Abstract
Three-dimensional light microscopy and three-dimensional electron microscopy (electron tomography) separately provide very powerful tools to study cellular structure and physiology, including the structure and physiology of mitochondria. Fluorescence microscopy allows one to study processes in live cells with specific labels and stains that follow the movement of labeled proteins and changes within cellular compartments but does not have sufficient resolution to define the ultrastructure of intracellular organelles such as mitochondria. Electron microscopy and electron tomography provide the highest resolution currently available to study mitochondrial ultrastructure but cannot follow processes in living cells. We describe the combination of these two techniques in which fluorescence confocal microscopy is used to study structural and physiologic changes in mitochondria within apoptotic HeLa cells to define the apoptotic timeframe. Cells can then be selected at various stages of the apoptotic timeframe for examination at higher resolution by electron microscopy and electron tomography. This is a form of “virtual” 4-dimensional electron microscopy that has revealed interesting structural changes in the mitochondria of HeLa cells during apoptosis. The same techniques can be applied, with modification, to study other dynamic processes within cells in other experimental contexts.
1. Introduction
Light microscopy (LM) can be used to visualize live cells in both 2-D and 3-D and to study changes during various cellular processes. Furthermore, fluorescence light microscopy provides powerful tools to monitor the movement of particular cellular components labeled with fluorophores and physiologic changes by fluorescent indicators. However, even the highest resolution light microscopy modalities do not deliver resolution sufficient to study mitochondrial fine structure. Transmission electron microscopy (TEM) and 3-D electron tomography (ET) generate the highest resolution 2-D and 3-D images of mitochondria currently available, providing a bridge in the meso-resolution range between light microscopy and techniques such as X-ray and electron crystallography and multidimensional NMR spectroscopy that can yield atomic models of macromolecules (Fig. 2.1). However, TEM and ET suffer from the fact that they can only image “dead” specimens and cannot be used to follow dynamic processes in mitochondria such as those that occur during apoptosis. Combining LM with TEM and ET provides the best of both worlds if one can use LM to monitor a dynamic cellular process that can then be halted at defined states by fixation followed by preparation of the identical cells or organelles for observation of ultrastructure by TEM and ET. By sampling different stages of a process based on changes observed by LM, one can add in some form the time dimension to the three spatial dimensions provided by ET, yielding a form of virtual 4-D electron microscopy. This is the subject of the next sections using the example of release of cytochrome c and loss of membrane potential from mitochondria during apoptosis to illustrate the utility of this approach, which can be applied, with appropriate modification, to many different systems. The techniques of LM, TEM, and even ET that we describe are quite conventional individually, but when combined to examine identical cells and subcellular organelles, they provide a very powerful multimode technique to characterize the dynamics of intracellular processes. We will illustrate the power of this approach by presenting our results on the structural changes in HeLa cell mitochondria during apoptosis as examples at each stage of the process.
Figure 2.1.
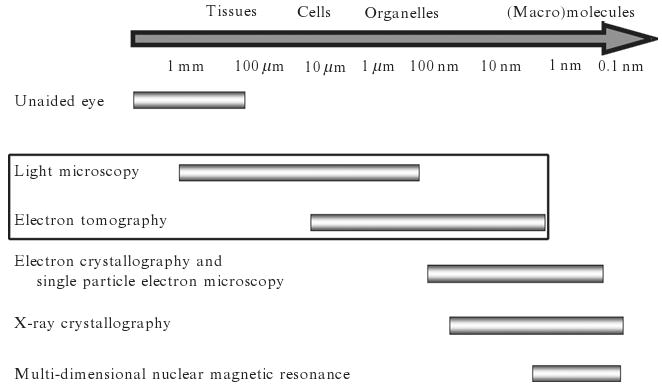
Resolution domains of common techniques used in structural biology research. Electron microscopy and tomography fills in the meso-resolution region between light microscopy and high-resolution techniques such as X-ray and electron crystallography and NMR spectroscopy.
2. Light Microscopy
2.1. Cell culture
In the following section we describe application of correlated light and electron microscopy and ET to characterize the ultrastructural changes in mitochondria during apoptosis in HeLa cells. However, the approach of this specific example can be applied to other cell culture models of other processes that can be characterized by LM, usually through the use of fluorescent labels and/or fluorescent indicators. We simultaneously monitored mitochondria within HeLa cells during apoptosis for cytochrome c release and inner mitochondrial membrane potential (ΔΨm) by use of confocal fluorescence microscopy of cells permanently transfected with fluorescent cytochrome c fusion proteins that were also labeled with TMRE, a membrane potential indicator (Goldstein et al., 2000, 2005; Sun et al., 2007). In some studies we have also used calcein-AM to load mitochondria with calcein, while quenching cytoplasmic calcein with CoCl2 to monitor the possible occurrence of a mitochondrial permeability transition (MPT) in which a large high-conductance channel opens in the mitochondrial inner membrane (Petronilli et al., 1999; Sun, 2007). By growing the cells in petri dishes that contain a glass coverslip with an etched grid (MatTek Corp. Ashland, MA), we can not only easily find and image by LM identical cells over an extended period of time, but we can also subsequently fix and embed the identical cells for examination by TEM and ET (Fig. 2.2). Using the grid to identify by confocal microscopy a field of cells at defined stages of apoptosis, we prepare cells for TEM by conventional methods of chemical fixation and then locate the identical cells during sectioning for TEM and ET.
Figure 2.2.

Correlated light and electron microscopy of HeLa cells at three stages of the apoptotic time frame after 16 h of exposure to 100 μM etoposide. Scale bars represent 25 μm in the fluorescent micrographs and low magnification TEMs and 500 nm in the high magnification TEMs. (A) Stage 1, Mitochondria retain cytochrome c as indicated by punctate FlAsH staining of Cyt. c-4CYS upper left and maintain a membrane potential indicated by punctate TMRE staining upper right. Lower left, Low magnification TEM of the same cells; lower right, high magnification TEM of normal mitochondria in one of the cells. (B) Stage 2, Mitochondria have released cytochrome c as indicated by diffuse FlAsH staining of Cyt. c-4CYS (upper left) but maintain a membrane potential indicated by punctate TMRE staining (upper right). Lower left, Low-magnification TEM of the same cells; lower right, high magnification TEM of a vesicular mitochondrion in one of the cells. (C) Stage 3, Mitochondria have released cytochrome c and have lost their membrane potential as indicated by diffuse FlAsH staining of Cyt. c-4CYS (upper left) and loss of TMRE staining (upper right). Lower left, Low-magnification TEM of the same cells; lower right, high magnification TEM of swollen/vesicular mitochondria in one of the cells.
In the example described in the following we used HeLa cells permanently transfected with a 13.3-kD cytochrome c fusion protein containing a short tetracysteine motif (Cyt. c-4CYS); however, one could monitor the movement of many other mitochondrial proteins in cells as long as they can be labeled for observation by LM. Similarly, the conditions of cell growth can be modified for other cell types and experimental protocols. The tetracysteine motif binds to the membrane permeable biarsenical fluorophores, FlAsH and ReAsH (Gaietta et al., 2002; Goldstein et al., 2005). In this example the cells were grown and maintained at 37° in Dulbecco's modified Eagles' medium (DMEM, Gibco) supplemented with 10% FBS, 2 mM l-glutamine, 200 mg/ml penicillin, and 100 mg/ml streptomycin sulfate in a humidified atmosphere of 5% CO2/95% air. Cells were sub-cultured 1:10 by incubating them in 0.25% trypsin (Gibco) while confluent and then resuspended in growth medium. Apoptosis was induced with 100 μM etoposide (Sigma) for 12 to 18 h. Cells can be treated for specific effects. For example, in some experiments zVAD-fmk (100 μM, Sigma) was added hours before the apoptotic stimulus to inhibit caspase activity or cyclosporine A was added to inhibit the MPT. FlAsH (250 nM) staining reveals the distribution of Cyt. c-4CYS, while TMRE (50 nM, Sigma) that partitions into those mitochondria that maintain a membrane potential was added to monitor the ΔΨm (Sun, 2007; Sun et al., 2007).
2.2. Confocal microscopy
Cell images were acquired by fluorescence confocal light microscopy with a Leica TCS SP2 inverted confocal microscope that uses monochromators with adjustable slits rather than filters allowing greater control in the selection of the wavelengths of light detected. This is advantageous when fluorophores in multiply labeled samples have overlapping emission spectra but is not essential, and a microscope that uses filters could also be used. FlAsH was excited with the 488 nm line from an argon laser attenuated to 35%, and TMRE was excited with a 543 nm line from a helium/neon laser attenuated to 34%. The detector slits of the confocal microscope were adjusted to detect FlAsH emission between 497 and 553 nm and TMRE emission between 555 and 620 nm. Apoptosis was initiated by the addition of etoposide (100 μM) that results in DNA fragmentation activating the p53 pathway. Western blot analysis of cytosolic and mitochondrial fractions indicated that half of cytochrome c had been released after 18 h, and previous studies have shown that cytochrome c release is asynchronous among cell populations. Once cytochrome c release begins within a HeLa cell, all mitochondria release all of their cytochrome c within a period of minutes (Goldstein et al., 2000, 2005). Thus, when we observed HeLa cell cultures 16 h after addition of etoposide, we found populations of cells at different stages of the apoptotic pathway, including many whose mitochondria had not yet released cytochrome c. Images in Fig. 2.2A were recorded 16 h after addition of etoposide to initiate apoptosis but before release of cytochrome c as shown by the distribution of Cyt. c-4CYS stained with FlAsH revealed in Fig. 2.2A (top left) that colocalized to mitochondria maintaining a membrane potential indicated by punctate TMRE staining in Fig. 2.2A (top right). In another field of cells recorded at a similar time point we observed cells whose mitochondria had released cytochrome c as shown by diffuse FlAsH staining but maintained a membrane potential indicated by punctate TMRE staining (Fig. 2.2B). Finally, the mitochondria in some cells had both released cytochrome c and had lost their membrane potential as indicated by diffuse FlAsH staining and loss of TMRE staining in Fig. 2.2C.
On the basis of the changes in fluorescence observed in HeLa cells expressing Cyt. c-4CYS labeled with FlAsH and TMRE, we identified three stages during apoptosis initiated by etoposide that define a new apoptotic time frame: Stage 1, Before release of cytochrome c and before loss of ΔΨm; Stage 2, After release of cytochrome c, but before loss of ΔΨm; and Stage 3, After both the release of cytochrome c and loss of ΔΨm.
We proceeded to study the ultrastructure of the mitochondria within the representative cells in these three stages by TEM and by ET of the identical cells that were characterized by fluorescence confocal microscopy.
2.3. Sample preparation for electron microscopy
2.3.1. Fixation
We found that the fixation, dehydration, and embedding protocol described below worked well with HeLa cells. Modifications or other protocols may be required in the case of other cell types. We generally use Durcupan as our embedding resin, because in our experience it has proven to provide the best results in ET of conventionally fixed and embedded samples; however, other resins may also be used.
After confocal imaging, the cells were fixed immediately by adding primary fixative at room temperature, followed by 1 h of incubation on ice. It is important to keep the samples cold at all times after primary fixation, until the 100% ethanol dehydration step, as described in the following. The fixative is composed of 2% paraformaldehyde and 2.5% glutaraldehyde in a 0.1 M sodium cacodylate, pH 7.4, buffer. After washing three times in an ice-cold 0.1 M sodium cacodylate buffer containing 3 μM calcium chloride for 3 min, the primary fixed cells were then incubated with 1% osmium tetroxide, 0.8% potassium ferrocyanide, and 3 μM calcium chloride in 0.1 M sodium cacodylate for 60 min on ice. After washing with distilled water three times for 3 min, the fixed cells were stained and stabilized in 2% uranyl acetate for 30 min on ice.
2.3.2. Dehydration, infiltration, and embedding
Cells were dehydrated in an ice-cold ethanol series of 20%, 50%, 70%, and 90% successively, on ice for 3 min each. The cells were then dehydrated at room temperature three times for 3 min each in 100% ethanol. Next, the cells were infiltrated in a mixture of 50% ethanol and 50% Durcupan ACM resin (Fluka) for 60 min with agitation at room temperature, followed by 100% Durcupan ACM twice for 1 h with agitation, after which the samples were placed in an oven to polymerize at 60 to 80° for at least 48 h.
2.4. Locating the identical cells
Once the samples were polymerized, the glass coverslip on the MatTek dish was peeled away from the bottom with a razor blade under the stereomicroscope. Careful insertion of the razor blade between the glass coverslip and the resin block allows air to go in and pop off the coverslip. The etched grid is reproduced on the sample block facilitating the identification of the appropriate grid location containing the same cells observed by confocal microscopy. The cell location is then circled according to the grid map with an indelible marker under a stereomicroscope, and the selected area is cut out and glued to a plain block for sectioning. Note that the surface of the block contains a single cell layer, so care must be taken when the block is trimmed and then sectioned.
2.4.1. Sectioning
The block surface was trimmed to as small an area of the sample as possible containing the cells of interest according to the grid map. Load the block into the holder of the microtome and bring the diamond knife close to the block. Because the sample thickness is only one cell layer, it is critical to make sure that the block surface is parallel to the diamond knife before sectioning. The gap between the sample surface and the knife-edge serves as a reference during adjustment, and once they are parallel to each other, the gap between the sample surface and the knife-edge will be identical in width all the way from left to right. When moving the block up and down, the gap will remain the same from the upper to lower edges of the surface. Sections were collected and post stained with 2% uranyl acetate and Sato lead (Perkins et al., 1997) before examination in an FEI Tecnai 12 TEM. Sections of different thicknesses may be collected from the same block; for example. thinner sections (ca. 80 nm) for standard TEM thin section imaging and thicker sections (200 to 500 nm) for ET.
2.5. Conventional TEM
TEM of thin sections revealed an interesting structural transformation during apoptosis in this system. Figure 2.2A–C contains low and high magnification TEM images of the identical cells imaged by confocal microscopy at each of the three stages that define the apoptotic time frame. Stage 1 cells whether untreated or treated but observed before changes detected by confocal microscopy showed only normal mitochondria (Fig. 2.2A, bottom row). On the other hand, in cells at Stage 2, which follows release of cytochrome c but precedes loss of ΔΨm, many mitochondria exhibit a markedly different ultrastructural morphology that we termed “vesicular” (Fig. 2.2B, bottom), although most mitochondria in Stage 2 cells appear normal. By Stage 3, cells have released cytochrome c, and lost ΔΨm and normal mitochondria are in the minority. Many mitochondria in Stage 3 cells exhibit the vesicular morphology and/or appear swollen as indicated by a rounded or distended appearance and lighter staining of the mitochondrial matrix (Sun et al., 2007). Characterization of these morphologic changes required the third spatial dimension provided by ET described next.
3. Electron Microscope Tomography
3.1. Introduction
3.1.1. Electron tomography
ET is based on a series of tilted images usually collected from a comparatively thick section or from isolated mitochondria (Frey et al., 2006). An ET reconstruction provides a 3-D density map that can be rotated and viewed from any angle and computationally “sliced” at voxel-thick intervals (a voxel or volume element is the 3-D analog of a pixel or picture element). The volume can be further segmented to model separately features of interest. These volumes can then be explored with sophisticated interactive 3-D rendering and visualization programs. The continual development of computational approaches for real-time interactive 3-D data processing and analyses aims at facilitating the generation of ET volumes by increasing automation of the processes. However, the volumes still need to be interpreted, and this requires substantial expert knowledge about mitochondrial structure and function. We will present a method that has worked well for our 3-D structural studies of mitochondria in conventionally fixed and embedded specimens. Other guides and resources can be found in these methods-oriented articles (Engelhardt, 2007; Frey et al., 2006; Marco et al., 2004; Marko and Hsieh, 2007; O'Toole et al., 2007). Figure 2.3 presents an outline of data collection, processing, segmentation, and visualization for ET.
Figure 2.3.
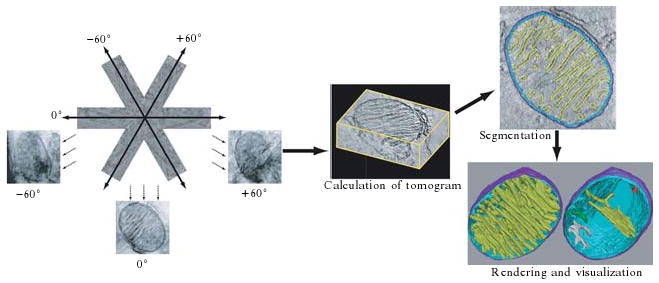
Schematic of the process required to generate a 3-D model by electron tomography. On the left collection of the tilt series from −60° to +60° at regular angular intervals is depicted with projection images of the −60°, 0°, and +60° tilts shown. In the center is a representation of the reconstructed volume containing part of a mitochondrion. Upper right shows segmentation of one slice of the tomogram by tracing the outer mitochondrial membrane in dark blue, the inner boundary membrane in turquoise, and the cristae membranes in yellow. Bottom right shows representations of two 3-D models based on segmentations of all of the sections of the tomogram. The model on the left shows the outer membrane in dark blue, the inner boundary membrane in turquoise, and all of the cristae in yellow. The model on the right is of the same mitochondrion but showing only four representative cristae in red, yellow, green, and grey from top to bottom.
3.2. Equipment and software requirements
3.2.1. Electron microscope
The minimum microscope requirement is a 120-kV TEM with a goniometer specimen holder that can tilt from 0° to ±60°. However, the ease of tilt series collection is governed by automation, and this requires computers that are now found on all modern TEMs. A eucentric goniometer that is computer controlled is advantageous because specimen movement and the focusing adjustment will be small during tilt series collection if the object is made eucentric. With a 120-kV microscope, however, one is limited in section thickness to approximately 200 nm, depending on the electron density of the sample, because thicker sections produce more inelastically and multiply-scattered electrons that add noise to the image. The noise is seen as blurring of features that can reduce resolution or even render the image unusable. Even so, research questions can still be addressed if all that is needed is a portion of the mitochondrial volume (e.g., crista junction or contact site analyses). More commonly used for ET of mitochondria are intermediate-voltage TEMs (IVEM) operating at 200 to 400 kV including energy-filtered TEMs (EFTEM) or high-voltage TEMs (HVEM) operating at 750 to 3000 kV. With IVEMs a section thickness of 500 nm is commonly used, and with the 3000 kV HVEM section thicknesses up to 5000 nm have been used. The increased accelerating voltages of these microscopes reduce the scattering cross section of the electron beam, permitting greater penetration of the thicker specimens with less inelastic and multiple scattering, thus improving image quality. The use of an energy filter also improves image quality by removing the lower energy inelastically scattered electrons that blur the image by chromatic aberration, because they have longer wavelengths than elastically scattered electrons and, therefore, have different focal planes. One must also remember that the path of the electron beam through slablike sections increases as the specimen is tilted. Decreased scattering cross section afforded at higher accelerating voltages becomes important when the specimen is tilted up to 60° or 70°, where path of the beam through the section doubles and triples, respectively.
3.2.2. Image recording
The use of a charge-coupled device (CCD) camera provides several advantages over the use of photographic film. (1) Direct digital recording eliminates the time spent for film development and subsequent digitization, thus increasing throughput. (2) Water contamination and vacuum pump-down in the TEM column are reduced because of the absence of photographic film. (3) CCD recording simplifies automated image collection. One advantage that photographic film still holds over CCD cameras is for large-area imaging needs (e.g., imaging long, clustered, or networked mitochondria). Film has a larger area than even the current high-end CCD cameras with 4k × 4k pixel detectors, such as the Gatan UltraScan™ 4000 and the Tietz TemCam F415. However, new generation CCD cameras have 8k × 8k pixel detectors, and this approaches the scan size of film that uses high-resolution film scanners (4000 dpi). Although scanning a tilt series recorded on film can take 3 to 6 h—longer than actually collecting the tilt series—an automatic scanning device was developed by Robert Glaeser (U. California, Berkeley) using a reasonably priced Nikon CoolScan (Typke et al., 2005).
3.2.2. Computer and software
Modern TEMs are equipped by the manufacturer with computers and software to automatically or semiautomatically acquire tilt series. For older TEMs, software for automatic tilt series acquisition is available from a few sources: TVIPS, www.tvips.com); SerialEM, http://bio3d.colorado.edu/; TOM, http://www.biochem.mpg.de/baumeister/; and Utrecht suite, www.bio.uu.nl/mcb/3dem/Electron_tomography.html.
With an automated system, a tilt series can be recorded within an hour or two once the microscope is properly aligned and the specimen selected.
After the tilt series has been collected, extensive image processing is required. Previously, ET image processing required a large workstation or even a supercomputer, but modern desktop computers, including clusters of such machines, are adequate for routine ET image processing. For large volumes, however, generating a reconstruction in under a day still requires parallelized code run on clusters or a supercomputer. Several complete software packages are available to process, display, and analyze ET data. Currently, the most used is the freeware IMOD software package (http://bio3d.colorado.edu/), and there is an active IMOD listserver group (imod@lists.Colorado.EDU) that diligently troubleshoots installation and software problems as they arise. Similar software packages have been developed and made available, including SPIRE (SPIDER) (www.wadsworth.org/spider_doc/); SUPRIM (http://ami.scripps.edu/software/suprim/); TOM (http://www.biochem.mpg.de/baumeister/tom_e/index.html); EM3D (http://em3d.stanford.edu/), TxBR (http://ncmir.ucsd.edu/); EMAN (http://blake.bcm.edu/EMAN/);BSOFT (http://lsbr.niams.nih.gov/bsoft/bsoft.html); IMAGIC (http://www.imagescience.de/); and XMIPP (http://xmipp.cnb.csic.es/).
Certain of these packages emphasize molecular ET, whereas others specialize in cellular ET. The packages that we have used the most for mitochondrial ET are IMOD and TxBR (Lawrence et al., 2006).
3.3. Tomographic data collection
Acquisition of high-quality images of mitochondria is the critical step to ensure a 3-D reconstruction at nanometer resolutions.
3.3.1. Considerations for tomography of mitochondria
The whole purpose of ET is to reconstruct volumes. It makes sense to aim for the thickest section that will provide the desired image quality and resolution. It is attractive to use sections thick enough (200 to 5000 nm) to contain a significant fraction of the organelle within the section volume. Threadlike mitochondria such as those found in axons can be as thin as 200 nm, but most mitochondria are thicker than this. Furthermore, even a thin mitochondrion may not be oriented parallel to and lie within a section. Yet, as pointed out previously, thicker sections produce more inelastically and multiply scattered electrons that degrade the image quality and resolution. It is also important to remember that the length of the beam path through a section increases twofold to threefold at tilt angles of 60° and 70°, respectively. Two strategies have been developed to perform ET on larger mitochondria without compromising resolution. One strategy is the use of energy-filtering TEMs and the other is to perform serial ET.
3.3.2. Energy-filtering and serial tomography
As the specimen thickness increases, the proportion of inelastically scattered electrons also increases (Bouwer et al., 2007). Energy filtering applied to ET is a way to image thicker specimens (>500 nm) by reducing or eliminating altogether the blurring from chromatic aberration that results from inelastic scattering. Energy filters operating in either “zeroloss” mode or “most-probable-loss” mode select electrons within a narrow energy range, minimizing chromatic aberration from electrons of different energy (wavelength) and yielding images with significantly higher signal-to-noise ratio (Bouwer et al., 2004, 2007). We have used the 300-kV EFTEM at the NCMIR to produce high-quality reconstructions of mitochondria in 1000-nm-thick sections.
The technique of serial sectioning can be combined with ET to derive a reconstruction of large volumes. Serial sections are cut through the sample block with a thickness determined by the desired resolution. Generally, for 120-kV TEMs the section thickness should be no greater than 200 nm, for 400-kV TEMs the section thickness should be no greater than 500 nm, and for 300-kV EFTEM no greater than 1000 nm. Tilt series are collected from the sliced mitochondrion found in each section, and ET reconstructions are computed for each tilt series, a single or double tilt series for each section. Afterward, the resulting series of volumes are aligned and combined to form a single, larger volume that contains the entire mitochondrion. The principal drawback to this approach is the loss of typically 25 to 40 nm of material at the edge of each section because of sectioning artifacts, which sometimes makes alignment, segmentation, and modeling across these gaps difficult. Extensive serial ET was recently used by Noske et al. to reconstruct two entire insulin-secreting beta cells including all of the mitochondria (Noske et al., 2007).
3.3.3. Estimation of tomographic resolution
In planning an ET experiment, one must first determine whether sufficient resolution is possible for a specimen of given thickness, tilt geometry, and angular range and increment. The desired resolution determines the magnification of the images in the tilt series and the number of images and the angular interval with which the tilt series is recorded. One must also understand that the resolution is not isotropic, because the resolution along the z-axis (the axis perpendicular to the section plane) will be lower as it is not possible to tilt the specimen to 90° with conventional TEM holders because of occlusion by the specimen holder or the specimen grid bars. The resulting “missing wedge” of information, the shape of the volume above and below the specimen into which it cannot be tilted, lowers the resolution in z by an elongation factor. The missing wedge can be reduced in volume to a “missing pyramid” by recording a double-tilt series, two tilt series around orthogonal tilt axes of the same specimen. At the NCMIR, a newly designed 360° total tilt ET specimen holder has been used to reconstruct muscle mitochondria with isotropic resolution. This holder will support cantilevered samples of either the pyramidal geometry, ideal for cells and tissue, or epoxy thread geometry, useful for isolated mitochondria or cells in suspension.
Obtaining an accurate estimate of resolution in ET reconstructions of mitochondria is difficult because each mitochondrion is unique, so the concept of statistical reproducibility used in single-particle analysis of molecules to calculate resolution is not applicable. It is reasonable to postulate that the resolution in ET should be governed by: (1) the thickness of the section, or more accurately, the thickness of the portions of heavy-metal staining and other electron-dense material that affects electron scattering, (2) the collection schema (e.g., single-tilt, double-tilt, or conical tilt), (3) the angular range and spacing of projection images in a tilt series, and (4) the signal-to-noise ratio in each image. Because the Crowther equation (Crowther et al., 1970), even modified for slab (section) geometry, often underestimates the resolution in ET reconstructions of biologic material, other constructs for estimating sufficient sampling of Fourier space are currently in use (Frank, 2006). A number of tomographers today use some form of the Fourier shell correlation (FSC) (Frangakis and Hegerl, 2001) and/or spectral signal-to-noise ratio (SSNR) (Penczek, 2002) to estimate the resolution in an ET reconstruction. Despite criticism by Penczek and Frank (in (Frank, 2006; pp 307–330), we have used the Bsoft program to estimate resolution in our reconstructions because it has been documented and is straightforward to use (Cardone et al., 2005; Heymann et al., 2008). A typical resolution range for our mitochondrial reconstructions is 5 to 10 nm.
3.3.4. Choice of pixel size and magnification
On the basis of the Shannon sampling theory, an image must be sampled at a frequency of at least twice the intended resolution. In practice, however, sampling at three or four times the desired resolution gives better image quality, especially when CCD cameras are used, in which case images are sometimes binned down by 2× by averaging 2 × 2 arrays of adjacent pixels to improve the signal-to-noise ratio before ET processing. Thus, if the desired resolution for the image were 5 nm, a pixel size of 1.25 nm or even finer would be appropriate. When a CCD camera is used, the magnification must be chosen to match the desired resolution with the spacing of the elements of the CCD array taking into account whether the data from adjacent elements will be binned in producing the final images. When film is used, the choice of magnification is less critical, because current film scanners can digitize at various resolutions up to approximately 4000 dpi, depending on the film scanner, and so there is a range of magnifications that could be adequate.
3.3.5. Size and number of fiducial gold particles
The images of the tilt series must be carefully aligned to a common origin and spatially adjusted to correct for possible distortions during data collection to produce a high-quality tomogram (see Section 4.1). For the best image alignment, we found that it is advisable to use 30 to 80 colloidal gold particles spread more-or-less uniformly across the image as fiducial markers. Gold particles should be applied on both sides of the section, because both IMOD and TxBR perform better if the particles used for alignment are not all on one side. The number of particles on each side need not be equal, but there should be at least four particles on a side for good triangulation. We apply different size gold particles to each side (e.g., 15-nm particles on one side and 20-nm particles on the other side), so that we can visually gauge whether there are enough particles on each side within the imaging area before collecting a tilt series. To correct for section bending or warping, one can use the “local” alignment option with IMOD (etomo). However, for proper correction, the number of fiducial markers needs to be greater—20 or more markers per side depending on the extent of the warping. A greater number of markers are required for the quadratic option (40 marker minimum) or the cubic option (60 marker minimum) in TxBR. These higher-order options provide a more accurate alignment and hence a higher quality reconstruction (Lawrence et al., 2006).
Problems arise if the density or size of gold particles is too low or too high. Generally, larger size gold particles are used for thicker sections because the increased electron density in the projection image can obscure smaller particles. If the density of gold particles is low, sections can be made more hydrophilic by glow-discharging or UV irradiation before applying the colloidal gold solution. Just as one can have too few gold particles, one can also have too many gold particles or clusters of particles, which would cover and interfere with the structures under investigation and create streaks that “bleed” into the tomogram. Thus, it is advisable to use the smallest size gold particles that will not be obscured by the electron-dense (stained) material, remembering that the distance between particle and nearby stained material changes and can overlap on tilting the specimen. The utility “ccderaser” in IMOD can be used to remove gold particles, especially those in clusters, from images before computing the reconstruction and thus minimize the streaking.
3.3.6. Orientation of mitochondrion
For single-tilt reconstructions, it is advantageous to orient elongated mitochondria with their long axis parallel to the tilt axis to enhance outer and inner boundary membrane visualization. For double-tilt reconstructions, the orientation of the long axis should be 45° from the tilt axis for each tilt series. Similarly, when working with mitochondria having a predominance of lamellar cristae (e.g., brown fat or muscle), the lamellae should be oriented along the tilt axis.
3.3.7. Pre-irradiating (“cooking”) the area of interest
In ET of conventionally fixed and embedded specimens, “cooking” the area of interest and the surrounding area before a tilt series is collected is practiced so that the greatest shrinkage and mass loss occurs before data collection. We use an automatic cooking sequence that irradiates the specimen at each tilt angle for a user-defined amount of time, usually 10 to 20 sec. It is important to note that for a double-tilt series, the cooking should be done for each tilt axis before starting the tilt series. Because the beam-induced mass loss and warping should be minimized between first and last images of a tilt series to maximize reconstruction quality, it is important to record a tilt series as quickly as feasible, paying attention to beam-induced specimen movement. Rapid image acquisition is facilitated by automated collection software. It is helpful to apply a thin film of carbon to both sides of the section, because the carbon film is conductive, adds stability, and reduces specimen charging. Once started, a tilt series should be completed with minimal interruptions because beam dynamics change with time and the effect is especially noticeable in images if the pause is overnight.
3.3.8. Single-tilt or double-tilt
With the double-tilt series protocol reconstructions are computed separately from separate tilt series taken about two orthogonal axes, and then the two reconstructions are aligned computationally to each other and combined to achieve a single reconstruction. This double-axis approach results in resolution that is more isotropic and is especially valuable in reconstructing membranes that are perpendicular to a tilt axis. Because of the advantages in reducing the missing data from a wedge to a pyramid with smaller volume, the double-tilt series collection schema is now the standard. With modern ET, there are only a few justifications for recording single-tilt series. They are: (1) if the specimen is fragile or cannot sustain the roughly double beam exposure required for double-tilt collection; (2) lack of time, this may be because the series is recorded on film with its inherently longer processing time, or because no automated collection is available; or (3) for the given specimen and research objectives a single-tilt series reconstruction will achieve the desired result. The last is justified when the membranes are heavily stained, which is how we commonly prepare mitochondrial samples, and the segmentation is done manually. Although fainter in specified orientations because of the missing wedge, the membranes are usually still visible and hence traceable. When automated segmentation is used, it makes sense to use double-tilt series to minimize “gaps” in the membrane opacity. Double-tilt ET, however, is not free from problems. The additional radiation used to collect the second tilt series induces further change (shrinkage and warping) in the specimen, so that the successive reconstructions are not from an identical specimen. Taking into account this change, pre- (TxBR) or post- (IMOD) reconstruction software is used to massage the two “halves” of the double-tilt data into a merged reconstruction.
3.3.9. Angular increment and tilt range
For ET of slablike sections, there is a limitation in the range of tilts that one can collect, generally ±60° to ±70°. This is due to several factors such as the specimen support grid or rod occluding the image at high tilt, the physical stops in the microscope that prevent the specimen rod from hitting pole pieces and apertures, and increased inelastic and multiple scattering, hence image degradation, resulting from the increased path of the beam at high tilt angles. Only a few TEMs have 180°-tilt ET specimen holders, and these require specimens with cylindrical or pyramidal geometry (Zhang et al., 1998). Typical angular increments used in collecting tilt series are either 1° or 2°. A double-tilt series with 1° increment between ±70° amounts to 282 images. Even with automation software, this is a lot of beam exposure for biologic samples. Usually mitochondrial membranes are well delineated for a double-tilt series with 2° increment between ±60°. It should be noted that the most commonly used reconstruction algorithm, weighted back-projection, requires a constant angular increment. Other schemas can sample Fourier space more evenly by use of a smaller increment for high tilts and a larger one for low tilts, following a formula. With these schemas, algorithms other than weighted back-projection are used to reconstruct the volume (e.g., ART or SIRT).
3.3.10. Microscope alignment, eucentricity, and focus
The TEM needs to be aligned well to acquire high-quality images. Smaller condenser apertures combined with larger spot sizes should be used to increase the spatial coherence of the electron beam while maintaining sufficient brightness. Condenser astigmatism should be corrected to minimize the ellipticity of the beam. Objective astigmatism should be corrected in the normal way and is facilitated with a CCD camera that allows calculation of the image Fourier transform whose noise spectrum is circularly symmetric when the objective lens astigmatism is corrected.
Before a tilt series is collected, the eucentric height for the object must be set in which the goniometer of the TEM is adjusted by moving the tilt axis up or down until it coincides with the plane of the object to allow eucentric tilting. This should be done at the magnification to be used for recording the tilt series. The most accurate way to set the eucentric height is to rotate the grid from ±60° and adjust the z-height so that the object at −60° is translated from the object at +60° only along the tilt axis. Computer control of the goniometer provides the greatest accuracy.
In acquiring a tilt series, an image is recorded, the goniometer is tilted by a defined increment, the object is recentered and refocused, and then another image is recorded. These steps are repeated until the full tilt range is covered. Recentering and refocusing are minimal when the eucentricity is accurate. It is important to recenter before refocusing, because recentering introduces slight beam-induced movement, and this movement along with residual stage movement usually dies down in the few seconds it takes to refocus. In this way, the next image is ready to be recorded immediately after focusing has been completed. All these steps can be done manually; however, the more automated these steps are the less operator time is required with less chance of error. With modern TEM's and CCD cameras, all of these steps can be accomplished automatically once the microscope is aligned.
3.4. Tomographic image processing
Image processing routines are constantly evolving, not only to add functionality but also to make procedures more user-transparent. This renders ET accessible to the nonexpert and as a result has led to the proliferation of publications reporting the use of ET, including those from scientists whose technical expertise is in areas other than electron microscopy and tomography.
3.4.1. Tilt series alignment
The alignment of the individual images of a tilt series is a critical step in obtaining high-quality reconstructions. The two most common alignment techniques are (1) cross-correlation alignment and (2) fiducial mark tracking, usually with colloidal gold particles applied to the section surfaces. The advantages of cross-correlation alignment are that it does not require application of gold particles and it is a fast procedure requiring little input from the user. Our experience, however, has shown that cross-correlation alignment is not capable of correcting image rotation or magnification changes between images of the tilt series to the same precision that fiducial tracking achieves. However, Engelhardt (Engelhardt, 2007) reports that second-generation “trifocal alignment” and third-generation “alignment without correspondence” methods, which are fully automatic alignment algorithms, perform well and are ready to replace fiducial tracking. To our knowledge, these have yet to be applied to mitochondrial ET.
The advantage of fiducial mark tracking is that in addition to alignment, the fiducial marks provide sufficient information to de-warp (correct image and specimen distortions) and to make higher order adjustments (e.g., quadratic or cubic alignments) in TxBR. Their success is based on tracking a large number of fiducial marks through the tilt series. Semiautomated fiducial placement has allowed the use of a relatively large number of markers (e.g., 40 markers tracked on 120 images amounts to the accurate placement of 4800 fiducials). We use IMOD to place 30 to 80 markers on the 0° untilted image. An automatic particle-tracking program is then used to track each marker through all of the images of the tilt series. The resulting dataset is then edited manually to correct any errors in the placement of the fiducials or to fill gaps in the dataset that occur when the tracking “loses” the particle, which can happen for high-tilts if the particle is obscured by an electron-dense region or if the area is warped. The positions of the fiducial markers are then used to refine tilt angles, image shifts and rotations, magnification changes, and beam and sample distortions. The resultant transforms are then applied to the tilt series stack of images to produce an aligned stack.
3.4.2. Tomographic reconstruction
The next step is to compute the ET reconstruction by use of the image densities in the aligned tilt series. This step is the most computationally intensive in the processing stream and can produce reconstruction files that are very large (>1 gigabyte). Even so, because of an increase in computer power, reconstructions are usually computed within a few hours. R*-weighted backprojection is the most popular method owing to its computational simplicity and predictable outcome. Both IMOD and TxBR use this method as default. Most alternative methods fall under “iterative techniques” (Frank, 2006) and maximum entropy methods (Engelhardt, 2007). The two most commonly used iterative techniques are the algebraic reconstruction technique (ART) and the simultaneous iterative reconstruction technique (SIRT), both of which rely on optimizing the reconstruction by iterative comparison between projections of the reconstruction and images of the aligned tilt series. Fourier and more general transform methods offer advantages of increased speed of reconstruction and corrections for multiple distortions arising from TEM imaging (Lawrence et al., 2006; Sandberg et al., 2003).
3.5. Visualization and measurements of volumes
Visualization from multiple vantage points is essential to resolve and interpret details in the 3-D configuration of mitochondria (Fig. 2.3). The first step in visualization is simply examining slices in arbitrary directions and making animations of slice fly-throughs. We use IMOD or Analyze (http://www.mayo.edu/bir/Software/Analyze/Analyze.html) for this. Another commonly used commercial visualization package is Amira (http://www.tgs.com), which is suitable for several steps in visualization, including the making of sophisticated movies. An advantage of ET is the capability to computationally section the reconstruction in slices as thin as 1 voxel in any orientation. This permits accurate measurements and analyses with the multipurpose freeware “ImageJ” (http://rsb.info.nih.gov/nih-image) or “homegrown” analysis software provided by several ET laboratories (e.g., Synuarea and Synuvolume [NCMIR]) that make surface area and volume measurements from segmented reconstructions.
3.5.1. Segmentation
The next step is usually manual segmentation of mitochondrial substructure (typically membranes) by use of traced contours, followed by surface rendering of the meshed stacked contours (Fig. 2.3). Once the contours of all the objects of interest are traced, the resulting model can then be viewed in several ways. The full model may be displayed to show all objects or individual objects can be turned on or off or rendered translucent to explore the shapes of features and the spatial distribution of a class of objects (e.g., crista junctions) and their relation to other objects in the volume. This functionality has been useful when examining the elaborate membrane systems of mitochondria, which can change significantly over short distances. For example, the models in Fig. 2.3 at the lower right are of the same mitochondrion; the model on the left shows the outer membrane in blue, the inner boundary membrane in turquoise, and all of the cristae in yellow, whereas the model on the right displays only four representative cristae in different colors. Comprehension of cristae shape and crista junction and contact site size and distribution have been enhanced by assigning color and transparency level to the different structural components. We use either IMOD or Xvoxtrace (NCMIR) for segmentation, and this step is rate-limiting in ET throughput, because usually more than 100 slices are present in the reconstruction and often more than 30 objects are segmented. However, because oversampling exceeds the resolution present in the reconstruction, typically not every slice needs to be traced. For features that do not change much from slice-to-slice, tracing every other or even every fourth slice often suffices. Because of the segmentation bottleneck, researchers are developing autosegmentation tools (Bartesaghi et al., 2005; Frank, 2006; Garduno et al., 2008; Jiang et al., 2004; Narasimha et al., 2008; Nguyen and Ji, 2008). However, to date, manual segmentation has proven superior to the currently available autosegmentation tools, except perhaps for the topologically simple mitochondrial outer membrane.
3.6. Movies, tele tomography, and databases
3.6.1. Movies
Movies illustrate the rich quantity of ET information with more scope and detail than can be presented conveniently in images and are portable across different software and hardware environments (see http://www.sci.sdsu.edu/TFrey/MitoMovie.htm). Effective movies can be composites of separate movie clips. For example, the first clip may be a fly-through of the slices of the reconstruction. The fly-through can be done in different (often perpendicular) directions to highlight changes in features on a length scale. The second clip could be the surface-rendered view of the fully segmented reconstruction. The segmented volume is ideal for rotations and zoom-ins to focus the attention on the features of interest. Subsequent clips could involve turning on or off objects or making some partially transparent while rotating and zooming; this is a powerful way to emphasize shapes and spatial relationships. The rotation or zoom-in could be paused at any stage to display annotation (e.g., text and arrows) to demonstrate features.
3.6.2. Tele tomography
Because the higher-voltage TEMs most commonly used for ET are expensive, relatively few are available. To make ET more accessible, tele tomography (https://telescience.ucsd.edu/) was developed for the remote operation of TEMs and for online image processing. All that is required is a computer with Internet access. In practice, one sends the sample to the facility supporting tele tomography. At a preappointed time, someone at that facility places the sample in the TEM and informs the sender that ET data collection can now be performed. The sender not only collects a tilt series but also routes the images to computers at the facility for remote image processing or alternately those at the sender's institution. A 400-kV TEM and a 300-kV EFTEM at the NCMIR, a 3-MV TEM in Osaka, Japan, and a 1.2-MV TEM in Seoul, Korea, are tele tomography capable.
3.6.3. Databases
Because of the explosion of the amount of ET data that are now being generated, dedicated databases have been established. These databases not only aid the organization of these data but also provide the capability for data mining. A database containing mitochondrial ET data deposited is the cell-centered database (CCDB) (http://ccdb.ucsd.edu/CCDBWebSite/index.html). Through the CCDB, researchers can access mitochondrial volume reconstructions, the original tilt series, the aligned tilt series, the segmented objects, and measurements. Data mining is made available by means of keyword searches and enhanced by linkages to other archival databases.
3.7. New developments for electron tomography of mitochondria in situ
Image enhancement being developed for mitochondrial ET involves anisotropic nonlinear diffusion and bilateral filtering. These computational techniques suppress the noise in reconstructions while preserving high-resolution structural features. We anticipate that these techniques will not only enhance the visualization of features but also aid autosegmentation of mitochondria, because the tracking of membranes will be less easily derailed by the noise. Because of the effort involved with generating ET data, sampling size is often low. The recent combination of stereology and electron tomography (Vanhecke et al., 2007) has opened the door for unbiased and precise quantitative analysis by use of the wealth of information in a few reconstructions. The parameters most easily measured with so-called quantitative ET are volumes, surfaces, lengths, and numbers.
3.8. Application to apoptotic HeLa cells
Our earlier work along with that of others using ET established the crista junction paradigm of mitochondrial structure in which the inner membrane has a more complicated conformation than that depicted in earlier models and popularized in textbooks. The crista junction paradigm describes the conformation of the inner membrane with two principal components, the inner boundary membrane closely apposed to the outer membrane and the crista membrane projecting into the matrix compartment of the mitochondrion and joined to the inner boundary membrane by tubular crista junctions of variable length as shown in Fig. 2.3 (lower right) (Perkins et al., 1997). This is believed to divide the inner membrane into two functional components with electron transport and energy transduction concentrated in the crista membrane and communication between the matrix and the cytosol through the inner boundary membrane at contact sites with the outer membrane (Frey and Mannella, 2000; Frey and Sun, 2008; Frey et al., 2002). Likewise, the space between the inner and outer mitochondrial membranes is now thought to comprise two separate compartments, the intermembrane space between the outer membrane and the inner boundary membrane and the intracristal spaces within the cristae, that only communicate through crista junctions (Scorrano et al., 2002; Yamaguchi et al., 2008; Zick et al., 2008).
The application of ET to the HeLa cells in Stages 1 to 3 of the apoptotic program determined by correlated LM proved invaluable in understanding the structural changes that had occurred and led to a topographical model for fission/fusion of the mitochondrial inner membrane. We were able to identify three mitochondrial ultrastructure classes: Normal mitochondria have lamellar cristae connected to the inner boundary membrane by means of crista junctions; vesicular mitochondria in which the inner membrane has fragmented into many separate vesicular matrix compartments, and swollen mitochondria containing swollen matrix compartments. Furthermore, we also observed hybrid structures that combined features of two of these classes: Normal/vesicular and vesicular/swollen. Three-dimensional models of fully segmented tomograms representing these five classes are shown in Fig. 2.4. These could be correlated to the stages of the apoptotic timeframe suggesting that the sequence of changes progressed from exclusively normal mitochondria in Stage 1 cells to a significant number of normal/vesicular and vesicular mitochondria in Stage 2 cells during and after release of cytochrome c to many vesicular/swollen and swollen mitochondria in Stage 3 cells. Significantly, a bare majority of mitochondria in Stage 2 cells maintained normal morphology, and even Stage 3 cells contained a significant number of mitochondria that appeared normal. This suggests that this structural transformation is not required for release of cytochrome c from the intracristal spaces. Indeed, this was confirmed when inhibition of caspases by z-VAD-fmk effectively prevented the structural transformation but did not prevent release of cytochrome c. We speculate that the transformation to vesicular mitochondria represents the changes in inner mitochondrial membrane topology required for mitochondrial fission that in this case occurs throughout the mitochondrial volume rather than just at the sites of fission (Frey et al., 2006; Sun et al., 2007). This provides an example of the power of combining fluorescence confocal LM with TEM and ET of identical cells to characterize the changes in ultrastructure that accompany a dynamic cellular process.
Figure 2.4.
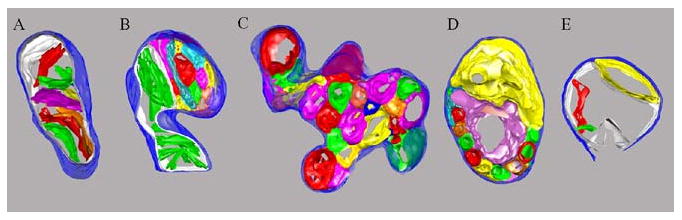
Three-dimensional models created from electron tomograms of mitochondria representing each of the structure classes observed in HeLa cells undergoing apoptosis induced by treatment with etoposide. The outer membrane is represented in dark blue and the inner boundary membrane, where present, in white. (A) Normal mitochondria contain only lamellar cristae represented in various colors connected to the inner boundary membrane by crista junctions. (B) Normal/vesicular mitochondria exhibit normal lamellar cristae connected to inner boundary membrane by means of crista junctions in one region (green cristae in the lower region) and separate vesicular matrix compartments (various colors in the upper region). (C) Vesicular mitochondria have only separate vesicular matrix compartments represented here in various colors in the absence of an inner boundary membrane. (D) Vesicular/swollen mitochondria have separate vesicular matrix compartments, and the large magenta and yellow compartments appear to be swollen. (E) Swollen mitochondria have normal and/or vesicular components that are swollen often to the point that the outer membrane is ruptured as seen at the bottom of this mitochondrion.
Acknowledgments
We acknowledge the help and support of our collaborators, Doug Green and Cristina Munoz-Pinedo at St. Jude Children's Research Hospital for providing HeLa cells transfected with cytochrome c fusion proteins and Mark Ellisman at the NCMIR/UCSD. We are grateful to Guido Gaietta (UCSD) for assistance with FlAsH staining and to Roger Y. Tsien (UCSD) for providing the FlAsH reagent. This work was supported by a Blasker Science and Technology Grant from the San Diego Foundation to TGF, NIH National Center for Research Resources Grant No. P41 RR004050 to Mark Ellisman, NIH Roadmap Grant GM72033 to Roger Y. Tsien and Mark Ellisman, and NIH grants ES010337 and DK54441.
References
- Bartesaghi A, Sapiro G, Subramaniam S. An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms. IEEE Trans Image Process. 2005;14:1314–1323. doi: 10.1109/TIP.2005.852467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwer JC, Mackey MR, Lawrence A, Deerinck TJ, Jones YZ, Terada M, Martone ME, Peltier S, Ellisman MH. Automated most-probable loss tomography of thick selectively stained biological specimens with quantitative measurement of resolution improvement. J Struct Biol. 2004;148:297–306. doi: 10.1016/j.jsb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Bouwer JC, Mackey MR, Lawrence A, Deerinck TJ, Jones YZ, Terada M, Martone ME, Peltier ST, Ellisman MH. The application of energy-filtered electron microscopy to tomography of thick, selectively stained biological samples. Methods Cell Biol. 2007;79:643–660. doi: 10.1016/S0091-679X(06)79026-4. [DOI] [PubMed] [Google Scholar]
- Cardone G, Grunewald K, Steven AC. A resolution criterion for electron tomography based on cross-validation. J Struct Biol. 2005;151:117–129. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Amos LA, Finch JT, De Rosier DJ, Klug A. Three dimensional reconstructions of spherical viruses by fourier synthesis from electron micrographs. Nature. 1970;226:421–425. doi: 10.1038/226421a0. [DOI] [PubMed] [Google Scholar]
- Engelhardt P. Three-dimensional reconstruction of chromosomes using electron tomography. Methods Mol Biol. 2007;b:365–385. doi: 10.1007/978-1-59745-294-6_18. [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Frank J. Electron Tomography. Springer; New York: 2006. [Google Scholar]
- Frey T, Renken C, Perkins G. Insight into mitochondrial structure and function from electron tomography. Biochim Biophys Acta. 2002;1555:196–203. doi: 10.1016/s0005-2728(02)00278-5. [DOI] [PubMed] [Google Scholar]
- Frey TG, Mannella CA. The internal structure of mitochondria. Trends Biochem Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- Frey TG, Perkins GA, Ellisman MH. Electron tomography of membrane-bound cellular organelles. Annu Rev Biophys Biomol Struct. 2006;35:199–224. doi: 10.1146/annurev.biophys.35.040405.102039. [DOI] [PubMed] [Google Scholar]
- Frey TG, Sun MG. Correlated light and electron microscopy illuminates the role of mitochondrial inner membrane remodeling during apoptosis. Biochim Biophys Acta. 2008;1777:847–852. doi: 10.1016/j.bbabio.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Garduno E, Wong-Barnum M, Volkmann N, Ellisman MH. Segmentation of electron tomographic data sets using fuzzy set theory principles. J Struct Biol. 2008;162:368–379. doi: 10.1016/j.jsb.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JC, Munoz-Pinedo C, Ricci JE, Adams SR, Kelekar A, Schuler M, Tsien RY, Green DR. Cytochrome c is released in a single step during apoptosis. Cell Death Differ. 2005;12:453–462. doi: 10.1038/sj.cdd.4401596. [DOI] [PubMed] [Google Scholar]
- Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol. 2008;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Ji Q, McEwen B. Model-based automated segmentation of kinetochore microtubule from electron tomography. Conf Proc IEEE Eng Med Biol Soc. 2004;3:1656–1659. doi: 10.1109/IEMBS.2004.1403500. [DOI] [PubMed] [Google Scholar]
- Lawrence A, Bouwer JC, Perkins G, Ellisman MH. Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J Struct Biol. 2006;154:144–167. doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Marco S, Boudier T, Messaoudi C, Rigaud JL. Electron tomography of biological samples. Biochemistry (Mosc) 2004;69:1219–1225. doi: 10.1007/s10541-005-0067-6. [DOI] [PubMed] [Google Scholar]
- Marko M, Hsieh CE. Three-dimensional cryotransmission electron microscopy of cells and organelles. Methods Mol Biol. 2007;369:407–429. doi: 10.1007/978-1-59745-294-6_20. [DOI] [PubMed] [Google Scholar]
- Narasimha R, Aganj I, Bennett AE, Borgnia MJ, Zabransky D, Sapiro G, McLaughlin SW, Milne JL, Subramaniam S. Evaluation of denoising algorithms for biological electron tomography. J Struct Biol. 2008;164:7–17. doi: 10.1016/j.jsb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Ji Q. Shape-driven three-dimensional watersnake segmentation of biological membranes in electron tomography. IEEE Trans Med Imaging. 2008;27:616–628. doi: 10.1109/TMI.2007.912390. [DOI] [PubMed] [Google Scholar]
- Noske AB, Costin AJ, Morgan GP, Marsh BJ. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J Struct Biol. 2007;161:298–313. doi: 10.1016/j.jsb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Giddings TH, Jr, Dutcher SK. Understanding microtubule organizing centers by comparing mutant and wild-type structures with electron tomography. Methods Cell Biol. 2007;79:125–143. doi: 10.1016/S0091-679X(06)79005-7. [DOI] [PubMed] [Google Scholar]
- Penczek PA. Three-dimensional spectral signal-to-noise ratio for a class of reconstruction algorithms. J Struct Biol. 2002;138:34–46. doi: 10.1016/s1047-8477(02)00033-3. [DOI] [PubMed] [Google Scholar]
- Perkins G, Renken C, Martone ME, Young SJ, Ellisman M, Frey TG. Electron tomography of neuronal mitochondria: Three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- Petronilli V, Miotto G, Canton M, Brini M, Colonna R, Bernardi P, Di Lisa F. Transient and long-lasting openings of the mitochondrial permeability transition pore can be monitored directly in intact cells by changes in mitochondrial calcein fluorescence. Biophys J. 1999;76:725–734. doi: 10.1016/S0006-3495(99)77239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg K, Mastronarde DN, Beylkin G. A fast reconstruction algorithm for electron microscope tomography. J Struct Biol. 2003;144:61–72. doi: 10.1016/j.jsb.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- Sun MG. Biology. Ph.D. San Diego State University; San Diego: 2007. Mitochondrial Structure During Apoptosis; p. 140. [Google Scholar]
- Sun MG, Williams J, Munoz-Pinedo C, Perkins GA, Brown JM, Ellisman MH, Green DR, Frey TG. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat Cell Biol. 2007;9:1057–1072. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- Typke D, Nordmeyer RA, Jones A, Lee J, Avila-Sakar A, Downing KH, Glaeser RM. High-throughput film-densitometry: An efficient approach to generate large data sets. J Struct Biol. 2005;149:17–29. doi: 10.1016/j.jsb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Vanhecke D, Studer D, Ochs M. Stereology meets electron tomography: Towards quantitative 3D electron microscopy. J Struct Biol. 2007;159:443–450. doi: 10.1016/j.jsb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Lartigue L, Perkins G, Scott RT, Dixit A, Kushnareva Y, Kuwana T, Ellisman MH, Newmeyer DD. Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol Cell. 2008;31:557–569. doi: 10.1016/j.molcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Takaoka A, Miyauchi K. A 360°-tilt specimen holder for electron tomography in an ultrahigh-voltage electron microscope. Rev Sci Instrum. 1998;69:4008–4009. [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]


