Abstract
The definition of European population genetic substructure and its application to understanding complex phenotypes is becoming increasingly important. In the current study using over 4000 subjects genotyped for 300 thousand SNPs we provide further insight into relationships among European population groups and identify sets of SNP ancestry informative markers (AIMs) for application in genetic studies. In general, the graphical description of these principal components analyses (PCA) of diverse European subjects showed a strong correspondence to the geographical relationships of specific countries or regions of origin. Clearer separation of different ethnic and regional populations was observed when northern and southern European groups were considered separately and the PCA results were influenced by the inclusion or exclusion of different self-identified population groups including Ashkenazi Jewish, Sardinian and Orcadian ethnic groups. SNP AIM sets were identified that could distinguish the regional and ethnic population groups. Moreover, the studies demonstrated that most allele frequency differences between different European groups could be effectively controlled in analyses using these AIM sets. The European substructure AIMs should be widely applicable to ongoing studies to confirm and delineate specific disease susceptibility candidate regions without the necessity to perform additional genome-wide SNP studies in additional subject sets.
Keywords: ancestry informative markers, population substructure, complex genetic disease, population stratification
INTRODUCTION
Over the last several years there has been substantial progress in using genotypes to ascertain population genetic substructure and apply this information to association testing [1–9]. These studies have been advanced by the availability of efficient platforms for genotyping several hundred thousand SNPs, increased efforts in sampling various population groups, and application of both highly supervised (clustering algorithms) and largely unsupervised methods for analyzing high dimensional data (i.e. genotypes in many individuals)[1,3,6]. The results of such studies in European and European American populations have enabled the description of relationships among various European ethnic groups and the ability to use these relationships to better inform association studies by accounting for population stratification differences between cases and control groups without the necessity to utilize family based methods such as transmission disequilibrium testing [10–13]. The current study was undertaken to further define the relationships among European population groups and to use this information in identifying and testing SNP sets enriched for population substructure information.
Our initial description of European population substructure primarily defined a single axis of variation (north/south) using seven thousand genome-wide SNPs [10]. Subsequently, studies by multiple groups, including our own, have utilized principal components analyses (PCA) or multidimensional scaling to further define European population substructure using several hundred thousand SNPs that are present in genome-wide panels [11–16]. These mathematical methods reduce higher dimensional data (each individual genotype) in to lower dimensions based on patterns of identity by descent relationships. In particular, a recent study using PCA of ~200,000 SNP genotypes has shown that with the exclusion of certain outliers there is a remarkable concordance between the first two principal components and geographical position of the country of origin for the included subject samples [15]. The current study uses different subjects including population groups that were not included in previous studies adds further support to this observation. In addition our study shows how inclusion or exclusion of particular ethnic groups and regions alter the PCA graphical description and evaluation of relationships among European populations. Furthermore, the current study shows that analyses of geographically or genotyped defined sub-regions (e.g. southern European populations) may allow a clearer evaluation of ancestry information and differences that may be important in evaluating association studies or defining homogeneous ethnic groups. Notably our analyses included Middle Eastern population groups. These population groups are more closely related to European populations than South Asian population groups [14,17] and are more closely related to southern European groups as shown here. In addition, individuals of origin in the Middle East or individuals with mixed European/Middle Eastern heritage may self-identify as European or non-Hispanic white for different sample set collections or demographic characterizations.
The practical aspects of applying population substructure to association testing have recently been reviewed [18,19]. For several different study designs the ability to define population substructure using specific smaller sets of SNPs rather than tens or hundreds of thousands of genome-wide SNPs may be advantageous in: 1) pre-selecting population members before genome-wide SNP studies, 2) limited testing of candidate SNPs, and 3) fine mapping critical chromosomal regions using additional samples not included in initial genome-wide scans. Previously, several groups have identified European ancestry informative markers (AIMs) that can partially address population stratification. In the current study, we have identified and examined SNP sets for ascertaining more subtle population stratification in both northern and southern European populations. The results provide strong support for the application of these SNP marker sets to genetic studies of European populations.
MATERIALS AND METHODS
Populations studied
For European substructure studies presented in Figures 1–3, the populations including those from the Human Genome Diversity Panel (HGDP), HapMap, the I-control database, Italian, Spanish, Swedish, and European Americans. The HGDP, HapMap, and I-control database genotypes were available from online databases. These included HapMap subjects (48 CEU), HGDP subjects (14 Orcadian, 28 Sardinian, 13 Northern Italian, 8 Tuscan) and 1488 subjects from Childrens Hospital of Philadelphia from the I-ControlDB (www.illumina.com/iControlDB, Illumina, San Diego, CA). Genotypes from other HGDP subjects (20 Druze, 23 Bedouin, 22 Palestinian, 13 Russian, 12 Basque, 12 Adygei) and 255 European American neurodevelopmental controls were obtained from the NIH Laboratory of Neurogenetics (http://neurogenetics.nia.nih.gov/paperdata/public/). The sample set also used 591 Swedish genotypes (collected by L.K.). Additional samples that were genotyped included 12 Spanish samples (collected by PV), and 1873 European Americans that were recruited as part of the New York Cancer Project (NYCP); a prospective longitudinal study[20]. For a subset of the NYCP participants complete 4 grandparent information was available and used in graphic depictions (see Figure 1 legend). All of the subjects (4446) were included in Figure 1. Figures 2 (185 subjects) and 3 (213 subjects) included only those individuals from specified groups as indicated in the legend.
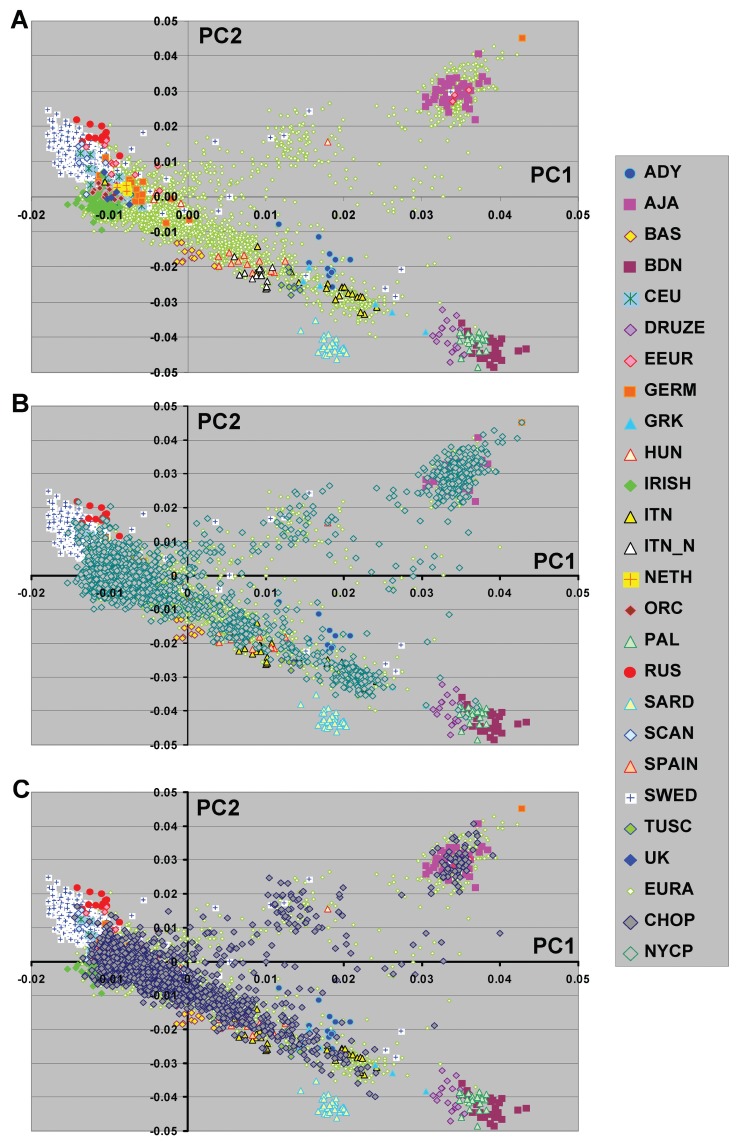
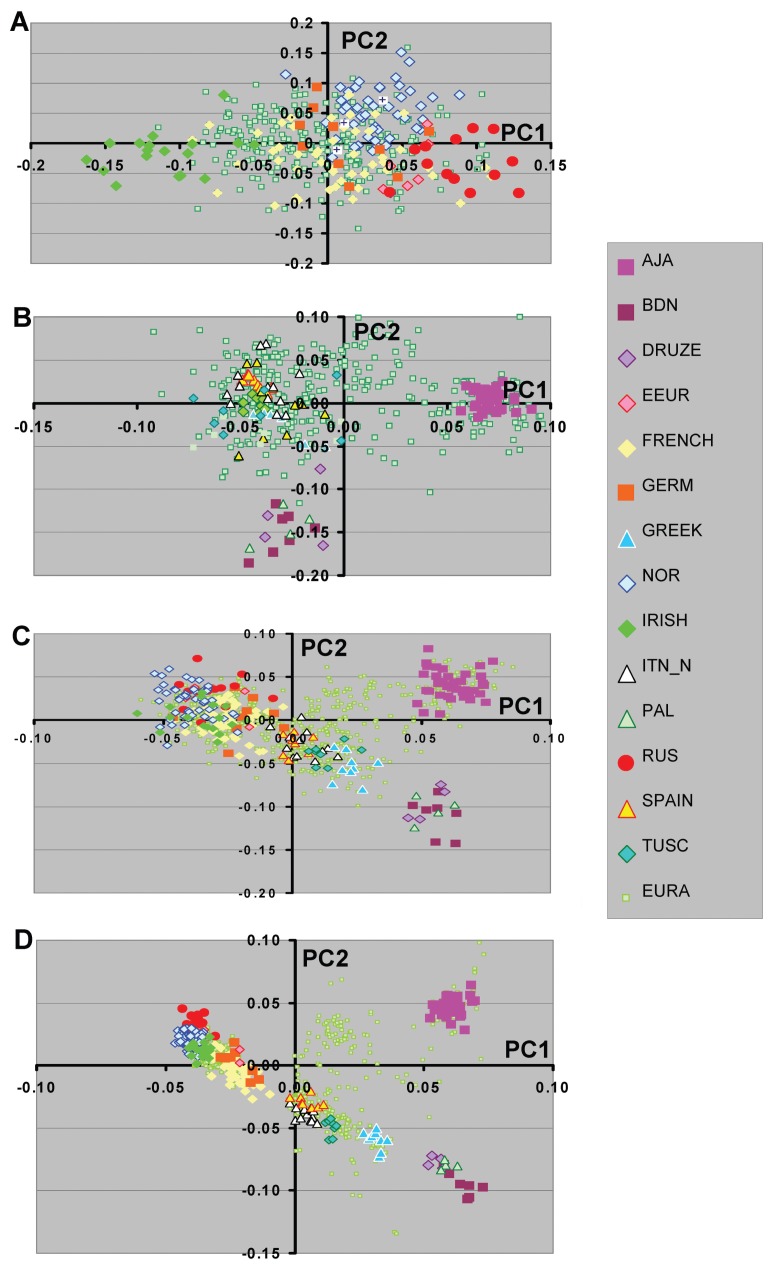
Figure 1.
Principal component analyses of substructure in a diverse set of subjects of European descent. Graphic representation of the first two PCs based on analysis with >250K SNPs are shown. Color code shows subgroup of subjects for each population group. The subjects included Adygei (ADY, 12 subjects), Ashkenazi Jewish American (AJA, 40 subjects), Basque (BAS, 12 subjects), Bedouin (BDN, 23 subjects), CEPH European American (CEU, 48), Druze (20 subjects), Eastern European American (EEUR, 11 subjects), German American (GERM, 17 subjects), Greek American (GRK, 7), Hungarian American (HUN, 4 ), IRISH (84 subjects), Italian American (ITN, 20 subjects), northern Italian (ITN_N, 13 subjects), Dutch American (NETH, 3 subjects), Orcadian (ORC, 14 subjects), Palestinian (PAL, 22 subjects), Russian (RUS, 13 subjects), Sardinian (SARD, 28 subjects), Scandinavian American (SCAN, 6 subjects ), Spanish (SPAIN, 12 subjects), Swedish (SWED, 591 subjects), Tuscany (TUSC, 8 subjects), and United Kingdom American (UK, 5 subjects). Each of the specific country or ethnic color coded origins had consistent 4 grandparent origin information. The total number of individuals in this analysis was 4446. In panel A European Americans (EURA) without 4 grandparental information are shown (contains both NYCP and CHOP). Panels B and C illustrate the distribution of the EURA from NYCP (1873 subjects) and CHOP (1488 subjects), respectively.
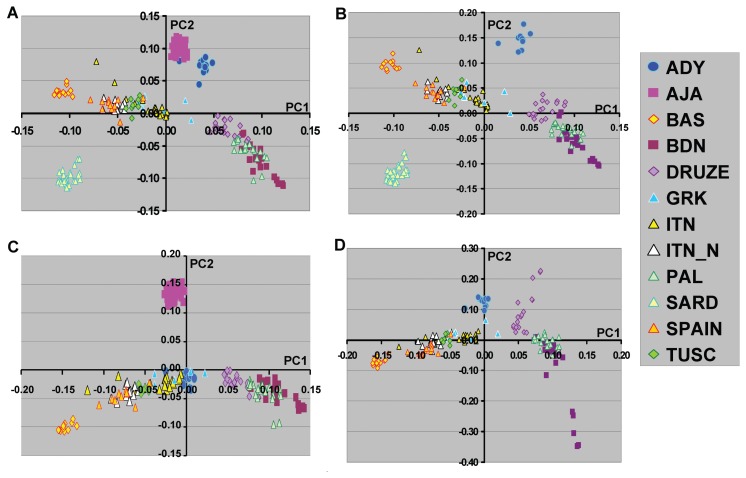
Figure 3.
Principal component analyses of Southern European populations. A, All subjects groups. B, PCA analysis without Ashkenazi Jewish American (AJA). C, PCA without the Sardinian (SARD) group. D, PCA without AJA and SARD. The subject numbers were the same as those indicated in Figure 1.
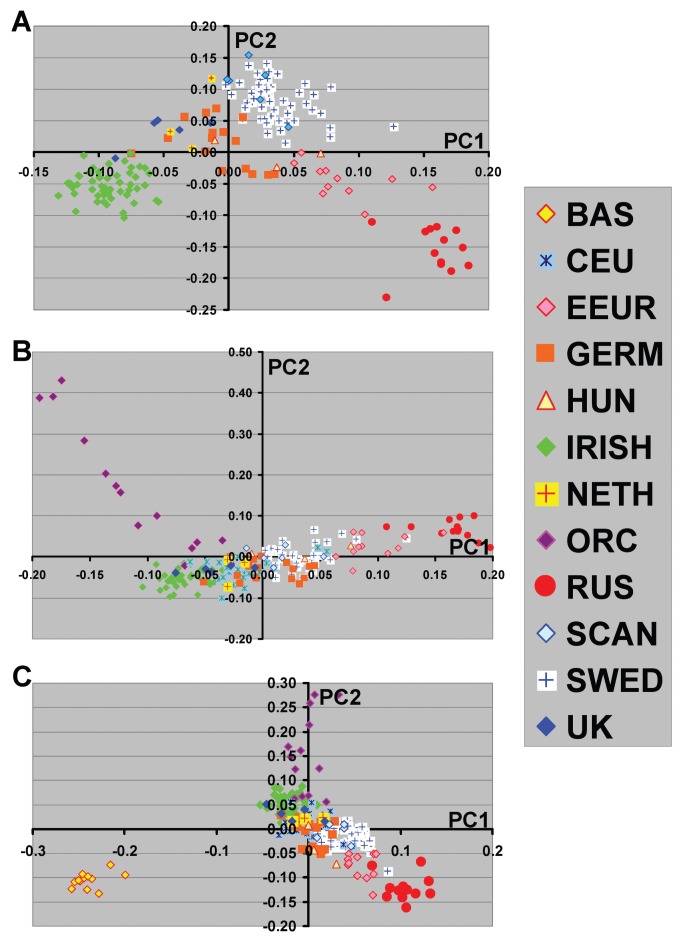
Figure 2.
Principal component analyses of Northern European populations. The color coded group membership is shown with the symbols corresponding to Figure 1 legend information. The subject sizes were as shown in Figure 1 with the exception of the Swedish group for which the sample size was reduced to 40 subjects. A, Northern European population groups without inclusion of Orcadian (ORC), CEU and Basque (BAS) subjects. B, and C, show PCA results when either ORC and CEU or BAS and CEU groups are added. Inclusion or exclusion of CEU did not affect the PCA pattern (data not shown).
For the testing of ESAIMs the study also included samples from 139 French (H-J.G.) and 240 Norwegian (collected by L.H.) subjects. Finally, genotypes from South Asian HGDP samples (7 Burusho and 15 Balochi) were used in some analyses and were obtained from NIH Laboratory of Neurogenetics (http://neurogenetics.nia.nih.gov/paperdata/public/)
For all European and European American subjects, blood cell samples were obtained from all individuals, according to protocols and informed-consent procedures approved by institutional review boards, and were labeled with an anonymous code number linked only to demographic information.
It should be noted that the current set of subjects has relatively few individuals of known Eastern European ancestry (11 subjects) and no subjects of known self identified ancestry from many specific European countries or ethnicity (e.g. southern Slavic population groups). However, the large number of subjects from both NYCP (1832 individuals) and CHOP (1487 individuals) are likely to provide good representation of most European population groups as well as individuals with mixed European heritage. For the NYCP participants, many have partial grandparent origin information and these show representation from all regions of Europe ([10,12], and unpublished information).
Genotyping
Genotyping was performed using a 300K Illumina array according to the Illumina Infinium 2 assay manual (Illumina, San Diego), as previously described [21].
Data Filters
SNPs and individual samples with less than 90% complete genotyping information from any data set were excluded from analyses. SNPs that showed extreme deviation from Hardy Weinberg equilibrium (p<0.00001) in individual population groups were also excluded from analysis. The relatively strict HW criterion was chosen to minimize genotyping error rates and potential artifacts introduced by differences between array lots and genotyping sources (i.e. data derived from multiple laboratories). These filters resulted in a total of 270 K autosomal SNPs that were used for these studies. In addition, individuals with evidence of >10 % contribution from other continents with the exception of the South Asian subjects were excluded from further study. This was performed using 128 continental AIMS[22]. Samples were also filtered for possible cryptic relationships using the PLINK program [6].
Statistical Analyses
Fst and Fis was determined using Genetix software[23] that applies the Weir and Cockerham algorithm [24]. A measure of informativeness for each SNP (In) was determined using an algorithm previously described [25]. Hardy-Weinberg equilibrium was determined using HelixTree 5.0.2 software (Golden Helix, Bozeman, MT, USA). Linkage disequilibrium (LD) was determined using Haploview.
Population structure was examined using STRUCTURE v2.1[1,9] using parameters and AIMs previously described [22]. Briefly, each analysis was performed without any prior population assignment and was performed at least 3 times with similar results using >200,000 replicates and >100,000 burn-in cycles under the admixture model. For all analyses reported, we used the “infer α” option with a separate α estimated for each population (where α is the Dirichlet parameter for degree of admixture). Runs were performed under the λ = 1 option where λ estimates the prior probability of the allele frequency and is based on the Dirichlet distribution of allele frequencies. This analysis was performed to exclude individuals with evidence of substantial continental admixture from Europe, Africa or the American continent (see Data Filters). STRUCTURE analysis was also used for dividing the European population groups into North European and South European population groups for the correlation studies using 192 ns-ESAIMS[12]. The STRUCTURE analysis of self-identified groups using these ESAIMs (Supplemental Figure 1) also corresponds to PCA results. For the correlation studies we used a cut-off of >0.8 membership in Pop 1 for North European population groups and < 0.8 membership in Pop1 (or >0.2 membership in Pop 2) for the South European populations.
PCA was performed using the EIGENSTRAT statistical package[3]. All analyses were performed after deleting the MHC region on chromosome 6, and known regions of common inversions on chromosome 8, 11 and 17, since regions of high linkage disequilibrium can overly influence PCA results [12].
Selection of European Substructure AIMs
Several strategies were investigated to identify SNPs for substructure information. These included using an algorithm for informativeness (In)[25], and SNP scores from PCA as well as using either the entire population studied or selecting particular disparate population groups. The most effective strategy in our hands was to combine set of markers including those that were previously selected for distinguishing population groups along a north/south axis [12] with additional SNP sets chosen using In values between particular northern European groups or particular southern European groups. For the northern ESAIMs we combined a previous set of SNPs [12] with additional SNP sets chosen from Irish/Swedish or Irish/central Europe analyses. For these groups the PCA results were used to select subjects that matched known 4 grandparent Irish, 4 grandparent central European (German and French). Each of these selections included a minimum of 200 subjects from each group. For the southern European ESAIMs we utilized AJA/Arab subjects for the SNP selection. For this selection the AJA included 200 subjects and the Arab subjects included 50 subjects (16 Druze, 16 Bedouin and 18 Palestinian). Finally, each SNP set was screened to remove any SNPs with strong LD (r2 >0.8) and those were not compatible with I-select (Illumina) genotyping platform. The ESAIMs are provided in Supplemental Table 1. Note that the EUROset1 SNPs contain 85% (1037/1212) of the ESAIM SNPs used in our previous studies [12].
All supplementary materials are available online at www.molmed.org
RESULTS
Population Differentiation among European Populations
To examine similarities and differences in population differentiation among European and closely related populations paired Fst values were determined between 18 population groups that were typed with genome-wide SNP arrays (see Methods). The studies included genotypes derived from HGDP[14], and samples collected in Europe and European American participants (see Methods). The Fst values (Table 1) were obtained using three random non-overlapping sets of 3500 SNPs distributed over the autosomal genome (minimum of 50 kb distance between SNPs). The small differences in these independent samplings (mean SD = 0.0009; median SD =0.0008) indicate that this approach resulted in good estimations of paired Fst values. In general, Fst values corresponded to geographical relationships with smaller values between population groups with origins in neighboring countries/regions (e.g. Tuscan/Greek, Fst = 0.001) compared with those from very different regions in Europe (e.g. Russian/Palestinian, Fst = 0.020) similar to previous studies [10]. Ashkenazi Jewish participants showed smaller paired Fst values with southern European populations (e.g. Ashkenazi/Italian, Fst =0.004) than with northern populations (e.g. Ashkenazi/Swedish, Fst = 0.0120). All of the intra-European paired Fst values were nearly an order of magnitude smaller than those observed in our previous studies when intercontinental groups (e.g. Europe and East Asia) are determined (e.g. Han Chinese/Swedish, Fst = 0.11) [17]. However, there is substantial overlap in Fst values comparing European and Middle East groups and those between European and South Asian population groups [17]. For example, the Balochi and Burusho (two HGDP ethnic group sampled in Pakistan) showed the following paired Fsts (Balochi/Palestinian, Fst = 0.016; Balochi/Swedish, Fst = 0.018; Burusho/Palestinian, Fst = 0.027; Burusho/Swedish, Fst = 0.029; Balochi/Burusho, Fst = 0.008). These paired Fsts overlapped with several inter-European Fsts (e.g. Palestinan/Swedish Fst = 0.019, Basque/Bedouin Fst = 0.020). Thus, this measurement of population differentiation does not appear to provide a clear grouping of different ethnic groups (see Discussion).
Table 1.
Paired Fst values for European Populations
| FstA | DRUZE | BDN | PAL | AJA | GRK | ITN | ADY | SPN | BAS | IRISH | GERM | EEURA | RUS | SWED | ORC | SARD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BDNB | 0.0072 | |||||||||||||||
| PAL | 0.0064 | 0.0056 | ||||||||||||||
| AJA | 0.0088 | 0.0108 | 0.0093 | |||||||||||||
| GRK | 0.0052 | 0.0064 | 0.0057 | 0.0042 | ||||||||||||
| ITN | 0.0057 | 0.0079 | 0.0064 | 0.0040 | −0.0001 | |||||||||||
| ADY | 0.0092 | 0.0123 | 0.0108 | 0.0107 | 0.0054 | 0.0067 | ||||||||||
| SPN | 0.0096 | 0.0103 | 0.0101 | 0.0056 | 0.0035 | 0.0010 | 0.0090 | |||||||||
| BAS | 0.0186 | 0.0204 | 0.0199 | 0.0144 | 0.0098 | 0.0084 | 0.0180 | 0.0060 | ||||||||
| IRISH | 0.0154 | 0.0187 | 0.0170 | 0.0109 | 0.0067 | 0.0048 | 0.0110 | 0.0037 | 0.0086 | |||||||
| GERM | 0.0121 | 0.0147 | 0.0136 | 0.0072 | 0.0039 | 0.0029 | 0.0089 | 0.0015 | 0.0079 | 0.0010 | ||||||
| EEURA | 0.0128 | 0.0149 | 0.0133 | 0.0068 | 0.0049 | 0.0040 | 0.0086 | 0.0033 | 0.0091 | 0.0034 | 0.0014 | |||||
| RUS | 0.0194 | 0.0211 | 0.0202 | 0.0137 | 0.0108 | 0.0088 | 0.0120 | 0.0079 | 0.0126 | 0.0038 | 0.0037 | 0.0029 | ||||
| SWED | 0.0167 | 0.0204 | 0.0191 | 0.0120 | 0.0084 | 0.0064 | 0.0117 | 0.0055 | 0.0100 | 0.0020 | 0.0007 | 0.0025 | 0.0036 | |||
| ORC | 0.0194 | 0.0212 | 0.0201 | 0.0146 | 0.0103 | 0.0080 | 0.0136 | 0.0063 | 0.0124 | 0.0039 | 0.0048 | 0.0055 | 0.0092 | 0.0046 | ||
| SARD | 0.0163 | 0.0183 | 0.0166 | 0.0131 | 0.0088 | 0.0072 | 0.0204 | 0.0071 | 0.0133 | 0.0140 | 0.0117 | 0.0132 | 0.0210 | 0.0155 | 0.0162 | |
| TUSC | 0.0086 | 0.0102 | 0.0096 | 0.0066 | 0.0005 | 0.0004 | 0.0094 | 0.0023 | 0.0084 | 0.0055 | 0.0032 | 0.0045 | 0.0108 | 0.0061 | 0.0098 | 0.0083 |
The Paired Fst value is the mean determined from three nonoverlapping sets of 3500 SNPs using the Weir and Cockerham algorithm [24]. Complete data including the standard deviation is provided in Supplemental Table 2.
Population groups included Druze, Bedouin (BDN), Palestinian (PAL), Ashkenazi Jewish American (AJA), Greek (GRK), Italian (ITN), Adygei (ADY), Spanish (SPN), Basque (BAS), IRISH, German (GERM), Eastern European (EEUR), Russian (RUS), Swedish (SWED), Orcadian (ORC), Sardinian (SARD), and Tuscan (TUSC).
Fis values were also determined for each of the population sample and did not indicate a strong inbreeding component for any of the tested sample groups with the exception of the Bedouin participants (Supplemental Table 3).
Principal Components Analyses Using >250K SNPs
To further explore the relationship among European population groups and examine population substructure PCA was performed using the genotype results from a set of ~300 thousand autosomal SNPs that was common to each of the populations examined. For most individuals with self-reported ethnic identities there was a general correspondence with the geographical location of origin (Figure 1A). For example the relationship of Italian groups and the subjects from the island country of Sardinia shows a striking resemblance to maps of Europe. In addition, genotypes from the same or related population groups typed in different laboratories showed similar PCA results (e.g. north Italian and Tuscan groups genotyped as part of HGDP overlapped with Italian American subjects).
For European American participants without self-identified ethnic affiliation there was a wide distribution in PC1 and PC2 that overlapped with those individuals of known ethnic affiliation or country of origin. These included subjects from two large publically available data sets (New York Cancer Project (NYCP) and Childrens Hospital of Philadelphia (CHOP) genotypes in I-control database (www.illumina.com/iControlDB, Illumina, San Diego, CA) (Figure 1B and Figure 1C). Not surprisingly, these sample sets from collection sites in the New York City region and Philadelphia also showed differences in their relative distribution pattern observed in principal component (PC)1 and PC2. This is particularly evident with regards both the Ashkenazi Jewish and Irish ethnic groups (Table 2).
Table 2.
Distribution of NYCP and CHOP European Subjects
| Presumed EthnicityA | PCA RangeB | Percent of SamplesC | ||
|---|---|---|---|---|
| PC1 | PC2 | NYCP | CHOP | |
| Ashkenazi | 0.030, 0.040 | 0.020, 0.040 | 13.0% | 4.5% |
| North European | −0.016, −0.005 | −0.010, 0.015 | 59.7% | 45.4% |
| Italian/Greek | −0.002, 0.025 | −0.035, −0.010 | 13.4% | 16.7% |
| Irish | −0.016, −0.010 | −0.010, 0.0001 | 14.0% | 3.8% |
Presumed ethnicity corresponding to the PCA results. The Irish group is a subset of the North European larger group.
The coordinate range for PC1 and PC2 for four different groups of subjects. The first value indicates the lower boundary and the second value the upper boundary for each PC range.
The percent of subjects is shown for NYCP (1832 subjects) and CHOP (1487 subjects).
PCA of Northern and Southern European Population Groups
Our previous studies showed that further elucidation of the relationships among Northern European individuals was possible when analyses were performed excluding subjects of southern European origin. For the current study, we next examined Northern and Southern population groups separately and the effect of including or excluding different ethnic groups (Figure 2 and Figure 3). We have included the Middle East groups in the analysis of Southern European population groups due to the close relationship of these population. For subjects of Northern European origin, the inclusion of Orcadian subjects showed a substantial change in the PC2 relationships in which the other Northern European populations were no longer separated on this axis (compare Figure 2A with Figure 2B). The pattern observed with the Orcadian individuals, also suggests that there may be ongoing admixture between this population and less isolated European populations. In addition, the inclusion of Basque subjects, a group that appeared intermediate between Northern and Southern European populations, changed both PC1 and PC2 results (Figure 2C). These differences in PCA results may be critical in assessing the ability of particular ancestry marker sets to define relevant population substructure for specific studies (see Discussion).
Similarly, studies were performed for the Southern European population groups (Figure 3). Here the inclusion or exclusion of the Basque population group had no effect on the PC1/PC2 graphical representation (data not shown). However, the inclusion or exclusion of Sardinian and Ashkenazi Jewish population groups shifted the position of the Adygei subjects. It is also worth noting that the inclusion of the Arab population groups results in larger separation between northern Italian and southern Italian (and/or Greek) subjects and suggests that inclusion of the Arab population genotypes may be useful in analyses of southern European population groups (data not shown).
PCA of European and South Asian Population Groups
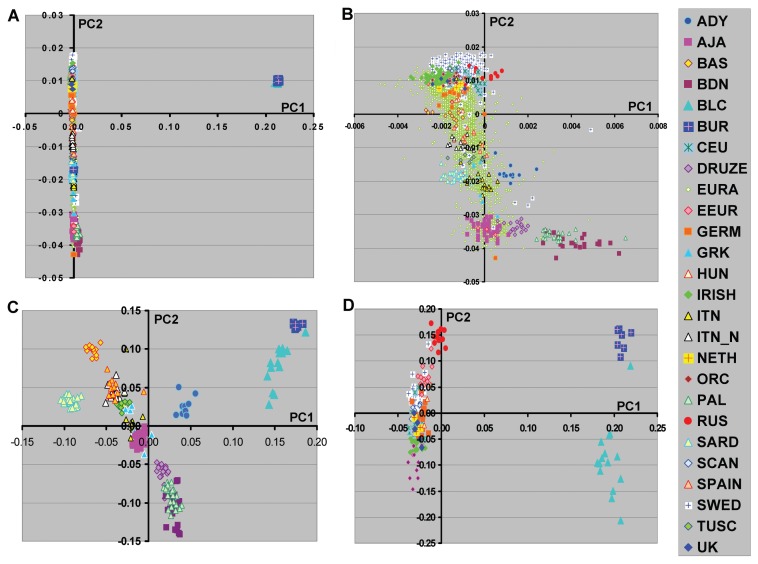
To further explore the relationship of population groups in Europe, we also performed PCA including the HGDP South Asian groups, Burusho and Balochi, These South Asian population groups showed relatively small population differentiation with continental European subjects ([14,17,26,27]. Thus inclusion of these populations might further suggest clines, contributions, or relationships between European and these neighboring geographic ethnic groups. PCA analyses showed that these South Asian groups were different from any of the European groups (Figure 4) consistent with previous studies [14,26,27]. This difference was less pronounced when only southern in contrast to northern European groups were examined (Figure 4C). Interestingly, the Adygei, a population from the Caucasus showed a closer relationship to these South Asian ethnic groups than other European groups, consistent with geographical relationships.
Figure 4.
PCA analyses of European population groups together with two South Asian groups. A, All European population groups together with Balochi (BLC, 15 subjects) and Burusho (BUR, 7 subjects). B, Expanded view of European populations from PCA shown in A. C, PCA results from southern European population groups analyzed together with BAL and BUR. D, PCA results from northern European population groups analyzed together with BAL and BUR.
Identification and testing of European Substructure Ancestry Informative Markers
AIMs that discern population substructure are likely to be useful in candidate gene, chromosomal position based association studies, and defining homogeneous subject sets [28]. Our previous studies identified two sets of European substructure AIMs (ESAIMs): ns-ESAIMs and north-ESAIMs. The ns-ESAIMs separated European populations on a single axis (north/south) that corresponds to the largest variation in European substructure in PCA (PC1). The north-ESAIMs provided distinction along an east west gradient when only northern European population groups were studied. The availability of genotypes from many additional European population groups as well as additional European American subjects provided an opportunity to ascertain and test sets of SNP AIMs that might improve population substructure analyses. In addition, for the current study we only included SNP AIMs compatible for typing on the Illumina Infinium platform.
For north-ESAIMs we selected additional SNPs by using a measurement of informativeness (In)[25] using subjects not used in our previous studies. These additional SNPs were selected from those with the greatest In determined from analyses of 1) Irish and Swedish genotypes and 2) French and Swedish genotypes (see Methods for details). These SNPs were combined with our previously identified north-ESAIMs and ~300 north/south-ESAIMs. To assess the potential usefulness of these AIMs we performed PCA using an independent set of samples. The results (Figure 5A) showed the clustering of self identified groups in PC1 and PC2.
Figure 5.
Ability of European Substructure AIMs to discern population substructure. A, PCA analysis of northern Europeans with North AIMs set 2. B, PCA analysis of Southern Europeans with South AIMs. C, PCA of all test population samples with EUROset2 ESAIMs. D, PCA of all test populations with 270 K SNPS. Note: The French subjects included only those identified as Northern European (~75% of subjects) and does not represent the diversity of all French subjects.
Similarly, for south-ESAIMs, we utilized combination of Arab ethnic groups (Druze, Palestinians and Bedouin) and Ashkenazi Jewish subjects to identify SNPs that could distinguish between these groups. These were combined with ~300 north/south AIMs that can separate Spanish, Italian, Greek and Ashkenazi Jewish ethnic groups [12]. The PCA results using independent sample (Figure 5B) showed the clustering of self identified groups.
We also used the combined set of SNPs to examine the northern and southern European populations together. The PCA results (Figure 5C) show very similar patterns to that observed with 270K SNPs (Figure 5D).
As a measure of the ability of the ESAIMs to accurately assess substructure we examined the correlation (r2) with 270K SNPs (Table 3). Northern and southern population groups were analyzed both separately and together, and the results were also compared with random marker sets. For each test population group (all, north and south subgroups), the complete ESAIM set containing 3519 SNPs (EUROset2 in Table 3) showed better correlation results than random sets of six thousand SNPs. Smaller marker sets (e.g. NORTHset2 and SOUTH) showed equivalent results when the test group was limited to either northern or southern European populations. In addition, although 3000 random SNPs showed high correlation in the entire test group (both north and south together) for PC1 and PC2, the correlations for the northern and southern groups were much weaker when considered separately.
Table 3.
Summary of Correlations for European AIMs
| All Test GroupA | NORTH Test | SOUTH Test | ||||
|---|---|---|---|---|---|---|
| Marker SetB | PC1 | PC2 | PC1 | PC2 | PC1 | PC2 |
| Random 3K | 0.92C | 0.60 | 0.20 | 0.01 | 0.78 | 0.04 |
| Random 6K | 0.96 | 0.77 | 0.55 | 0.05 | 0.93 | 0.36 |
| Random 12K | 0.98 | 0.88 | 0.76 | 0.08 | 0.96 | 0.72 |
| Random 24K | 0.99 | 0.95 | 0.88 | 0.37 | 0.98 | 0.87 |
| Ranodm 48K | 1.00 | 0.97 | 0.95 | 0.63 | 0.99 | 0.88 |
| EUROset1 (2795) | 0.95 | 0.79 | 0.54 | 0.01 | 0.92 | 0.45 |
| EUROset2 (3519) | 0.96 | 0.81 | 0.57 | 0.19 | 0.92 | 0.55 |
| NORTHset1 (2034) | Na | Na | 0.53 | 0.02 | na | Na |
| NORTHset2 (2762) | Na | Na | 0.56 | 0.19 | na | Na |
| SOUTH (1078) | Na | Na | Na | Na | 0.87 | 0.40 |
The “All” Test Group included 789 European subjects and did not include any subjects used in AIMs selection (see Methods). The NORTH (378 subjects) and South (411 subjects) were subsets of the “ALL Test Group” determined from STRUCTURE results (see Methods).
The random marker results are the mean from three independently selected marker groups for each category (3K, 6K, 12K, 24K and 48K). The EUROset2 contained EUROset1 markers. Similarly, the NORTHset2 contained the NORTHset1 SNPs. There was a small overlap of SNPs in the NORTH and SOUTH SNP set (see Materials and Methods and Supplemental Table 1 for Details).
All correlations are shown as r2 values with PCA using 270K SNPs.
Performance of ESAIMs in Controlling for Population Substructure
To further assess the potential value and performance of ESAIMs we examined SNPs with large (10–25%) allele frequency differences between different European subgroups. The different subgroups were selected based on PCA results and included both individuals with self-identified ethnic affiliation and those without known self-identity information (see Supplemental Figure 2). This procedure allowed the separation of five distinct subgroups of individuals that correspond to different regions or ethnic groups. These included French, Irish/United Kingdom, Scandinavian, Ashkenazi, and other southern European including Italian and Greek (OSOUTH). We then identified those SNPs with the largest allele frequency difference between different pairs of subject groups. From these SNPs we selected test SNPs for which the significant differences (in case/control association tests) between groups would most likely be due to differences in population substructure. This was determined by EIGENSTRAT[3] analyses using 270K SNP set (see Methods) and included markers for which EIGENSTRAT corrected p values were substantially different from baseline. We then examined the ability of different ESAIMs and random AIMs to adjust for population substructure differences that could account for the allele frequency differences of the test SNPs by examining Armitage χ2 p values before and after EIGENSTRAT correction based on PCA (Table 4).
Table 4.
PCA Correction for Ancestry Using ESAIMs
| SNP | Chr | Mb | p-valueA | 270KB | EUROset1C | EUROset2 | North | South | Ran 3K | Ran 6K | Ran 12K | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASH/OSOUTHD | rs6587597 | 1 | 150 | 8.2E-11E | 2.5E-01 | 6.5E-01 | 2.9E-01 | 1.5E-02 | 5.8E-01 | 2.2E-01 | 3.2E-01 | 1.7E-01 |
| ASH/OSOUTH | rs1400452F | 2 | 18 | 3.0E-11 | 1.3E-03 | 1.8E-03 | 9.1E-04 | 2.5E-06 | 1.1E-04 | 8.6E-03 | 4.3E-04 | 2.1E-02 |
| ASH/OSOUTH | rs4859259 | 3 | 184 | 2.2E-10 | 5.3E-01 | 7.5E-01 | 7.5E-01 | 4.4E-01 | 3.7E-01 | 2.8E-02 | 1.1E-01 | 5.8E-01 |
| ASH/OSOUTH | rs7456425 | 7 | 81 | 4.7E-09 | 7.5E-01 | 7.5E-01 | 6.5E-01 | 6.1E-02 | 8.3E-02 | 5.1E-02 | 5.3E-01 | 6.5E-01 |
| ASH/OSOUTH | rs7139066 | 12 | 26 | 1.3E-10 | 6.9E-02 | 3.4E-01 | 2.5E-01 | 1.4E-02 | 4.0E-02 | 2.5E-01 | 1.6E-02 | 2.9E-01 |
| OSOUTH/FRENCH | rs6723108 | 2 | 135 | 5.2E-11 | 6.5E-01 | 2.5E-01 | 4.6E-02 | 5.1E-02 | 7.8E-02 | 4.4E-03 | 1.1E-01 | 2.4E-01 |
| OSOUTH/FRENCH | rs1584930 | 3 | 95 | 1.3E-07 | 3.4E-01 | 7.7E-03 | 2.4E-02 | 6.1E-02 | 4.5E-04 | 1.6E-02 | 1.3E-03 | 4.7E-03 |
| OSOUTH/FRENCH | rs10515893 | 5 | 165 | 2.9E-08 | 2.4E-01 | 5.1E-02 | 1.7E-01 | 4.8E-02 | 8.2E-03 | 3.1E-04 | 1.4E-02 | 3.6E-02 |
| OSOUTH/FRENCH | rs2072633 | 6 | 32 | 2.9E-08 | 1.3E-01 | 4.2E-03 | 6.2E-03 | 1.1E-03 | 1.1E-02 | 5.6E-04 | 7.8E-02 | 8.9E-02 |
| OSOUTH/FRENCH | rs11663558 | 18 | 19 | 5.9E-08 | 1.0E+00 | 1.6E-01 | 1.8E-01 | 6.5E-02 | 1.6E-02 | 8.9E-02 | 8.9E-02 | 1.7E-01 |
| FRENCH/SCAN | rs4954564 | 2 | 137 | <E-12 | 3.2E-01 | 3.7E-04 | 5.8E-01 | 4.8E-01 | 4.9E-07 | 1.6E-04 | 5.0E-04 | 1.0E-02 |
| FRENCH/SCAN | rs2856718 | 6 | 33 | 3.5E-12 | 3.7E-01 | 2.2E-03 | 2.3E-02 | 1.4E-02 | 4.6E-06 | 1.5E-05 | 7.7E-03 | 1.5E-02 |
| FRENCH/SCAN | rs11038910 | 11 | 47 | 4.7E-09 | 1.1E-01 | 4.1E-04 | 5.2E-03 | 6.2E-03 | 2.3E-04 | 9.7E-05 | 1.9E-02 | 1.0E+00 |
| FRENCH/SCAN | rs1146904 | 13 | 76 | 2.2E-08 | 6.5E-02 | 1.7E-02 | 4.9E-03 | 2.2E-03 | 4.1E-04 | 7.4E-07 | 2.3E-02 | 2.1E-02 |
| FRENCH/SCAN | rs11631323F | 15 | 96 | 1.7E-09 | 1.0E+00 | 3.8E-03 | 2.1E-02 | 9.6E-03 | 8.7E-05 | 2.2E-03 | 1.1E-01 | 1.8E-01 |
| SCAN/IRISHUK | rs10517522 | 4 | 39 | 3.9E-08 | 1.3E-02 | 3.0E-02 | 3.2E-02 | 9.1E-03 | 5.7E-07 | 1.3E-05 | 4.8E-04 | 6.5E-02 |
| SCAN/IRISHUK | rs1265048 | 6 | 31 | 6.9E-08 | 1.8E-02 | 3.0E-03 | 2.5E-02 | 5.8E-03 | 1.8E-06 | 8.3E-05 | 1.8E-02 | 6.9E-03 |
| SCAN/IRISHUK | rs511512 | 13 | 76 | 8.3E-10 | 2.7E-02 | 4.7E-03 | 6.5E-03 | 8.2E-03 | 3.3E-08 | 2.7E-08 | 1.9E-08 | 3.3E-04 |
| SCAN/IRISHUK | rs255052 | 16 | 67 | 1.5E-08 | 1.7E-01 | 1.1E-01 | 4.4E-01 | 9.4E-02 | 1.3E-07 | 5.0E-04 | 1.8E-01 | 5.4E-02 |
| SCAN/IRISHUK | rs9303363 | 17 | 50 | 1.0E-08 | 1.8E-01 | 3.4E-02 | 6.9E-03 | 3.4E-03 | 1.5E-07 | 6.9E-03 | 6.5E-03 | 4.8E-02 |
| FRENCH/IRISHUK | rs6723108 | 2 | 135 | 2.2E-10 | 5.6E-04 | 8.9E-08 | 4.8E-01 | 6.5E-01 | 4.2E-10 | 4.0E-10 | 5.1E-06 | 8.2E-04 |
| FRENCH/IRISHUK | rs1965299F | 4 | 117 | 1.2E-07 | 2.8E-04 | 9.2E-05 | 1.1E-04 | 1.8E-05 | 5.4E-07 | 3.4E-07 | 6.3E-05 | 1.7E-03 |
| FRENCH/IRISHUK | rs12498670 | 4 | 178 | 4.9E-07 | 1.5E-03 | 2.9E-05 | 3.9E-05 | 3.7E-05 | 3.2E-07 | 8.7E-07 | 7.4E-05 | 5.6E-04 |
| FRENCH/IRISHUK | rs3135029 | 6 | 33 | 7.4E-07 | 7.4E-02 | 1.5E-03 | 1.4E-02 | 2.6E-03 | 1.6E-04 | 4.1E-06 | 9.2E-05 | 1.9E-03 |
| FRENCH/IRISHUK | rs11645416 | 16 | 50 | 6.5E-08 | 3.5E-04 | 2.9E-05 | 1.5E-03 | 2.0E-04 | 1.9E-07 | 8.9E-08 | 1.5E-05 | 9.1E-04 |
The p value was determined by the Armitage Chi2 test using the first population group as “case” and the second group as “control”.
The adjusted p value based on correction for PCA using EIGENSTRAT software is shown for 270K SNPs and each of the AIMs and random marker sets. The EUROset1 and EUROset2 are as defined in Table 2 and Supplemental Table 3.
The EUROset1 contains 1037 of 1212 ESAIMs previously reported [12] and an additional 1757 SNPs (see Methods).
The five different population groups were defined by PCA (see methods): Ashkenazi (ASH) 88 individuals, Other South (OSOUTH) 210 individuals, French 444 individuals, and Irish United Kingdom (IRISHUK) 328 individuals.
Bold p values highlight p values <10−4.
These SNPs were isolated significant values. For each of the other SNPs chosen, multiple closely linked SNPs (within 50 Kb) showed similar p values.
For certain population pairs (e.g. ASH/OSOUTH and OSOUTH/FRENCH) most of the ESAIM sets and even 3K random SNPs performed well (Table 4). For example in the ASH/OSOUTH population pairing, the rs4859259 that showed an initial uncorrected p value of 2.2E-10. After correction with any of the SNP sets only very modest or insignificant p values were observed. However, for the other population pairs the 3K random SNPs performed poorly in controlling for population structure. For one particular paired population group (FRENCH/IRISHUK) the most comprehensive ESAIMs (EUROset2) containing 3519 selected SNPs showed substantially better ability to control for population substructure than 3K or 6K random SNP sets. It is also notable that the EUROset2 SNPs overall performed better than the EUROset1 SNPs that include both the majority of our previous set of north-ESAIMs [12] as well as additional SNPs (see Materials and Methods). For example, EUROset1 failed to show an appropriate correction for rs6723108 in the FRENCH/IRSHUK population pair (initial p value = 2.2E-10, EUROset1 corrected p value = 8.9E-08) where as when using the EUROset2 markers the p value was adjusted (EUROset2 corrected p value = 4.8E-01).
Overall, the EUROset2 SNPs appeared to be comparable to 12K random SNP sets and only moderately less effective than the 270K SNPs. These data together with the PCA correlation studies support the use of the EUROset2 SNPs in future studies. The ESAIMs are provided in Supplemental Table 1.
DISCUSSION
The current study extends the definition of European population substructure and the relationships among diverse European ethnic groups. The results are generally consistent with the observation that the largest variations in PCA (those graphically depicted in PC1/PC2) correspond to geographical relationships. This result is also consistent with our recent studies of East Asia population substructure [29]. While perhaps surprising this result is consistent with migration (demic expansion) and geographic physical boundaries being the most critical or common factor in determining genotypic patterns in population groups within continents.
The current study extends the analysis of European population genetic structure to include additional southern European groups and Arab populations. Even within Italy, the relative position of northern Italians compared with subjects from Tuscany is consistent with the general geographic correspondence of PCA results.. Interestingly, the majority of Italian Americans (NYCP 4 grandparent defined) appear to derive from southern Italy and overlap with subjects of Greek heritage. Both of these observations are consistent with previous historical information [30,31].
Possible exceptions to this observation of geographic correspondence include the Ashkenazi Jewish population. While the Ashkenazi are clearly of southern origin based on both PCA and STRUCTURE studies, in our analyses of diverse European populations (Figure 1), this group appears to have a unique genotypic pattern that may not reflect geographic origins. Furthermore, the inclusion or exclusion of particular ethnic groups (i.e. Ashkenazi Jewish, and Sardinian for southern European, and Orcadian for Northern European) shifted the relationships in PCA when southern or northern Europeans were examined separately. Similarly, the inclusion of South Asian populations (Figure 4C) changes the relationships of the population groups with the Ashkenazi Jewish population appearing in the center of a presumed southern European cline. These findings are consistent with our previous observations [12], and show that PCA results are highly dependent on which population groups are included in the analysis. Thus, there should be some caution in interpreting these results and other results from similar analytic methods with respect to ascribing origins of particular ethnic groups. It is worth mentioning that two of the afore noted groups in our analyses, Sardinian and Orcadian, are either island populations suggesting that relative isolation of particular groups may underlie some of the observed results. However, it should also be noted that the Bedouin population group with high inbreeding (Supplemental Table 3), is not represented differently in PCA than other Arab groups without evidence in high inbreeding.
The differences observed between measures of population differentiation and analyses based on either model dependent clustering or PCA are also noteworthy. Although a common measurement of population differentiation (Fst) has been used in measurements of “distance/years” between population groups [32] the paired Fst values do not necessarily correspond to a measurement of the differences in genotypic patterns. This is illustrated in the current study with respect to the relationship of South Asian population groups and European groups. Although paired Fst values overlap between intra-European and inter-European/South Asian groups, PCA shows that the largest axis of variation unambiguously separates European and South Asian groups. Presumably this is of practical importance since it is the patterns of genotypic variation that are critical in causing false positives (and perhaps false negatives) due to population stratification differences between case and control groups.
The current study focuses on the PC1 and PC2 results in analyses of European substructure. Additional PC’s do show additional substructure, however, the amount of variation is relatively small compared to PC1 and PC2 when northern and southern European populations are considered separately (Supplemental Figure 3). In addition, it should be noted that the SNPs included in any of the population substructure studies excluded regions of high linkage disequilibrium and the ESAIMs were selected to exclude SNPs with r2>0.8.
An important aspect of the current study was the identification and characterization of AIMs applicable to association studies in European and European American populations. The current availability of several thousand genotypes from a single platform (Illumina in the current study) enabled a more extensive selection of AIMs and the ability to test the performance of these selected markers in different sample sets. With respect to northern European populations, a direct comparison with our previous north-ESAIM set was not possible due to the absence of a portion of these SNPs in our current dataset. However, the currently recommended EUROset2 SNPs performed better than the EUROset1 SNPs containing 85% the north ESAIMs (Table 4).
Our strategy in selecting markers was based on using In[25] differences between paired population groups that showed large separations in PC1 and PC2 in either southern European, northern European, or combined analyses. Alternate approaches including the use of PCA SNP scores did not result in ESAIM sets of comparable ability to discern European substructure (data not shown).
In the current study two analyses were presented that we believe demonstrate the ability and usefulness of these ESAIMs to ascertain and correct for population substructure. It should be acknowledged that one of these criteria (correlation with “whole genome” PCA results in PC1 and PC2) will depend in part on the diversity of the individuals and perhaps whether a particular population group is included within the sample set (i.e. PC1 and PC2 are not necessarily fixed as discussed above). The inclusion of large numbers of samples from collections in NYC and Philadelphia that have a broad distribution of ethnic origin, albeit as defined by PCA, suggests that these markers will be of particular value in studies where ethnic origin matching by self-reporting is not available or unreliable. However, the applicability of these or other ESAIM sets may vary depending on whether the current sample set is reasonably inclusive of subjects in a particular study. Our assessment of the ability of the marker sets to account for allele frequency differences between large European/European American subgroups supports the practical application of these ESAIMs (Table 4) and suggests that these ESAIMs will address population substructure in most if not all European/European American datasets. In conclusion, we believe that the current ESAIMs (EUROset2) will have wide applicability to complex genetic studies within European populations.
Supplemental Data
Acknowledgments
We thank Stephen Johnson and Robert Lundsten for informatics support on the New York Cancer Project samples. We also thank Anthony Liew and Houman Khalili for expert assistance with genotyping. We thank the volunteers from the different populations for donating blood samples. This work was supported by NIH grants DK071185, AR050267, and AR44422.
Footnotes
DISCLOSURE
The authors have declared that no competing interests exist.
REFERENCES
- 1.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 4.Satten GA, Flanders WD, Yang Q. Accounting for unmeasured population substructure in case-control studies of genetic association using a novel latent-class model. Am J Hum Genet. 2001;68:466–477. doi: 10.1086/318195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoggart CJ, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson KJ, Belkhir K. A Bayesian approach to the identification of panmictic populations and the assignment of individuals. Genet Res. 2001;78:59–77. doi: 10.1017/s001667230100502x. [DOI] [PubMed] [Google Scholar]
- 8.Epstein MP, Allen AS, Satten GA. A simple and improved correction for population stratification in case-control studies. Am J Hum Genet. 2007;80:921–930. doi: 10.1086/516842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seldin MF, et al. European Population Substructure: Clustering of Northern and Southern Populations. PLoS Genetics. 2006;2:1339–1351. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauchet M, et al. Measuring European population stratification with microarray genotype data. Am J Hum Genet. 2007;80:948–956. doi: 10.1086/513477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AL, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 15.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–103. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lao O, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 17.Nassir R, et al. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC Genet. 2009;10:39. doi: 10.1186/1471-2156-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York Cancer project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–310. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosoy R, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX, software under WindowsTM for the genetic of populations. 4.02 ed. Montpellier, France: Laboratory Genome, Populations, Interactions CNRS UMR 5000, University of Montpellier II; 2001. [Google Scholar]
- 24.Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 27.Yang N, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 28.Seldin MF, Price AL. Application of ancestry informative markers to association studies in European Americans. PLoS Genet. 2008;4:e5. doi: 10.1371/journal.pgen.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian C, et al. Analysis of East Asia genetic substructure using genome-wide SNP arrays. PLoS ONE. 2008;3:e3862. doi: 10.1371/journal.pone.0003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangione J, Morreale B. LA Storia: Five Centuries of the Italian American Experience. New York City: HarperCollins; 1993. [Google Scholar]
- 31.Woodhead AG. Greeks in the west. New York City: Praeger; 1962. [Google Scholar]
- 32.Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. xiii. Princeton, N.J: Princeton University Press; 1996. p. 413. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.