Abstract
Background
Single nucleotide polymorphisms (SNPs) in matrix metalloproteinase (MMP) genes may be associated with myocardial infarction (MI) and coronary artery disease (CAD), but studies of multiple MMP genes and their tissue inhibitors (TIMPs) are scarce. Further, differentiation of predictive ability by endpoint (MI vs. CAD) has not been addressed. This study evaluated the association with MI of SNPs in genes encoding MMPs 1, 2, 3, and 9 and TIMPs 1, 2, and 3.
Methods
Genotypes of patients (N=5,148) with MI (n=1,693) and angiographically-defined CAD (=1 lesion of =70% stenosis, n=1,967) were compared to MI-free (n=3,455) and non-CAD patients (n=1,122), respectively. Due to linkage disequilibrium, MMP-1 and MMP-3 SNPs (chromosome 11) were combined, as were the two MMP-9 SNPs.
Results
For MI, only MMP-9 group CT/RQ (OR=1.25, p=0.007 vs. wild-type CC/RR) had greater MI risk, with TT/QQ having a weak trend (OR=1.43, p=0.10). These findings remained (CT/RQ) or were strengthened (TT/QQ) after full adjustment. For CAD, association was found for MMP-1/-3 groups 2G1G/6A6A (OR=1.45, p=0.022), 2G1G/6A5A (OR=1.49, p=0.001), 2G1G/5A5A (OR=1.64, p=0.003), and 1G1G/5A5A (OR=1.35, p=0.035) compared to wild-type.
Conclusions
Composite MMP-9 genotypes but not other SNPs were associated with MI, while MMP-1/-3 genotypes were CAD-associated. The largest MMP/TIMP gene study to date, this study suggests care in selection and definition of clinical phenotypes. Further, this suggests that the evaluated SNPs only approximately account for intra-genic variation in these genes and that comprehensive evaluation of all variation in these genes should better elucidate associations with MI and CAD phenotypes.
Background
Coronary artery disease (CAD) is a complex, multi-factorial, chronic, and highly prevalent vascular disorder that often leads to acute myocardial infarction (MI).(1) A multitude of CAD risk factors are known, each of which may have environmental and genetic components.(2) A variety of evidence, including that an individual’s family history of early cardiovascular disease is an independent risk factor for CAD and MI (3,4), further suggest genetic underpinnings of other undiscovered factors.
While chronic CAD can lead to full obstruction of a coronary artery, acute closure and MI are usually precipitated by the rupture or erosion of an unstable plaque with provocation of an occlusive thrombus, regardless of the degree of prior narrowing.(5–7) Matrix metalloproteinases (MMPs) are implicated in the degradation of the extracellular matrix of coronary plaque, including thinning of the fibrous cap.(6,8) As the cap thins, the plaque becomes increasingly unstable and prone to rupture, possibly stimulating plaque enlargement and often resulting in acute MI. MMPs are suppressed by tissue inhibitor metalloproteinases (TIMPs).(6,9) While not associated with traditional cardiac risk factors, MMPs and TIMPs represent a biologic pathway that could importantly modulate the risk of acute MI.
Variation in genes encoding MMPs and TIMPs may be associated with cardiovascular disease (10–17), but few studies have examined more than one gene or even more than one SNP.(18–25) This study evaluated the association of SNPs in the genes encoding MMP-1, MMP-2, MMP-3, MMP-9, TIMP-1, TIMP-2, and TIMP-3 with MI among patients undergoing coronary angiography.
Methods
Patient Population
Patients included in this study were drawn from the cardiac catheterization registry of the Intermountain Heart Collaborative Study. This registry includes patients undergoing coronary angiography at tertiary-care hospitals within Utah-based Intermountain Healthcare. Individuals included in this study were evaluated by coronary angiography and donated a DNA sample between August, 1994, and June, 2001. This study was approved by the LDS Hospital Institutional Review Board.
Genetic Variables
MMP genes evaluated in this study included MMP-1 or collagenase I (promoter insertion-deletion SNP rs1799750 [−1607 1G/2G]), MMP-2 or gelatinase A (promoter SNP rs243865 [C-1360T]), MMP-3 or stromelysin I (promoter insertion-deletion SNP rs3025058 [−1613 5A/6A]), MMP-9 or gelatinase B (two SNPs: promoter SNP rs3918242 [C-1562T] and exon 6 SNP rs2664538 [R279Q with G→A]). TIMP genes included in the study were TIMP-1 (exon 5 SNP rs4898 [F124F with T→C]) on the X chromosome, TIMP-2 (promoter SNP rs8179093 [A-596C]), and TIMP-3 (exon 3 SNP rs9862 [H83H with C→T]). SNPs were chosen as those that were the most studied in cardiovascular disease and/or the most common (for MMP-9, the two SNPs were used due to potential LD and joint effect). Genotyping of all SNPs was performed using 5′ exonuclease (Taqman®) chemistry on the ABI Prism® 7000 (Applied Biosystems, Foster City, CA). All assays were validated by direct sequencing using Big Dye v 3.1 terminator chemistry (Applied Biosystems).
Covariables
Covariable data were collected by physician- and patient-report. Covariables included patient age, sex, ethnicity, smoking, hypertension, hyperlipidemia, diabetes, family history of early coronary disease, and high-sensitivity C-reactive protein (hsCRP). Smoking was defined as positive for current smoking or a >10 pack-year history. Hypertension, hyperlipidemia, and diabetes were determined by physician-report based on clinical and laboratory findings and/or current medication therapy, including that hypertension was defined as systolic blood pressure =140 mmHg or diastolic blood pressure =90 mmHg; hyperlipidemia as fasting total cholesterol =200 mg/dL or low-density lipoprotein cholesterol (LDL-C) =130 mg/dL; and diabetes as a fasting glucose =126 mg/dL. Patient-reported family history was present if a first-order relative suffered cardiovascular death, MI, or coronary revascularization before age 55 (males) or 65 (females). Measurement of hsCRP was performed using an immunoturbidimetric method (Roche Diagnostics Corporation).
Primary Study Endpoint: MI
MI events were defined by enzyme measurement or ECG from Intermountain’s electronic record repository. An MI was defined based on biomarker evidence (a CK-MB >6 mg/dl and a CK-MB index >3%) supplemented by ECG criteria and clinical symptoms and signs. Non-MI controls included patients with no CAD and MI-free CAD patients. All patients were included in MI association analyses (N=5,148), with n=1,693 MI patients and n=3,455 MI-free patients.
Secondary Endpoint
Angiographically-determined CAD was defined by physician report from standard coronary angiography. Control patients (n=1,122) free from CAD (i.e., no or minimal [<10%] stenosis) and case patients (n=1,967) with significant CAD (i.e., =1 lesion of ≥70% stenosis) but no MI were evaluated as the CAD phenotype (patients with maximum of 10–69% stenosis were excluded as indeterminate). Since the clinical decision to treat is made based on the presence of flow-limiting lesions, stenoses of 70–100% were considered the most clinically-relevant CAD phenotype.
Statistical Considerations
Due to LD, MMP-1 and MMP-3 SNPs (in the MMP cluster of chromosome 11q21-23) were combined a priori in one variable (MMP-1/-3), as were the two MMP-9 SNPs. For MMP-1 and MMP-3 SNPs, LD was found with D′=0.50 and r2=0.24, while MMP-9 SNPs had D′=0.99 and r2=0.29. Because it is located on the X chromosome, all analyses of TIMP-1 were sex-stratified.
Analyses of SNP associations to MI were performed by the chi-square test. Overall and subpopulation (by sex, smoking, diabetes) analyses were performed. For genetic variables with significant or suggestive associations, multivariable logistic regression was used to calculate adjusted odds ratios (ORs) and 95% confidence intervals (CI) after forced entry of covariables (i.e., age, sex, and cardiac risk factors). Multiplicative MMP × TIMP regression interaction variables tested SNP interactions. Additional models adjusted for hsCRP. Secondary endpoint analyses evaluated SNP associations to CAD status.
Statistical analyses were performed with SPSS (v.15.0, SPSS Inc., Chicago, IL). Associations of inferred haplotypes to MI and CAD were evaluated using SimHap. With Bonferroni correction, two-tailed p values were considered significant at p=0.00833 (correction for multiple comparisons of the six SNP variables), with p=0.05 designated as a suggestive association.
Results
Overall findings for clinical covariables and associations of these variables with MI and CAD are shown in Table 1. All covariables were associated with CAD and most with MI, excepting age and hypertension status. MMP-9 composite genotypes were exceptionally uncommon for CT/RR (n=3) and TT/RQ (n=1), and no patient had TT/RR genotype.
Table 1.
Patient characteristics by MI and CAD status.
| Characteristic | Overall (N=5,148) | MI (n=1,693) | no MI (n=3,455) | CAD* (n=1,967) | no CAD (n=1,122) |
|---|---|---|---|---|---|
| Age (years) | 63±12 | 64±12 | 63±12 | 65±11 | 58±13‡ |
| Sex (male) | 69% | 74% | 66%‡ | 76% | 49%‡ |
| Hypertension | 57% | 59% | 57% | 61% | 46%‡ |
| Hyperlipidemia | 53% | 57% | 51%‡ | 62% | 31%‡ |
| Diabetes | 19% | 21% | 18%† | 22% | 11%‡ |
| Smoking | 23% | 27% | 21%‡ | 25% | 16%‡ |
| Family History | 39% | 42% | 38%† | 43% | 31%‡ |
| hsCRP (mg/L)§ | 2.95 | 5.89 | 2.32‡ | 3.17 | 2.62‡ |
| Ethnicity | |||||
| Asian | 0.9% | 1.3% | 0.7%† | 0.4% | 1.1%† |
| African | 0.6% | 0.7% | 0.5% | 0.5% | 0.6% |
| Hispanic | 2% | 2.3% | 1.7% | 1% | 3%† |
| Native Am. | 0.5% | 0.7% | 0.4% | 0.3% | 0.6% |
| Caucasian | 92% | 92% | 93%¶ | 94% | 90%¶ |
| Other | 0.8% | 0.9% | 0.7% | 0.7% | 0.7% |
| Unknown | 3% | 3% | 3% | 3% | 4%† |
Patients (n=2,059) with MI or with indeterminate CAD (stenoses 10–69%) were excluded from CAD analyses;
p<0.05 and
p<0.001 for comparisons of MI vs. no MI, or CAD vs. no CAD;
N=3,314 for available hsCRP measurements;
referent. Age is expressed as the mean±standard deviation and hsCRP as the median.
Myocardial Infarction
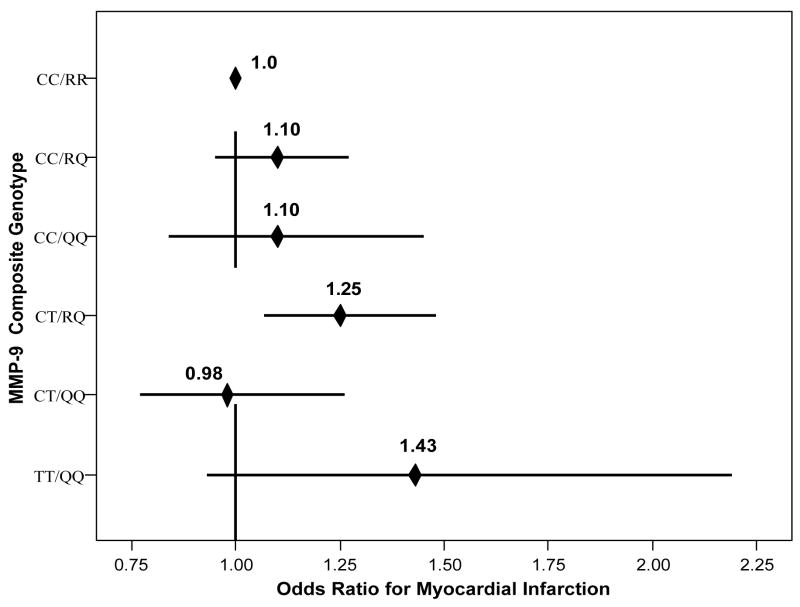
Univariable analysis showed that MMP-9 composite genotypes were associated with MI (Table 2), with significance for CT/RQ genotype (OR=1.25, CI=[1.07,1.48], p=0.007) compared to CC/RR. TT/QQ had increased MI risk (Figure 1), but this was not significant (OR=1.43, CI=[0.93,2.19], p=0.10). No other SNPs were associated with MI (Table 2, Table 3), and no interactions between MMP SNPs and their respective TIMP SNPs were found. Adjustment for age, sex, and risk factors strengthened these associations (Table 4, Model A). Adjustment for hsCRP (Table 4, Model B) retained the elevated risk for CT/RQ, while significance and greatly elevated risk were found for TT/QQ (OR=2.23, p=0.006).
Table 2.
Percentages of genotypes within MI- and CAD-defined groups.
| MI | no MI (p-value*) | CAD | no CAD (p-value*) | |
|---|---|---|---|---|
| MMP | n=1,693 | 3,455 | 1,967 | 1,122 |
| MMP-1/MMP-3 | ||||
| 2G2G/6A6A | 12% | 13% (-----)† | 12% | 16% (-----)† |
| 2G1G/6A6A | 10% | 10% (0.81) | 10% | 9% (0.022) |
| 1G1G/6A6A | 2% | 2% (0.22) | 1% | 2% (0.66) |
| 2G2G/6A5A | 8% | 9% (0.99) | 8% | 9% (0.33) |
| 2G1G/6A5A | 32% | 31% (0.33) | 32% | 28% (0.001)‡ |
| 1G1G/6A5A | 11% | 11% (0.52) | 10% | 12% (0.43) |
| 2G2G/5A5A | 1% | 1% (0.87) | 1% | 1% (0.88) |
| 2G1G/5A5A | 9% | 10% (0.55) | 11% | 8% (0.003)‡ |
| 1G1G/5A5A | 15% | 15% (0.69) | 15% | 14% (0.035) |
| MMP-9 C-1562T/R279Q | ||||
| CC/RR | 40% | 43 (-----)† | 43% | 42% (-----)† |
| CC/RQ | 28% | 27% (0.19) | 27% | 26% (0.62) |
| CC/QQ | 5% | 5% (0.48) | 4% | 5% (0.87) |
| CT/RR | 0.06% | 0.06% § | 0.07% | 0% § |
| CT/RQ | 19% | 16% (0.007)‡ | 16% | 17% (0.84) |
| CT/QQ | 6% | 7% (0.90) | 7% | 7% (0.52) |
| TT/RR | 0% | 0% § | 0% | 0% § |
| TT/RQ | 0% | 0.03% § | 0.03% | 0% § |
| TT/QQ | 2% | 1% (0.10) | 2% | 2% (0.70) |
| MMP-2 C-1360T | ||||
| CC | 57% | 56% (-----)† | 56% | 56% (-----)† |
| CT | 38% | 38% (0.56) | 38% | 39% (0.90) |
| TT | 6% | 6% (0.87) | 6% | 5% (0.93) |
| TIMP-1 F124F (T→C) | ||||
| Female | ||||
| TT | 27% | 31% (-----)† | 30% | 32% (-----)† |
| TC | 52% | 51% (0.21) | 51% | 50% (0.44) |
| CC | 22% | 19% (0.08) | 19% | 18% (0.49) |
| Male | ||||
| T | 54% | 55% (-----)† | 55% | 56% (-----)† |
| C | 46% | 45% (0.42) | 45% | 44% (0.58) |
| TIMP-2 A-596C | ||||
| AA | 38% | 38% (-----)† | 38% | 38% (-----)† |
| AC | 47% | 47% (0.90) | 48% | 45% (0.49) |
| CC | 15% | 15% (0.90) | 14% | 17% (0.08) |
| TIMP-3 H83H (C→T) | ||||
| CC | 26% | 24% (-----)† | 25% | 22% (-----)† |
| CT | 49% | 51% (0.16) | 50% | 53% (0.07) |
| TT | 25% | 25% (0.54) | 25% | 25% (0.45) |
Unadjusted;
Referent;
Significant after correction for multiple testing;
Not tested.
Figure 1.
Plot of the relative odds for the univariable association of MMP-9 composite genotypes to MI.
Table 3.
Percentages of genotypes in individual SNPs for post hoc comparisons of MMP-1, MMP-3, and MMP-9 within MI- and CAD-defined groups.
| MMP | MI | no MI (p-value*) | CAD | no CAD (p-value*) |
|---|---|---|---|---|
| MMP-1 | ||||
| 2G2G | 22% | 23% (-----)† | 21% | 26% (-----)† |
| 2G1G | 50% | 50% (0.56) | 52% | 46% (<0.001) |
| 1G1G | 28% | 27% (0.47) | 26% | 28% (0.22) |
| MMP-3 | ||||
| 6A6A | 24% | 25% (-----)† | 23% | 27% (-----)† |
| 6A5A | 52% | 50% (0.45) | 50% | 48% (0.033) |
| 5A5A | 24% | 26% (0.65) | 27% | 24% (0.012) |
| MMP-9 C-1562T | ||||
| CC | 72% | 75% (-----)† | 75% | 74% (-----)† |
| CT | 26% | 24% (0.06) | 23% | 24% (0.54) |
| TT | 2.1% | 1.6% (0.19) | 2% | 2% (0.89) |
| MMP-9 R279Q | ||||
| RR | 40% | 43% (-----)† | 43% | 42% (-----)† |
| RQ | 47% | 44% (0.026) | 44% | 44% (0.58) |
| 14% | 13% (0.33) | 13% | 14% (0.30) | |
Unadjusted;
Referent.
Table 4.
Regression models for MMP-9 and MI with adjustment for (Model A): age, sex, ethnicity, and cardiac risk factors (hypertension, hyperlipidemia, diabetes, smoking, family history), and (Model B): age, sex, ethnicity, cardiac risk factors, and hsCRP (i.e., Model A + hsCRP).
| Composite Genotypes for C-1562T/R279Q |
Model A | Model B |
|---|---|---|
| OR (95% CI), p-value | OR (95% CI), p-value | |
| CC/RR | 1.0, ----- | 1.0, ----- |
| CC/RQ | 1.12 (0.97, 1.29), 0.13 | 1.13 (0.94, 1.36), 0.20 |
| CC/QQ | 1.08 (0.82, 1.43), 0.60 | 0.99 (0.69, 1.43), 0.96 |
| CT/RQ | 1.27 (1.08, 1.49), 0.005* | 1.32 (1.07, 1.63), 0.009 |
| CT/QQ | 0.98 (0.76, 1.26), 0.87 | 0.99 (0.73, 1.35), 0.99 |
| TT/QQ | 1.48 (0.96, 2.28), 0.08 | 2.23 (1.29, 3.87), 0.004* |
Significant after correction for multiple testing.
Post hoc analysis of MMP-9 SNPs separately (Table 3) showed weak associations for C-1562T (vs. CC, CT: OR=1.14, p=0.06; TT: OR=1.33, p=0.19) and R279Q (vs. RR, RQ: OR=1.15, p=0.026; QQ: OR=1.09, p=0.33).
Analyses of inferred haplotypes showed weak association of the MMP-9 TQ haplotype to MI (haplotypic OR=1.10 [0.98, 1.24], p=0.09) but no association for MMP-1/MMP-3 haplotypes.
Sex strata
Among females, effects were increased for MMP-9 CT/RQ (OR=1.61, p=0.002) and TT/QQ (OR=1.64, p=0.19). For males, MMP-9 results were not as strong as overall for CT/RQ (OR=1.13, p=0.23) or TT/QQ (OR=1.37, p=0.25); however, interaction of MMP-9 and TIMP-1 was suggested for males carrying the variant TIMP-1 C genotype and MMP-9 CT/RQ (OR=1.44, p-interaction=0.021), CT/QQ (OR=1.34, p-interaction=0.029), and TT/QQ (OR=1.70, p-interaction=0.44). No interaction of MMP-9 with sex was found (all p-interaction>0.05).
Smoking strata
Among non-smokers, effect sizes for MMP-9 genotypes were similar to overall (data not shown). Among smokers, though, MMP-9 and TIMP-1 genotypes had considerably elevated effect sizes. For female smokers, TIMP-1 was significant (vs. TT, TC: OR=2.62, p=0.013; CC: OR=3.36, p=0.0079) and MMP-9 had altered risk levels by genotype (OR=0.90, 2.27, 1.51, 1.31, 3.10 for CC/RQ, CC/QQ, CT/RQ, CT/QQ, and TT/QQ, respectively). Male smokers had suggestive association for TIMP-1 (vs. T, C: OR=1.34, p=0.028) and MMP-9 (OR=1.28, 1.19, 1.34, 1.71 [p=0.047], 2.94 [p=0.041], respectively); further, interaction of TIMP-1 with MMP-9 was suggested.
Diabetes strata
Among non-diabetic patients, the MI association of MMP-9 was strengthened, while MMP-9 had a weaker and non-significant effect among diabetics; no other gene was associated with MI based among diabetic status (data not shown).
Secondary Endpoints
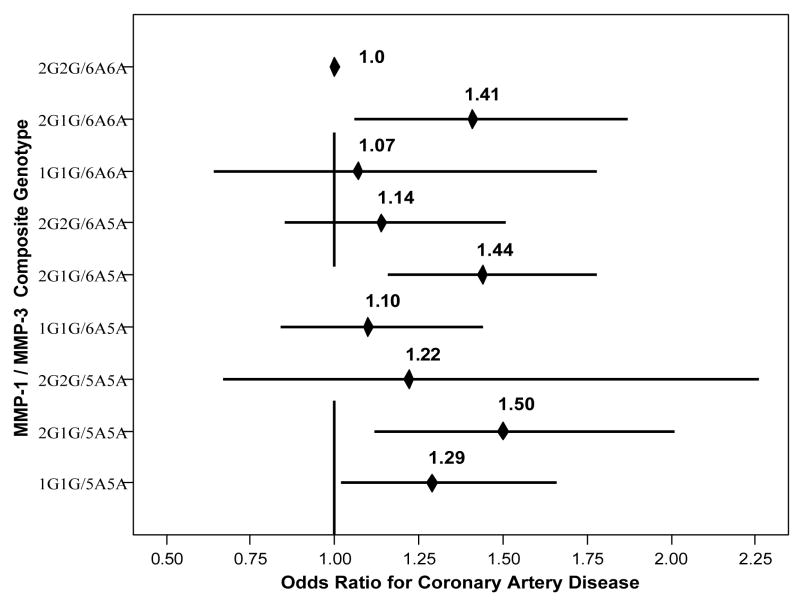
Univariable analyses (Table 2) showed that MMP-1/-3 was associated with CAD (Figure 2), including (compared to 2G2G/6A6A) composite genotypes 2G1G/6A6A (OR=1.45, CI=[1.06,1.99], p=0.022), 2G1G/6A5A (OR=1.49, CI=[1.16,1.90], p=0.001), 2G1G/5A5A (OR=1.64, CI=[1.19,2.26], p=0.003), and 1G1G/5A5A (OR=1.35, CI=[1.02,1.79], p=0.035). Suggestive association was found for TIMP-2 (vs. AA, AC: OR=1.06, p=0.49; CC: OR=0.82, p=0.075). No interactions were found between the MMP SNPs and their respective TIMPs. Adjustment for covariables, including hsCRP, had only modest effects on these associations although associations became suggestive (Table 5). No interaction of sex with MMP-1/MMP-3 or the individual SNPs was found.
Figure 2.
Plot of the relative odds for the univariable association of MMP-1/MMP-3 composite genotypes to CAD.
Table 5.
Regression models for MMP-1/MMP-3 combined genotypes in association with CAD vs. No-CAD (excluding MI patients) with adjustment for (Model A): age, sex, and ethnicity, and (Model B): age, sex, ethnicity, cardiac risk factors (hypertension, hyperlipidemia, diabetes, smoking, family history), and hsCRP (i.e., Model A + cardiac risk factors + hsCRP).
| Composite Genotypes for MMP-1/MMP-3 |
Model A | Model B |
|---|---|---|
| OR (95% CI), p-value | OR (95% CI), p-value | |
| 2G2G/6A6A | 1.0, ----- | 1.0, ----- |
| 2G1G/6A6A | 1.16 (0.79, 1.71), 0.45 | 1.33 (0.88, 2.02), 0.18 |
| 1G1G/6A6A | 1.06 (0.51, 2.22), 0.87 | 1.15 (0.51, 2.61), 0.74 |
| 2G2G/6A5A | 1.26 (0.85, 1.88), 0.26 | 1.22 (0.80, 1.87), 0.35 |
| 2G1G/6A5A | 1.29 (0.96, 1.75), 0.10 | 1.26 (0.92, 1.74), 0.16 |
| 1G1G/6A5A | 0.92 (0.64, 1.35), 0.68 | 0.90 (0.60, 1.34), 0.60 |
| 2G2G/6A5A | 0.71 (0.31, 1.62), 0.42 | 0.70 (0.29, 1.70), 0.43 |
| 2G1G/6A5A | 1.44 (0.97, 2.13), 0.07 | 1.46 (0.96, 2.22), 0.08 |
| 1G1G/6A5A | 1.21 (0.85, 1.71), 0.29 | 1.21 (0.83, 1.75), 0.32 |
Inferred haplotypes of MMP-1/MMP-3 showed potential associations for the 2G5A haplotype (haplotypic OR=1.20 [1.01, 1.42], p=0.032) and the 1G5A haplotype (haplotypic OR=1.17 [1.03, 1.32], p=0.012), but not for MMP-9 haplotypes.
Post hoc analysis of MMP-1 and MMP-3 SNPs separately for CAD (Table 3) found significance for the MMP-1 heterozygote only (vs. 2G2G, 2G1G: OR=1.40, p<0.001; 1G1G: OR=1.14, p=0.22) and a suggestive association for MMP-3 (vs. 6A6A, 6A5A: OR=1.22, p=0.033; 5A5A: OR=1.31, p=0.012).
Results of SNP comparisons between MI cases and non-MI CAD controls (excluding non-CAD controls) were similar to the overall MI findings for MMP-9 (Table 6) and for the other SNPs (data not shown); for MI cases compared to non-MI non-CAD controls (excluding CAD controls), results were also similar to overall although the effect sizes of MMP-1/-3 genotypes were markedly higher (data not shown).
Table 6. MI patients vs. MI-free CAD controls.
Regression model for MMP-9 composite genotypes in association with MI after adjustment (compare to Table 4, model A).
| MMP-9 genotype | OR (95% CI), p-value |
|---|---|
| CC/RR | 1.0, ----- |
| CC/RQ | 1.08 (0.92, 1.26), 0.38 |
| CC/QQ | 1.21 (0.88, 1.67), 0.23 |
| CT/RQ | 1.30 (1.08, 1.56), 0.006* |
| CT/QQ | 0.96 (0.73, 1.27), 0.80 |
| TT/QQ | 1.37 (0.84, 2.25), 0.21 |
Significant after correction for multiple testing.
Discussion
Previous studies have examined MI and CAD associations of SNPs in the genes for MMP-1, MMP-2, MMP-3, and MMP-9.(10–24) Many of these evaluated only the MMP-3 5A/6A SNP (10–14) or a SNP in another gene (15–17). Several have evaluated multiple MMP or TIMP SNPs.(18–25) Some of the studies failed to replicate prior findings (15,16,18–25), perhaps because of differences in study endpoints, small population sizes, differing study designs (e.g., cohort vs. matched controls), failure to account for epistasis (gene interactions: e.g., MMP-1 and MMP-3), and unmeasured intra-genic allelic heterogeneity.
Phenotype Selection and Definition
The most studied MMP SNP is the MMP-3 gene 5A/6A variant (10–14,18,19,21–23) and only a few have not found a disease association (19). While all studies of MMP-3 evaluated the 5A/6A promoter SNP, only two have evaluated other MMP-3 SNPs and in those the 5A/6A SNP was as good or better at risk stratification than the others.(22,23) Prior studies of MMP-3 have found increased risk associated with both the 5A (11,13,14,18,21,22) and the 6A (10,12,13,23,26,27) allele, or no association (19), but the phenotypes studied therein included MI (11,14,21–23,27), CAD presence (23), CAD progression (10,12), coronary aneurysm (18), patient-reported symptoms and CAD history (19), cardiac events (13), and CAD score (14).
The largest population evaluated for MMP genes to date, this study did not find an association of composite MMP-1/-3 genotypes with MI, or of individual SNPs in post hoc analyses. MMP-1/-3, though, was associated with the secondary endpoint of CAD. MMP-9, the second most studied MMP gene (15–17,20), was associated with MI but not CAD. In much smaller prior studies, the MMP-9 C-1562T promoter SNP (15–17,20) and the R279Q SNP (20) were found to be associated with CAD (15,17,20) but not MI (15,16).
Because MMPs are implicated in plaque rupture (6,8), this study evaluated MI as the primary endpoint. Phenotypic differences may account in part for the failure of the present study to detect the same associations as prior studies. Since the current study utilized the most relevant clinical definitions of MI and CAD (and studied >5,000 patients), but results differed from some prior studies, this points toward the careful selection of phenotype as a study endpoint for each candidate gene.
Because the definition of clinical phenotypes may be of substantive importance for detection of genetic associations, this study attempted to test association of SNPs to MI events. Genes involved in plaque destabilization and MI may not be the same genes that initiate or enlarge a coronary lesion, thus non-MI controls (with or without CAD) were compared with MI cases to test associations to the MI event. As a separate, secondary evaluation, CAD presence was compared (with exclusion of MI patients) with coronary stenosis-free controls.
Genetic and Allelic Heterogeneity
The MMP-9 C-1562T and R279Q SNPs were in moderate LD as measured by r2 and in strong LD when haplotype construction was considered. As previously described (20), three of the nine possible composite genotypes were essentially missing, perhaps due to selection against the T-R haplotype prior to birth via non-CAD mechanisms or because most patients with the T-R haplotype do not survive an MI long enough for admission to a hospital. These interesting points, the large sample size, and the significant (p<0.008) stratification of risk by MMP-9 and MMP-1/-3 composite genotypes suggest that multiple SNP analysis provides greater ability to decipher genetic effects. The strong associations for composite genotype variables (i.e., unphased haplotypes) suggest either that haplotypes underlie the associations or that the SNPs evaluated herein may imperfectly account for one or more unmeasured causal SNPs in each gene that are in LD with the tested SNPs. It may also be that important SNP interactions exist within the genes.
Recent attempts at high-resolution evaluation of MMP-1 and MMP-3 genes (23,24) corroborated prior MMP-3 findings but only found haplotype effects in MMP-1 and no association for the 1G/2G SNP. Those two studies, though, were most interesting because of the state-of-the-art genetic epidemiologic methods that were attempted. Such methodology comprehensively evaluates all variation within each candidate gene to better elucidate associations through the use of tagging SNPs and tagging SNP haplotypes.(28) Tagging SNPs account for the variation within a gene while reducing genotyping costs and the need for statistical correction for multiple comparisons.
The findings of the current study and prior work on MMP and TIMP genes suggest that each should be comprehensively evaluated. This includes the genes not showing associations because their findings may have resulted from a failure to detect association (e.g., due to use of the wrong SNP). It also includes the MMP-1 and MMP-3 genes because limited diversity (only 60 [24] and 40 [23] chromosomes, respectively) was used to discover SNPs in the two high-resolution studies, they did not use tagging SNPs, and they did not correct for multiple comparisons. Other cardiovascular studies have recently used an incarnation of this methodology but only investigated clinical phenotypes in afterthought.(29,30)
Finally, prior studies of MMP and TIMP genes studied populations of around one hundred (10,17,25), multiple hundreds (11,12,13,16,18,19,21), between 1,000–1,700 participants.(14,15,20,22–24,27) While the current study of >5,000 patients did not replicate all of those prior studies’ findings, it improved on their sample sizes by 3- to 50-fold. Evaluation of many thousands of patients, as in this study, should help to reduce the variability between study findings. The exception to those smaller sample sizes was a study by Yamada (26) wherein the MMP-3 6A variant was found to predict elevated MI risk among females (no non-MI CAD patients were included), but stark differences (perhaps due to ethnic composition) in allele frequencies and potential differences in LD structure may account for differences in the current study’s findings.
Limitations
This study may be limited by biases arising in observational investigation, including that patients may have been at higher CAD risk than the general population. Several traditional risk factors, though, were adjusted for in multivariable analyses. The study may also be limited because the ethnic composition was primarily Caucasian and the results may or may not apply to other ethnic groups.
While no cellular expression evaluations were performed for the MMP or TIMP variants herein, prior evaluations have demonstrated decreased transcription in MMP-1 among 1G variant carriers (19,24), higher promoter activity in MMP-3 for the 5A variant (11), and greater transcription in MMP-9 for the T allele of C-1562T and the Q variant of R279Q.(15) This may suggest an explanation for the MI association of MMP-9 variants found in this study and presents an intriguing combination of effect from MMP-1/MMP-3 variants.
A strength of the study is that the actual coronary status of each patient was known from the gold-standard test (i.e., angiography), thus eliminating the phenotypic uncertainty inherent in many population-based genetic studies where the study endpoint is a surrogate CAD measure (e.g., symptoms) or only acute events are measured.
Conclusions
This study found associations of MMP-9 composite genotypes with MI and also suggested associations of MMP-1 and MMP-3 genotypes with CAD. This builds on prior reports from a multitude of disparate association studies by evaluating a much larger population (by far the largest to date for MMP genes) and multiple MMP and TIMP genes related by a common biologic pathway. The differing findings for MI and CAD endpoints suggest that selection and definition of phenotypes should be standardized. The study findings also suggest that the evaluated SNPs only approximately account for intra-genic variation in these MMP and TIMP genes and that future research should focus on complete characterization of the genetic variation within each of these genes, with subsequent testing for associations with MI and/or CAD.
Acknowledgments
This study was funded by American Heart Association Western States Affiliate Fellowship 0415023Y (B.D.H.), and by NIH grants CA098364 (N.J.C.) and HL071878 (J.L.A. & J.F.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340(2):15–26. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 2.Wilson PW, D’Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–47. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 3.Nora JJ, Lortscher RH, Spangler RD, et al. Genetic epidemiologic study of early-onset ischemic heart disease. Circulation. 1980;61:503–8. doi: 10.1161/01.cir.61.3.503. [DOI] [PubMed] [Google Scholar]
- 4.Barrett-Connor E, Khaw KT. Family history of heart attack as an independent predictor of death due to cardiovascular disease. Circulation. 1984;69:1065–9. doi: 10.1161/01.cir.69.6.1065. [DOI] [PubMed] [Google Scholar]
- 5.van der Wal AC, Becker AE, van der Luce CM, et al. Site of intimal rupture or erosion of thrombosis in coronary atherosclerotic plaques is characterized by an inflammatory process irrespective of the dominant plaque morphology. Circulation. 1994;89:36–44. doi: 10.1161/01.cir.89.1.36. [DOI] [PubMed] [Google Scholar]
- 6.Libby P. Coronary artery injury and the biology of atherosclerosis: inflammation, thrombosis, and stabilization. Am J Cardiol. 2000;86(suppl):3J–9J. doi: 10.1016/s0002-9149(00)01339-4. [DOI] [PubMed] [Google Scholar]
- 7.Libby P. What have we learned about the biology of atherosclerosis? The role of inflammation. Am J Cardiol. 2001;88(suppl):3J–6J. doi: 10.1016/s0002-9149(01)01879-3. [DOI] [PubMed] [Google Scholar]
- 8.Galis ZS, Sukhova GK, Lark MW, et al. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1994;94:2493–2503. doi: 10.1172/JCI117619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orbe J, Fernandez L, Rodriguez JA, et al. Different expression of MMPs/TIMP-1 in human atherosclerotic lesions. Relation to plaque features and vascular bed. Atherosclerosis. 2003;170:269–76. doi: 10.1016/s0021-9150(03)00251-x. [DOI] [PubMed] [Google Scholar]
- 10.Ye S, Watts GF, Mandalia S, et al. Preliminary report: genetic variation in the human stromelysin promoter is associated with progression of coronary atherosclerosis. Br Heart J. 1995;73:209–15. doi: 10.1136/hrt.73.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terashima M, Akita H, Kanazawa J, et al. Stromelysin promoter 5A/6A polymorphism is associated with acute myocardial infarction. Circulation. 1999;99:2717–9. doi: 10.1161/01.cir.99.21.2717. [DOI] [PubMed] [Google Scholar]
- 12.Humphries SE, Luong L, Talmud PJ, et al. The 5A/6A polymorphism of the stromelysin-1 (MMP-3) gene predicts progression of angiographically determined coronary artery disease in men in the LOCAT gemfibrozil study. Atherosclerosis. 1998;139:49–56. doi: 10.1016/s0021-9150(98)00053-7. [DOI] [PubMed] [Google Scholar]
- 13.de Maat MPM, Jukema JW, Ye S, et al. Effect of the stromelysin-1 promoter on efficacy of pravastatin in coronary atherosclerosis and restenosis. Am J Cardiol. 1999;83:852–6. doi: 10.1016/s0002-9149(98)01073-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu PY, Li YH, Chan SH, et al. Genotype-phenotype association of matrix metalloproteinase-3 polymorphism and its synergistic effect with smoking on the occurrence of acute coronary syndrome. Am J Cardiol. 2006;98:1012–7. doi: 10.1016/j.amjcard.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Ye S, Herrmann SM, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99:1788–94. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Warzecha D, Wilcken D, et al. Polymorphism in the gelatinase B gene and the severity of coronary arterial stenosis. Clin Sci (Lond) 2001;101:87–92. [PubMed] [Google Scholar]
- 17.Cho HJ, Chae IH, Park KW, et al. Functional polymorphism in the promoter region of the gelatinase B gene in relation to coronary artery disease and restenosis after percutaneous coronary intervention. J Hum Genet. 2002;47:88–91. doi: 10.1007/s100380200006. [DOI] [PubMed] [Google Scholar]
- 18.Lamblin N, Bauters C, Hermant X, et al. Polymorphisms in the promoter regions of MMP-2, MMP-3, MMP-9, and MMP-12 genes as determinants of anuerysmal coronary artery disease. J Am Coll Cardiol. 2002;40:43–8. doi: 10.1016/s0735-1097(02)01909-5. [DOI] [PubMed] [Google Scholar]
- 19.Ye S, Gale CR, Martyn CN. Variation in the matrix metalloproteinase-1 gene and risk of coronary heart disease. Eur Heart J. 2003;24:1668–71. doi: 10.1016/s0195-668x(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 20.Morgan AR, Zhang B, Tapper W, et al. Haplotypic analysis of the MMP-9 gene in relation to coronary artery disease. J Mol Med. 2003;81:321–6. doi: 10.1007/s00109-003-0441-z. [DOI] [PubMed] [Google Scholar]
- 21.Nojiri T, Morita H, Imai Y, et al. Genetic variations of matrix metalloproteinase-1 and –3 promoter regions and their associations with susceptibility to myocardial infarction in Japanese. Int J Cardiol. 2003;92:181–6. doi: 10.1016/s0167-5273(03)00100-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Huang J, Chen J, et al. Haplotype analysis of the matrix metalloproteinase 3 gene and myocardial infarction in a Chinese Han population. The Beijing atherosclerosis study. Thromb Haemost. 2004;92:867–73. doi: 10.1160/TH04-03-0192. [DOI] [PubMed] [Google Scholar]
- 23.Beyzade S, Zhang S, Wong Y, et al. Influences of matrix metalloproteinase-3 gene variation on extent of coronary atherosclerosis and risk of myocardial infarction. J Am Coll Cardiol. 2003;41:2130–7. doi: 10.1016/s0735-1097(03)00482-0. [DOI] [PubMed] [Google Scholar]
- 24.Pearce E, Tregouet DA, Samnegard A, et al. Haplotype effect of the matrix metalloproteinase-1 gene on risk of myocardial infarction. Circ Res. 2005;97:1070–6. doi: 10.1161/01.RES.0000189302.03303.11. [DOI] [PubMed] [Google Scholar]
- 25.Krex D, Rohl H, Konig IR, et al. Tissue inhibitor of metalloproteinases-1, -2, and -3 polymorphisms in a white population with intracranial aneurysms. Stroke. 2003;34:2817–21. doi: 10.1161/01.STR.0000099966.51485.5F. [DOI] [PubMed] [Google Scholar]
- 26.Yamada Y, Izawa H, Ichihara S, et al. Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med. 2002;347:1916–23. doi: 10.1056/NEJMoa021445. [DOI] [PubMed] [Google Scholar]
- 27.Hirashiki A, Yamada Y, Murase Y, et al. Association of gene polymorphisms with coronary artery disease in low-or high-risk subjects defined by conventional risk factors. J Am Coll Cardiol. 2003;42:1429–37. doi: 10.1016/s0735-1097(03)01062-3. [DOI] [PubMed] [Google Scholar]
- 28.Horne BD, Carlquist JF, Cannon-Albright LA, et al. High-resolution characterization of linkage disequilibrium structure and selection of tagging single nucleotide polymorphisms: application to the cholesteryl ester transfer protein gene. Ann Hum Genet. 2006;70:524–35. doi: 10.1111/j.1469-1809.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 29.Kathiresan S, Larson MG, Vasan RS, et al. Contribution of clinical correlates and 13 C-reactive protein gene polymorphisms to interindividual variability in serum C-reactive protein level. Circulation. 2006;113:1415–23. doi: 10.1161/CIRCULATIONAHA.105.591271. [DOI] [PubMed] [Google Scholar]
- 30.Crawford DC, Sanders CL, Qin X, et al. Genetic variation is associated with C-reactive protein levels in the third National Health and Nutrition Examination Survey. Circulation. 2006;114:2458–65. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]