7), CtBP1 and 2 are able to regulate numerous genes with dependence on NADH concentration and thus the energy and redox state of the cell. Intriguingly, CtBP1 transcriptionally represses the SirT1 histone deacetylase when NADH is elevated (8), possibly regulating SirT1 energy-restricted longevity in some species. E-cadherin is repressed by CtBP (9–11) during hypoxia-induced tumor metastasis, where hypoxia significantly increases cellular NADH (9). Of note, there is a report that hypoxia stimulates adrenal output of corticosterone secretion and steroidogenic acute regulatory protein and peripheral benzodiazepine receptor in rat pups (12).
Although CtBP represses transcription of nuclear receptor target genes (13–19), CtBP1 has only been shown to interact with SF-1 (20). Bartholin et al. (13) showed that the functional CtBP interaction motif of a retinoid X receptor-interacting corepressor is required for acute repression of a retinoic acid-inducible reporter and retinoic acid-induced teratogenesis. CtBP1 and 2 reversibly interact with and reinforce the corepressor function of RIP140 (140-kDa receptor interacting protein) (14, 15, 17, 18), which in a ligand-dependent manner interacts with nuclear receptors (21, 22), including SF-1 (23), via one or more of several NR boxes (LXX(L/M)L motifs). CtBP1 also interacts with a novel estrogen receptor corepressor, ZNF366 (16), which, like RIP140, conserves the CtBP PXDLS interaction motif. CtBP1 and 2 have also been characterized in a large complex that includes histone H3 K9 deacetylase and methyltransferase activities of HDAC2 and G9a, respectively, and smaller, possibly heteromeric complexes of CtBP1 and 2 of unknown function (24). A related complex that relies on H3 K4 demethylase (LSD1) function and a structural factor, CoREST (corepressor for element-1-silencing transcription factor) (19, 25), represses growth hormone (GH) transcription, and complex members are up-regulated by estradiol in estrogen-receptor positive cells (19). In the same study, selective RNA interference of individual CoREST complex members and corresponding derepression of GH transcription suggested that CtBP contributes to repression of GH via the steady state CoREST complex, which is tethered to the GH promoter via a DNA-binding factor that recognizes a response element near a thyroid hormone receptor-binding site. This collection of studies indicates that CtBP coordinates the acute switch from nuclear receptor coactivator-associated chromatin structure to transcriptionally inactive chromatin. To date, we have shown that SF-1 dissociates from the CYP17 promoter during CtBP recruitment, which precedes a temporary return of the chromatin to a histone-enriched state, indicated by a more than 6-fold increase in histone H2B occupancy (20). This event sequence occurs during the course of ACTH/cAMP signaling and requires PKA.
Post-translational modification affects CtBP interactions and integrates activation of signaling pathways into repression of nuclear CtBP function (10, 18, 26). When acetylated, the CtBP-interacting motif loses the ability to bind interaction partners and to repress transcription (18). CtBP2 is acetylated on an isoform-specific nuclear localization sequence (26). CtBP1 is phosphorylated by p21-activated kinase (PAK) 1 during signaling through the epidermal growth factor receptor, stimulating nuclear export of CtBP1 and down-regulating its dehydrogenase activity (10). Notably, PAK-recognized serine-containing motifs (RXXS) are conserved in both CtBP1 and 2. In contrast to CtBP2, nuclear predominance of the closely related CtBP1 short isoform varies with cell type and binding partner expression levels (11, 27). NADH-dependent heterodimerization with CtBP2 is thought to increase the nuclear concentration of CtBP1, which lacks a nuclear localization sequence (27). For these reasons, modification of interaction surfaces of any CtBP1-binding partner has the potential to affect CtBP1 compartmentalization (11). Based on the role of post-translational modification in regulating CtBP function, we hypothesized that cAMP/PKA signaling regulates CYP17 transcription rate in part by dictating timing of CtBP-mediated chromatin remodeling during cycles of transcription (20). The aim of the present study was to identify the mechanism by which PKA modulates CtBP repressor function during ACTH/cAMP-dependent CYP17 transcription.
EXPERIMENTAL PROCEDURES
Reagents
Dibutryl 3′-5′-cyclic adenosine monophosphate (Bt2cAMP) was obtained from Sigma. α-Amanitin, H-89, and the catalytic subunit of PKA were obtained from EMD Biosciences, Inc. (La Jolla, CA). 5,6-Carboxy-2′,7′-dichloro-dihydrofluoroscein diacetate was purchased from Invitrogen. Active PAK6 and anti-SF-1 were obtained from Millipore (Billerica, MA). Anti-CtBP1 and 2 were purchased from BD Biosciences (Palo Alto, CA).
Cell Culture
H295R adrenocortical cells (28, 29) were generously donated by Dr. William E. Rainey (Medical College of Georgia, Augusta, GA) and cultured in Dulbecco’s modified Eagle’s/F-12 medium (Invitrogen) supplemented with 10% Nu-Serum I (BD Biosciences), 1% ITS Plus (BD Biosciences), antibiotics, and as indicated, sodium pyruvate (Mediatech, Herndon, VA).
Confocal Microscopy and Time Lapse Imaging
A Carl Zeiss LSM 510 (Germany) was used to image live and fixed cells. 3-μm cross-sections were imaged via excitation with 351- (UV) or 543-nm laser, and emission at 385–470 and 560 nm recorded, respectively, for endogenous autofluorescent pyridine nucleotide and fixed secondary antibody conjugated to DyLight 549 (Pierce). Regions of interest for nuclear and cytoplasmic CtBP1 and 2 were defined in the Zeiss image analysis software, and the background-subtracted absolute and relative intensities for time points were then calculated.
In Vitro Kinase Assay
Purified wild type or mutant immobilized receptor was incubated with recombinant, active ERK2, PAK1, PAK6, or the catalytic subunit of PKA and 1 μCi of [γ-32P]ATP (MP Biomedicals, Solon, OH) in an assay buffer optimal for the kinase being tested for 1 h at 30 °C. The protein-bound beads were washed twice with radioimmune precipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 2 mM sodium fluoride, 2 mM EDTA, 0.1% SDS, and protease inhibitor mixture I (EMD) with 500 μM 4-(2-aminoethyl) benzene-sulfonyl fluoride, 150 nM aprotinin, 1 μM E-64, 0.5 mM EDTA, and 1 μM leupeptin) and three times with phosphate-buffered saline and resuspended in SDS-PAGE gel loading buffer, and the proteins were resolved on 10% acrylamide gels. Dried Coomassie-stained gels were exposed to a PhosphorImager screen, and phosphorylated proteins were imaged on a Fluorescence/Phospho-Imager (Fuji Film). Assay buffer for reactions containing PAK6 contained 45 mM HEPES, pH 7.9, 10 mM MgCl2, 2 mM MnCl2, and 0.2 mM dithiothreitol, whereas PKA reaction buffer contained 20 mM Tris-Cl, pH 7.5, 100 mM NaCl, 12 mM MgCl2, and 1 mM dithiothreitol. Positive reactions were obtained for each kinase/buffer system in reactions with myelin basic protein (Sigma) or CREB PKA target fragment, CREBtide (EMD).
Coimmunoprecipitation and Western Blotting
H295R cells were cultured in 100-mm dishes, and cytoplasmic and nuclear fractions were obtained using the NE-PER kit (Pierce), or whole cell extracts were prepared in radioimmune precipitation assay buffer. For exogenous CtBP expression, 5 μg of pFH.CtBP1 or 2 (WT or mutant), and/or 5 μg of pACT.CtBP1 were transfected 60 h before harvest. For kinetics by coimmunoprecipitation (coIP), 100-μg nuclear extracts or for other experiments, an equal fraction or total sample of whole cell lysates were pre-cleared for 45 min at 4 °C and then incubated overnight with protein A/G beads (Santa Cruz Biotechnology, Santa Cruz, CA) and 4 μg of anti-CtBP1 (BD Biosciences), 3 μg of anti-VP16 or 2 μg of GCN5 (Santa Cruz), 2 μg of anti-FLAG (Sigma), or 2 μg of anti-SF-1 (Millipore). The beads were washed two times in radioimmune precipitation assay buffer with protease inhibitors (EMD) and twice in phosphate-buffered saline, then boiled in SDS-PAGE buffer, and separated in 10% gels before transfer to polyvinylidene difluoride membrane (Millipore) and Western blotting with anti-CtBP2. The blots were developed with ECF (Amersham Biosciences), imaged with a FLA-3000 (Fuji Film), and then quantified using Fuji Image Gauge v3.0 software.
Plasmids and Transfection
pRC-CMV.CtBP1, pFH.CtBP2, and pFH.CtBP1.NLS were kindly donated by Dr. G. Chinnadurai (Institute for Molecular Virology, Saint Louis University School of Medicine, St. Louis, MO). CtBP1 and 2 were sub-cloned into pET42 vector (Novagen, La Jolla, CA). GCN5 (pOZ-N.PCAF-B) was generously provided by Dr. Yoshihiro Nakatani (Dana Farber Cancer Institute, Boston, MA). pBind.GCN5, pBind.SF-1, and pAct.CtBP1 were subcloned and described in a previous study (20). Reporter assays were performed in 24-well plates, and transfections relied on per well combinations of the following plasmids, unless otherwise indicated: pGL3.CYP17 2×57 or −300 (150 ng), pCR3.1.SF-1 (30 ng), and pRC-CMV.CtBP1 (30 ng).
Mutagenesis
Phosphorylation sites and dimerization interface sites of CtBP1 and 2 were mutated using the Stratagene QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by sequencing.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as previously described (20). Briefly, H295R cells were synchronized by incubating with 2.5 μM α-amanitin for 2 h. Some cells were treated with 1 mM Bt2cAMP and/or 5 mM sodium pyruvate for time periods ranging from 15 min to 4 h. Following cross-linking with formaldehyde, treated cells and paired controls were washed twice in phosphate-buffered saline and harvested into radioimmune precipitation assay buffer containing protease inhibitor mixture I (EMD). The lysates were then sonicated to obtain optimal DNA fragment lengths of 100–1000 bp followed by centrifugation for 15 min at 4 °C. Fifty μl of supernatant was retained as input. Purified chromatin was precleared with 1 μg of rabbit or mouse IgG and immunoprecipitated overnight at 4 °C on a tube rotator using 5 μg of anti-SF-1 (Millipore), or anti-CtBP1 or 2 (BD Biosciences) and protein A/G plus (Santa Cruz). The immobilized protein-DNA complexes were subjected to a series of 5-min washes, cross-links were reversed, and DNA was purified by phenol: chloroform extraction and ethanol precipitation. Real time PCR was carried out using the iTaq SYBR Green Supermix with ROX (Bio-Rad), as previously published (20), with primers amplifying the region of the CYP17 promoter from position −104 to +43: 5′-GGC TGG GCT CCA GGA GAA TCT TTC TTC CAC-3′, and reverse 5′-CGG CAG GCA AGA TAG ACA GCA GTG GAG TAG-3′.
Biomolecular Simulation
Recreation of the CtBP1 homodimer (3) was performed manually to generate the symmetry of the homodimer as displayed in the publication by Kumar et al. (3), and the structures were merged in a Swiss PDB Viewer 3.7 (30) and energy-minimized. CtBP2 was threaded through one of the CtBP1 monomers in this structure, and the resulting CtBP2 monomer was refined using the SWISS-MODEL service (30). Following energy minimization of the resulting heterodimer, peptides comprising CtBP1 residues 133–145 and/or CtBP2 residues 139–151 were saved as an isolated structure, maintaining dimeric contacts. In silico mutagenesis was carried out in a Swiss PDB Viewer followed by 30 cycles of energy minimization in vacuo, during which the total system free energy was determined to plateau within 0.1% of the energy of the previous calculation for at least five cycles of minimization.
Statistics
One-sample t tests were performed in GraphPad Prism 5.00 (GraphPad Software, Inc., San Diego, CA). Significant difference from a compared value was defined as p < 0.05.
RESULTS
ACTH and cAMP Induce Rapid Accumulation of Reduced Pyridine Nucleotides via PKA Activation
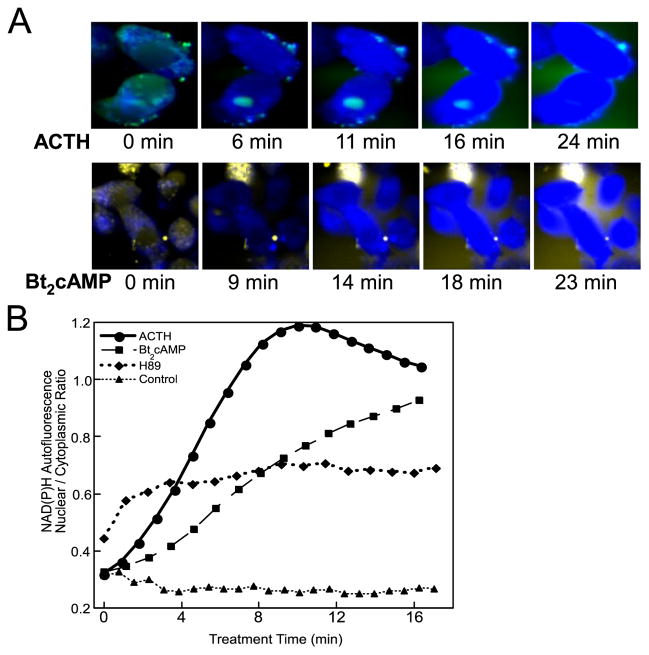
It is established that ACTH induces accumulation of reduced NADPH in the adrenal cortex (31, 32) and that NADPH is the major provider of reducing equivalents to steroidogenic (33) and other pathways relying on cytochrome P-450 enzymes (34, 35). In the adrenal cortex, PKA specifically induces metabolic flux through the pentose phosphate pathway (31, 36), reducing NADP+. Based on these previous studies, we first verified that ACTH and dibutryl cAMP (Bt2cAMP) stimulate accumulation of reduced pyridine nucleotide in cultured H295R human adrenocortical cells by measuring the accumulation of NAD(P)H autofluorescence using confocal microscopy (Fig. 1A). Increases in NAD(P)H autofluorescence typically peaked at ~16-fold of the levels in cells prior to stimulation, whereas unstimulated cells did not show an increase in cytoplasmic NAD(P)H autofluorescence for up to 60 min (data not shown). The overall ratio of oxidative:reductive reactions in treated cells as measured by dichlorofluorescein, a reactive oxygen species indicator, indicated that cells favored the loss of oxidative radicals during acute ACTH/cAMP stimulation (Fig. 1A).
FIGURE 1.
A, NAD(P)H confocal autofluorescence during ACTH/cAMP treatment of H295R cells. Reduced pyridine nucleotide autofluorescence (blue) with UV laser stimulation was recorded at times following the addition of 100 nM ACTH or 1 mM Bt2cAMP. Reactive oxygen indicator, dichlorofluorescein diacetate, was imaged separately (green or yellow). B, static nuclear or cytoplasmic regions were defined as regions of interest and the signal ratio of nuclear/cytoplasmic autofluorescence was calculated and averaged for 5–9 cells under each experimental condition. Pretreatment with H-89 (10 μM) was carried out for 20 h before ACTH stimulation and measurement.
We next asked whether ACTH/cAMP stimulates differential pyridine nucleotide reduction in distinct subcellular compartments by defining nuclear and cytoplasmic regions of each cell in the above time courses and calculating the average ratio of nuclear to cytoplasmic NAD(P)H in the time courses shown in Fig. 1B. Nuclear levels of NAD(P)H, which are initially one-third to one-half of cytoplasmic levels, increase at a faster rate than cytoplasmic levels but ultimately reach similarly elevated levels. PKA inhibition with 10 μM H-89 severely blunted increased NAD(P)H and prevented nucleocytoplasmic redistribution (Fig. 1B). We conclude that ACTH/cAMP activates NAD(P)+ reduction and redistribution in H295R cells primarily, although not exclusively, via PKA-stimulated metabolism.
Pyridine Nucleotides Activate Nuclear-Cytoplasmic CtBP1/CtBP2 Shuttling
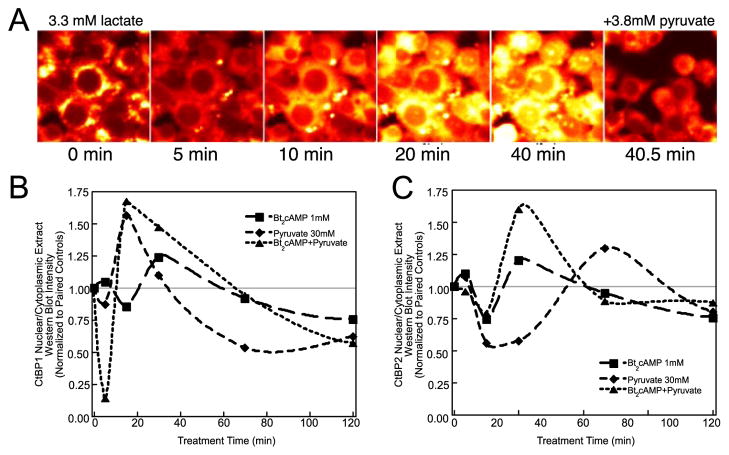
NADH, the established cofactor for the CtBP1 dehydrogenase (3, 6) involved in its homodimerization (37), could be influenced by ACTH/cAMP along with NADPH. By serving as a substrate for lactate dehydrogenase, pyruvate depletes NADH, thereby maintaining a high NAD+/NADH ratio (9, 38). To validate the use of pyruvate for the purpose of manipulating NAD(P)H, we confirmed that exogenous lactate rapidly (within 15–30 min) stimulates NAD(P)H autofluorescence in H295R cells and that application of a slightly higher concentration of pyruvate more rapidly (within 1 min) quenches this signal below initial levels (Fig. 2A).
FIGURE 2. Pyridine nucleotide metabolism affects CtBP1 and 2 localization, SF-1-mediated CYP17 DNA binding, and transcription in H295R cells.
A, confocal measurement of NAD(P)H autofluorescence during a treatment time course following addition of 3.3 mM sodium lactate and then immediately following 3.8 mM sodium pyruvate addition. Some cells were detached during addition of pyruvate. B and C, CtBP1 and 2 in nuclear and cytoplasmic extracts from stimulated H295R cells. Following the indicated treatment with 1 mM Bt2cAMP and/or 5 mM pyruvate-supplemented medium and fractionation, 25 μg of protein from nuclear or cytoplasmic extracts was analyzed by SDS-PAGE and Western blotting of CtBP1 (B) or CtBP2 (C). Western blots of 25 μg of nuclear or cytoplasmic H295R cellular extracts were analyzed with paired controls, and the ratio of nuclear to cytoplasmic CtBP1 from Western blot densitometry normalized to this ratio in paired controls was calculated and plotted as a function of time. The experiment was performed twice in triplicate. Pyruvate treatments in these panels and other figures are expressed as amount of sodium pyruvate supplementing the 2 mM standard medium concentration.
The ability of CtBP1 to interact with other cellular proteins is affected by NADH binding (6, 37). In addition, CtBP1 partner switching is known to affect its subcellular localization (11, 27). Thus, we asked whether nuclear/cytoplasmic shuttling of CtBP1 and 2 occurs in response to ACTH/cAMP-evoked NAD(P)H changes. We also tested the effect of NAD(P)H depletion by incubating cells with excess pyruvate. H295R cells were treated with Bt2cAMP or pyruvate for times ranging from 5 to 120 min and then fractionated into nuclear and cytoplasmic extracts for analysis by Western blotting. Intriguingly, both CtBP1 and 2 were found to oscillate between nucleus and cytoplasm (Fig. 2, B and C). CtBP2 oscillation in response to Bt2cAMP matched that of CtBP1 during this time period. Pyruvate also stimulates CtBP1 and 2 shuttling, indicating a role for ACTH-induced modulation of pyridine nucleotide metabolism in the oscillatory fluxes of CtBP proteins across the nuclear envelope.
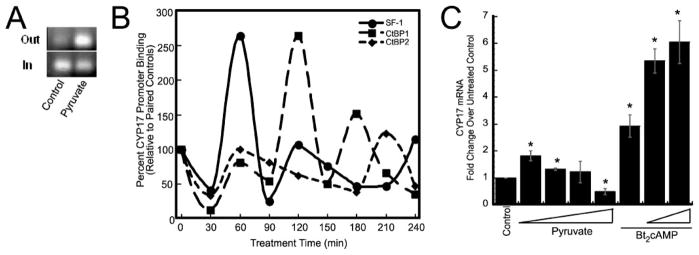
Pyruvate Modulates SF-1 and CtBP1 Binding to the CYP17 Promoter
As discussed earlier, CYP17 activation by Bt2cAMP involves both SF-1 and CtBP (20). To determine whether pyruvate can affect SF-1 binding to the CYP17 promoter, we performed ChIP of the promoter using SF-1 antibody (20) after 12 h of exposure to 30 mM pyruvate. Excess pyruvate increased SF-1 binding to the promoter (Fig. 3A), consistent with either stalled binding or enhanced transcription. We asked whether oscillation in the kinetics of the CtBP proteins and SF-1 on the CYP17 promoter (20), corresponds with nuclear-cytoplasmic oscillation of CtBP. Endogenous SF-1, CtBP1, and CtBP2 binding to this segment of the CYP17 promoter, −104/+43, was examined at half-hour intervals following synchronization with α-amanitin and then treatment with 5 mM pyruvate above standard concentration (Fig. 3B). CtBP2 oscillation on the promoter corresponds with bulk nuclear/cytoplasmic flux of this protein seen in cells treated with 30 mM pyruvate, waning with nuclear export at 30 min of pyruvate stimulation and returning by 60 min, consistent with a return of CtBP2 to the nucleus (Figs. 2C and 3B). However, CtBP1 binding to the CYP17 promoter at 120 min does not correspond with nuclear enrichment of this protein.
FIGURE 3.
A, ChIP measuring SF-1 binding to CYP17 −104/+43 following 12 h of treatment with 30 mM pyruvate. B, temporal ChIP of SF-1, CtBP1, and CtBP2 measuring SF-1 binding to CYP17 −104/+43 following treatments with 5 mM pyruvate for indicated times. Output is normalized to paired controls. C, quantitative reverse transcription-PCR of CYP17 normalized to actin and expressed as fold of controls was calculated for mRNA from H295R cells treated with medium supplemented with 0, 2, 5, 15, or 30 mM pyruvate, as indicated for 18 h. The experiment was repeated four times in quadruplicate, except for cotreatment data, from one experiment. *, significant difference from control.
Stalling of SF-1 on the CYP17 promoter is expected to repress CYP17 mRNA accumulation, whereas cycling of SF-1 and coregulators is consistent with transcription. We determined that pyruvate stimulates endogenous adrenal cortex CYP17 transcription in the absence or presence of ACTH/cAMP signaling (Fig. 3C), consistent with pyruvate promoting cyclic binding of SF-1 and CtBPs to the CYP17 promoter (Fig. 3A).
PKA Phosphorylates CtBP1 Dehydrogenase Domain Distal from the NADH-binding Site
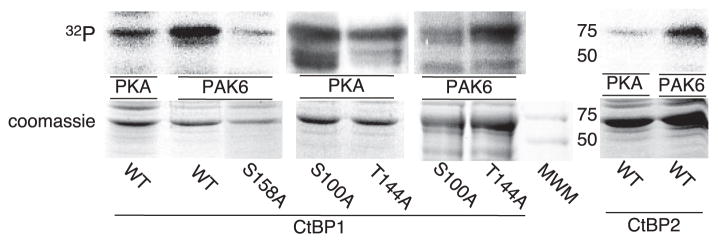
Having identified CtBP1 as key for basal repression of CYP17 transcription (20), we postulated that activation of the ACTH pathway may play a role in attenuating the repressor function of the CtBP proteins. CtBP1 is phosphorylated by PAK1 at Ser158, a modification that may decrease CtBP1 dehydrogenase activity in the homodimer (10). We predicted that post-translational modification by PKA (or PAK) could alter CtBP corepressor function. Using bacterially expressed GST-CtBP1 and 2 nuclear isoforms, we found that PKA phosphorylates both CtBP1 and CtBP2 in vitro (Fig. 4). Although PAK1 has been found to phosphorylate CtBP1 (10), we found that PAK6 phosphorylates both CtBP1 and CtBP2 (Fig. 4). Consistent with the findings of Barnes et al. (10), mutation of Ser158 to alanine substantially reduced the amount of radiolabeled phosphate incorporated into CtBP1 (Fig. 4), although the same mutation in a parallel assay did not decrease PKA phosphorylation (data not shown). Therefore, we conclude that PKA and PAK6 target unique sites in CtBP1.
FIGURE 4. PAK6 and PKA phosphorylation of CtBP1 and 2.
An in vitro kinase assay was performed with GST-CtBP1 (WT or S158A), GST-CtBP2 (WT) expressed in E. coli, bound to glutathione beads, and incubated with active PAK6 or the catalytic subunit of PKA, and [γ-32P]ATP. Coomassie staining (loading) and autoradiogram (32P labeling) of the GST-CtBP1 or 2 are shown as labeled. The same assay was also performed with S100A and T144A CtBP1 mutants bound to nickel affinity beads. A molecular weight marker (MWM) is shown on Coomassie staining panel, and the weights of standards (in kDa) are denoted.
Thr144, a consensus PKA (RXT motif) and possible minor PAK (RXXT motif) phosphorylation site, is conserved in both CtBP1 and 2. This site is on the NADH-dependent CtBP1 homodimerization interface (3), on a surface of the dehydrogenase domain opposite to the NADH-binding site and is distal to a hydrophobic cleft on the substrate-binding domain that enables binding of some CtBP partner proteins (39). Ser100 in CtBP1 is in a PAK motif that is exposed to the NADH-binding site on the substrate-binding domain and is in direct van der Waals contact with NADH in the crystal structure (3). In vitro kinase assays using a T144A mutant exhibited a decrease in the amount of radiolabeled 32P incorporated into CtBP1 when compared with WT or S100A when PKA was the kinase. Conversely, PAK6 was still able to phosphorylate the T144A mutant while exhibiting a decreased phosphorylation of the S100A mutant (Fig. 4). We conclude that CtBP1 is differentially targeted by PKA and PAK6 and that Thr144 is a PKA target, whereas Ser158 and Ser100 are PAK6 targets.
The CtBP Helical Bend Is a Phosphorylation-dependent Heterodimerization Motif
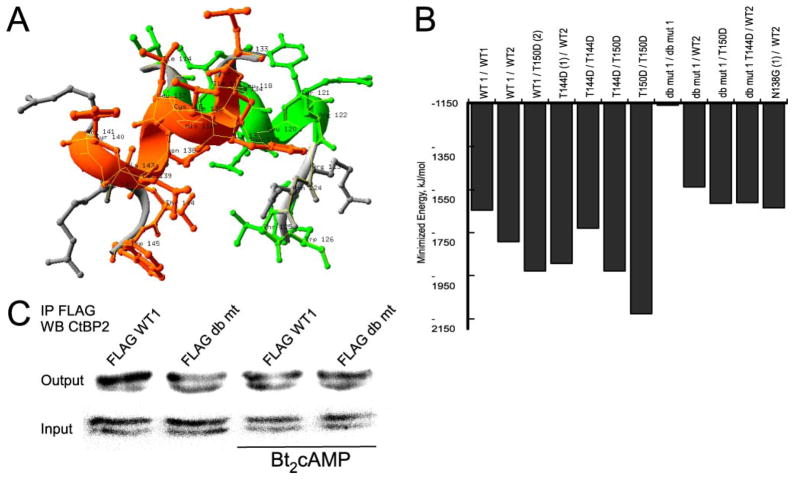
Thr144 in CtBP1 and Thr150 in CtBP2 are located at the dimerization interface, so we further examined dimerization regions of both proteins. By manually docking and energy minimizing a CtBP1 homodimer from the available monomeric structure (3), CtBP2 was modeled using one of the CtBP1 monomers as a threading template, and the regions of interest from contacting monomers were excised as shown in Fig. 5A. We then mutated select residues in silico to the lowest scoring rotamer using a Swiss PDB Viewer (30) and energy-minimized resulting structures 30 times (Fig. 5B). We found that T144D (CtBP1) and T150D (CtBP2) stabilized dimerization at the helical bend via hydrogen bonding with the backbone at Asn138. We also noted that a N138G mutant or a C134A/N138G double mutant drastically increased the predicted free energy of dimerization, consistent with a loss of dimerization potential. These calculations suggest that the helical bend of CtBP1 and 2 is a heterodimerization motif and that the specific conformation required for dimerization is favored by phosphorylation of Thr144 and disfavored by mutation of the helical bend motif, at Cys134 and Asn138. For reference, this motif is conserved as CXXXNXYRRXT and occurs at a bend in an otherwise continuous helix between the two arginines in dimeric crystal structures of CtBP1 (3) and CtBP2 (Protein Data Bank entry 2OME). To test this model experimentally, we performed coIP of WT FLAG-tagged CtBP1 and C134A/N138G (double mutant) with endogenous CtBP2 and found that the double mutant exhibits decreased binding to CtBP2, both in the presence and absence of Bt2cAMP (Fig. 5C).
FIGURE 5. The dimerization motifs of CtBP1 and 2.
A, peptides of CtBP1 and CtBP2 that form the core of the interface between the two proteins oriented in a heterodimer, created as described under “Experimental Procedures.” B, minimized energy of the two peptide system with CtBP1- or CtBP2-specific residues or mutations interfering with packing of the peptides or improving their interaction (T144D, mimicking phosphorylation). C, coIP of WT or C134A/N138G double mutant FLAG-CtBP1 with endogenous CtBP2 from H295R whole cell lysates. Cells treated with Bt2cAMP for 90 min show a 28% decrease in levels of endogenous CtBP2. IP, immunoprecipitation.
PKA-catalyzed Phosphorylation of T144 Regulates CtBP Partnering
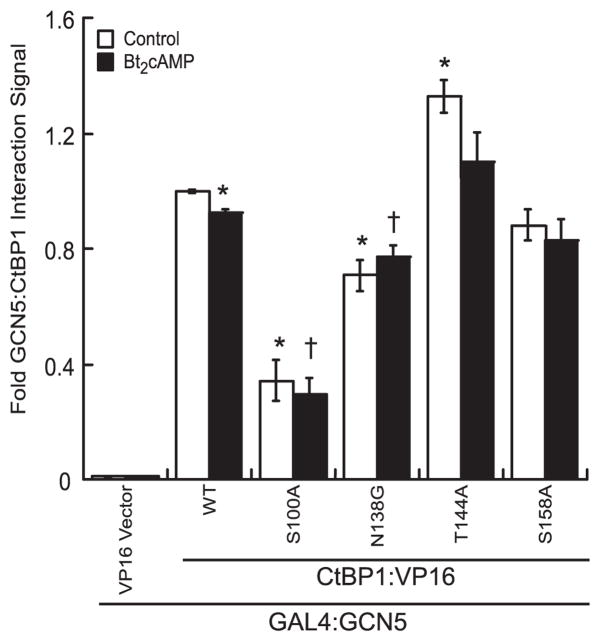
We have previously found that the coactivator GCN5 preferentially interacts with the monomeric, NADH binding-deficient CtBP1 G183V mutant (20). This G183V mutant of CtBP1 has also been shown to exhibit increased affinity for the coactivator p300 when compared with WT, supporting a role for NADH-sensitive CtBP1 sequestration of bromodomain containing acetyltransferases (37). We hypothesized that phosphorylation of CtBP1 by PKA at Thr144 disfavors GCN5 interaction with CtBP1 by inducing dimerization of CtBP1 and CtBP2 (20, 37). The hinge created in the N138G mutant could mimic the helical bend induced by phosphorylation (Fig. 5B). Additional mutants were also made to test the role of confirmed PAK6 targets Ser100 and Ser158. To determine the effect of these CtBP1 mutants on the ability of the corepressor to interact with GCN5, we carried out mammalian two-hybrid assays using GAL4:GCN5 and WT or mutant VP16:CtBP1. GAL4:GCN5 interacts strongly with VP16:CtBP1 and, significantly, loses a modest albeit significant 8% of interaction strength upon stimulation with Bt2cAMP (Fig. 6). Mutation of the PKA phosphorylation site increases interaction, whereas the N138G mutant significantly disfavored interaction with GCN5. Effects of mutation of helical bend arginines on the interaction are provided in supplemental Fig. S1A. Alanine substitution of the PAK6-targeted site S100 severely compromises GCN5 interaction, perhaps because a decrease in side chain volume encourages more stable binding of pyridine nucleotide cofactor to CtBP1, which we have established is likely to disfavor interaction with GCN5 (20). The other PAK target, Ser158, had no significant effect on GCN5 interaction in the S158A mutant.
FIGURE 6. CtBP1 dimerization motif mutations affect GCN5/CtBP interaction.
A, mammalian two-hybrid interaction of GAL4.GCN5 and VP16.CtBP1 (WT or mutants) in H295R cells in the presence or absence of Bt2cAMP is shown ± S.E. pG5 reporter, GAL4/GCN5, and VP16/CtBP1 plasmids were transfected in the ratio 50 ng:50 ng:50 ng; n = 3; n = 2. The symbols indicate significant difference from control (*) or Bt2cAMP stimulated (†) interaction with WT CtBP1.
SF-1/CtBP1 Interaction and CYP17 Induction Respond to CtBP1 and CtBP2 Dimerization Interface Mutations
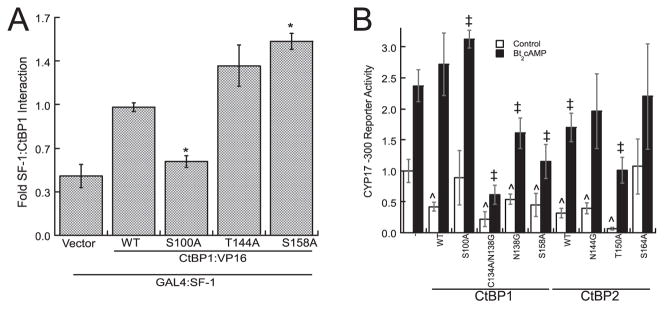
Because CtBP1 also interacts with SF-1 (20), we next determined whether phosphorylation of CtBP1 at Thr144 modulates interaction with the receptor. As shown in Fig. 7A, the S158A mutant strengthened the SF-1/CtBP1 interaction compared with WT. However T144A did not affect interaction, suggesting that the absence of Thr144 phosphorylation does not affect the interaction with SF-1. The S100A mutation weakened interaction signal to background levels, similar to the effect on GAL4/GCN5. These data suggest that phosphorylation of CtBP1 at distinct sites differentially affects the interaction between SF-1 and CtBP1 versus GCN5 and CtBP1. It is likely that arginines define the PKA phosphorylation site at Thr144. CtBP1 helical bend arginine charge neutralization had no significant effect on SF-1 interaction, whereas the R141E charge reversal did significantly increase SF-1/CtBP1 interaction (supplemental Fig. S1B).
FIGURE 7. Interaction of SF-1 with CtBP1 helical bend mutants.
A, mammalian two-hybrid SF-1/CtBP interaction with WT CtBP1 or mutants near the helical bend were measured. The plasmids were transfected as in Fig. 6, the experiments were performed in triplicate, and the results are displayed with S.E. *, differs significantly from basal SF-1/WT CtBP1 signal. B, CYP17 −300 reporter (125 ng) activity was measured in the presence of SF-1 (25 ng), and WT or mutant CtBP1 or homologous site mutants of CtBP2 (25 ng). Symbols indicate significant difference from basal (^) or Bt2cAMP-stimulated (‡)SF-1-induced reporter activity in the absence of over- expressed CtBP.
We have previously shown that CtBP1 represses CYP17 reporter gene activity in H295R cells (20). To test whether CtBP interaction mutants affect CYP17 transcription, we examined the effect of the mutants on reporter gene activation. N138G and C134A/N138G significantly inhibited Bt2cAMP-mediated activation (Fig. 7B), consistent with a requirement for CtBP1 dimerization in the loss of CtBP1-mediated repression of CYP17. S158A CtBP1 repressed CYP17 reporter more effectively than WT, whereas S100A lost the ability to repress the reporter.
CtBP2 partnering with CtBP1 is predicted to be energetically favorable to CtBP1 homodimerization (Fig. 5B). Therefore, we used the reporter gene assay to determine effects of mutating residues in the dimerization domain of CtBP2 on CYP17 transcription. CtBP2 overexpression significantly represses CYP17. Mutation of CtBP2 Thr150 (CtBP1 Thr144), which we predicted facilitates CtBP1/CtBP2 heterodimerization (Fig. 5B), strongly attenuates CYP17 transcription. Mutation of the CtBP2 residue Ser164 (CtBP1 Ser158) reverses repression. Our findings indicate that CtBP1/CtBP2 heterodimerization may be required in efficient CYP17 transcription.
The effects of CtBP1 and 2 arginine mutations of the dimerization interfaces are shown in supplemental Fig. S1C. The R142A mutation, which significantly strengthened5 GCN5/CtBP1 interaction (Fig. 6), also significantly enhances repression of CYP17 during Bt2cAMP stimulation.
Endogenous Interactions of CtBP1 and 2 with SF-1 and GCN5 Are Sensitive to Bt2cAMP and Pyruvate
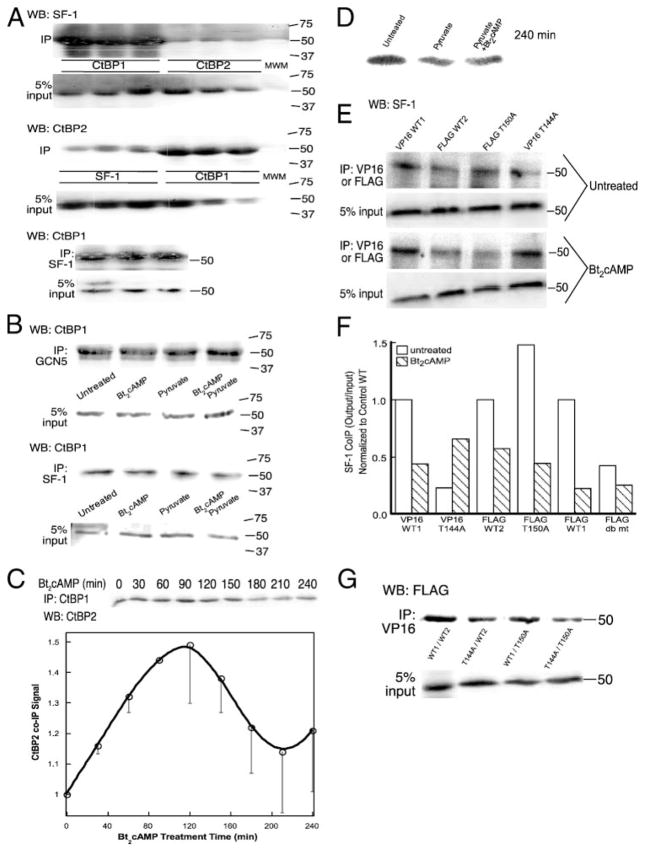
Our data suggest that ACTH/cAMP-stimulated phosphorylation promotes CtBP1 partner switching and the nuclear export of CtBP1/CtBP2 heterodimers. To further examine this phosphorylation-induced partner shuffling, we carried out coIP assays for endogenous CtBP1, 2, and SF-1 interactions in H295R cells and found that CtBP1, but not CtBP2, strongly interacts with SF-1, whereas CtBP2 prefers to interact with CtBP1 rather than SF-1 (Fig. 8A). Bt2cAMP and pyruvate decreased interaction of CtBP1 with SF-1, whereas only Bt2cAMP decreased the GCN5/CtBP1 interaction (Fig. 8B). Consistent with previous findings implicating NADH binding in CtBP1 function (20, 37), Bt2cAMP and pyruvate additively impacted the SF-1/CtBP1 interaction.
FIGURE 8. CtBP1 interaction with CtBP2 and SF-1 in H295R extracts.
A, coIP with anti-CtBP1, anti-CtBP2, or anti-SF-1 antibodies was performed on H295R whole cell lysates. Shown are output (IP) and 5% of the input in replicates of three samples isolated from untreated cells. Top panel, Western blot (WB) for SF-1 and IP for CtBP1 and CtBP2. Middle panel, WB for CtBP2 and IP for SF-1 or CtBP1. Bottom panel, WB for CtBP1 and IP for SF-1. Molecular weight marker (MWM) is shown, and the weights of standards (in kDa) are denoted. B, coIP of GCN5 or SF-1 with CtBP1 was performed on whole cell lysates following the indicated treatments for 90 min: control, 1 mM Bt2cAMP and/or 5 mM pyruvate. The molecular weights are denoted in kDa. C, excess protein A/G beads and CtBP1 antibody were incubated with 70 μg of H295R nuclear extracts taken following the indicated Bt2cAMP treatments. Western blots of washed beads were quantified via densitometry for CtBP2 signal and are expressed as coIP output. D, CtBP1 from cells treated as indicated for 4 h was immunoprecipitated from nuclear extracts, and interaction with CtBP2 was quantified by Western blotting of washed outputs. E, Western blots of GCN5 following coIP of VP16-CtBP1 or FLAG-CtBP2 from H295R whole cell lysates. The molecular weights are denoted in kDa. F, quantification of coIP results, including coIP of SF-1 with FLAG-CtBP1 not shown in E. G, Western blots of tagged CtBP2 following coIP of VP16-CtBP1 in whole cell lysates of transfected cells. The molecular weights are denoted in kDa.
Kinetics of Endogenous Nuclear CtBP Heterodimerization in Response to Bt2cAMP
To further explore the roles of ACTH/cAMP-induced pyridine nucleotide accumulation and PKA-catalyzed phosphorylation in CtBP1/CtBP2 heterodimerization, we quantified Bt2cAMP-stimulated interaction kinetics between CtBP proteins in coIP experiments using the nuclear fraction of H295R cells. As shown in Fig. 8C, CtBP1 and 2 interaction increases in nuclei within 30 min, peaks at 90–120 min, and proceeds to decay after 120 min, concomitant with flux of both proteins into the nucleus by 30 min (Fig. 2, B and C). Conditions favoring CtBP heterodimerization remain for up to 120 min, although this does not maintain positive flux of CtBP1 or 2 into the nucleus throughout this period (Fig. 2, B and C). We next determined the effect of excess pyruvate on CtBP1/CtBP2 interaction, repeating coIP experiments using lysates isolated from cells treated with 30 mM pyruvate. Pyruvate decreased the CtBP1/CtBP2 interaction by 25 and 19% in the absence and presence of Bt2cAMP, respectively (Fig. 8D).
SF-1 coIP with a tagged CtBP1 T144A mutant also revealed a 77% decrease in binding compared with WT, whereas Bt2cAMP increased interaction only with the phospho-deficient mutant by 3-fold (Fig. 8, E and F). In contrast, although basal CtBP2 interaction was much weaker, T150A mutation increased SF-1 binding 48% compared with WT CtBP2, and Bt2cAMP still decreased interaction with this mutant by more than two-thirds.
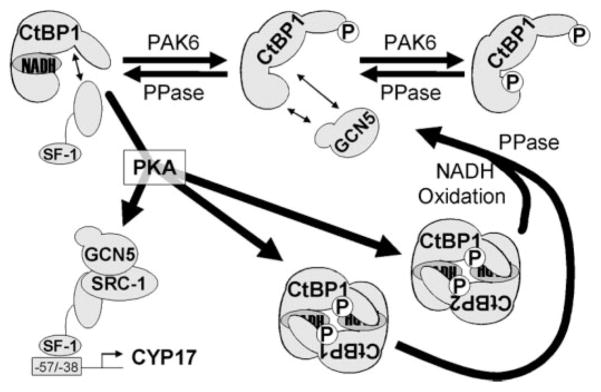
A Model of CtBP-mediated CYP17 Repression and Relief by Kinase Signaling
Bt2cAMP induces CtBP1 and 2 heterodimerization in the nucleus (Fig. 8C). We asked what the combined effect of CtBP1 Thr144 and CtBP2 Thr150 mutation is on their heterodimerization by performing coIP of tagged WT or T144 CtBP1 and found that the T144A mutation weakened CtBP1/CtBP2 heterodimerization (Fig. 8G). Further mutation of the corresponding PKA phosphorylation site in CtBP2 (Thr150) also resulted in decreased interaction, supporting our hypothesis that phosphorylation of CtBP proteins in response to ACTH signaling promotes partner switching and CYP17 induction.
Our results indicate that CtBP switches partners and subcellular localization concomitantly and that CtBP heterodimers form in response to Bt2cAMP. Therefore, we propose a model of allosteric changes in CtBP1 that control the nuclear concentration of monomer available for regulating exchange of CYP17 coactivators from their active complexes containing SF-1 and GCN5. Nuclear export of CtBP heterodimers prevails in response to ACTH/cAMP and allows induction of CYP17 transcription (Fig. 9).
FIGURE 9. Working model of CYP17 induction by PKA.
PAK6 and NADH differentially affect the ability of SF-1 and GCN5 to interact with CtBP1. PKA phosphorylates CtBP1 (and possibly 2), causing oligomerization of CtBP proteins in the nucleus. Reversal of PKA phosphorylation at Thr144 or Thr150 and/or oxidative conditions may prevail as cytochrome P-450-mediated steroidogenesis proceeds and may cause the system to revert to its initial state.
DISCUSSION
The array of pleiotropic effects of PKA induction by ACTH/cAMP in the adrenal cortex includes pyridine nucleotide accumulation and a multifaceted, complex mechanism of transcriptional induction. We sought to identify a mechanism of PKA-induced adrenal cortex transcription that corresponds with the induced formation of SF-1-coactivator complexes (20). We have shown here that reduced pyridine nucleotide can accumulate rapidly in the nucleus in response to PKA activation (Fig. 1), and pyridine nucleotide manipulation causes shuttling of CtBP1 and 2 across the nuclear envelope (Fig. 2) and dynamic CYP17 promoter binding (Fig. 3). Significantly, PKA directly phosphorylates CtBP1 and 2 (Fig. 4), and phosphorylation of the target site at Thr144 in CtBP1 has the capacity to improve the stability of the CtBP dimer (Fig. 5B). GCN5 and SF-1 interactions with CtBP1 are sensitive to Thr144 mutation, as well as mutation of other CtBP dimerization motif residues. We conclude from these results that catalyzed phosphorylation of Thr144 and the increase in nuclear pyridine nucleotides, probably NADH, stimulate CYP17 transcription promoting dissociation of CtBP from SF-1-coactivator complexes.
CtBP1 and 2 repress transcription of numerous genes via participation in master regulatory complexes of transcription corepressors (19, 24) that promote chromatin compaction via association with histone deacetylase and methyltransferase enzymes while also potentially repressing transcription in general by sequestering histone acetyltransferases, required for formation of heterochromatin, via a conserved interaction motif in bromodomains (37). The specificity of CtBP repression, however, is conferred by specific interactions with trans factors and depends on the trophic status of a cell and NAD+/NADH ratio. CtBP1 interacts with a growing list of gene-specific DNA-binding factors (8, 9, 16, 40), to which we add the nuclear receptor SF-1. The divergent effects of mutagenesis of the dimerization motif on the interaction with SF-1 (Figs. 6A and 7E) versus the interaction with GCN5 (Fig. 5), as well as of mutating the Rossman fold (20) suggest that conditions favoring SF-1 interaction are unique from the ones favoring GCN5 (Fig. 9). The exception, however, is that both this SF-1 interaction and the GCN5 interaction with CtBP proteins are reduced by Bt2cAMP treatment (Fig. 8B), in a mechanism that requires Thr144 (Figs. 6 and 8, E and F).
PAK phosphorylation site residues (CtBP1 S158 or CtBP2 S164) are at extreme ends of the CtBP heterodimer interaction interface and affect the ability of Bt2cAMP to induce CYP17 transcription (Fig. 7B and supplemental Fig. S1C). The location of both Ser100, juxtaposed to NADH in the catalytic domains of the homodimeric crystal structure (3), and Ser158, at the ends of the dimerization interface closest to the NADH binding sites (10), support a role for PAK-mediated exclusion of NADH from monomers assembling into oligomeric CtBP during the shift to activation of CYP17 transcription.
Ser158 phosphorylation of CtBP1 has been shown to inactivate dehydrogenase activity (10) consistent with a direct role of CtBP dehydrogenase activity in modulating target gene repression or activation. Thus, dehydrogenase activity of CtBP1 likely affects tighter binding to SF-1 in the absence of PAK signaling, such that the CtBP1 S158A mutation prevents Bt2cAMP derepression of CYP17, whereas, in contrast, mutation of the cognate residue in CtBP2, S164A, causes CtBP2 to lose basal repressive capacity (supplemental Fig. S1C).
Consistent with the role of PAKs in modulating CtBP dehydrogenase activity, CtBP1 Ser100 is a PAK6 target (Fig. 4), and its phosphorylation is predicted to sterically interfere with NADH binding to CtBP1. It is particularly interesting that mutation of this site decreases two-hybrid interactions with bothGCN5 and SF-1(Figs. 6 and 7A). Our data showing PAK site effects on Bt2cAMP induction of CYP17 suggest that activation of PAK isoform(s) is linked to PKA in the adrenal cortex. PAK signaling, classically downstream of serum growth factors and upstream of ERK signaling, could thus link the activation of ACTH/PKA signaling and ERK pathways during steroidogenic gene derepression. Progressive phosphorylation or dephosphorylation of PAK sites may induce multiple modes of preferential CtBP partner binding (Fig. 9). Interestingly, we have found that PAK6 is acutely phosphorylated and active in response to ACTH/cAMP signaling.3
In summary, we have shown that CtBP is a PKA target at a unique motif that is important for CtBP dimerization. Mutation of the dimerization motif as well as PAK6 and PKA phosphorylation targets, as well as modulation of the availability of reduced pyridine nucleotide, each affect the ability of CtBP proteins to repress CYP17 in human adrenal cortex cells. These conditions affect a switch in binding partners for CtBP1 and 2, in particular facilitating assembly of CtBP1-CtBP2 heteromeric complexes, which appear to be required for efficient CYP17 transcription.
Supplementary Material
Acknowledgments
We thank Dr. G. Chinnadurai (Washington University, St. Louis, MO) for the generous gift of CtBP1 and 2 plasmids, Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute, Boston, MA) for the donation of the GCN5 plasmid, Dr. Donghui Li of the Sewer Lab for technical assistance, and Srinath Jagarlapudi and Natasha Lucki for critical reading of the manuscript.
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The abbreviations used are: CtBP, carboxyl-terminal binding protein; ACTH, adrenocorticotropin; PKA, cAMP-dependent protein kinase; Bt2cAMP, dibutryl cAMP; PAK, p21-activated kinase; coIP, coimmunoprecipitation; ChIP, chromatin immunoprecipitation; SF-1, steroidogenic factor-1; GH, growth hormone; ERK, extracellular signal-regulated kinase; WT, wild type.
E. B. Dammer and M. B. Sewer, unpublished observation.
This work was supported by NIGMS, National Institutes of Health Grant GM073241.
References
- 1.Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Proc Natl Acad Sci U S A. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian P, Zhao LJ, Chinnadurai G. FEBS Lett. 2003;537:157–160. doi: 10.1016/s0014-5793(03)00119-4. [DOI] [PubMed] [Google Scholar]
- 3.Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- 4.Achouri Y, Noël G, Van Schaftingen E. Biochem Biophys Res Commun. 2007;352:903–906. doi: 10.1016/j.bbrc.2006.11.111. [DOI] [PubMed] [Google Scholar]
- 5.Fjeld C, Birdsong WT, Goodman RH. Proc Natl Acad Sci U S A. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, Piston DW, Goodman RH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 7.Thio SSC, Bonventre JV, Hsu SIH. Nucleic Acids Res. 2004;32:1836–1847. doi: 10.1093/nar/gkh344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhang Q, Wang SY, Nottke ACN, Rocheleau JV, Piston DW, Goodman RH. Proc Natl Acad Sci U S A. 2006;103:9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Li F, Kumar R. Nat Struct Biol. 2003;10:622–628. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- 11.Alpatov R, Munguba C, Caton P, Joo JH, Shi Y, Shi Y, Hunt ME, Sugrue SP. Mol Cell Biol. 2004;24:10223–10235. doi: 10.1128/MCB.24.23.10223-10235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raff H, Hong JJ, Oaks MK, Widmaier EP. Am J Physiol. 2003;284:R78–R85. doi: 10.1152/ajpregu.00501.2002. [DOI] [PubMed] [Google Scholar]
- 13.Bartholin L, Powers SE, Melhuish TA, Lasse S, Weinstein M, Wotton D. Mol Cell Biol. 2006;26:990–1001. doi: 10.1128/MCB.26.3.990-1001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castet A, Boulahtouf A, Versini G, Bonnet S, Augereau P, Vignon F, Khochbin S, Jalaguier S, Cavaillès V. Nucleic Acids Res. 2004;32:1957–1966. doi: 10.1093/nar/gkh524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christian M, Tullet JMA, Parker MG. J Biol Chem. 2004;279:15645–15651. doi: 10.1074/jbc.M313906200. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Garcia J, Periyasamy M, Thomas RS, Christian M, Leao M, Jat P, Kindle KB, Heery DM, Parker MG, Buluwela L, Kamalati T, Ali S. Nucleic Acids Res. 2006;34:6126–6136. doi: 10.1093/nar/gkl875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tazawa H, Osman W, Shoji Y, Treuter E, Gustafsson JA, Zilliacus J. Mol Cell Biol. 2003;23:4187–4198. doi: 10.1128/MCB.23.12.4187-4198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vo N, Fjeld C, Goodman RH. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 20.Dammer EB, Leon A, Sewer MB. Mol Endocrinol. 2007;21:415–438. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- 21.Wei LN, Farooqui M, Hu X. J Biol Chem. 2001;276:16107–16112. doi: 10.1074/jbc.M010185200. [DOI] [PubMed] [Google Scholar]
- 22.Cavailles V, Dauvois S, Danielian PS, Parker MG. Proc Natl Acad Sci U S A. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellgren G, Børud B, Hoang T, Yri OE, Fladeby C, Liena EA, Lund J. Mol Cell Endocrinol. 2003;203:91–103. doi: 10.1016/s0303-7207(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Sawada J-i, Sui G, Affar EB, Whetstine JR, Lan F, Ogawa H, Luke MPS, Nakatani Y, Shi Y. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. J Biol Chem. 2006;281:4183–4189. doi: 10.1074/jbc.M509051200. [DOI] [PubMed] [Google Scholar]
- 27.Verger A, Quinlan KGR, Crofts LA, Spanò S, Corda D, Kable EPW, Braet F, Crossley M. Mol Cell Biol. 2006;26:4882–4894. doi: 10.1128/MCB.02402-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staels B, Hum DW, Miller WL. Mol Endocrinol. 1993;7:423–433. doi: 10.1210/mend.7.3.8387159. [DOI] [PubMed] [Google Scholar]
- 29.Rainey WE, Bird IM, Mason JI. Mol Cell Endocrinol. 1994;99:R17–R20. doi: 10.1016/0303-7207(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 30.Guex N, Peitsch M. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 31.Frederiks WM, Kummerlin IPED, Bosch KS, Vreeling-Sindelarova H, Jonker A, Van Noorden CJF. J Histochem Cytochem. 2007;55:975–980. doi: 10.1369/jhc.7A7222.2007. [DOI] [PubMed] [Google Scholar]
- 32.Shorin IP, Shershnev VN, Iakobson GS. Biull Eksp Biol Med. 1976;81:173–175. [PubMed] [Google Scholar]
- 33.Saad M, Monte-Alegre S, Saad S. Horm Res. 1991;35:1–3. doi: 10.1159/000181866. [DOI] [PubMed] [Google Scholar]
- 34.Feo F, Ruggiu M, Lenzerini L, Garcea R, Daino L, Frassetto S, Addis V, Gaspa L, Pascale R. Int J Cancer. 1987;39:560–564. doi: 10.1002/ijc.2910390504. [DOI] [PubMed] [Google Scholar]
- 35.Pascale R, Ruggiu M, Simile M, Daino L, Vannini G, Seddaiu M, Satta G, Feo F. Res Commun Chem Pathol Pharmacol. 1990;69:361–364. [PubMed] [Google Scholar]
- 36.Costa Rosa LFBP, Curi R, Murphy C, Newsholme P. Biochem J. 1995;310:709–714. doi: 10.1042/bj3100709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, Cho EJ, Kim ST, Youn HD. Nat Struct Mol Biol. 2005;12:423–428. doi: 10.1038/nsmb924. [DOI] [PubMed] [Google Scholar]
- 38.Williamson DH, Lund P, Krebs HA. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan KGR, Verger A, Kwok A, Lee SHY, Perdomo J, Nardini M, Bolognesi M, Crossley M. Mol Cell Biol. 2006;26:8202–8213. doi: 10.1128/MCB.00445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan KGR, Verger A, Yaswen P, Crossley M. Biochim Biophys Acta. 2007;1775:333–340. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.