Summary
Insulator sequences help to organize the genome into discrete functional regions by preventing inappropriate cross-regulation. This is thought to be mediated in part through associations with other insulators located elsewhere in the genome. Enhancers that normally drive Drosophila even skipped (eve) expression are located closer to the TER94 transcription start site than to that of eve. We discovered that the region between these genes has enhancer-blocking activity, and that this insulator region also mediates homing of P-element transgenes to the eve-TER94 genomic neighborhood. Localization of these activities to within 0.6 kb failed to separate them. Importantly, homed transgenic promoters respond to endogenous eve enhancers from great distances, and this long-range communication depends on the homing/insulator region, which we call Homie. We also find that the eve promoter contributes to long-distance communication. However, even the basal hsp70 promoter can communicate with eve enhancers across distances of several megabases, when the communication is mediated by Homie. These studies show that, while Homie blocks enhancer-promoter communication at short range, it facilitates long-range communication between distant genomic regions, possibly by organizing a large chromosomal loop between endogenous and transgenic Homies.
Keywords: Insulator, Homing, Enhancer blocking, Epigenetics, Even skipped, Drosophila
INTRODUCTION
Sequences that help to organize chromatin into functional domains can have a profound influence on gene regulation. Enhancers are capable of activating transcription across tens or hundreds of kilobases (kb) along a chromosome. Paradoxically, gene-rich genomic regions contain many genes within that distance, without known functional crosstalk. Insulator sequences have been identified that can prevent inappropriate enhancer-promoter (E-P) communication, helping to resolve this apparent paradox (Bushey et al., 2008; Dorman et al., 2007; Gaszner and Felsenfeld, 2006; Valenzuela and Kamakaka, 2006). Insulators are typically found between genes, or within complex loci such as the bithorax complex (BX-C) (Maeda and Karch, 2006; Maeda and Karch, 2007), where they act in combination with other sequences to orchestrate complex regulatory programs during development.
Underlying mechanisms appear to involve the formation of loops, possibly organizing chromatin into functionally isolated domains (Bushey et al., 2008; Dorman et al., 2007; Gaszner and Felsenfeld, 2006; Valenzuela and Kamakaka, 2006). The scs and scs′ insulators (Udvardy et al., 1985) are each bound by distinct protein complexes (Gaszner et al., 1999; Hart et al., 1997; Zhao et al., 1995) that interact with each other, resulting in a chromosomal loop that encompasses the 87A7 hsp70 genes (Blanton et al., 2003). The gypsy transposon exhibits enhancer-blocking activity (Geyer and Corces, 1992; Geyer et al., 1988; Modolell et al., 1983; Peifer and Bender, 1988) that requires the Suppressor of Hairy wing protein (Parnell et al., 2006; Ramos et al., 2006; Spana et al., 1988), as well as CP190 and Mod(mdg)4, which form a complex (Gause et al., 2001; Ghosh et al., 2001; Pai et al., 2004). Vertebrate insulators often bind the CTCF protein (Bell and Felsenfeld, 2000; Bell et al., 1999; Hark et al., 2000; Kanduri et al., 2000), and CTCF interacts with cohesins (Parelho et al., 2008; Wendt et al., 2008), an interaction that correlates with its enhancer-blocking activity.
Insulators can cooperate with other regulatory sequences. In the BX-C, gene activities in early embryos are differentially regulated, and these patterns of gene activity are maintained through Polycomb- and Trithorax-response elements (PREs and TREs, respectively) (Maeda and Karch, 2006). The regulatory regions Mcp (Busturia et al., 1997; Karch et al., 1994; Muller et al., 1999), Fab-7 (Gyurkovics et al., 1990; Karch et al., 1994; Mihaly et al., 1997) and Fab-8 (Barges et al., 2000; Zhou et al., 1999) each contain closely linked PRE/TREs and insulators. CTCF binds to Mcp and Fab-8 (Holohan et al., 2007), whereas Fab-7 binds other factors (Aoki et al., 2008; Schweinsberg and Schedl, 2004). CTCF can facilitate repressive interactions between an insulator and a promoter that involve Polycomb group (PcG) complexes (Li et al., 2008). In addition, promoter targeting sequences can overcome insulator activity to maintain an active state (Zhou and Levine, 1999), and promoter specificity sequences, such as the promoter-tethering element in Abdominal-B (Akbari et al., 2008), facilitate specific E-P interactions. Loop formation by insulators (Cleard et al., 2006) and PRE/TREs (Lanzuolo et al., 2007) may be an essential component of maintaining proper gene expression through development. The DNA-binding GAGA factor, which binds to many PRE/TREs, can also contribute to enhancer blocking (Belozerov et al., 2003; Ohtsuki and Levine, 1998; Schweinsberg et al., 2004; Schweinsberg and Schedl, 2004). Loop attachment sites may block the propagation along the DNA of chromatin modifications or protein complexes that enhance transcription (Bushey et al., 2008; Dorman et al., 2007; Gaszner and Felsenfeld, 2006; Valenzuela and Kamakaka, 2006). Much remains to be discovered concerning how chromosomal architecture affects gene expression, and how regulatory elements that control this architecture carry out their functions.
In Drosophila, it has been observed that some sequences cause transgenes to insert non-randomly in the genome, near the site of origin of those sequences. This transgene homing has been observed for regions of engrailed (Hama et al., 1990; Kassis, 2002; Kassis et al., 1992), linotte (Taillebourg and Dura, 1999), also known as derailed, and the BX-C (Bender and Hudson, 2000). The BX-C homing element may contain an insulator that separates two enhancer regions (Bender and Hudson, 2000), whereas for engrailed, the homing element is associated with a PRE (Kwon et al., 2009). Homed reporter transgenes have been seen to communicate with enhancers from the endogenous locus across several other genes (Devido et al., 2008; Hama et al., 1990; Kassis et al., 1992; Kwon et al., 2009).
The Drosophila even skipped (eve) locus has been particularly well characterized (Fujioka et al., 1999; Goto et al., 1989; Harding et al., 1989; Sackerson et al., 1999; Small et al., 1992; Small et al., 1996), including the identification of a PRE at its 3′ end (Fujioka et al., 2008; Oktaba et al., 2008). The 3′-adjacent gene, TER94 (Leon and McKearin, 1999; Pinter et al., 1998; Ruden et al., 2000), is expressed in the syncytial blastoderm and, by embryonic stage 11, throughout the central nervous system (CNS; this study and Berkeley Drosophila Genome Project) (Tomancak et al., 2002). Several of the eve enhancers are close to TER94, yet TER94 is not expressed in an eve pattern (or vice versa). We found that the region between these genes has enhancer-blocking activity. The same region also mediates transgene homing, and homed transgenes communicate with the endogenous eve enhancers. Long-range E-P communication occurs from as far away as 3300 kb. This E-P communication requires the insulator/homing region in the transgene, but not the PRE. Thus, regulatory interactions between enhancers and promoters can occur between linearly distant genomic regions, and such interactions can be mediated by sequences with insulator properties that also mediate transgene homing.
MATERIALS AND METHODS
Plasmid construction and transgenic analysis
All sequence coordinates in this study are relative to the transcription start site of eve (+1) (Frasch et al., 1988), unless otherwise stated. Details of eZ, hZ, eZ46-15W and eZAR-MeW constructs, and derivatives of pCfhL, are available on request. For the ΦC31-RMCE (Bateman et al., 2006) attP target plasmid, two attP sequences derived from pUAST-P2 (Bateman et al., 2006) were inserted (in opposite orientations) into eZRR11K, flanking eve-lacZ and mini-white. To replace transgenic attP-flanked targets, various donor regions (described in the Results section) were cloned into attBΔ2 (Fujioka et al., 2008), a modified piB-GFP plasmid (Bateman et al., 2006). RMCE events were identified by loss of mini-white-dependent eye color, and confirmed by PCR.
P-element insertion sites were identified by inverse PCR (Ochman et al., 1988; Huang et al., 2000) and homed sites were confirmed using one PCR primer from the flanking genomic sequence and one from within the P-element. Transgenesis (Fujioka et al., 2000; Rubin and Spradling, 1982), in situ hybridization and antibody staining (Fujioka et al., 1999) were performed as previously described.
RESULTS
Enhancer blocking by the eve-TER94 border region
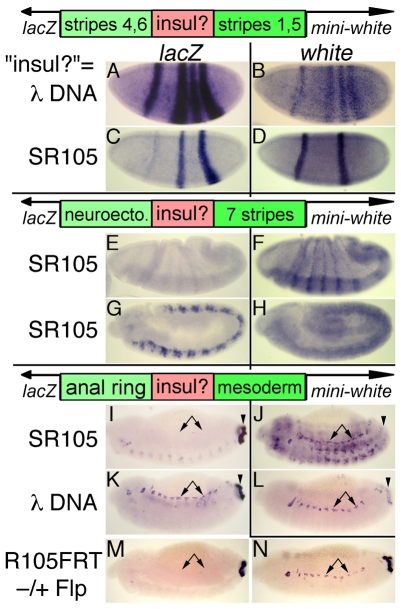
Although some eve enhancers are closer to the TER94 promoter than to the eve promoter, the expression patterns of the two genes are distinctive (Fig. 1). TER94 is expressed ubiquitously in embryos, whereas eve is expressed in a discrete pattern in several tissues. Based on this, we tested whether the border region acts as an enhancer-blocking insulator. We began by examining the region from +8.4 to +10.5 kb (SR105) in three enhancer-blocking assay constructs. One of these constructs (eZ46-15W) contains eve early-stripe enhancers (for stripes 4+6, 1 and 5), an eve-promoter-lacZ reporter gene (eZ), and the mini-white gene (W), with the two promoters divergently transcribed. The stripe 4+6 enhancer is proximal to the eve-promoter-lacZ reporter, and the stripe 1 and 5 enhancers are proximal to mini-white. Transgenes carrying this construct showed expression of both reporter genes in all four stripes, even when a 2 kb stretch of phage λ DNA was inserted between the two stripe enhancers (Fig. 2A,B). By contrast, when SR105 was placed between the enhancers, lacZ was expressed strongly in stripes 4 and 6, but only very weakly (five out of seven lines) or not at all (two out of seven lines) in stripes 1 and 5, whereas mini-white was expressed in the complementary pattern (Fig. 2C,D), showing that SR105 has enhancer-blocking activity. Some variation in blocking activity with the site of insertion is expected based on studies with other insulators (Belozerov et al., 2003; Majumder and Cai, 2003).
Fig. 1.
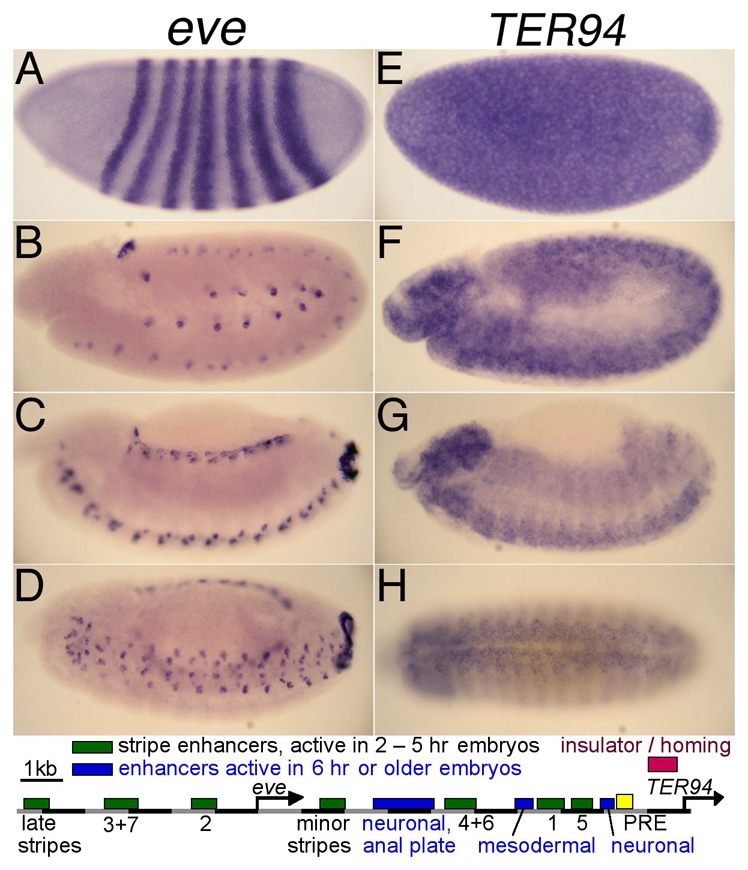
The eve and TER94 expression patterns are distinct. Staged Drosophila embryos were subjected to in situ hybridization against eve (A-D) or TER94 (E-H). Stage 4 (A,E) and stage 11 (B,F) embryos, and lateral (C,G) and ventral (D,H) views of stage 13 embryos. Bottom panel: map of the eve locus and 3′ border region. The start sites and direction of transcription of eve and TER94 are shown as arrows. Green and blue boxes, eve enhancers; yellow box, a PRE (Fujioka et al., 2008); red box, the core insulator/homing element.
Fig. 2.
The region between eve and TER94 has enhancer-blocking activity. Top diagram: enhancer-blocking test vector (eZ46-15W) for A-D. The tested sequence (red box marked `insul?') is flanked by two distinct enhancers (green boxes), upstream of divergently transcribed reporter genes (lacZ and mini-white). Middle (pCfhL) and lower (eZAR-MeW) diagrams, test vectors for E-H and I-N, respectively, have the same design, with different enhancers. The `insul?' test sequence is shown on the left for each row: either λ phage DNA (negative control), SR105 or R105 (the latter flanked by FRT sites). SR105 and R105 were in the same orientation relative to the direction of transcription of mini-white as they normally are relative to that of eve (and TER94). Transgenic embryos carrying these constructs were subjected to in situ hybridization against lacZ or white. (A-D) Stage 5 embryos carrying eZ46-15W. (E-H) Stage 6 (E,F) or stage 11 (G,H) embryos carrying pCfhL. (I-N) Stage 13 embryos carrying eZAR-MeW. Arrows and arrowheads indicate positions of mesodermal and AR expression, respectively. The lateral ectopic expression in J is not relevant for this assay, and is possibly due to a position effect. With R105 flanked by FRT sites as `insul?', lacZ was driven only by the proximal AR enhancer (M). When R105 was removed by FLP-mediated recombination in vivo, activation by the distal mesodermal enhancer is seen (N, arrows).
Next, we tested whether this enhancer-blocking region works with heterologous elements, using a standard vector (pCfhL). This vector consists of the fushi tarazu (ftz) neuronal enhancer proximal to a heat shock promoter-lacZ reporter gene, and the ftz 7-stripe element proximal to mini-white (Hagstrom et al., 1996). When either SR105 (three lines) or a smaller element of 1.3 kb (R105, from +9.2 to +10.5 kb; eight lines) was inserted between the two enhancers, each activated the proximal reporter gene much more strongly than the distal one (Fig. 2E-H). Again, we observed some minor variation in the strength of enhancer blocking with the site of insertion (not shown), but in each case, the element specifically reduced expression driven by the distal enhancer. Thus, this enhancer-blocking insulator can function with heterologous E-P combinations.
Finally, we tested enhancer-blocking activity at later embryonic stages using a construct (eZAR-MeW) with the eve anal plate ring (AR) enhancer proximal to the same eve-promoter-lacZ reporter (eZ) described above for eZ46-15W, and the eve mesodermal enhancer proximal to mini-white. When SR105 was placed between these two enhancers, lacZ was expressed in the AR but not the mesoderm (Fig. 2I), whereas mini-white was expressed in the mesoderm but not the AR (Fig. 2J). By contrast, when λ DNA was inserted between the two enhancers, both reporters were expressed in both patterns (Fig. 2K,L). Thus, the directional enhancer-blocking activity of SR105 is also functional at later stages of embryogenesis.
Enhancer blocking depends on the continued presence of the element
When either SR105 (data not shown) or the smaller R105 sequence (Fig. 2M) was flanked by FLP recombination target (FRT) sites (Golic and Lindquist, 1989) in the eZAR-MeW construct, enhancer-blocking activity was the same as for SR105. After subsequent removal of the element through FLP-mediated recombination, lacZ expression was observed in both tissues (Fig. 2N). This rules out the possibility that the apparent enhancer blocking is due to transgenes targeted to genomic sites that inactivate one reporter gene but not the other.
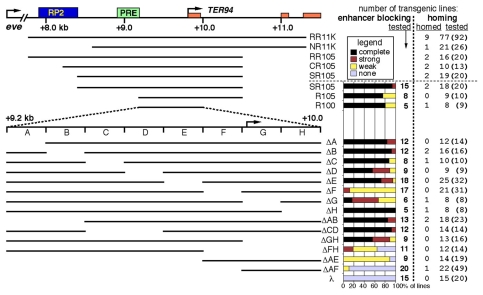
Enhancer blocking conferred by a 600 bp element
We dissected the insulator first of all by deleting from each end. The region from +9.2 to +10.0 kb (Fig. 3, R100) retained activity. Then, internal deletions of ∼100 bp were made (Fig. 3). We saw three distinct levels of activity. Whereas some regions blocked E-P communication completely in both directions, others blocked the interaction between the AR enhancer and mini-white completely, while only partially blocking lacZ expression in the mesoderm. This partial blocking could be further quantified based on the developmental stage at which mesodermal β-galactosidase (β-gal) accumulated to detectable levels. Strong blocking caused a delay until embryonic stages 13-14, whereas weak blocking allowed an earlier appearance of mesodermal β-gal (at stages 11-12). Furthermore, there was some variation in activity with the site of insertion, as detailed in Fig. 3.
Fig. 3.
Enhancer-blocking and homing activity conferred by the eve-TER94 border region. Diagrams on the left represent the eve-TER94 border region (top), and subregions (line diagrams below) tested for enhancer-blocking and homing activity. Distances (in kb) are relative to the eve transcription start site. Green box, a PRE; orange boxes, TER94 exons; blue box, RP2 enhancer. R100 was subdivided into eight ∼100 bp regions (A-H), and these were deleted individually or in combination, as indicated. Columns on the right list the number of independent transgenic insertion lines tested for either enhancer-blocking activity or homing, as well as the number of lines found to be homed. Enhancer blocking: below the dotted line, the region diagrammed on the left was present in an eZAR-MeW transgene, analyzed as shown in Fig. 2I-N. Colored bars indicate the percentage of lines showing enhancer blocking, with either complete, strong, or weak activity (see key). Those with `complete' activity showed full enhancer blocking in both directions, i.e. neither lacZ nor mini-white expression driven by the distal enhancer was detected. Those lines in which lacZ expression driven by the distal (mesodermal) enhancer led to detectable lacZ levels only at later stages (embryonic stages 13-14) and showed no mini-white expression driven by the distal (AR) enhancer were considered to have `strong' activity. Those in which mesodermal lacZ accumulated to detectable levels by stage 11-12 but showed no AR mini-white expression were counted as having `weak' activity. Those that showed both early mesodermal lacZ and AR mini-white expression were considered to have no activity (`none'). In four of the SR105 lines (out of 15), the element was between the eve-promoter-lacZ reporter and the two enhancers. Each of these lines showed no lacZ expression, which was counted as complete enhancer blocking (mini-white was driven by both enhancers, as expected). Homing: under `homed' is the number of independent insertions carrying the region on the left that were within 200 kb of eve. Under `tested' is the number of lines stained for β-gal expression; the total number of lines established is in parentheses. Above the dotted line, the region diagrammed on the left was present upstream of an eve-promoter-lacZ reporter in a P-element transgene, except for 11 of the 19 SR105 lines, which carried the hsp70-lacZ reporter (one of each was homed). Most of the homed lines were discovered to be homed because they expressed β-gal in an eve pattern. The locations of these and of a subset of the `non-homed' insertions were determined by inverse PCR and sequencing. Overall, 57 eZ, 7 hsp70-lacZ and 77 eZAR-MeW insertions that were found by genetic mapping to be on the second chromosome were localized in this way (listed in Table S1 in the supplementary material). As not all of the second chromosome lines were localized by inverse PCR, the number of homed lines might be an underestimate, i.e. there might be additional homed lines among those that do not express β-gal in an eve-like pattern. In most cases, untested lines were not stained because they were not healthy. In other cases, a subset of healthy lines was chosen at random for analysis. Details of insulator/homing regions in eZ constructs: in most cases, the 5′ end of the region (i.e. the end that is normally closer to eve) was oriented towards the eve-lacZ reporter; exceptions are noted below. RR11K lines (which contain eZ constructs) include both attP lines (4 out of 49 established lines found to be homed) and non-attP lines (5/43 homed). The non-attP RR11K lines include: the wild-type element in the orientation described above (3/10 homed); the wild-type element in the opposite orientation (0/5 homed); and modified elements with sequence alterations within the PRE, which is not required for either enhancer blocking or homing (Fujioka et al., 2008) (2/28 homed). NR11K includes lines with the region in the normal orientation (0/9 homed) and lines with the inverted orientation (1/17 homed). RR105 include a homed transgene with an eGFP reporter, which expresses GFP in an eve-like pattern (1/10 homed).
Deletion of subregions A-E (see Fig. 3) individually caused little or no reduction in activity. By contrast, deletion of all five subregions together strongly reduced activity (ΔAE, Fig. 3). Deletion of region F (ΔF) resulted in only weak activity in most lines. Nonetheless, two non-overlapping regions, one containing region F (ΔAE) and the other not (ΔFH), both showed partial activity (Fig. 3). Thus, multiple small regions contribute to activity.
R100 contains the TER94 start site and, with some other insulators, a region near a transcription start site is needed for enhancer blocking (Avramova and Tikhonov, 1999; Kellum and Schedl, 1992; Kuhn et al., 2004). Here, subregion G contains the start site, and although it might contribute to enhancer blocking, it is not required (ΔGH, Fig. 3). ΔGH ends 45 bp upstream of the TER94 start site, yet retains strong activity. Thus, although it seems likely that the functional region overlaps with TER94 regulatory sequences, transcription initiation within the insulator is apparently not required for enhancer blocking. This conclusion is reinforced by the fact that subregion A-F (ΔGH), which does not include the TER94 start site, retains considerably more activity than subregion F-H (ΔAE), which spans the start site, and includes more than 100 bp of upstream sequence.
The 3′ end of the eve locus contains a homing element; homed transgenes engage in long-range enhancer-promoter communication
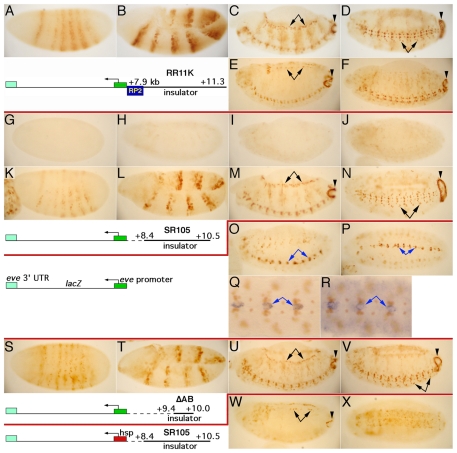
While investigating activities in the border region between eve and TER94, we created a transgene carrying sequences from +7.9 to +11.3 kb driving the eve-promoter-lacZ reporter (RR11K, Fig. 3). This region contains a single eve enhancer (Fujioka et al., 1999), the eve 3′ PRE (Fujioka et al., 2008) and the 5′ portion of TER94 (FlyBase) (Tweedie et al., 2009). Even though this construct contains only one eve enhancer, which is active only in the RP2 and a/pCC cells of the CNS, many lines carrying it nonetheless showed β-gal expression in a full eve pattern (Fig. 4A-D). Inverse PCR revealed that these transgenes had inserted in the chromosomal neighborhood of the eve locus (within 130 kb of eve, green arrowheads in Fig. 5A). We see this phenomenon only when transgenes are inserted in the eve neighborhood. Thus, the transgenic promoter is communicating with endogenous eve enhancers over large distances, across a number of other genes (Fig. 5A). The eve locus has been analyzed for enhancer activities in our lab using hundreds of transgenic lines with the same reporter (Fig. 4, middle diagram), but we have never before observed this phenomenon. This suggested that both homing activity and long-range E-P communication were conferred by the region from +7.9 to +11.3 kb, which also contains the insulator.
Fig. 4.
The eve-TER94 insulator is required for, and the eve promoter contributes to, long-range E-P communication. Staged embryos were subjected to anti-β-gal staining, except Q and R, which show anti-Eve staining (orange) with either lacZ (Q) or CG1623 (R) RNA in situ hybridization (blue). (A-D) RR11K-I (see Table S1 in the supplementary material; diagram below A,B) inserted in the third exon of TER94. Note eve-like stripe (A,B), mesodermal (arrows in C), AR (arrowheads in C and D) and CNS (D) expression. Expression in cells near the midline is from the RP2+a/pCC enhancer (transgenic and/or endogenous), and expression away from the midline, indicated by arrows in D, is from the endogenous eve U/CQ and EL enhancers (compare with Fig. 1C,D). (E) attP-eZRR11K-II, inserted at -1652 kb (relative to the eve start site; see Table S1 in the supplementary material). Note that although AR expression is robust (arrowhead), mesodermal expression is weaker (arrows; it becomes clearly detectable only at this slightly later stage relative to C,M and U). (F) attP-eZRR11K-NN, inserted at -3322 kb (see Table S1 in the supplementary material). Note that AR expression is robust (arrowhead) but that, unlike in D, off-midline CNS expression is not clearly detectable. (G-J) SR105 with the eve-promoter-lacZ reporter (diagram below K,L) inserted on chromosome 1. Note that there is no eve-like expression, showing that the eve insulator and promoter do not have the ability to drive expression unless they communicate with endogenous enhancers. (K-N) The original attP-RR11K insertion at -142 kb (attP-eZRR11K-X, which showed indistinguishable expression) was replaced by SR105 with the eve-promoter-lacZ reporter (diagram below K,L). This and other replacements also differed from the original insertion in not having the mini-white gene. Note the eve-like expression, as seen in A-D. (O-R) The same attP insertion was replaced by the eve-promoter-lacZ reporter without a homing/insulator element (diagram to the left of Q). Note the lack of eve-like expression, showing that the insulator is required for long-range E-P communication. Instead, lateral and midline CNS expression (blue arrows) in non-eve-expressing cells is seen. This was confirmed by double staining for Eve protein and lacZ RNA (Q, shown at higher magnification). The same cells express the gene into which the transgene is inserted (R), showing that without the insulator, the reporter acts as a typical enhancer trap. (S-V) The same attP insertion was replaced by the eve-promoter-lacZ reporter with the 600 bp ΔAB region (diagram below S,T; see Fig. 3). Note that expression is like that in A-D and K-N, showing that this minimal insulator supports long-range E-P communication. (W,X) The same -142 kb attP insertion was replaced by SR105 with the hsp70-lacZ reporter (diagram to the lower left of W). Note that expression in the mesoderm (arrows) and AR (arrowhead) is weaker, and that expression in the CNS (X) is similar to that of TER94 (compare with Fig. 1H). Early stripe expression was barely detectable (not shown).
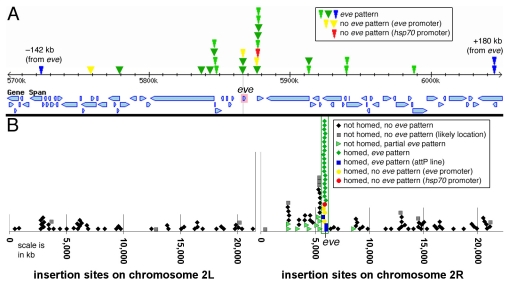
Fig. 5.
Insertion sites of transgenes carrying the insulator/homing region. (A) Genetic map covering 350 kb of chromosome 2R, from 5700 to 6050 kb. The transcription start site of eve (at 5867 kb on this scale) is marked with a vertical gray line. Transcription units (as seen in FlyBase) are shown as blue boxes. Insertion sites of homed lines are indicated by narrow arrowheads for eZ and hsp70-lacZ constructs (above the dotted line in Fig. 3), and wide arrowheads for eZAR-MeW constructs (below the dotted line in Fig. 3). Colors indicate the following: green, transgenes expressing β-gal in an essentially complete eve pattern; blue, attP RR11K insertions that also show an eve-like pattern; red, an SR105 transgene with an hsp70-lacZ reporter not expressed in an eve-like pattern; yellow (near the red arrowhead), CR105 with an eve-promoter-lacZ reporter that gave expression only in the AR; yellow (just upstream of the eve start site), an attP-RR11K insertion that does not give an eve-like β-gal expression. (B) Distribution of insertion sites of eZ and eZAR-MeW transgenes on the second chromosome. The region shown in A is bracketed by two vertical green lines. Note that insertion sites are concentrated near the eve locus, and transgenes that communicate with endogenous eve (green arrowheads) are up to 3300 kb away. Most other insertion lines within this region (all but three of the black diamonds) carry the AR and mesodermal eve enhancers (eZAR-MeW), obscuring potential long-range interactions (see text). Gray squares correspond to the most likely locations of transgenes where inverse PCR-derived sequences gave somewhat ambiguous results (see Table S1 in the supplementary material).
The homing/insulator element can mediate strong E-P communication from over 170 kb away
To further investigate long-range E-P communication, lacZ reporter-carrying transgenes without eve stripe, mesodermal or AR enhancers were used (listed above the dashed line in Fig. 3). Out of 171 lines obtained with these constructs, 143 were examined for an eve-like pattern throughout embryogenesis, and 13 showed such a pattern (Fig. 4A-D). All 13 were localized by inverse PCR to between ∼140 kb upstream and 180 kb downstream of eve (Fig. 5A, narrow green and blue arrowheads). We refer to lines inserted within this neighborhood of eve as `homed'. An additional 44 of these lines that were inserted on the second chromosome but did not show an eve-like pattern were localized by inverse PCR, and three of them were found to be homed (narrow yellow and red arrowheads, Fig. 5A). One of these showed β-gal expression only in the AR (CR105), whereas the other two showed no eve-like expression. One of these two carried the hsp70 basal promoter-lacZ reporter (SR105), and the other carried eve-promoter-lacZ (RR11K).
In the 13 homed lines that show an eve-like lacZ pattern, expression begins around stage 5, with the early eve stripe pattern, and is followed by later eve-like expression in both the mesoderm and AR (Fig. 4A-D). Endogenous eve is also expressed in RP2, a/pCC, CQ/U and EL neurons. Although most of these transgenic insertions (RR11K and RR105; Fig. 3) carry the RP2+a/pCC enhancer (so that expression in these neurons cannot be attributed to the endogenous enhancer), even those that do not carry this enhancer (NR11K, CR105 and SR105; Fig. 3) express β-gal in these neurons. Furthermore, expression of β-gal is also seen in CQ/U and EL neurons, which is attributable only to communication with endogenous eve enhancers. This expression is often weaker and delayed in its appearance relative to endogenous eve expression. Overall, most transgenic insertions within the `homed' region communicate with all of the endogenous eve enhancers, with some variation in the strength of the interaction. All of those insertions that communicate carry the eve-promoter-lacZ reporter, whereas the one that carries hsp70-lacZ (described above) does not. This suggested that promoter specificity might contribute to long-range E-P communication, an idea that we test below.
Transgene clustering outside the `homed' region
The tendency of the homing element to induce transgene insertion in a larger region of chromosome 2R might be observed as an increased frequency of insertion on the entire second chromosome. Based on a random sampling of transgenes not carrying this region from previous studies in our laboratory, the second chromosome insertion frequency is 42% (based on 485 lines). By contrast, even when homed lines are excluded, 47% of transgenes carrying the homing region inserted on the second chromosome (211 out of 446 lines obtained). As can be seen in Fig. 5B, many of these insertions occurred just centromere proximal from the `homed' region, and there are also a number of other regions where several insertion sites are clustered. Within these clusters on the second chromosome, there are five regions (outside the `homed' region) where two or more insertions occurred within 2.5 kb, suggesting that the homing element might be tethering to regions other than the eve-TER94 locus. It will be interesting to determine whether these regions harbor insulators.
It is also noteworthy that within the `homed' region, four insertions are within 2 kb of each other near the promoter of the Mef2 gene, ∼20 kb upstream of the eve promoter (Fig. 5A; see Table S1 in the supplementary material). Such regions might bind protein complexes that interact with the homing element, facilitating transgene insertion and also organizing chromosomal architecture in developing tissues.
The homing/insulator element can mediate E-P communication from 3300 kb away
In addition to homed insertions, there is also clustering of insertions in a larger region of chromosome 2R, mostly centromere proximal from eve. Within this larger region, many lines showed communication with the endogenous eve AR and mesodermal enhancers (Fig. 5B). Out of the aforementioned 143 lines examined for eve-like β-gal expression, ten that were not homed (green triangles, Fig. 5B) showed AR expression (Fig. 4E,F), and in some cases eve-like mesodermal expression (Fig. 4E), but not stripe or CNS (CQ/U and EL neuronal) expression. Two of these lines carried hsp70-lacZ, and eight carried eve-promoter-lacZ. At least nine of these ten lines were inserted on chromosome 2R, within 3400 kb of eve (but outside the `homed' region); the other line could not be localized by inverse PCR. Among these 143 lines, three others were also found to be within this distance of eve, but did not show β-gal expression with any aspect of the eve pattern. Insertions on chromosomes 1, 2L and 3 were also examined, and none showed eve-like expression (Fig. 4G-J).
The other insertions in this region (shown in Fig. 5B as black diamonds) carried the AR and mesodermal enhancers (in the context of the eZAR-MeW enhancer-blocking construct), preventing the determination of whether they communicate with the endogenous eve enhancers. Conversely, the possibility of long-range E-P communication resulted in ambiguity as to whether transgenic eve-promoter-lacZ reporters inserted in this part of the genome were driven by transgenic or endogenous enhancers. Therefore, these lines were not considered in the analysis of enhancer blocking.
The insulator mediates homing
The transgenes constructed for the dissection of enhancer-blocking activity were also tested for homing activity and long-range E-P communication (Figs 3 and 5). We found that there is a close correlation among the required regions for these activities. Thus, the 600 bp ΔAB construct, which retains clear enhancer-blocking activity, is also sufficient for homing and long-range E-P communication, as two homed lines were obtained with this construct (out of 18 tested, Fig. 3), and each showed communication with the endogenous eve enhancers (Fig. 4S-V). Furthermore, out of 210 transgenes tested for homing that carry all or part of the R100 insulator (R100 and those below it in Fig. 3), nine (4.3%) were homed.
These data show that the eve 3′ border region confers three distinct activities, enhancer blocking, homing and long-range E-P communication. The analysis described below further addresses the extent to which these activities are related.
The homing/insulator element mediates long-range E-P communication
To analyze long-range E-P communication further, we used ΦC31 recombinase-mediated cassette exchange (RMCE). In this system, an attB-carrying insertion plasmid can replace an existing attP insertion in a target line, allowing modified elements to be compared in the same chromosomal environment (Bateman et al., 2006; Groth et al., 2004). To create homed target sites, an attP cassette carrying the eve-promoter-lacZ reporter and the homing region from +7.9 to +11.3 kb (RR11K; top diagram in Fig. 4) was inserted by conventional P-element transformation. Two resulting lines inserted at -142 and +180 kb (Fig. 5A, blue arrowheads) were used to dissect the requirements for long-range E-P communication. These lines showed β-gal expression in an eve-like pattern throughout embryogenesis (similar to Fig. 4K-N; data not shown). When each of these insertions was replaced by one that lacked the homing/insulator element, β-gal expression in an eve-like pattern was completely lost (Fig. 4O,P). Instead, expression was observed in non-eve-expressing cells of the CNS, suggesting that the transgenic reporter is responding to non-eve enhancers near the insertion site. This was verified for both insertion sites (Fig. 4Q,R; see Fig. S1 in the supplementary material; data not shown). Thus, the transgenic enhancer-blocking element is required for long-range communication with endogenous eve enhancers.
We next tested whether smaller regions with insulator activity support long-range E-P communication. When we exchanged in the SR105 insulator/homing region, the eve-like β-gal pattern was maintained (Fig. 4K-N). In this version of RMCE, either direction of insertion is possible (Bateman et al., 2006). Although one direction of the modified insertion at -142 kb showed increased ectopic β-gal expression in some cells (see Fig. S1 in the supplementary material), in no case were the eve aspects of pattern affected. Thus, SR105 fully supports long-range E-P communication. We also exchanged both insertions with one carrying the minimal 600 bp insulator (ΔAB, Fig. 3). The β-gal pattern was not affected (Fig. 4S-V; see Fig. S1 in the supplementary material), showing that the minimal insulator is sufficient for long-range E-P communication.
Promoter specificity in long-range communication with eve enhancers
To test whether the transgenic eve promoter contributes to long-range E-P communication, a cassette carrying hsp70-lacZ (along with the SR105 insulator/homing element) was exchanged into both attP target sites. Compared with the same cassette carrying eve-promoter-lacZ, the early striped pattern was severely weakened (see Fig. S1 in the supplementary material; data not shown), and AR and mesoderm expression were delayed and at a reduced intensity (Fig. 4W; see Fig. S1 in the supplementary material). Moreover, in the CNS, β-gal was expressed ubiquitously, which is similar to the expression of TER94 (Fig. 4X, compare with Fig. 1H). These data indicate that, although the insulator/homing region is required for long-range E-P communication, the transgenic eve promoter also contributes to the efficient communication with endogenous eve enhancers. These data further suggest that the eve promoter communicates preferentially with eve enhancers over those of TER94. However, as described above, we have also seen that this basal hsp70 promoter can communicate with the endogenous eve AR and mesodermal enhancers over much greater distances (Fig. 4E,F), when present in a transgene with the insulator/homing element. Thus, the insulator/homing element is the primary determinant of long-range E-P communication, and the eve promoter contributes to its strength and enhancer preference.
DISCUSSION
An insulator separates eve and TER94
Some of the eve enhancers are close to the TER94 promoter, yet they do not activate TER94. Although TER94 is expressed nearly ubiquitously in embryos, it is expressed only at a low level in the mesoderm and anal plate, where eve expression is high in a subset of cells, making it unlikely that eve enhancers acting on TER94 would be masked by this expression (Fig. 1). Therefore, something isolates TER94 from eve enhancers (and probably vice versa). Indeed, the region between the 3′-most eve regulatory element, a PRE, and the TER94 transcription start site has the properties of an enhancer-blocking insulator (Figs 2 and 3). It exhibits directional enhancer blocking in transgenes carrying eve enhancers in combination with either the eve promoter region or heterologous promoters, as well as between heterologous enhancers and promoters.
The role of the TER94 promoter in enhancer blocking
We dissected this insulator region in the context of transgenes carrying two different enhancers between divergently transcribed reporter genes (Fig. 2I-N). Some deletion mutants were still able to block the AR enhancer from activating the mini-white reporter, while allowing the eve mesodermal enhancer to activate the eve-promoter-lacZ reporter across the mutant insulator (Fig. 3). This might result from a relatively weak interaction between the eve AR enhancer and the heterologous mini-white promoter, which suggests a degree of specificity of eve enhancers for their cognate promoter. This mechanism also contributes to long-range E-P communication mediated by the insulator, as discussed below. Furthermore, the recently discovered presence of an insulator at the 3′ end of mini-white (Chetverina et al., 2008) might contribute to stronger enhancer blocking in this direction.
We first narrowed enhancer-blocking activity down to an 800 bp sequence (R100, Fig. 3) that spans the 5′ end of TER94. Further dissection showed that the start site of TER94 is not required (ΔGH, Fig. 3). This makes it unlikely that transcriptional interference (Martianov et al., 2007; Mazo et al., 2007) makes a strong contribution to our results, although it could be significant in some cases, such as for ΔF, which retains the TER94 start site. Notably, region F, extending from ∼150 to 45 bp upstream of this start site, seems particularly important for enhancer blocking. A similar situation pertains to the well-studied insulators scs and scs′ (Avramova and Tikhonov, 1999; Geyer, 1997; Kellum and Schedl, 1992; Kuhn et al., 2004). Perhaps some promoter regions induce a chromatin configuration that blocks the progression of activating complexes or chromatin modifications, through which enhancers communicate with target promoters.
The eve-TER94 insulator and homing activity
The region between eve and TER94 also induces transgene homing. About 7% of transgenes carrying this region (27 out of 380 lines tested) inserted within 180 kb of eve. Among 27 homed lines, eight inserted within 1.5 kb of the endogenous insulator (Fig. 5), suggesting that homing involves direct tethering, possibly through a homophilic protein complex formed on the element in the germline, where transgenic insertion occurs. We call the responsible element Homie, for homing insulator at eve.
Although it is more difficult to dissect the region required for homing than it is to dissect the region required for enhancer blocking (due to the number of transgenic insertions required to validate a negative result), there is a clear correlation between these activities. Of the 210 transgenes tested for homing that carry all or part of the 800 bp R100 insulator (Fig. 3), nine of them (4.3%) were homed, even though the `homed' region is less than 0.4% of the genome. Protein-protein interactions among insulators, when they occur in the germline, might lead to transgene homing.
The eve PRE and homing activity
In previous studies of the eve 3′ region, we produced hundreds of lines that carried the eve PRE, yet we did not observe homing. Therefore, the eve PRE is not sufficient for homing. Furthermore, as the minimal homing element does not contain the PRE, this PRE is not required for either homing activity or long-range E-P communication. However, the engrailed homing region has PRE activity (Kwon et al., 2009), indicating that some PREs may engage in homotypic interactions that facilitate homing. Consistent with this, long-range interactions among PREs were seen in the BX-C (Lanzuolo et al., 2007). Furthermore, the engrailed PRE may also facilitate long-distance E-P communication (Devido et al., 2008).
An extensive genomic region is a target for Homie-mediated homing and very long-range E-P communication
The eve-promoter-lacZ reporter in a homed transgene is usually expressed in a full eve pattern, showing communication with all of the endogenous eve enhancers from as far away as 180 kb, and across a number of other genes (Fig. 4; Fig. 5A). Beyond the homing target region, there is a tendency for Homie-carrying transgenes to insert on chromosome 2R, particularly centromere proximal from eve (Fig. 5B; additionally, Table S1 in the supplementary material lists the locations of all these mapped transgenic inserts). We have not referred to these insertions as `homed', mainly to distinguish them from transgenes that pick up a full eve pattern of expression. However, they usually (9 out of 12) pick up a partial eve pattern. Intriguingly, Homie-carrying transgenes inserted as far as 3300 kb away (Fig. 5B; see Table S1 in the supplementary material), are capable of interacting with the endogenous eve AR and mesodermal enhancers (Fig. 4E,F). Previous indications of long-range E-P interactions mediated by transgenic insulators have come from the genetic and phenotypic analysis of transvection (Kravchenko et al., 2005) and related regulatory interactions (Hendrickson and Sakonju, 1995; Hopmann et al., 1995; Sipos et al., 1998).
We directly tested the requirement for Homie in long-range E-P communication using ΦC31-RMCE to compare transgenes with and without this region at the same chromosomal insertion site. Removal of Homie resulted in complete loss of the eve pattern. The same results were obtained at two different landing sites, at opposite ends of the homing region (Fig. 5A, blue arrowheads). Communication of distant `shadow' enhancers with promoters across several intervening genes has recently been proposed, based upon bioinformatics-based identification of functionally conserved enhancer regions with no other apparent target promoters (Hong et al., 2008). Our results suggest that for such distant enhancers to communicate effectively, they may need promoter-targeting and/or promoter-tethering sequences (Akbari et al., 2008; Zhou and Levine, 1999), and that some of these sequences might also act as insulators, generating a chromosomal architecture that facilitates functionally important interactions while preventing deleterious ones.
How does Homie mediate such long-range E-P communication? Both preferential insertion and the ability to pick up a partial eve pattern from long range could be explained by a homologous tethering mechanism (Bantignies et al., 2003; Vazquez et al., 2006), if we assume that this region of 2R is in relative proximity to the eve locus within a chromosome territory (Cremer and Cremer, 2001), both in the germline and in the developing AR and mesoderm. Homologous tethering might stabilize a functional E-P interaction, which in turn might facilitate transcription initiation through a combination of mechanisms, including targeting to regions of active transcription within the nucleus (de Laat and Grosveld, 2003; Fraser, 2006; Simonis and de Laat, 2008).
Promoter specificity in long-range E-P communication
We used RMCE to test the role of promoter specificity in long-range communication. Exchanging a basal hsp70 promoter for the eve promoter caused a complete loss of communication with some endogenous eve enhancers but not others. The communication that remained was with the AR and mesodermal enhancers, the same ones that often communicate with either the eve or hsp70 promoters in transgenes inserted up to 3300 kb away (Fig. 5B; see Table S1 in the supplementary material). The ability of these enhancers to communicate at a much longer range than others might indicate relatively stable E-P interactions that can survive entropic forces tending to randomize their positions in the nucleus. Alternatively, the interactions of these enhancers might be specifically facilitated by Homie.
Another indication of the effects of promoter specificity in long-range E-P communication is that when the eve promoter was replaced by that of hsp70, β-gal reporter expression in the CNS changed from an eve-like pattern to one similar to that of TER94 (Fig. 4X). Although it is possible that this TER94-like expression is driven by enhancers located near the insertion site, it is clear that which enhancers are targeted by the transgenic promoter depends in part on promoter specificity. Similar influences have recently been found on E-P communication at the engrailed locus (Kwon et al., 2009).
Reconciling insulator with long-range facilitator functions
How can Homie act as an insulator and also mediate long-range communication? The key may lie in the details of the resulting chromosomal architecture. Precedence for this idea comes from the phenomenon of insulator bypass, in which the enhancer-blocking activity of a single insulator can be negated by placing a second insulator between the enhancer and promoter (Cai and Shen, 2001; Muravyova et al., 2001). This phenomenon is consistent with data from those homed insertions that lie just downstream of endogenous Homie. In these cases, both the transgenic and endogenous Homies are interposed between the lacZ reporter and the endogenous enhancers that drive its expression. Our data also show that the apparent bypass of endogenous Homie does not require that transgenic Homie lies between the interacting enhancer and promoter. In one case, the transgenic promoter lies between the two Homies, with the interacting enhancers on the outside. We propose that Homie has directionality, so that the two copies of Homie line up in parallel with each other within a wall-like structure. In the cases where both Homies are between the interacting enhancer and promoter, the Homies are inverted in orientation, whereas in the other case they are in the same orientation. In both cases, their lining up in parallel would tend to place the interacting enhancer and promoter on the same side of this wall-like structure, facilitating their communication. By contrast, a single copy of Homie would tend to block communication between sequences on either side, by placing them on opposite sides of the structure. Similar effects of insulator directionality have been seen for the Fab-8 and Mcp insulators (Kyrchanova et al., 2007; Kyrchanova et al., 2008).
Insulator directionality in facilitating long-range communication
In most homed lines, we did not see mini-white expression in an eve pattern. This might be due to the mini-white promoter being relatively weak and/or less compatible with eve enhancers than is the eve promoter, or even the hsp70 promoter, which also often picked up AR or mesodermal enhancer activity from great distances (facilitated by Homie). Intriguingly, however, although in most of the transgenes carrying Homie its 5′ end was oriented toward the lacZ reporter, in one line (inserted at +46 kb), this orientation was reversed, and in that line mini-white was expressed in the eve pattern. Thus, it is possible that Homie directionality, through the mechanism described above for insulator bypass, might play a role in determining whether or not a weak E-P interaction is facilitated.
What is the normal function of Homie?
There are two likely possibilities for how Homie functions in the regulation of eve and TER94. The first is that it simply prevents eve enhancers from activating TER94, and also prevents eve from being expressed broadly in the CNS like TER94, which would probably cause mis-specification of neurons (Broihier and Skeath, 2002). Another, not mutually exclusive, possibility is that Homie works in conjunction with the nearby PRE to orchestrate functionally appropriate chromosomal architectures during development. Known insulators in the BX-C are each situated near a PRE (Maeda and Karch, 2006), and these PRE-insulator regions interact with promoters in several contexts (Cleard et al., 2006; Lanzuolo et al., 2007). Our data suggest a similar interaction with the eve promoter region, based on the fact that three of our homed lines are inserted within the eve promoter region. Such an interaction might help enhancers from the 3′ end of the eve locus communicate with the eve promoter, while also preventing inappropriate interaction with TER94 enhancers. One motivation for such a model is that in mutants for the PcG gene polyhomeotic, eve is ectopically expressed throughout the CNS (Smouse et al., 1988), which is reminiscent of normal TER94 expression. Thus a loss of PcG repression, acting through the PRE, might disrupt the normal insulator function that prevents inappropriate activation of eve. This suggests that the functions of the PRE and Homie are coordinated during development, allowing the PRE to maintain either an activated or repressed state of eve in different cells (Fujioka et al., 2008), while maintaining the functional isolation of eve from TER94.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/18/3077/DC1
Supplementary Material
We thank Galina Yusibova and Jian Zhou for excellent technical assistance, Michele P. Calos, C.-Ting Wu and Jack R. Bateman for ΦC31 system reagents, Paul Schedl for pCfhL, and Judy Kassis for helpful discussions and for comments on the manuscript. This work was supported by NSF-MCB0818118 and NIH-R01GM050231 awards to J.B.J. and M.F. Deposited in PMC for release after 12 months.
References
- Akbari, O. S., Bae, E., Johnsen, H., Villaluz, A., Wong, D. and Drewell, R. A. (2008). A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development 135, 123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, T., Schweinsberg, S., Manasson, J. and Schedl, P. (2008). A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila. Mol. Cell. Biol. 28, 1047-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, Z. and Tikhonov, A. (1999). Are scs and scs' `neutral' chromatin domain boundaries of the locus? Trends Genet. 15, 138-139. [DOI] [PubMed] [Google Scholar]
- Bantignies, F., Grimaud, C., Lavrov, S., Gabut, M. and Cavalli, G. (2003). Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17, 2406-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges, S., Mihaly, J., Galloni, M., Hagstrom, K., Muller, M., Shanower, G., Schedl, P., Gyurkovics, H. and Karch, F. (2000). The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127, 779-790. [DOI] [PubMed] [Google Scholar]
- Bateman, J. R., Lee, A. M. and Wu, C. T. (2006). Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173, 769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A. C. and Felsenfeld, G. (2000). Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482-485. [DOI] [PubMed] [Google Scholar]
- Bell, A. C., West, A. G. and Felsenfeld, G. (1999). The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387-396. [DOI] [PubMed] [Google Scholar]
- Belozerov, V. E., Majumder, P., Shen, P. and Cai, H. N. (2003). A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 22, 3113-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, W. and Hudson, A. (2000). P element homing to the Drosophila bithorax complex. Development 127, 3981-3992. [DOI] [PubMed] [Google Scholar]
- Blanton, J., Gaszner, M. and Schedl, P. (2003). Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17, 664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broihier, H. T. and Skeath, J. B. (2002). Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron 35, 39-50. [DOI] [PubMed] [Google Scholar]
- Bushey, A. M., Dorman, E. R. and Corces, V. G. (2008). Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia, A., Wightman, C. D. and Sakonju, S. (1997). A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 124, 4343-4350. [DOI] [PubMed] [Google Scholar]
- Cai, H. N. and Shen, P. (2001). Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291, 493-495. [DOI] [PubMed] [Google Scholar]
- Chetverina, D., Savitskaya, E., Maksimenko, O., Melnikova, L., Zaytseva, O., Parshikov, A., Galkin, A. V. and Georgiev, P. (2008). Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 36, 929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard, F., Moshkin, Y., Karch, F. and Maeda, R. K. (2006). Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38, 931-935. [DOI] [PubMed] [Google Scholar]
- Cremer, T. and Cremer, C. (2001). Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292-301. [DOI] [PubMed] [Google Scholar]
- de Laat, W. and Grosveld, F. (2003). Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11, 447-459. [DOI] [PubMed] [Google Scholar]
- Devido, S. K., Kwon, D., Brown, J. L. and Kassis, J. A. (2008). The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development 135, 669-676. [DOI] [PubMed] [Google Scholar]
- Dorman, E. R., Bushey, A. M. and Corces, V. G. (2007). The role of insulator elements in large-scale chromatin structure in interphase. Semin. Cell Dev. Biol. 18, 682-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch, M., Warrior, R., Tugwood, J. and Levine, M. (1988). Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev. 2, 1824-1838. [DOI] [PubMed] [Google Scholar]
- Fraser, P. (2006). Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 16, 490-495. [DOI] [PubMed] [Google Scholar]
- Fujioka, M., Emi-Sarker, Y., Yusibova, G. L., Goto, T. and Jaynes, J. B. (1999). Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126, 2527-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, M., Jaynes, J. B., Bejsovec, A. and Weir, M. (2000). Production of transgenic Drosophila. Methods Mol. Biol. 136, 353-363. [DOI] [PubMed] [Google Scholar]
- Fujioka, M., Yusibova, G. L., Zhou, J. and Jaynes, J. B. (2008). The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development 135, 4131-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner, M. and Felsenfeld, G. (2006). Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7, 703-713. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., Vazquez, J. and Schedl, P. (1999). The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13, 2098-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause, M., Morcillo, P. and Dorsett, D. (2001). Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21, 4807-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. K. (1997). The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev. 7, 242-248. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K. and Corces, V. G. (1992). DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6, 1865-1873. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., Green, M. M. and Corces, V. G. (1988). Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc. Natl. Acad. Sci. USA 85, 8593-8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, D., Gerasimova, T. I. and Corces, V. G. (2001). Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20, 2518-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G. and Lindquist, S. (1989). The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59, 499-509. [DOI] [PubMed] [Google Scholar]
- Goto, T., Macdonald, P. and Maniatis, T. (1989). Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57, 413-422. [DOI] [PubMed] [Google Scholar]
- Groth, A. C., Fish, M., Nusse, R. and Calos, M. P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics, H., Gausz, J., Kummer, J. and Karch, F. (1990). A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9, 2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom, K., Muller, M. and Schedl, P. (1996). Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10, 3202-3215. [DOI] [PubMed] [Google Scholar]
- Hama, C., Ali, Z. and Kornberg, T. B. (1990). Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 4, 1079-1093. [DOI] [PubMed] [Google Scholar]
- Harding, K., Hoey, T., Warrior, R. and Levine, M. (1989). Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 8, 1205-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M. and Tilghman, S. M. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486-489. [DOI] [PubMed] [Google Scholar]
- Hart, C. M., Zhao, K. and Laemmli, U. K. (1997). The scs' boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17, 999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, J. E. and Sakonju, S. (1995). Cis and trans interactions between the iab regulatory regions and abdominal-A and Abdominal-B in Drosophila melanogaster. Genetics 139, 835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan, E. E., Kwong, C., Adryan, B., Bartkuhn, M., Herold, M., Renkawitz, R., Russell, S. and White, R. (2007). CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, J. W., Hendrix, D. A. and Levine, M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science 321, 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann, R., Duncan, D. and Duncan, I. (1995). Transvection in the iab-5,6,7 region of the bithorax complex of Drosophila: homology independent interactions in trans. Genetics 139, 815-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. M., Rehm, E. J. and Rubin, G. M. (2000). Recovery of DNA sequences flanking P-element insertions: inverse PCR and plasmid rescue. In Drosophila Protocols (ed. W. Sullivan, M. Ashburner and R. S. Hawley), pp. 429-437. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [DOI] [PubMed]
- Kanduri, C., Pant, V., Loukinov, D., Pugacheva, E., Qi, C. F., Wolffe, A., Ohlsson, R. and Lobanenkov, V. V. (2000). Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10, 853-856. [DOI] [PubMed] [Google Scholar]
- Karch, F., Galloni, M., Sipos, L., Gausz, J., Gyurkovics, H. and Schedl, P. (1994). Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22, 3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis, J. A. (2002). Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46, 421-438. [DOI] [PubMed] [Google Scholar]
- Kassis, J. A., Noll, E., VanSickle, E. P., Odenwald, W. F. and Perrimon, N. (1992). Altering the insertional specificity of a Drosophila transposable element. Proc. Natl. Acad. Sci. USA 89, 1919-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum, R. and Schedl, P. (1992). A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12, 2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko, E., Savitskaya, E., Kravchuk, O., Parshikov, A., Georgiev, P. and Savitsky, M. (2005). Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 25, 9283-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, E. J., Hart, C. M. and Geyer, P. K. (2004). Studies of the role of the Drosophila scs and scs' insulators in defining boundaries of a chromosome puff. Mol. Cell. Biol. 24, 1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, D., Mucci, D., Langlais, K. K., Americo, J. L., DeVido, S. K., Cheng, Y. and Kassis, J. A. (2009). Enhancer-promoter communication at the Drosophila engrailed locus. Development 136, 3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova, O., Toshchakov, S., Parshikov, A. and Georgiev, P. (2007). Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 27, 3035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova, O., Toshchakov, S., Podstreshnaya, Y., Parshikov, A. and Georgiev, P. (2008). Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 28, 4188-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo, C., Roure, V., Dekker, J., Bantignies, F. and Orlando, V. (2007). Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 9, 1167-1174. [DOI] [PubMed] [Google Scholar]
- Leon, A. and McKearin, D. (1999). Identification of TER94, an AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Mol. Biol. Cell 10, 3825-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Hu, J. F., Qiu, X., Ling, J., Chen, H., Wang, S., Hou, A., Vu, T. H. and Hoffman, A. R. (2008). CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 28, 6473-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, R. K. and Karch, F. (2006). The ABC of the BX-C: the bithorax complex explained. Development 133, 1413-1422. [DOI] [PubMed] [Google Scholar]
- Maeda, R. K. and Karch, F. (2007). Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 17, 394-399. [DOI] [PubMed] [Google Scholar]
- Majumder, P. and Cai, H. N. (2003). The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 100, 5223-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov, I., Ramadass, A., Serra Barros, A., Chow, N. and Akoulitchev, A. (2007). Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666-670. [DOI] [PubMed] [Google Scholar]
- Mazo, A., Hodgson, J. W., Petruk, S., Sedkov, Y. and Brock, H. W. (2007). Transcriptional interference: an unexpected layer of complexity in gene regulation. J. Cell Sci. 120, 2755-2761. [DOI] [PubMed] [Google Scholar]
- Mihaly, J., Hogga, I., Gausz, J., Gyurkovics, H. and Karch, F. (1997). In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124, 1809-1820. [DOI] [PubMed] [Google Scholar]
- Modolell, J., Bender, W. and Meselson, M. (1983). Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc. Natl. Acad. Sci. USA 80, 1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, M., Hagstrom, K., Gyurkovics, H., Pirrotta, V. and Schedl, P. (1999). The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153, 1333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova, E., Golovnin, A., Gracheva, E., Parshikov, A., Belenkaya, T., Pirrotta, V. and Georgiev, P. (2001). Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291, 495-498. [DOI] [PubMed] [Google Scholar]
- Ochman, H., Gerber, A. S. and Hartl, D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki, S. and Levine, M. (1998). GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 12, 3325-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba, K., Gutierrez, L., Gagneur, J., Girardot, C., Sengupta, A. K., Furlong, E. E. and Muller, J. (2008). Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15, 877-889. [DOI] [PubMed] [Google Scholar]
- Pai, C. Y., Lei, E. P., Ghosh, D. and Corces, V. G. (2004). The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16, 737-748. [DOI] [PubMed] [Google Scholar]
- Parelho, V., Hadjur, S., Spivakov, M., Leleu, M., Sauer, S., Gregson, H. C., Jarmuz, A., Canzonetta, C., Webster, Z., Nesterova, T. et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422-433. [DOI] [PubMed] [Google Scholar]
- Parnell, T. J., Kuhn, E. J., Gilmore, B. L., Helou, C., Wold, M. S. and Geyer, P. K. (2006). Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 26, 5983-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer, M. and Bender, W. (1988). Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc. Natl. Acad. Sci. USA 85, 9650-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter, M., Jekely, G., Szepesi, R. J., Farkas, A., Theopold, U., Meyer, H. E., Lindholm, D., Nassel, D. R., Hultmark, D. and Friedrich, P. (1998). TER94, a Drosophila homolog of the membrane fusion protein CDC48/p97, is accumulated in nonproliferating cells: in the reproductive organs and in the brain of the imago. Insect Biochem. Mol. Biol. 28, 91-98. [DOI] [PubMed] [Google Scholar]
- Ramos, E., Ghosh, D., Baxter, E. and Corces, V. G. (2006). Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172, 2337-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M. and Spradling, A. C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348-353. [DOI] [PubMed] [Google Scholar]
- Ruden, D. M., Sollars, V., Wang, X., Mori, D., Alterman, M. and Lu, X. (2000). Membrane fusion proteins are required for oskar mRNA localization in the Drosophila egg chamber. Dev. Biol. 218, 314-325. [DOI] [PubMed] [Google Scholar]
- Sackerson, C., Fujioka, M. and Goto, T. (1999). The even-skipped locus is contained in a 16-kb chromatin domain. Dev. Biol. 211, 39-52. [DOI] [PubMed] [Google Scholar]
- Schweinsberg, S. E. and Schedl, P. (2004). Developmental modulation of Fab-7 boundary function. Development 131, 4743-4749. [DOI] [PubMed] [Google Scholar]
- Schweinsberg, S., Hagstrom, K., Gohl, D., Schedl, P., Kumar, R. P., Mishra, R. and Karch, F. (2004). The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168, 1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis, M. and de Laat, W. (2008). FISH-eyed and genome-wide views on the spatial organisation of gene expression. Biochim. Biophys. Acta 1783, 2052-2060. [DOI] [PubMed] [Google Scholar]
- Sipos, L., Mihaly, J., Karch, F., Schedl, P., Gausz, J. and Gyurkovics, H. (1998). Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 149, 1031-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, S., Blair, A. and Levine, M. (1992). Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, S., Blair, A. and Levine, M. (1996). Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 175, 314-324. [DOI] [PubMed] [Google Scholar]
- Smouse, D., Goodman, C., Mahowald, A. and Perrimon, N. (1988). polyhomeotic: a gene required for the embryonic development of axon pathways in the central nervous system of Drosophila. Genes Dev. 2, 830-842. [DOI] [PubMed] [Google Scholar]
- Spana, C., Harrison, D. A. and Corces, V. G. (1988). The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2, 1414-1423. [DOI] [PubMed] [Google Scholar]
- Taillebourg, E. and Dura, J. M. (1999). A novel mechanism for P element homing in Drosophila. Proc. Natl. Acad. Sci. USA 96, 6856-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak, P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S. E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S. E. et al. (2002). Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3, 0088.1-0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie, S., Ashburner, M., Falls, K., Leyland, P., McQuilton, P., Marygold, S., Millburn, G., Osumi-Sutherland, D., Schroeder, A., Seal, R. et al. (2009). FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37, D567-D570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy, A., Maine, E. and Schedl, P. (1985). The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185, 341-358. [DOI] [PubMed] [Google Scholar]
- Valenzuela, L. and Kamakaka, R. T. (2006). Chromatin insulators. Annu. Rev. Genet. 40, 107-138. [DOI] [PubMed] [Google Scholar]
- Vazquez, J., Muller, M., Pirrotta, V. and Sedat, J. W. (2006). The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol. Biol. Cell 17, 2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt, K. S., Yoshida, K., Itoh, T., Bando, M., Koch, B., Schirghuber, E., Tsutsumi, S., Nagae, G., Ishihara, K., Mishiro, T. et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796-801. [DOI] [PubMed] [Google Scholar]
- Zhao, K., Hart, C. M. and Laemmli, U. K. (1995). Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81, 879-889. [DOI] [PubMed] [Google Scholar]
- Zhou, J. and Levine, M. (1999). A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell 99, 567-575. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Ashe, H., Burks, C. and Levine, M. (1999). Characterization of the transvection mediating region of the Abdominal-B locus in Drosophila. Development 126, 3057-3065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.