SUMMARY
The development of effective malaria vaccines may be hindered by extensive genetic diversity in the surface proteins being employed as vaccine antigens. Understanding of the extent and dynamics of genetic diversity in vaccine antigens is needed to guide rational vaccine design and to interpret the results of vaccine efficacy trials conducted in malaria endemic areas. Molecular epidemiological, population genetic, and structural approaches are being employed to try to identify immunologically relevant polymorphism in vaccine antigens. The results of these studies will inform choices of which alleles to include in multivalent or chimeric vaccines; however, additional molecular and immuno-epidemiological studies in a variety of geographic locations will be necessary for these approaches to succeed. Alternative means of overcoming antigenic diversity are also being explored, including boosting responses to critical conserved regions of current vaccine antigens, identifying new, more conserved and less immunodominant antigens, and developing whole-organism vaccines. Continued creative application and integration of tools from multiple disciplines, including epidemiology, immunology, molecular biology, and evolutionary genetics and genomics, will likely be required to develop broadly protective vaccines against Plasmodium and other antigenically complex pathogens.
Keywords: Plasmodium falciparum, Malaria vaccines, Genetic diversity, Vaccine escape, Molecular epidemiology
INTRODUCTION
The complicated biology of Plasmodium falciparum has presented major obstacles to the development of effective malaria vaccines. As it passes through the stages of its life cycle, the malaria parasite expresses different, stage-specific antigens, each stimulating a specific immune response. Adding further complexity, P. falciparum has a long evolutionary history with its human host and exhibits extensive genetic diversity, particularly in the surface antigens that have been under prolonged selective pressure by the human immune response and that have been the main targets of subunit vaccines (1). Moreover, the parasite continues to evolve through mutation and sexual recombination in response to drugs and other malaria interventions, providing a moving target for these interventions. When malaria vaccines are deployed, “vaccine resistant malaria” can be expected to emerge and threaten vaccine efficacy just as drug resistance has compromised the efficacy of the drugs used to prevent and treat malaria.
Genetic variability in protective antigens has posed a challenge for the development of vaccines against other pathogens including bacteria (e.g. Streptococcus pneumoniae) and rapidly evolving viruses (e.g. HIV, Hepatitis C virus, and influenza virus). Because of the large amount of genetic variation in these pathogens, vaccines directed against them can only feasibly target a subset of the genetic variants circulating in a population, which, depending on the level of vaccine coverage, could lead to an increased frequency of variants not targeted by the vaccine. This phenomenon has been observed with pneumococcal vaccines; higher rates of carriage and invasive disease due to existing non-vaccine serotypes followed the introduction of the seven-valent conjugate pneumococcal vaccine (PCV7) (2,3), and new recombinant forms of the bacterium capable of vaccine escape are emerging (4). Malaria vaccine deployment is likely to result in similar selection of vaccine-resistant variants.
To date, only a few malaria vaccine trials have been large enough to be able to detect vaccine-induced selection. Results of a Phase 2 trial of a multi-antigen blood stage vaccine in Papua New Guinea suggested possible selection for clinical infections with non-vaccine type parasites in vaccinated individuals (5). In an efficacy trial of a pre-erythrocytic vaccine (RTS,S/AS02A) in Mozambique, there was no evidence of selection for divergent genotypes of the T-cell epitope regions of the circumsporozoite gene in symptomatic or asymptomatic malaria infections, but multiplicity of infection was reduced in asymptomatic infections in vaccinated individuals compared to control individuals (6). Measures of allele-specific efficacy from trials of MSP-1 and AMA-1 vaccines that had no overall efficacy will be available soon and should indicate whether vaccine escape contributed to these failures (7,8). Factors that might hinder the ability to detect vaccine-induced selection in vaccine trials include low initial frequency of the vaccine target allele(s) requiring large sample sizes to detect a reduction in frequency, and the difficulty of detecting selective effects against backgrounds of both extensive natural diversity in the vaccine antigen and high levels of naturally acquired immunity in the study population. Vaccine-induced selection may be more readily apparent in trials conducted in relatively non-immune individuals such as young infants or in populations with lower rates of malaria transmission and immunity.
To develop strategies to overcome the extensive genetic diversity in P. falciparum and design broadly protective vaccines, it is helpful first to understand the distribution and natural dynamics of vaccine antigen polymorphisms in endemic populations where diversity is driven by naturally acquired immunity. In clinical trials of malaria vaccines, allele-specific efficacy should be measured as a key study endpoint, and this information used to inform subsequent vaccine design and testing. In this review, we discuss the extent of diversity present in some of the leading P. falciparum vaccine candidate antigens, approaches to identify the diversity most relevant to vaccine escape and cross-protection, and the importance of conducting molecular epidemiological studies prior to development and testing of vaccines.
GENETIC DIVERSITY IN THE LEADING VACCINE ANTIGENS
Merozoite surface protein 1
MSP-1 is the major protein on the surface of the blood stage of the parasite. It is synthesized as a 190kDa precursor, which undergoes proteolytic cleavage into four fragments that remain on the merozoite surface as a glycosylphosphatidylinositol-anchored complex. Before erythrocyte invasion, the entire MSP-1 complex is shed, except for the C-terminal 19kDa (MSP-119), which remains on the surface as the merozoite enters the erythrocyte (9). MSP-119 contains two epidermal growth factor (EGF)-like domains, which are thought to have an important function in erythrocyte invasion (10). Naturally acquired antibodies to MSP-119 can inhibit erythrocyte invasion by preventing the secondary processing that releases this fragment from the rest of the MSP-1 complex (9,11,12), and are associated with protection from clinical malaria in field studies (13–18).
The sequence of the msp-1 gene can be organized into 17 blocks based on sequence variability (19,20). Most of the sequence in MSP-1 groups into two distinct allele families (20), with the exception of Block 2, which is a repetitive region that consists of four allele families (19,21). Block 17 contains MSP-119, which has been the focus of malaria vaccine development because of its highly conserved sequence and hypothesized critical function. However, even this region contains at least six nonsynonymous single nucleotide polymorphisms (SNPs). Studies have demonstrated cross-reactive antibody responses between MSP-119 haplotypes (22,23), but also some differential recognition (22–25), particularly of polymorphic amino acids within the second EGF-like domain (22,23).
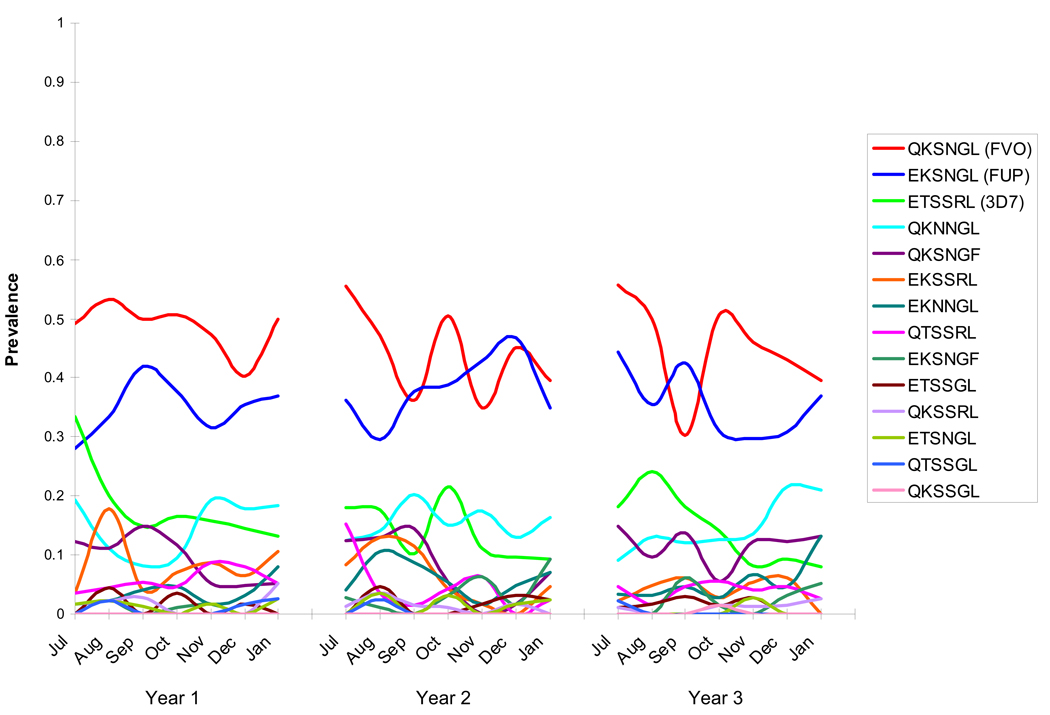
The frequency of MSP-119 haplotypes (based on the six polymorphic sites) was examined over three years in a cohort of children and young adults studied at a vaccine testing site in Mali (26). Parasites with MSP-119 amino acid sequences corresponding to the FVO (QKSNGL) and FUP (EKSNGL) strains of P. falciparum were most prevalent in all three years of the study (Figure 1) and in all age groups, while parasites with sequences corresponding to the 3D7 (ETSSRL) strain were less prevalent. Parasites corresponding to the FUP (EKSNGL) strain also have the highest frequency in Vietnam (27), Kenya (28), and Thailand (29). If immunity elicited by an MSP-119-based vaccine is allele-specific, a vaccine derived from either the FVO or FUP strain might have better initial efficacy than a vaccine derived from the leading vaccine strain 3D7 (30) in settings where these MSP-119 haplotypes predominate and haplotypes corresponding to 3D7 are less frequent (26–29,31).
Figure 1. Prevalence of MSP-119 haplotypes over three years in Bandiagara, Mali.
Parasites with haplotypes corresponding to the FVO and FUP strains of P. falciparum predominated while the vaccine strain 3D7 was at low frequency throughout three consecutive malaria transmission seasons at a malaria vaccine testing site in Mali (26).
Apical Membrane Antigen 1
AMA-1 is also being targeted for malaria vaccines against the blood stage of the parasite. This 83 kDa protein contains three major domains stabilized by eight disulfide bonds (32) and is synthesized during the late intraerthrocytic phase. The protein is initially present in the apical complex of the merozoite (33), and then processed to a 66kDa form that moves to the surface as the merozoite is released from the infected erythrocyte (34). AMA-1 is thought to play a role in erythrocyte invasion through either reorienting the merozoite after initial contact with the erythrocyte or through initiating the junctional contact between the merozoite and host cell (35). Proteomic studies have shown that AMA-1 is also expressed in the sporozoite stage (36), suggesting that it may play a similar role in hepatocyte invasion (37).
All of the sequence diversity in the gene encoding AMA-1 is in the form of SNPs; there are no repeat regions as there are in other vaccine candidate antigens (e.g. MSP-1 and circumsporozoite protein (CSP)). Most of these SNPs are located in domain I. A recent analysis of 355 ama-1 sequences from GenBank (including sequences from the following references: (33,38–45)) showed 32 polymorphic amino acid positions in domain I, 11 in domain II, and 9 in domain III, excluding polymorphisms observed in only one sequence (46). These polymorphisms are located predominantly on one face of the AMA-1 molecule, with most polymorphic residues situated near a hydrophobic trough that is hypothesized to be a receptor binding site between AMA-1 and associated proteins that form the machinery for erythrocyte invasion (Figure 2)(47,48). In ongoing work, more than 200 unique AMA-1 haplotypes have been identified among just over 500 samples collected in a single Malian town (unpublished data). No single haplotype predominates, with all haplotypes having a frequency less than 4%, and approximately 3% of haplotypes having an amino acid exactly matching that of the 3D7 strain of P. falciparum. To date, no AMA-1 haplotypes have been found that have an amino acid sequence exactly matching that of the FVO strain.
Figure 2. Polymorphic residues in AMA-1.
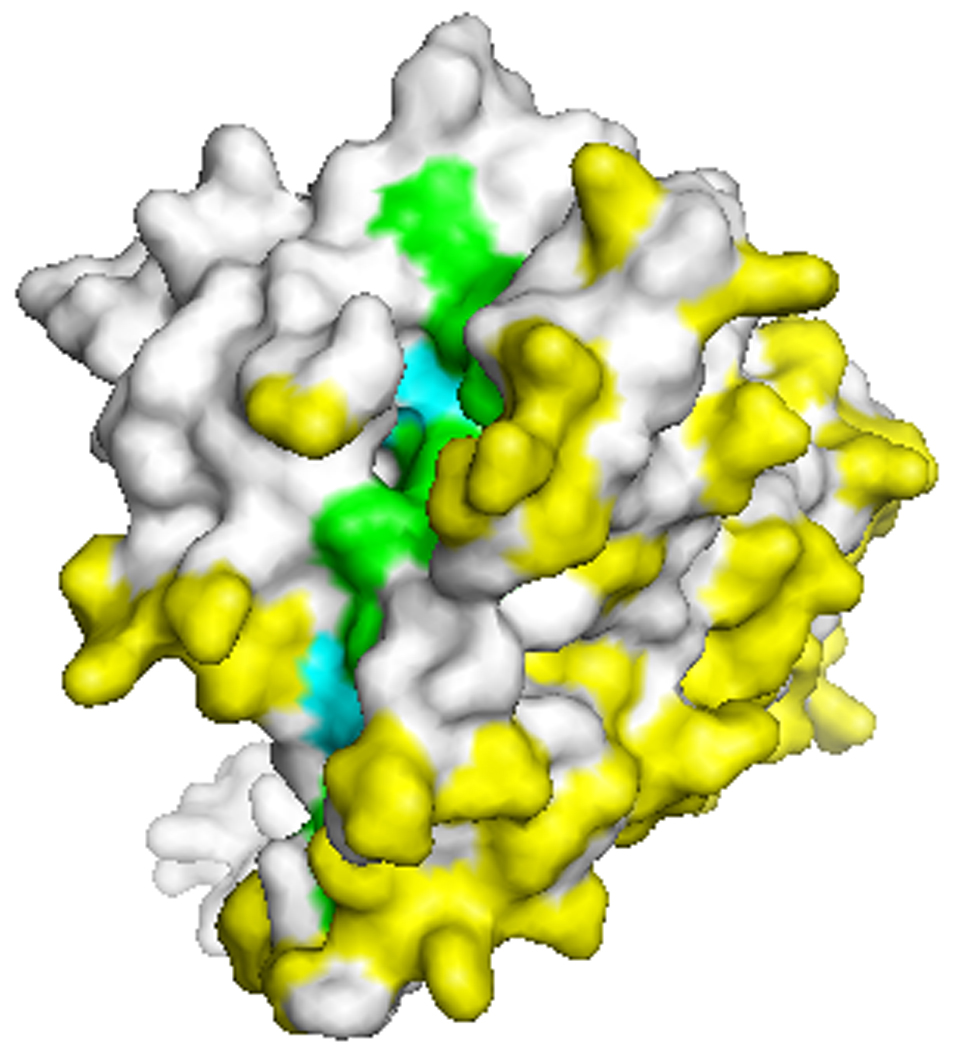
View of the top of the AMA-1 molecule showing the hydrophobic trough (green and blue residues) and the polymorphic face of the protein. Yellow and blue residues indicate polymorphic amino acid positions. Many polymorphic residues cluster around the hydrophobic trough hypothesized to be a receptor binding pocket (47,48). Crystal structure was kindly provided by Adrian Batchelor.
People living in malaria endemic areas produce antibodies to AMA-1 (49–53). These antibodies are capable of inhibiting erythrocyte invasion in vitro (54) and are associated with protection in field studies (51). Several studies in animal models have shown significant allele-specificity in the inhibitory activity of anti-AMA-1 antibodies (54–56), and these results have been corroborated by subsequent allelic exchange experiments (57,58). The specificity of one inhibitory monoclonal antibody was shown to depend on the amino acid present at position 197, the most polymorphic site on the molecule, with six possible amino acids. In malaria-exposed individuals from Papua New Guinea, antibodies against conserved regions of AMA-1 were predominant in most individuals; however, most also had a smaller fraction of allele-specific antibodies (49). For a comprehensive review of AMA-1 as a vaccine candidate, see (59).
Circumsporozoite Protein
CSP, the predominant protein on the surface of the sporozoite, is the most thoroughly studied preerythrocytic vaccine candidate, and is the target of the RTS,S/AS02A vaccine, which has demonstrated efficacy against uncomplicated and severe clinical malaria (60). This 58 kDa protein is thought to have an important function in sporozoite development and motility (61).
The gene encoding CSP contains a central repetitive region flanked on each side by a non-repetitive region (62). The tandem repeats in the central part of the molecule consist of four amino acids each, most with the amino acid sequence NANP and a minority with NVDP or NVNP (62–64). The repeats are species-specific and have been shown to elicit antibody responses (65,66). An average of 41 repeats per allele (range 37–49) was observed in a sample of 48 complete CSP sequences from various geographic regions (63).
On either side of the repeat region are two conserved regions: region 1 (R1) near the N-terminus and region 2-plus (R2) near the C-terminus of CSP. R2 serves as the parasite’s hepatocyte-binding ligand (67), and R1 and R2 are conserved among all malaria parasites (62,67,68). There is, however, some polymorphism in the 5’ and 3’ ends of the csp gene, and in particular, in the regions coding for the Th2R and Th3R T-cell epitopes (31,63,64,69–80). As with most polymorphic loci in P. falciparum, the extent of diversity within these T-cell regions varies with the level of transmission in different endemic areas. In order of increasing levels of malaria transmission, four Th2R–Th3R haplotypes were observed in Peru (n=139) (31), 20 in Vietnam (n=142), and 24 haplotypes were observed in a smaller set of samples from The Gambia (n=44) (80). Preliminary data from a small sample set in Mali (n=50) have shown 15 SNPs in Th2R and seven SNPs in Th3R, and 28 Th2R–Th3R haplotypes (K. Gandhi, unpublished data), comparable to the number of SNPs observed in The Gambia (18 polymorphisms in the two regions combined) (80). Previous studies of CSP diversity have indicated that the predominant CSP alleles differ according to geographic region (31,63,70,72,74,76). If vaccine-induced immunity is allele-specific, such regional differences in haplotype frequency might translate into regional differences in vaccine efficacy, with important implications for both vaccine testing and eventual deployment. Some immunological studies have suggested that the polymorphisms in the T-cell regions of CSP can affect T-cell recognition and HLA binding (81–84). However, no evidence of selection of non-vaccine alleles in the CSP T-cell regions was detected in a Phase 2 pediatric efficacy trial of the RTS,S/AS02A vaccine, which is based on the 3D7 CSP sequence (6). While this finding supports the idea that genetic diversity in CSP does not compromise the efficacy of RTS,S/AS02A, it is possible that modest or transient selective effects could have been missed. Substantial proportions (44% in Cohort 1 and 26% in Cohort 2) of infections containing multiple alleles were excluded from the molecular analyses (6). Survival analyses of time to the first infection (or first clinical episode) with non-vaccine strain CSP might have detected time-dependent selective effects. Moreover, if RTS,S selection operates via immune responses directed against the CSP repeat region, this study, which examined only polymorphism in T cell epitopes, would not have detected it. Lastly, a weak selection signal might have been obscured by naturally acquired immune responses in these children who were repeatedly exposed to malaria infection. To determine whether the allelic diversity of CSP is compromising RTS,S efficacy, further investigation of the specificity of RTS,S-induced T-cell responses, as well as of the role of antibody responses induced by the repetitive regions of the vaccine construct, is warranted both in the recently started Phase 3 trial of RTS,S/AS01 and in studies of the vaccine in young infants who lack acquired immunity. Molecular analyses from this trial may also shed light on the finding of reduced multiplicity of infection in vaccinated individuals and whether reduced complexity of blood-stage infections contributes to protective immunity (85).
Other candidate antigens
Much less is known about diversity in other vaccine candidate antigens such as merozoite surface protein 3 (MSP-3), serine repeat antigen 5 (SERA5), the liver stage antigens (e.g. LSA-1 and LSA-3), and sexual stage antigens (e.g. Pfs48/45).
MSP-3 and SERA5 are blood-stage vaccine candidates that have recently entered clinical testing. Alleles of the gene encoding MSP-3 group into two major allelic types (86) and contain polymorphism both in the form of indels and SNPs (1,87,88). A recent evaluation of 101 MSP-3 sequences showed extensive polymorphism, with 358 polymorphic sites (out of 1063 total nucleotide sites) including 91 SNPs outside indels, 36 SNPs within indels, and 231 other indels. Among 51 Nigerian isolates there were 41 distinct MSP-3 alleles, and 20 alleles among 50 Thai isolates (88). The first efficacy trial of an MSP-3 vaccine, presently underway in Mali, may show whether this extreme degree of polymorphism results in allele-restricted efficacy.
The gene encoding SERA5 contains four exons, with most sequence diversity observed in exon II (89,90). Most of the diversity in exon II is in the form of indels, and amino acid substitutions were also observed at 17 positions (1.7kb evaluated) (89).
LSA-1 and LSA-3 are pre-erythrocytic candidates being targeted for CTL-inducing subunit malaria vaccines. The genes encoding these antigens both contain repetitive and non-repetitive regions, with the majority of T-cell epitopes located within non-repetitive regions. An alignment of 71 sequences of the N-terminal non-repetitive region of LSA-1 from Brazil, Papua New Guinea, Kenya, and Malaysia showed 11 non-synonymous substitutions and 1 synonymous substitution (91). Two of these non-synonymous substitutions were also observed in an alignment of 137 isolates from Peru (31). In the 1204 amino acids comprising the non-repetitive regions of LSA-3, 7 amino acid substitutions were detected between the K1 and 3D7 strains (92). No amino acid substitutions were found in the HLA-B53 restricted la90 CTL epitope of LSA-3 (93). LSA-3 may also be expressed in the blood stages of the parasite life cycle, potentially allowing LSA-3-based vaccines to target multiple life stages (94).
Pfs48/45 is a sexual stage antigen synthesized by gametocytes and expressed on the surface of gametes and zygotes (95). This transmission-blocking vaccine candidate antigen tends to be less polymorphic than most other erythocytic and pre-erythrocytic antigens, and has relatively low non-synonymous polymorphism (1,96,97). Among 44 sequences sampled from diverse geographic regions, 23 out of 449 amino acid positions were polymorphic, with Kenyan isolates showing more diversity than comparable isolates from Thailand, India, and Venezuela (96).
As vaccine candidates based on these polymorphic antigens progress in clinical testing, it will be important to conduct larger surveys of genetic diversity in endemic areas where malaria vaccines will be tested and later deployed. It will also be crucial to include allele-specific efficacy as a key efficacy endpoint and to power trials sufficiently to detect selection of non-vaccine variants.
Positive natural selection on malaria vaccine antigens
It has long been hypothesized that the extensive polymorphism present in some malaria antigens may be the result of positive natural selection acting to maintain genetic diversity and allow the parasite to escape attack by the human immune system. Indeed, evidence for selection has been reported for all three leading vaccine antigens, MSP-1, AMA-1 and CSP.
In the case of MSP-1, the region of the protein under the strongest selection is the N-terminal block 2 repetitive region (98); however, there is also evidence for positive selection acting on MSP-119 (99,100), the C-terminal portion of the molecule that has been included in MSP-1-based vaccines. A recent comparison of MSP-142 in P. vivax and P. falciparum provided evidence that the polymorphism in MSP-119 is under positive selection, and unlike the N-terminal regions of the molecule, which have been proposed to be under balancing selection, this analysis suggested that MSP-119 is under directional selection (99). Polymorphism in AMA-1 also appears to be maintained by positive natural selection in the form of balancing selection, particularly for domain I (38,41,42,45,101) and domain III (45,101). The evidence for positive natural selection acting on the polymorphic T-cell regions of CSP is mixed, with some studies providing evidence for selection (1,102) and others finding no such evidence (80). These conflicting results may be due to differences in sample size, sampled populations, or the use of different tests to estimate departures from neutrality.
The rationale for using population genetics analyses to pinpoint the specific parts of malaria antigens under the greatest positive natural selection is based on the desire to identify the polymorphisms that are the most immunologically relevant in hopes of gaining information that can guide vaccine design and provide insight into the evolution of antigen proteins. Recently, more direct means have been used to hone in on immunologically relevant antigen polymorphism, namely, novel molecular epidemiological approaches that assess changes in parasite genotypes as risk factors for malaria infection and disease in semi-immune populations.
NARROWING THE FOCUS ON IMMUNOLOGICALLY RELEVANT DIVERSITY
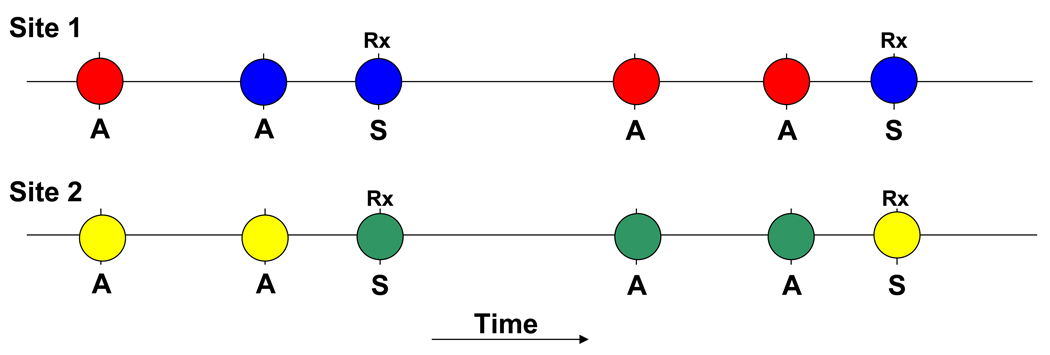
Since diversity itself is not necessarily an indication of immune selection, it is important to understand which polymorphisms are important for immune protection, as this information will inform choices about which alleles to include in multivalent or chimeric vaccines. Using samples collected from participants in a prospective cohort study of malaria incidence conducted over three years at a vaccine-testing site in Mali (103), the within-host dynamics of MSP-119 polymorphisms were examined (26). Study participants contributed blood samples every month and at every clinical episode of malaria. On average, each participant was infected with P. falciparum (detected by PCR) at 14 time points over the course of three transmission seasons. The six polymorphic nucleotides in MSP-119 were genotyped in samples corresponding to consecutive infections, and statistical analyses were performed to estimate the association between change in the predominant amino acid at each polymorphic site between consecutive episodes, and clinical outcome, including the development of clinical symptoms and the time to next clinical infection. The concepts underlying these analyses are shown in Figure 3, which shows the hypothetical results of follow up of one individual over the course of the transmission season at two polymorphic sites of a vaccine antigen. The hypothesis is that because of allele-specific immunity, individuals are more likely to get sick when they are infected with a parasite that is different from the preceding one in immunologically important regions of the molecule. Results of this type of analysis of samples from the Malian cohort suggested that changes in amino acids at MSP-119 positions 1691, 1700, and 1701, but not at the other three polymorphic residues, were associated with individuals becoming symptomatic, suggesting that these amino acid positions may be particularly important in determining allele-specificity of anti-MSP-119 immune responses. These results are consistent with previous studies of anti-MSP-119 immunity that have indicated some differential recognition of MSP-119 haplotypes based on polymorphic amino acids in the second EGF-like domain (22,23). However, coordinated assessment of both parasite genotypes and allele-specific antibody responses will be required to confirm the importance of these residues in determining the dynamics of MSP-119 diversity, since other factors, including immune responses to other antigens, could influence the dynamics. If the amino acids present at these positions do define MSP-119 “serotypes”, then this greatly reduces the amount of genetic diversity vaccine developers would need to take into account in developing a polyvalent MSP-1-based vaccine (Figure 4) (26).
Figure 3. Analysis of within-host dynamics of vaccine antigen alleles to identify immunologically relevant polymorphisms.
Circles represent time points when parasites are detected within infected individuals. “A” represents asymptomatic time points, and “S” represents symptomatic time points when individuals are treated (“Rx”). Different colored circles indicate different alleles at each polymorphic site. In this hypothetical example, at site 1, an allele change occurs both when the individual goes from being asymptomatic to symptomatic and when they have consecutive asymptomatic infections. However, at site 2, allele changes occur when the individual goes from being asymptomatic to symptomatic but not when they remain asymptomatic at consecutive time points. If consecutive infections from multiple individuals are compared, it can be determined whether there is an association between allele changes at certain positions and the development of clinical malaria.
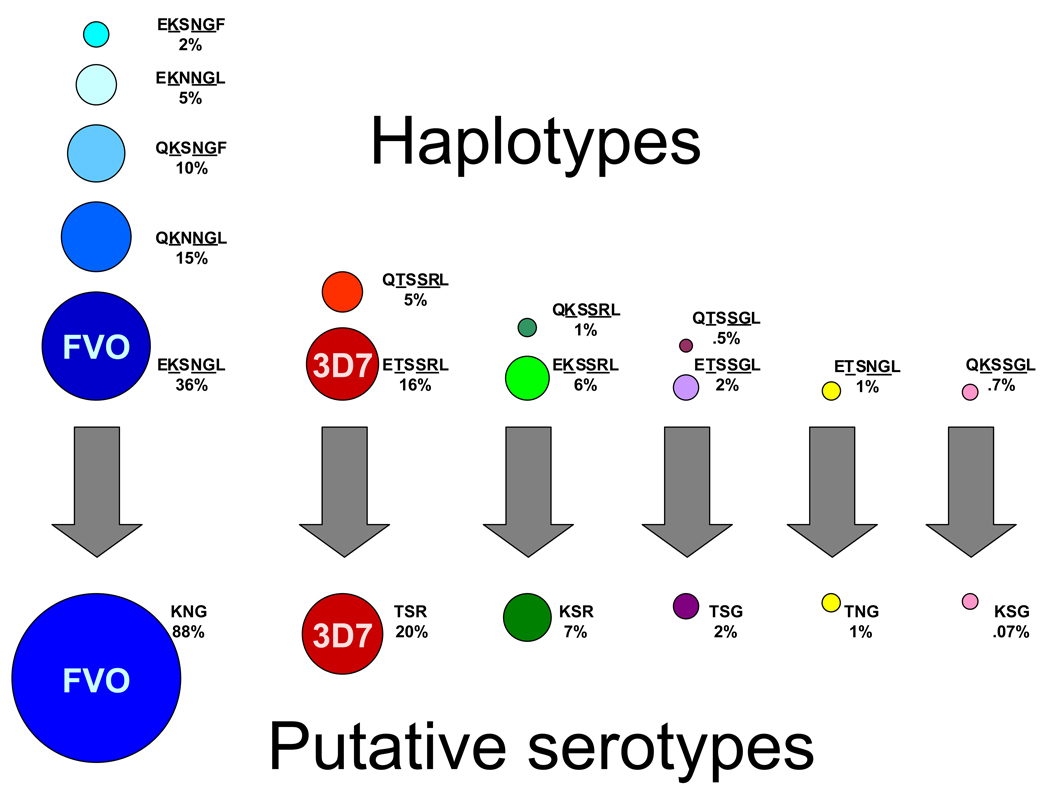
Figure 4. Grouping MSP-119 haplotypes into “serotypes”.
In the top half of the figure, circles represent 14 MSP-119 haplotypes (based on six polymorphic sites), with circle area proportional to the prevalence of the haplotypes among infections in a Malian cohort. These haplotypes can be grouped into six putative “serotypes” (lower part of figure) based only on amino acids at positions 1691, 1700, and 1701. Amino acid changes at these three sites were associated with the development of clinical malaria in individuals in the cohort. FVO and 3D7 haplotypes and putative serotypes correspond to these two leading vaccine strains. Parasites with residues KNG, respectively, at these putatively important sites were observed in nearly 90% of infections, suggesting that MSP-1 corresponding to the FVO vaccine strain would have been the best initial choice for inclusion in a vaccine (26).
Narrowing the focus on the most relevant diversity is even more important for an extremely polymorphic protein like AMA-1. Solving the crystal structure of AMA-1 has greatly aided this process (47), and a recent study exploited this development to map clusters of antigenic escape residues on the AMA-1 structure (57). Chimeric proteins were created and specific clusters of residues within each of the three domains (and subclusters of domain I) were switched from FVO type to 3D7 type (e.g. a chimera consisting of the FVO AMA-1 protein with a 3D7 sequence at domain I). The chimeras were then used to determine the inhibitory contribution of clusters of polymorphic residues, based on their ability to deplete strain-specific antibodies and reverse parasite growth inhibition. These allelic exchange experiments showed that a cluster of residues surrounding the hydrophobic pocket in domain I had the greatest effect on antigenic escape, with a cluster of polymorphisms in domain II also demonstrating a significant effect. The authors suggest that this information can be used to group malaria parasites into inhibitory groups based on whether or not they share amino acids in immunologically relevant regions of the protein, or that chimeric proteins themselves could be used as vaccine constructs that elicit an immune response that covers multiple parasite strains (57). Similar diversity-covering approaches are being undertaken by others (46).
Another group used a Bayesian clustering algorithm, originally designed to infer population structure using data from multiple, unlinked, neutral loci, to categorize AMA-1 sequences into groups of similar haplotypes (39). One hundred seven unique haplotypes were identified from among full-length AMA-1 sequences from 158 parasite isolates, of which 97 were from cloned parasites from different regions of the world and 61 were from three time points in Mali. Using the program STRUCTURE, the 107 haplotypes grouped into six haplotype groups. These groups were represented in all of the geographic regions sampled and at all time points. Antibodies isolated from rabbits vaccinated with recombinant AMA-1 based on the 3D7 sequence inhibited invasion by parasites from the same haplotype group significantly more than parasites from other haplotype groups, suggesting that this approach may define immunologically relevant groups (39).
Using a similar approach to that used to assess the within-host dynamics of MSP-119, ongoing work is assessing how individual polymorphisms, clusters of polymorphisms, and haplotype groups in AMA-1 relate to the development of clinical symptoms in individuals over time. If the results of in vitro growth inhibition assays are borne out by these studies of natural selection of AMA-1 variants under immune pressure in vivo, this will strengthen the rationale for considering such evidence of allele-specific immunity in vaccine design.
ROLE OF MOLECULAR EPIDEMIOLOGICAL STUDIES
Vaccine testing
Clinical trials of vaccine efficacy provide valuable opportunities to understand how polymorphism in vaccine antigens affects the immune response to these antigens and vice versa, which in turn will inform efforts to optimize vaccine design. However, to accurately interpret the results of efficacy trials of malaria subunit vaccines and to estimate allele-specific efficacy, it is important to know the frequency of the target allele(s) in the population where the vaccine is being tested. Without this knowledge, it is not possible to distinguish between a vaccine that is simply ineffective and one that may have high allele-specific efficacy against a target allele that is present at a low frequency. For example, the FMP1 vaccine antigen is based on the C-terminal 42kDa of MSP-1 from the 3D7 strain of P. falciparum (30). This vaccine failed to show clinical efficacy in a Phase 2 pediatric trial conducted in Kenya (8). Previous small studies of MSP-119 diversity in Kenya found the 3D7 allele at frequencies of no more than 10–25% (28,104), which might account for failure of a 3D7-based vaccine to offer significant protection. Further examination of MSP-119 genotypes and allele-specific immune responses in vaccinated and control individuals from this trial may demonstrate whether the lack of efficacy was due to allele-specific immunity.
Developing analytical plans to estimate allele-specific efficacy in clinical trials will be challenging. Careful thought needs to be given to how to assess allele-specific effects in studies using different types of primary study end points (e.g. parasite density, incidence of clinical malaria, or time to first infection), and how to address mixed-allele infections. The frequency of target alleles should be taken into account when calculating sample sizes, since the number of subjects required to detect allele-specific efficacy is greater than that required to detect overall efficacy. Methods used to assess allele-specific efficacy of vaccines against other genetically diverse pathogens (e.g. HIV) might be adapted to analyze data from malaria vaccine trials (105). Such analyses may prevent prematurely abandoning vaccine candidates that provide allele-specific protection and that could potentially be improved by including additional target alleles.
Vaccine design
Molecular epidemiological studies not only allow estimates of vaccine efficacy to be understood in the context of vaccine antigen allele frequencies, they also provide data to aid the rational design of malaria vaccines. Having this kind of information from endemic areas where vaccines will be tested and deployed will permit vaccine developers to choose target strains based on the frequencies of the parasite alleles circulating in the population, rather than using such data retrospectively to explain the failure of specific vaccine candidates.
If antigenic escape and selection for non-vaccine genotypes occurs with monovalent vaccine candidates and/or if these constructs do not provide broad enough coverage, it may be necessary to develop polyvalent malaria vaccines, as has been done for influenza A and pneumococcus. This would increase the cost of developing and manufacturing vaccines, and some have suggested that such polyvalent vaccines may not perform any better than their monovalent counterparts (106). Alternatively, it may be possible to select or engineer vaccine antigens that are more cross-protective. For example, 355 AMA-1 sequences from GenBank were aligned to determine the location and extent of polymorphism in the protein and identify any linkage between specific residues, and this information was used to design three artificial AMA-1 constructs that share conserved amino acids while covering the greatest possible amount of polymorphism (46). Combined, these constructs incorporate, on average, 97% of the variability observed in the sampled sequences. Rabbits vaccinated with a combination of these artificial AMA-1 proteins were shown to induce functional immune responses against three diverse AMA-1 alleles that were comparable to responses generated by vaccination with monovalent homologous alleles (46). As described above, data obtained from molecular epidemiological studies combined with population genetic analyses and/or analyses of protein structure can be used to identify subsets of residues (26,57) that may be particularly important to take into consideration in engineering such artificial antigens, ensuring that the most important residues for protection are represented.
In the case of extremely polymorphic proteins like AMA-1, it may be desirable to avoid polymorphism altogether and to engineer vaccine constructs that boost the immune response to protective epitopes in conserved regions of these proteins. At least one invasion-inhibitory epitope has been identified in the domain II loop on the non-polymorphic face of AMA-1 (107). If subunit vaccines based on conserved epitopes can divert the immune response away from highly polymorphic regions, they may be able to induce strain-transcending immunity. Alternatively, genomic and proteomic approaches are being used to identify new vaccine targets that are not immunodominant and likely to be more conserved than the current highly immunodominant and polymorphic candidates (108–110). Novel approaches that integrate proteomics with careful epidemiological studies may be particularly fruitful. For example, sera from individuals who are resistant and susceptible to malaria can be screened against arrays of parasite proteins (111) to identify antigens that are most important to clinical protection.
Continued molecular monitoring after vaccine deployment
As vaccines are introduced, parasite populations should continue to be monitored to detect subsequent changes that could affect vaccine efficacy. Today it is hard to imagine periodic re-design of malaria vaccines as is done annually now with influenza vaccines, or geographically tailored vaccines based on molecular epidemiology. However, as illustrated by the exponential pace of the genomic revolution, what is feasible and affordable can change quickly, and technological advances in vaccine design and production might eventually make these seemingly fantastic strategies possible.
GENETIC DIVERSITY AND WHOLE-ORGANISM VACCINES
Recombinant DNA vaccines seemingly offer the promise of overcoming genetic diversity because, at least in theory, they are relatively simple to design and modify and have the potential to elicit protective immune responses against multiple different antigens (and different variants of antigens) from different life stages of the parasite (112). The possibility of targeting several parasite antigens simultaneously also served as the impetus for development of other multi-stage multi-antigen malaria vaccines, including viral-vectored vaccines (113) and synthetic antigens containing multiple parasite proteins (114). So far none of these vaccines has progressed very far in the clinical pipeline.
Whole-organism vaccines represent another promising albeit controversial alternative to subunit vaccines that has the potential to overcome the problem of vaccine resistance (115). This approach has been proposed for both pre-erythrocytic (116) and blood stages (117) of the parasite life cycle, and a pre-erythrocytic whole-organism vaccine is presently being evaluated in a clinical trial at the U.S. Navy Medical Research Center and the University of Maryland’s Center for Vaccine Development. It is hoped that this approach might mitigate the problem of parasite genetic diversity by generating immunity to a range of parasite antigens (115); however, it is not known whether there will be enough redundancy in the immune response to overcome diversity in individual antigens.
Whole-organism sporozoite vaccines have been developed and used in East Africa to prevent bovine fever caused by the apicomplexan tick-borne parasite Theileria parva. While a three-strain cocktail vaccine has been used in hopes of avoiding vaccine escape, the success of this strategy is uncertain, and the genetic basis of any allele-specific protection has not been elucidated (118).
Since the basis of protective immunity elicited by whole-organism vaccines is not known, it will be crucial to look for evidence of selection, both in known antigens and genome-wide, in the context of clinical efficacy trials in malaria endemic areas. Prior to such trials, sequencing of P. falciparum genomes from diverse geographic locations, and particularly from local vaccine testing sites, will help inform choices about which strain(s) to include in a whole-organism vaccine, should genetic diversity pose a problem, and will also provide important information about parasite population genetics, including population structure, diversity, and linkage disequilibrium, that will inform genome-wide analyses to detect regions of the genome associated with vaccine escape. The success of these investigations will require a multi-disciplinary approach incorporating expertise in clinical trials, molecular epidemiology, bioinformatics, and genomics, and will also depend on the ability to overcome technical hurdles such as polyclonal malaria infections and large-scale collection of parasite material suitable for direct application to genome-wide genotyping and current and next-generation sequencing platforms.
CONCLUSIONS
As the call for malaria eradication is taken up (119,120), protection conferred by a successful malaria vaccine will be critical to supplement waning natural immunity. The emergence and spread of “vaccine resistant malaria” could be as fatal to the success of the incipient global eradication campaign as drug resistant malaria was to the first one. The malaria parasite has had centuries to evolve mechanisms for evading the human immune response, including vast amounts of diversity in the immunodominant surface proteins targeted by most current vaccines. This diversity provides the fuel for selection of parasites able to escape the effects of vaccines that are based on a limited number of parasite alleles.
Molecular epidemiological studies provide information about the frequency and dynamics of vaccine antigen polymorphisms that can be used to make informed decisions about which parasite alleles to include in vaccine formulations, and to evaluate accurately the efficacy of vaccines tested in malaria endemic areas. Molecular data obtained in well-planned longitudinal studies and clinical trials can be used to understand how the dynamics of antigen diversity correlate with clinical outcomes in individuals. This new approach, guided by knowledge of the structure of the target proteins, the level and specificity of the immune response, and the evolution of the parasite population, can be used to identify the diversity most relevant to vaccine escape and cross-protection, and thus reduce the amount of diversity that needs to be taken into account when designing polyvalent vaccines. Alternative means to overcome antigenic diversity, including boosting responses to critical conserved regions of known antigens, identification of more conserved and less immunodominant antigens, and development of whole-organism vaccines, are also being explored. Creative approaches that integrate tools from multiple disciplines will continue to be required to understand the mechanisms behind the interaction between the human immune system and the malaria parasite and to apply this knowledge to the development of broadly protective vaccines against P. falciparum and other human malaria species.
ACKNOWLEDGMENTS
The authors and their work are supported by the Howard Hughes Medical Institute, by a Distinguished Clinical Scientist Award from the Doris Duke Charitable Foundation, and by U.S. National Institutes of Health grants K12RR023250 (Multidisciplinary Clinical Research Career Development Award), U19AI065683 from the National Institute of Allergy and Infectious Disease, and D43TW01589 from the Fogarty International Center, and contract W81XWH-06-1-0427 from the U.S. Department of Defense and the U.S. Agency for International Development’s Malaria Vaccine Development Program. We thank Adrian Batchelor for sharing the crystal structure of AMA-1, Kavita Gandhi for sharing preliminary genotyping data and for helpful comments on the manuscript, and Ogobara Doumbo, Mahamadou Thera, Amed Ouattara and others from the Malaria Research and Training Center in Mali for collaborating on molecular epidemiological studies and vaccine trials.
Footnotes
Disclosures: None
REFERENCES
- 1.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149(1):189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall B, McEllistrem MC, Gertz RE, Jr, Wedel S, Boxrud DJ, Gonzalez AL, Medina MJ, Pai R, Thompson TA, Harrison LH, McGee L, Whitney CG. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J Clin Microbiol. 2006;44(3):999–1017. doi: 10.1128/JCM.44.3.999-1017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez BE, Hulten KG, Lamberth L, Kaplan SL, Mason EO., Jr Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr Infect Dis J. 2006;25(4):301–305. doi: 10.1097/01.inf.0000207484.52850.38. [DOI] [PubMed] [Google Scholar]
- 4.Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3(11):168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, Rare L, Baisor M, Lorry K, Brown GV, Pye D, Irving DO, Smith TA, Beck HP, Alpers MP. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1-2b trial in Papua New Guinea. J Infect Dis. 2002;185(6):820–827. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- 6.Enosse S, Dobano C, Quelhas D, Aponte JJ, Lievens M, Leach A, Sacarlal J, Greenwood B, Milman J, Dubovsky F, Cohen J, Thompson R, Ballou WR, Alonso PL, Conway DJ, Sutherland CJ. RTS,S/AS02A Malaria Vaccine Does Not Induce Parasite CSP T Cell Epitope Selection and Reduces Multiplicity of Infection. PLoS Clin Trials. 2006;1(1):e5. doi: 10.1371/journal.pctr.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dicko A. Randomized, Controlled, Phase 2 Study of the Safety and Immunogenicity of AMA1-C1/Alhydrogel Vaccine for Plasmodium falciparum Malaria in Children in Bancoumana, Mali. Symposium on Recent Clinical Trials of Apical Membrane Antigen 1, a Leading Blood-Stage Vaccine Candidate for Plasmodium falciparum Malaria. Am J Trop Med Hyg. 2007;77 5 Suppl:144. [Google Scholar]
- 8.Ogutu BR. A Randomized, Controlled, Efficacy Trial of the FMP1/AS02A Plasmodium falciparium Malaria Vaccine in Young Western Kenyan Children. American Society of Tropical Medicine and Hygiene Annual Meeting. Symposium on Recent Results of Phase I and Phase II Clinical Trials of Three Candidate Malaria Antigens. Am J Trop Med Hyg. 2006;75 5 Supp:180. [Google Scholar]
- 9.Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion- inhibiting antibodies. J Exp Med. 1990;172(1):379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holder AA, Blackman MJ, Burghaus PA, Chappel JA, Ling IT, McCallum-Deighton N, Shai S. A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz. 1992;87 Suppl 3:37–42. doi: 10.1590/s0074-02761992000700004. [DOI] [PubMed] [Google Scholar]
- 11.Nwuba RI, Sodeinde O, Anumudu CI, Omosun YO, Odaibo AB, Holder AA, Nwagwu M. The human immune response to Plasmodium falciparum includes both antibodies that inhibit Merozoite Surface Protein 1 secondary processing and blocking antibodies. Infection and Immunity. 2002;70(9):5328–5331. doi: 10.1128/IAI.70.9.5328-5331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patino JAG, Holder AA, McBride JS, Blackman MJ. Antibodies that inhibit malaria Merozoite Surface Protein-1 processing and erythrocyte invasion are blocked by naturally acquired human antibodies. J Exp Med. 1997;186(10):1689–1699. doi: 10.1084/jem.186.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branch OH, Udhayakumar V, Hightower AW, Oloo AJ, Hawley WA, Nahlen BL, Bloland PB, Kaslow DC, Lal AA. A longitudinal investigation of IgG and IgM antibody responses to the merozoite surface protein-1 19-kiloDalton domain of Plasmodium falciparum in pregnant women and infants: associations with febrile illness, parasitemia, and anemia. Am J Trop Med Hyg. 1998;58(2):211–219. doi: 10.4269/ajtmh.1998.58.211. [DOI] [PubMed] [Google Scholar]
- 14.Branch OH, Oloo AJ, Nahlen BL, Kaslow D, Lal AA. Anti-merozoite surface protein-1 19-kDa IgG in mother-infant pairs naturally exposed to Plasmodium falciparum: subclass analysis with age, exposure to asexual parasitemia, and protection against malaria. V. The Asembo Bay Cohort Project. J Infect Dis. 2000;181(5):1746–1752. doi: 10.1086/315424. [DOI] [PubMed] [Google Scholar]
- 15.Egan AF, Morris J, Barnish G, Allen S, Greenwood BM, Kaslow DC, Holder AA, Riley EM. Clinical immunity to Plasmodium falciparum malaria is associated with serum antibodies to the 19-kDa C-terminal fragment of the merozoite surface antigen, PfMSP-1. J Infect Dis. 1996;173(3):765–769. doi: 10.1093/infdis/173.3.765. [DOI] [PubMed] [Google Scholar]
- 16.John CC, O'Donnell RA, Sumba PO, Moormann AM, Koning-Ward TF, King CL, Kazura JW, Crabb BS. Evidence that invasion-inhibitory antibodies specific for the 19-kDa fragment of merozoite surface protein-1 (MSP-1 19) can play a protective role against blood-stage Plasmodium falciparum infection in individuals in a malaria endemic area of Africa. J Immunol. 2004;173(1):666–672. doi: 10.4049/jimmunol.173.1.666. [DOI] [PubMed] [Google Scholar]
- 17.Okech BA, Corran PH, Todd J, Joynson-Hicks A, Uthaipibull C, Egwan TG, Holder AA, Riley EM. Fine specificity of serum antibodies to Plasmodium falciparum merozoite surface protein, PfMSP-1(19), predicts protection from malaria infection and high-density parasitemia. Infection and Immunity. 2004;72(3):1557–1567. doi: 10.1128/IAI.72.3.1557-1567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley EM, Allen SJ, Wheeler JG, Blackman MJ, Bennett S, Takacs B, Schonfeld HJ, Holder AA, Greenwood BM. Naturally acquired cellular and humoral immune responses to the major merozoite surface antigen (PfMSP1) of Plasmodium falciparum are associated with reduced malaria morbidity. Parasite Immunol. 1992;14(3):321–337. doi: 10.1111/j.1365-3024.1992.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1) Mol Biochem Parasitol. 1993;59(1):1–14. doi: 10.1016/0166-6851(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 20.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195(2):273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 21.Takala S, Branch O, Escalante AA, Kariuki S, Wootton J, Lal AA. Evidence for intragenic recombination in Plasmodium falciparum: identification of a novel allele family in block 2 of merozoite surface protein-1: Asembo Bay Area Cohort Project XIV. Mol Biochem Parasitol. 2002;125(1–2):163–171. doi: 10.1016/s0166-6851(02)00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan AF, Chappel JA, Burghaus PA, Morris JS, McBride JS, Holder AA, Kaslow DC, Riley EM. Serum antibodies from malaria-exposed people recognize conserved epitopes formed by the two epidermal growth factor motifs of MSP1(19), the carboxy-terminal fragment of the major merozoite surface protein of Plasmodium falciparum. Infect Immun. 1995;63(2):456–466. doi: 10.1128/iai.63.2.456-466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi YP, Sayed U, Qari SH, Roberts JM, Udhayakumar V, Oloo AJ, Hawley WA, Kaslow DC, Nahlen BL, Lal AA. Natural immune response to the C-terminal 19-kilodalton domain of Plasmodium falciparum merozoite surface protein 1. Infect Immun. 1996;64(7):2716–2723. doi: 10.1128/iai.64.7.2716-2723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diallo TO, Spiegel A, Diouf A, Perraut R, Kaslow DC, Garraud O. Short report: IgG1/IgG3 antibody responses to various analogs of recombinant ypfmsp119--a study in immune adults living in areas of Plasmodium falciparum transmission. Am J Trop Med Hyg. 2001;64(3–4):204–206. doi: 10.4269/ajtmh.2001.64.204. [DOI] [PubMed] [Google Scholar]
- 25.Udhayakumar V, Anyona D, Kariuki S, Shi YP, Bloland PB, Branch OH, Weiss W, Nahlen BL, Kaslow DC, Lal AA. Identification of T and B cell epitopes recognized by humans in the C- terminal 42-kDa domain of the Plasmodium falciparum merozoite surface protein (MSP)-1. J Immunol. 1995;154(11):6022–6030. [PubMed] [Google Scholar]
- 26.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, Kone AK, Traore K, Ouattara A, Djimde AA, Sehdev PS, Lyke KE, Diallo DA, Doumbo OK, Plowe CV. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4(3):e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003;304:65–75. doi: 10.1016/s0378-1119(02)01180-0. [DOI] [PubMed] [Google Scholar]
- 28.Qari SH, Shi YP, Goldman IF, Nahlen BL, Tibayrenc M, Lal AA. Predicted and observed alleles of Plasmodium falciparum merozoite surface protein-1 (MSP-1), a potential malaria vaccine antigen. Mol Biochem Parasitol. 1998;92(2):241–252. doi: 10.1016/s0166-6851(98)00010-3. [DOI] [PubMed] [Google Scholar]
- 29.Sakihama N, Kimura M, Hirayama K, Kanda T, Na-Bangchang K, Jongwutiwes S, Conway D, Tanabe K. Allelic recombination and linkage disequilibrium within Msp-1 of Plasmodium falciparum, the malignant human malaria parasite. Gene. 1999;230(1):47–54. doi: 10.1016/s0378-1119(99)00069-4. [DOI] [PubMed] [Google Scholar]
- 30.Angov E, Aufiero BM, Turgeon AM, Van Handenhove M, Ockenhouse CF, Kester KE, Walsh DS, McBride JS, Dubois MC, Cohen J, Haynes JD, Eckels KH, Heppner DG, Ballou WR, Diggs CL, Lyon JA. Development and pre-clinical analysis of a Plasmodium falciparum Merozoite Surface Protein-1(42) malaria vaccine. Mol Biochem Parasitol. 2003;128(2):195–204. doi: 10.1016/s0166-6851(03)00077-x. [DOI] [PubMed] [Google Scholar]
- 31.Chenet SM, Branch OH, Escalante AA, Lucas CM, Bacon DJ. Genetic diversity of vaccine candidate antigens in Plasmodium falciparum isolates from the Amazon basin of Peru. Malar J. 2008;7:93. doi: 10.1186/1475-2875-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. The disulfide bond structure of Plasmodium apical membrane antigen-1. J Biol Chem. 1996;271(46):29446–29452. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- 33.Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol Cell Biol. 1989;9(7):3151–3154. doi: 10.1128/mcb.9.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narum DL, Thomas AW. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1994;67(1):59–68. doi: 10.1016/0166-6851(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect Immun. 2004;72(1):154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419(6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 37.Silvie O, Franetich JF, Charrin S, Mueller MS, Siau A, Bodescot M, Rubinstein E, Hannoun L, Charoenvit Y, Kocken CH, Thomas AW, Van Gemert GJ, Sauerwein RW, Blackman MJ, Anders RF, Pluschke G, Mazier D. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem. 2004;279(10):9490–9496. doi: 10.1074/jbc.M311331200. [DOI] [PubMed] [Google Scholar]
- 38.Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect Immun. 2003;71(3):1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, Diemert D, Long C, Zhou H, Miura K, Ouattara A, Dolo A, Doumbo O, Su XZ, Miller L. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc Natl Acad Sci U S A. 2008;105(22):7857–7862. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisen DP, Marshall VM, Billman-Jacobe H, Coppel RL. A Plasmodium falciparum apical membrane antigen-1 (AMA-1) gene apparently generated by intragenic recombination. Mol Biochem Parasitol. 1999;100(2):243–246. doi: 10.1016/s0166-6851(99)00054-7. [DOI] [PubMed] [Google Scholar]
- 41.Escalante AA, Grebert HM, Chaiyaroj SC, Magris M, Biswas S, Nahlen BL, Lal AA. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2001;113(2):279–287. doi: 10.1016/s0166-6851(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 42.Garg S, Alam MT, Das MK, Dev V, Kumar A, Dash AP, Sharma YD. Sequence diversity and natural selection at domain I of the apical membrane antigen 1 among Indian Plasmodium falciparum populations. Malar J. 2007;6:154. doi: 10.1186/1475-2875-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kocken CH, Narum1 DL, Massougbodji A, Ayivi B, Dubbeld MA, van der Wel A, Conway DJ, Sanni A, Thomas AW. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol Biochem Parasitol. 2000;109(2):147–156. doi: 10.1016/s0166-6851(00)00250-4. [DOI] [PubMed] [Google Scholar]
- 44.Marshall VM, Zhang L, Anders RF, Coppel RL. Diversity of the vaccine candidate AMA-1 of Plasmodium falciparum. Mol Biochem Parasitol. 1996;77(1):109–113. doi: 10.1016/0166-6851(96)02583-2. [DOI] [PubMed] [Google Scholar]
- 45.Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165(2):555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun. 2008;76(6):2660–2670. doi: 10.1128/IAI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci U S A. 2005;102(36):12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins CR, Withers-Martinez C, Hackett F, Blackman MJ. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog. 2009;5(1) doi: 10.1371/journal.ppat.1000273. e1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortes A, Mellombo M, Masciantonio R, Murphy VJ, Reeder JC, Anders RF. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect Immun. 2005;73(1):422–430. doi: 10.1128/IAI.73.1.422-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mount AM, Mwapasa V, Elliott SR, Beeson JG, Tadesse E, Lema VM, Molyneux ME, Meshnick SR, Rogerson SJ. Impairment of humoral immunity to Plasmodium falciparum malaria in pregnancy by HIV infection. Lancet. 2004;363(9424):1860–1867. doi: 10.1016/S0140-6736(04)16354-X. [DOI] [PubMed] [Google Scholar]
- 51.Polley SD, Mwangi T, Kocken CH, Thomas AW, Dutta S, Lanar DE, Remarque E, Ross A, Williams TN, Mwambingu G, Lowe B, Conway DJ, Marsh K. Human antibodies to recombinant protein constructs of Plasmodium falciparum Apical Membrane Antigen 1 (AMA1) and their associations with protection from malaria. Vaccine. 2004;23(5):718–728. doi: 10.1016/j.vaccine.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 52.Thomas AW, Trape JF, Rogier C, Goncalves A, Rosario VE, Narum DL. High prevalence of natural antibodies against Plasmodium falciparum 83- kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture- enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am J Trop Med Hyg. 1994;51(6):730–740. doi: 10.4269/ajtmh.1994.51.730. [DOI] [PubMed] [Google Scholar]
- 53.Udhayakumar V, Kariuki S, Kolczack M, Girma M, Roberts JM, Oloo AJ, Nahlen BL, Lal AA. Longitudinal study of natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in a holoendemic region of malaria in western Kenya: Asembo Bay Cohort Project VIII. Am J Trop Med Hyg. 2001;65(2):100–107. doi: 10.4269/ajtmh.2001.65.100. [DOI] [PubMed] [Google Scholar]
- 54.Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect Immun. 2001;69(5):3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, Long CA, Miller LH, Stowers AW. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70(12):6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kocken CH, Withers-Martinez C, Dubbeld MA, van der Wel A, Hackett F, Valderrama A, Blackman MJ, Thomas AW. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect Immun. 2002;70(8):4471–4476. doi: 10.1128/IAI.70.8.4471-4476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc Natl Acad Sci U S A. 2007;104(30):12488–12493. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman AF, Batchelor A. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol. 2004;52(1):159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 59.Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24(2):74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Alonso PL, Sacarlal J, Aponte JJ, Leach A, Macete E, Milman J, Mandomando I, Spiessens B, Guinovart C, Espasa M, Bassat Q, Aide P, Ofori-Anyinam O, Navia MM, Corachan S, Ceuppens M, Dubois MC, Demoitie MA, Dubovsky F, Menendez C, Tornieporth N, Ballou WR, Thompson R, Cohen J. Efficacy of the RTS,S/AS02A vaccine against Plasmodium falciparum infection and disease in young African children: randomised controlled trial. Lancet. 2004;364(9443):1411–1420. doi: 10.1016/S0140-6736(04)17223-1. [DOI] [PubMed] [Google Scholar]
- 61.Beier JC, Vanderburg JP. Sporogonic Development in the Mosquito.1998. In: Sherman IW, editor. Malaria Parasite Biology, Pathogenesis, and Protection. Washington, DC: ASM Press; 1998. [Google Scholar]
- 62.Dame JB, Williams JL, McCutchan TF, Weber JL, Wirtz RA, Hockmeyer WT, Maloy WL, Haynes JD, Schneider I, Roberts D, et al. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225(4662):593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 63.Escalante AA, Grebert HM, Isea R, Goldman IF, Basco L, Magris M, Biswas S, Kariuki S, Lal AA. A study of genetic diversity in the gene encoding the circumsporozoite protein (CSP) of Plasmodium falciparum from different transmission areas--XVI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;125(1–2):83–90. doi: 10.1016/s0166-6851(02)00216-5. [DOI] [PubMed] [Google Scholar]
- 64.Rich SM, Hudson RR, Ayala FJ. Plasmodium falciparum antigenic diversity: evidence of clonal population structure. Proc Natl Acad Sci USA. 1997;94(24):13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ballou WR, Rothbard J, Wirtz RA, Gordon DM, Williams JS, Gore RW, Schneider I, Hollingdale MR, Beaudoin RL, Maloy WL, et al. Immunogenicity of synthetic peptides from circumsporozoite protein of Plasmodium falciparum. Science. 1985;228(4702):996–999. doi: 10.1126/science.2988126. [DOI] [PubMed] [Google Scholar]
- 66.Zhou Z, Xiao L, Branch OH, Kariuki S, Nahlen BL, Lal AA. Antibody responses to repetitive epitopes of the circumsporozoite protein, liver stage antigen-1, and merozoite surface protein-2 in infants residing in a Plasmodium falciparum-hyperendemic area of western Kenya. XIII. Asembo Bay Cohort Project. Am J Trop Med Hyg. 2002;66(1):7–12. doi: 10.4269/ajtmh.2002.66.7. [DOI] [PubMed] [Google Scholar]
- 67.Sinnis P, Clavijo P, Fenyo D, Chait BT, Cerami C, Nussenzweig V. Structural and functional properties of region II-plus of the malaria circumsporozoite protein. J Exp Med. 1994;180(1):297–306. doi: 10.1084/jem.180.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffman SL, Miller LH. Perspectives on Malaria Vaccine Development.1996. In: Hoffman SL, editor. Malaria Vaccine Development: A Multi-Immune Response Approach. Washington, DC: ASM Press; 1996. [Google Scholar]
- 69.Aidoo M, Udhayakumar V. Field studies of cytotoxic T lymphocytes in malaria infections: implications for malaria vaccine development. Parasitol Today. 2000;16(2):50–56. doi: 10.1016/s0169-4758(99)01592-6. [DOI] [PubMed] [Google Scholar]
- 70.Alloueche A, Silveira H, Conway DJ, Bojang K, Doherty T, Cohen J, Pinder M, Greenwood BM. High-throughput sequence typing of T-cell epitope polymorphisms in Plasmodium falciparum circumsporozoite protein. Mol Biochem Parasitol. 2000;106:273–282. doi: 10.1016/s0166-6851(99)00221-2. [DOI] [PubMed] [Google Scholar]
- 71.de la Cruz VF, Lal AA, McCutchan TF. Sequence variation in putative functional domains of the circumsporozoite protein of Plasmodium falciparum. Implications for vaccine development. J Biol Chem. 1987;262(25):11935–11939. [PubMed] [Google Scholar]
- 72.Doolan DL, Saul AL, Good ML. Geographically restricted heterogeneity of the Plasmodium falciparum CSP: relevance for vaccine development. Infect Immun. 1992;60:675–682. doi: 10.1128/iai.60.2.675-682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Good MF, Pombo D, Quakyi IA, Riley EM, Houghten RA, Menon A, Alling DW, Berzofsky JA, Miller LH. Human T-cell recognition of the circumsporozoite protein of Plasmodium falciparum: immunodominant T-cell domains map to the polymorphic regions of the molecule. Proc Natl Acad Sci U S A. 1988;85(4):1199–1203. doi: 10.1073/pnas.85.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jalloh A, van, Thien H, Ferreira MU, Ohashi J, Matsuoka H, Kanbe T, Kikuchi A, Kawamoto F. Sequence variation in the T-cell epitopes of the Plasmodium falciparum circumsporozoite protein among field isolates is temporally stable: a 5-year longitudinal study in southern Vietnam. J Clin Microbiol. 2006;44(4):1229–1235. doi: 10.1128/JCM.44.4.1229-1235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jongwutiwes S, Tanabe K, Hughes MK, Kanbara H, Hughes AL. Allelic variation in the circumsporozoite protein of Plasmodium falciparum from Thai field isolates. Am J Trop Med Hyg. 1994;51:659–668. doi: 10.4269/ajtmh.1994.51.659. [DOI] [PubMed] [Google Scholar]
- 76.Kumkhaek C, Phra-Ek K, Renia L, Singhasivanon P, Looareesuwan S, Hirunpetcharat C, White NJ, Brockman A, Gruner AC, Lebrun N, Alloueche A, Nosten F, Khusmith S, Snounou G. Are extensive T cell epitope polymorphisms in the Plasmodium falciparum circumsporozoite antigen, a leading sporozoite vaccine candidate, selected by immune pressure? J Immunol. 2005;175(6):3935–3939. doi: 10.4049/jimmunol.175.6.3935. [DOI] [PubMed] [Google Scholar]
- 77.Lockyer M, Marsh K, Newbold CI. Wild isolates of Plasmodium falciparum show extensive polymorphisms in T cell epitopes of the circumsporozoite protein. Mol Biochem Parasitol. 1989;37:275–280. doi: 10.1016/0166-6851(89)90159-x. [DOI] [PubMed] [Google Scholar]
- 78.Lockyer MJ, Schwarz RT. Strain variation in the circumsporozoite protein gene of Plasmodium falciparum. Mol Biochem Parasitol. 1987;22(1):101–108. doi: 10.1016/0166-6851(87)90073-9. [DOI] [PubMed] [Google Scholar]
- 79.Shi YP, Alpers MP, Povoa MM, Lal AA. Diversity in the immunodominant determinants of the circumsporozoite protein of Plasmodium falciparum parasites from malaria-endemic regions of Papua New Guinea and Brazil. Am J Trop Med Hyg. 1992;47:844–851. doi: 10.4269/ajtmh.1992.47.844. [DOI] [PubMed] [Google Scholar]
- 80.Weedall GD, Preston BM, Thomas AW, Sutherland CJ, Conway DJ. Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int J Parasitol. 2007;37(1):77–85. doi: 10.1016/j.ijpara.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 81.de la Cruz VF, Maloy WL, Miller LH, Lal AA, Good MF, McCutchan TF. Lack of cross-reactivity between variant T cell determinants from malaria circumsporozoite protein. J Immunol. 1988;141(7):2456–2460. [PubMed] [Google Scholar]
- 82.Guttinger M, Caspers P, Takacs B, Trzeciak A, Gillessen D, Pink JR, Sinigaglia F. Human T cells recognize polymorphic and non-polymorphic regions of the Plasmodium falciparum circumsporozoite protein. Embo J. 1988;7(8):2555–2558. doi: 10.1002/j.1460-2075.1988.tb03104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Udhayakumar V, Ongecha JM, Shi YP, Aidoo M, Orago AS, Oloo AJ, Hawley WA, Nahlen BL, Hoffman SL, Weiss WR, Lal AA. Cytotoxic T cell reactivity and HLA-B35 binding of the variant Plasmodium falciparum circumsporozoite protein CD8+ CTL epitope in naturally exposed Kenyan adults. Eur J Immunol. 1997;27(8):1952–1957. doi: 10.1002/eji.1830270819. [DOI] [PubMed] [Google Scholar]
- 84.Zevering Y, Khamboonruang C, Good MF. Natural amino acid polymorphisms of the circumsporozoite protein of Plasmodium falciparum abrogate specific human CD4+ T cell responsiveness. Eur J Immunol. 1994;24(6):1418–1425. doi: 10.1002/eji.1830240627. [DOI] [PubMed] [Google Scholar]
- 85.Sutherland CJ, Drakeley CJ, Schellenberg D. How is childhood development of immunity to Plasmodium falciparum enhanced by certain antimalarial interventions? Malar J. 2007;6:161. doi: 10.1186/1475-2875-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okenu DM, Thomas AW, Conway DJ. Allelic lineages of the merozoite surface protein 3 gene in Plasmodium reichenowi and Plasmodium falciparum. Mol Biochem Parasitol. 2000;109(2):185–188. doi: 10.1016/s0166-6851(00)00245-0. [DOI] [PubMed] [Google Scholar]
- 87.Huber W, Felger I, Matile H, Lipps HJ, Steiger S, Beck HP. Limited sequence polymorphism in the Plasmodium falciparum merozoite surface protein 3. Mol Biochem Parasitol. 1997;87(2):231–234. doi: 10.1016/s0166-6851(97)00067-4. [DOI] [PubMed] [Google Scholar]
- 88.Polley SD, Tetteh KK, Lloyd JM, Akpogheneta OJ, Greenwood BM, Bojang KA, Conway DJ. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007;195(2):279–287. doi: 10.1086/509806. [DOI] [PubMed] [Google Scholar]
- 89.Morimatsu K, Morikawa T, Tanabe K, Bzik DJ, Horii T. Sequence diversity in the amino-terminal 47 kDa fragment of the Plasmodium falciparum serine repeat antigen. Mol Biochem Parasitol. 1997;86(2):249–254. doi: 10.1016/s0166-6851(97)00038-8. [DOI] [PubMed] [Google Scholar]
- 90.Safitri I, Jalloh A, Tantular IS, Pusarawati S, Win TT, Liu Q, Ferreira MU, Dachlan YP, Horii T, Kawamoto F. Sequence diversity in the amino-terminal region of the malaria-vaccine candidate serine repeat antigen in natural Plasmodium falciparum populations. Parasitol Int. 2003;52(2):117–131. doi: 10.1016/s1383-5769(02)00088-0. [DOI] [PubMed] [Google Scholar]
- 91.Ravichandran M, Doolan DL, Cox-Singh J, Hoffman SL, Singh B. Research note: HLA degenerate T-cell epitopes from Plasmodium falciparum liver stage-specific antigen 1 (LSA-1) are highly conserved in isolates from geographically distinct areas. Parasite Immunol. 2000;22(9):469–473. doi: 10.1046/j.1365-3024.2000.00324.x. [DOI] [PubMed] [Google Scholar]
- 92.Daubersies P, Thomas AW, Millet P, Brahimi K, Langermans JA, Ollomo B, BenMohamed L, Slierendregt B, Eling W, Van, Belkum A, Dubreuil G, Meis JF, Guerin-Marchand C, Cayphas S, Cohen J, Gras-Masse H, Druilhe P. Protection against Plasmodium falciparum malaria in chimpanzees by immunization with the conserved pre-erythrocytic liver-stage antigen 3. Nat Med. 2000;6(11):1258–1263. doi: 10.1038/81366. [DOI] [PubMed] [Google Scholar]
- 93.Aidoo M, Lalvani A, Gilbert SC, Hu JT, Daubersies P, Hurt N, Whittle HC, Druihle P, Hill AV. Cytotoxic T-lymphocyte epitopes for HLA-B53 and other HLA types in the malaria vaccine candidate liver-stage antigen 3. Infect Immun. 2000;68(1):227–232. doi: 10.1128/iai.68.1.227-232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moyano EM, Gonzalez LM, Arahuetes S, Benito A. Liver stage antigen 3 isolated from a cDNA library of Plasmodium falciparum erythrocytic stages. Parasitol Res. 2007;102(1):111–115. doi: 10.1007/s00436-007-0737-y. [DOI] [PubMed] [Google Scholar]
- 95.Kumar N, Carter R. Biosynthesis of the target antigens of antibodies blocking transmission of Plasmodium falciparum. Mol Biochem Parasitol. 1984;13(3):333–342. doi: 10.1016/0166-6851(84)90124-5. [DOI] [PubMed] [Google Scholar]
- 96.Escalante AA, Grebert HM, Chaiyaroj SC, Riggione F, Biswas S, Nahlen BL, Lal AA. Polymorphism in the gene encoding the Pfs48/45 antigen of Plasmodium falciparum. XI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;119(1):17–22. doi: 10.1016/s0166-6851(01)00386-3. [DOI] [PubMed] [Google Scholar]
- 97.Kocken CH, Milek RL, Lensen TH, Kaslow DC, Schoenmakers JG, Konings RN. Minimal variation in the transmission-blocking vaccine candidate Pfs48/45 of the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1995;69(1):115–118. doi: 10.1016/0166-6851(94)00193-q. [DOI] [PubMed] [Google Scholar]
- 98.Conway DJ, Cavanagh DR, Tanabe K, Roper C, Mikes ZS, Sakihama N, Bojang KA, Oduola AMJ, Kremsner PG, Arnot DE, Greenwood BM, McBride JS. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med. 2000;6(6):689–692. doi: 10.1038/76272. [DOI] [PubMed] [Google Scholar]
- 99.Pacheco MA, Poe AC, Collins WE, Lal AA, Tanabe K, Kariuki SK, Udhayakumar V, Escalante AA. A comparative study of the genetic diversity of the 42kDa fragment of the merozoite surface protein 1 in Plasmodium falciparum and P. vivax. Infect Genet Evol. 2007;7(2):180–187. doi: 10.1016/j.meegid.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanabe K, Sakihama N, Nakamura Y, Kaneko O, Kimura M, Ferreira MU, Hirayama K. Selection and genetic drift of polymorphisms within the merozoite surface protein-1 gene of Plasmodium falciparum 5. Gene. 2000;241(2):325–331. doi: 10.1016/s0378-1119(99)00472-2. [DOI] [PubMed] [Google Scholar]
- 101.Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158(4):1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hughes AL. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics. 1991;127(2):345–353. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coulibaly D, Diallo DA, Thera MA, Dicko A, Guindo AB, Kone AK, Cissoko Y, Coulibaly S, Djimde A, Lyke K, Doumbo OK, Plowe CV. Impact of preseason treatment on incidence of falciparum malaria and parasite density at a site for testing malaria vaccines in Bandiagara, Mali. Am J Trop Med Hyg. 2002;67(6):604–610. doi: 10.4269/ajtmh.2002.67.604. [DOI] [PubMed] [Google Scholar]
- 104.Dent AE, Yohn CT, Zimmerman PA, Vulule J, Kazura JW, Moormann AM. A polymerase chain reaction/ligase detection reaction fluorescent microsphere assay to determine Plasmodium falciparum MSP-119 haplotypes. Am J Trop Med Hyg. 2007;77(2):250–255. [PMC free article] [PubMed] [Google Scholar]
- 105.Gilbert PB. Interpretability and robustness of sieve analysis models for assessing HIV strain variations in vaccine efficacy. Stat Med. 2001;20(2):263–279. doi: 10.1002/1097-0258(20010130)20:2<263::aid-sim660>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 106.Barclay VC, Chan BH, Anders RF, Read AF. Mixed allele malaria vaccines: Host protection and within-host selection. Vaccine. 2008 doi: 10.1016/j.vaccine.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Collins CR, Withers-Martinez C, Bentley GA, Batchelor AH, Thomas AW, Blackman MJ. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J Biol Chem. 2007;282(10):7431–7441. doi: 10.1074/jbc.M610562200. [DOI] [PubMed] [Google Scholar]
- 108.Chaudhuri R, Ahmed S, Ansari FA, Singh HV, Ramachandran S. MalVac: database of malarial vaccine candidates. Malar J. 2008;7:184. doi: 10.1186/1475-2875-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kanoi BN, Egwang TG. New concepts in vaccine development in malaria. Curr Opin Infect Dis. 2007;20(3):311–316. doi: 10.1097/QCO.0b013e32816b5cc2. [DOI] [PubMed] [Google Scholar]
- 110.Scarselli M, Giuliani MM, du-Bobie J, Pizza M, Rappuoli R. The impact of genomics on vaccine design. Trends Biotechnol. 2005;23(2):84–91. doi: 10.1016/j.tibtech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 111.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008 doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doolan DL, Hoffman SL. DNA-based vaccines against malaria: status and promise of the Multi-Stage Malaria DNA Vaccine Operation. Int J Parasitol. 2001;31(8):753–762. doi: 10.1016/s0020-7519(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 113.Tine JA, Lanar DE, Smith DM, Wellde BT, Schultheiss P, Ware LA, Kauffman EB, Wirtz RA, De, Taisne C, Hui GS, Chang SP, Church P, Hollingdale MR, Kaslow DC, Hoffman S, Guito KP, Ballou WR, Sadoff JC, Paoletti E. NYVAC-Pf7: a poxvirus-vectored, multiantigen, multistage vaccine candidate for Plasmodium falciparum malaria. Infect Immun. 1996;64(9):3833–3844. doi: 10.1128/iai.64.9.3833-3844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Z, Todd CW, Wohlhueter RM, Price A, Xiao L, Schnake P, Bonner PC, Martin AM, Goldman IF, De La V, Udhayakumar V, Lal AA. Development, characterization and immunogenicity of a multi-stage, multi-valent Plasmodium falciparum vaccine antigen (FALVAC-1A) expressed in Escherichia coli. Hum Vaccin. 2006;2(1):14–23. doi: 10.4161/hv.2.1.2437. [DOI] [PubMed] [Google Scholar]
- 115.Wykes M, Good MF. A case for whole-parasite malaria vaccines. Int J Parasitol. 2007;37(7):705–712. doi: 10.1016/j.ijpara.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 116.Luke TC, Hoffman SL. Rationale and plans for developing a non-replicating, metabolically active, radiation-attenuated Plasmodium falciparum sporozoite vaccine. J Exp Biol. 2003;206(Pt 21):3803–3808. doi: 10.1242/jeb.00644. [DOI] [PubMed] [Google Scholar]
- 117.Pombo DJ, Lawrence G, Hirunpetcharat C, Rzepczyk C, Bryden M, Cloonan N, Anderson K, Mahakunkijcharoen Y, Martin LB, Wilson D, Elliott S, Elliott S, Eisen DP, Weinberg JB, Saul A, Good MF. Immunity to malaria after administration of ultra-low doses of red cells infected with Plasmodium falciparum. Lancet. 2002;360(9333):610–617. doi: 10.1016/S0140-6736(02)09784-2. [DOI] [PubMed] [Google Scholar]
- 118.Uilenberg G, Silayo RS, Mpangala C, Tondeur W, Tatchell RJ, Sanga HJ. Studies on Theileriidae (Sporozoa) in Tanzania. X. A large-scale field trial on immunization against cattle Theileriosis. Tropenmed Parasitol. 1977;28(4):499–506. [PubMed] [Google Scholar]
- 119.Roberts L, Enserink M, Malaria Did they really say … eradication? Science. 2007;318(5856):1544–1545. doi: 10.1126/science.318.5856.1544. [DOI] [PubMed] [Google Scholar]
- 120.Tanner M, DE, Savigny D. Malaria eradication back on the table. Bull World Health Organ. 2008;86(2):82. doi: 10.2471/BLT.07.050633. [DOI] [PMC free article] [PubMed] [Google Scholar]