Abstract
Many human cancers show constitutive or amplified expression of the transcriptional regulator and oncoprotein Myc, making Myc a potential target for therapeutic intervention. Here we report the down-regulation of Myc activity by reducing the availability of Max, the essential dimerization partner of Myc. Max is expressed constitutively and can form unstable homodimers. We have isolated stabilizers of the Max homodimer by applying virtual ligand screening (VLS) to identify specific binding pockets for small molecule interactors. Candidate compounds found by VLS were screened by fluorescence resonance energy transfer, and from these screens emerged a potent, specific stabilizer of the Max homodimer. In vitro binding assays demonstrated that the stabilizer enhances the formation of the Max-Max homodimer and interferes with the heterodimerization of Myc and Max in a dose-dependent manner. Furthermore, this compound interferes with Myc-induced oncogenic transformation, Myc-dependent cell growth, and Myc-mediated transcriptional activation. The Max-Max stabilizer can be considered a lead compound for the development of inhibitors of the Myc network.
The transcriptional regulator Myc shows gain of function in a large variety of human cancers (Nesbit et al., 1999; Lutz et al., 2002), and increased Myc activity is correlated with poor prognosis (Adler et al., 2006). Myc is widely expressed in proliferating cells and down-regulated in differentiated cells (Eilers, 1999).
Myc mediates progression of the cell cycle by functioning as a transcriptional activator (Lüscher, 2001). Myc belongs to a network of basic helix-loop-helix leucine zipper (bHLHLZ) transcription factors that can activate or repress transcrpition as heterodimers with a single member of the same protein family, the Max protein (Blackwood and Eisenman, 1991; Lüscher, 2001). Interaction of the Myc-Max heterodimer with DNA at a consensus “E-box” binding site leads to the recruitment of additional transcriptional activators via the transactivation domain of Myc (Blackwood and Eisenman, 1991; McMahon et al., 1998).
Unlike Myc, the Max protein can homodimerize in vitro and in vivo (Blackwood and Eisenman, 1991; Blackwood et al., 1992). Max homodimers are less stable than Myc-Max heterodimers or other heterodimers of the Myc network (Fieber et al., 2001). The reduced stability of the Max homodimer results from a packing defect at its protein-protein interface (Nair and Burley, 2003). At physiological levels, Max homodimers fail to regulate transcription, but Max overexpression can lead to reporter gene repression (Kretzner et al., 1992; Yin et al., 1998). Overexpressed Max reduces Myc-induced carcinogenesis (Cogliati et al., 1993; Lindeman et al., 1995). In human cancer, higher Max levels are associated with a better prognosis (Yuza et al., 1999).
Small molecule inhibitors of Myc-Max dimerization have been identified (Berg et al., 2002; Yin et al., 2003; Xu et al., 2006; Follis et al., 2008). These inhibitors reduce Myc-induced DNA binding, transcriptional activation and oncogenic transformation.
An effective route to predicting inhibitors of protein-protein interactions is virtual ligand screening (VLS) (Brooijmans and Kuntz, 2003). The AutoDock Software suite has been used successfully to find inhibitors from chemical databases (Li et al., 2004; Dickerson et al., 2005; Rogers et al., 2006). The accuracy of VLS is limited by the structural information for the protein target. The discovery of inhibitors of protein-protein interactions is facilitated by a well-defined binding cavity at the protein-protein interface where a small molecule can compete with protein association.
The Myc protein is only partially structured in its uncomplexed form (Fieber et al., 2001), whereas the Myc-Max and Max-Max dimers are highly structured (Nair and Burley, 2003). The dimer structures are therefore more promising in silico docking targets for small-molecule interactions. We hypothesized that the docked molecules would most likely stabilize the bHLHLZ dimers, and that stabilization of the Max homodimer would reduce the availability of Max to heterodimerize with Myc and with other proteins in the network. This could result in a down-regulation of the entire Myc network. In cancer cells that overexpress Myc, the Myc-Max heterodimer may be inhibited preferentially compared with the other dimers of the network that are expressed at much lower levels. Because the packing defect of the Max homodimer is unique, we argued that this unstable dimer could be an excellent target for specific small molecule interactions.
Small molecule intervention in protein-protein interaction is usually aimed at inhibiting the association of the protein-protein partner (Berg, 2003). As an indirect way of interfering with the formation of the Myc-Max target dimer, we propose stabilization of the competing Max homodimer. Tying up Max in homodimer structures should result in the inhibition of Myc function in a cellular environment.
We present here the identification of stabilizers of the Max homodimer by VLS. To our knowledge, this is the first computational study to screen a large database of compounds over an entire structural element. The results focus on a lead molecule that is active as inhibitor of Myc-mediated transcription and oncogenic transformation.
Materials and Methods
Chemical Libraries. Compounds were selected from the National Cancer Institute (NCI) Diversity Set (Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis). For VLS AutoDock analyses, the Diversity Set was used in pdbq format, which includes Gasteiger partial charges and fully rotatable chemical bonds. The library represents the chemical diversity of 140,000 compounds available from the NCI and is restricted to compounds that have few rotatable chemical bonds, which is correlated with increased oral bioavailability (Veber et al., 2002). Of the 1990 compounds in the Diversity Set, 1668 could be modeled with accurate geometry and charges and were used in the VLS calculations. Molecules predicted to interact with the Myc-Max and/or Max-Max dimers were ordered from the NCI program and used in subsequent studies. The control Myc-Max dimerization inhibitor (Z,E)-5-(4-ethylbenzylidine)-2-thioxothiazolidin-4-one was generated by the lab of Ed Prochownik and obtained from Calbiochem (San Diego, CA). It was described in Yin et al. (2003) and is referred to here as compound ID 10058-F4.
Molecular Modeling. The AutoDock 3.0.5 software suite (Morris et al., 1998) was used to dock compounds to Myc-Max and Max-Max dimers (Protein Data Bank codes 1nkp and 1an2). The protein structures were processed with the graphical user interface AutoDock-Tools to prepare subsequent files for use in AutoDock. “Kollman” partial charges and Stouten solvation parameters were calculated for the pdb files and used to calculate energy maps for all relevant atom types using AutoGrid. The energy grids were sized at 60 × 126 × 60 Å, with the long axis parallel to the major axes of the Myc-Max and Max-Max structures, and had a grid spacing of 1 Å. Compounds were docked using the Lamarckian Genetic-Algorithm Local-Search algorithm in AutoDock, which evaluates a population of possible ligand states and retains those with the lowest predicted docking energies to be used in subsequent generations of ligand populations. The top docking solutions were also subject to mutations—or random changes—in the docking solution, as well as crossover. A stochastic local search was applied to these solutions to ensure local minimization. A generation population of 200 ligand states, a mutation rate of 0.02, a crossover rate of 0.8, and a local search probability of 0.06 were used in the docking calculations.
Because of the large size of the grids used in the docking calculations, 108 energy evaluations were used per docking calculation. Docking calculations were run on a cluster of 256 2.4-GHz dual processor Intel XEON CPUs. Eight separate docking calculations were used for each compound. Results from all eight runs from each compound were clustered based on their root-mean-square distance from each other to gauge the convergence of the docking calculations. At 100 million evaluations, more than 98% of the docking results had root-mean-square distance clusters, indicating that the positions of the eight separate docking runs were converging toward the same solution. The lowest docking energy and predicted free binding energy for each compound from the eight docking runs were retained. These energies were used to sort the VLS results and are the values used throughout the text. The predicted aqueous partition coefficient was calculated for all compounds using the XLOGP program (Wang et al., 1997) to filter away compounds with a low predicted solubility.
Clustering Analysis. Docking results were clustered based on their predicted binding location. The compound with the lowest predicted energy was used to seed each cluster. Other compounds within 10 Å of the seed compound center of gravity were added to that cluster. Of the remaining compounds, the one with the lowest energy was used to seed the next cluster, and the process was repeated until all compounds belonged to a cluster. Clusters were visually inspected for overlap because of differences in compound size, and those related by the Max-Max pseudo-symmetry were merged. Binding sites identified were similar to those discovered by the flood fill method for determining protein binding pockets (Beuscher et al., 2005), which directly searches the AutoGrid maps for contiguous regions of highly negative intermolecular binding potential.
Fluorescence Resonance Energy Transfer. Expression and purification of recombinant proteins Max, MycCFP, MaxCFP, and MaxYFP have been described previously (Berg et al., 2002). The bHLHLZ domains of Myc and of Max fused to the C terminus of cyan fluorescent protein (CFP) were allowed to dimerize in H2O with the bHLHLZ domain of Max fused to yellow fluorescent protein (YFP) protein in the presence or absence of test compounds at room temperature for 5 min. All proteins were used at 85 nM concentration. Fluorescence intensities were measured between 460 and 570 nm using a spectrofluorometer (LS 50B; PerkinElmer Life and Analytical Sciences, Waltham, MA). FRET, measured as the ratio of fluorescence intensity I525/I475, defined the degree of dimerization. In the absence of compound, Myc-Max and Max-Max FRET ratios were set to 1.0 (Table 1). In the presence of compound, stabilization was defined as an increase in the FRET ratio, and inhibition was defined as a decrease in the FRET ratio. Ratios were normalized to those in the absence of compound to define percentage stabilization or percentage inhibition.
TABLE 1.
FRET stabilization of Max-Max homodimerization by NCI compounds
In silico docking cluster and binding energy against the Max homodimer are shown for each compound, as are the measured FRET dimerization ratios against the Max-Max and Myc-Max homodimers. Cluster refers to predicted interaction with binding sites in Fig. 1A. FRET data are measured as the ratio I525/I475. Ratios are normalized against Max-Max or Myc-Max dimerization in the absence of compound, set independently to 1.00.
| Compound | Cluster | Binding Energy | Max-Max | Myc-Max |
|---|---|---|---|---|
| kcal/mol | ||||
| No compound | 1.00 ± 0.00 | 1.00 ± 0.00 | ||
| 5 μM | ||||
| NSC39863 | 3 | −9.3 | 1.87 ± 0.28 | 1.10 ± 0.75 |
| NSC13728 | 3 | −9.4 | 1.31 ± 0.07 | 0.97 ± 0.01 |
| NSC601364 | 1 | −10.2 | 1.18 ± 0.25 | 0.98 ± 0.13 |
| 10 μM | ||||
| NSC2979 | 1 | −8.2 | 1.49 ± 0.57 | 0.97 ± 0.07 |
| NSC40837 | 1 | −9.4 | 1.49 ± 0.08 | 0.94 ± 0.03 |
| NSC7616 | 1 | −9.6 | 1.47 ± 0.03 | 0.93 ± 0.03 |
| NSC683770 | 3 | −8.5 | 1.41 ± 0.47 | 0.96 ± 0.10 |
| NSC292213 | 2 | −7.5 | 1.37 ± 0.52 | 1.06 ± 0.17 |
| NSC30188 | 1 | −7.9 | 1.29 ± 0.07 | 0.98 ± 0.05 |
| NSC66207 | 1 | −10.6 | 1.26 ± 0.31 | 0.95 ± 0.09 |
| NSC93354 | 3 | −8.6 | 1.25 ± 0.44 | 0.95 ± 0.17 |
| NSC610938 | 2 | −8.3 | 1.19 ± 0.32 | 0.95 ± 0.11 |
| NSC299137 | 3 | −8.5 | 1.13 ± 0.16 | 0.86 ± 0.06 |
Electrophoretic Mobility Shift Assay. The bHLHLZ domains of Myc or Max were used in a 25-μl total volume of 25 mM Tris, pH 8.0, 50 mM KCl, 6.25 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 1.3 mM dithiothreitol, and 10 μg/ml poly(dI-dC). Dimethyl sulfoxide (DMSO) was used in place of compound and bovine serum albumin in place of MycCFP or MaxCFP proteins in control reactions. For Myc-Max and Max-Max dimerization, each dimer partner was added to a concentration of 85 nM. Reactions were incubated for 15 min at room temperature. A [32P]dCTP-labeled double-stranded DNA oligonucleotide with the sequence GATCAGTTGACCACGTGGTCTGGG, containing the consensus Myc network E-box binding site (Berg et al., 2002), was added to 25,000 cpm per reaction, and samples were incubated for an additional 15 min at room temperature. Samples were loaded onto a 6.6% native acrylamide gel and run at 170 V for 2 h in 0.5× Tris-borate EDTA (45 mM Tris-borate, 1 mM EDTA) running buffer. Gels were dried and exposed to film (BioMax XAR; Carestream Health, Rochester, NY).
Coimmunoprecipitation. MCF7-35IM cells were cultured for 72 h in the presence of 1 μg/ml doxycycline to induce the expression of the exogenous Myc (Venditti et al., 2002). Cells were then given fresh media and fresh doxycycline for another 24 h. Cells were lysed in a buffer containing 1% Nonidet P-40, 10% glycerol, 20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 1 mM dithiothreitol (DTT), 50 mM β-glycerophosphate, 1 mM Na3VO4, and 1× Complete protease inhibitor mixture (Roche, Indianapolis, IN). Fifteen hundred micrograms of total protein were incubated with NSC13728 at 0 (DMSO only), 50, 100, and 150 μM, respectively; then, 30 μl of rabbit anti-Max polyclonal antibody (C-124; Santa Cruz Biotechnology, CA) was added, and the mixture was incubated at 4°C overnight. Then, 40 μl of Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were added and incubated at 4°C for 6 h. After three washes with the lysis buffer, the beads were treated with 40 μl of 2× protein sample buffer. Bound proteins were separated in a 4-to-20% Tris-glycine SDS-PAGE gel (Invitrogen, Carlsbad, CA), then probed with mouse anti-Myc monoclonal antibody (C-33; Santa Cruz Biotechnology) or goat anti-Max antibody (Imgenex, San Diego, CA).
Enzyme-Linked Immunosorbent Assay. Max (300 ng/μl, in 1× PBS) was absorbed onto an ELISA plate (Nalge Nunc International, Rochester, NY) at 37°C for 1 h. After three washes with 1× PBS buffer, the plate was blocked with 3% nonfat milk in 1× PBS at 37°C for 1 h and washed three times with 1× PBS buffer. MycCFP (6.5 ng/μl, in 1× PBS) was incubated with NSC13728 at 0, 20, 40, and 60 μM, respectively, for 0.5 h at room temperature. The mixture was added to the plate and incubated at 37°C for 1 h. Rabbit polyclonal anti-Max antibody (Santa Cruz Biotechnology) diluted 1:1000 in 3% milk/1× PBS was used directly to detect Max in a parallel ELISA assay to confirm equal coating of Max in the wells. After three washes with 1× PBS, 1:10,000 diluted anti-GFP horseradish peroxidase-conjugated antibody (Novus Biologicals, Littleton, CO), or 1:10,000 diluted goat anti-rabbit horseradish peroxidase-conjugated antibody (Pierce, Rockford, IL) in 3% milk/1× PBS, was added. After 1-h incubation at 37°C and three washes, 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate solution (Calbiochem) was added and incubated for 5 min at room temperature for color development, then 100 μl of 2 M H2SO4 was added to stop the reaction. The absorbance at 450 nm was read with a microplate reader (SpectraMax 250; Molecular Devices, Sunnyvale, CA).
Analysis of Max and Myc Interaction by Surface Plasmon Resonance. The interaction between Myc and Max was analyzed using surface plasmon resonance (SPR) on a Biacore 3000 instrument (Biacore Ab, Uppsala, Sweden) at 25°C. Max was directly immobilized onto a CM5 sensor chip (BIAcore AB, Uppsala, Sweden) and targeted to the 8000 resonance unit setting using standard N-hydroxysuccinimide/ethyl diethyl carbodiimide coupling methodology (amine coupling kit; BIAcore AB). Analyte MycCFP or CFP alone prepared in Max buffer (200 mM HEPES, pH 7.0, 500 mM KCl, 30 mM MgCl2, 2 mM DTT, and 10 mM EDTA) at the concentration of 100 nM was injected over the Max-coated chip surface for 10 min, respectively, and was allowed to dissociate in the buffer for 5 min. All measurements were conducted in triplicate with Max buffer at a flow rate of 10 μl/min, and included double-referenced controls (i.e., with an in-line blank reference flow cell and blank buffer injections). A positive binding was defined as the SPR response at the end of dissociation phase being equal or greater than 10 S.D. of response for the buffer-only control. The chip surface was regenerated with 10 mM glycine-HCl, pH 1.7, for 30 s before proceeding to the next round of analysis.
Analytical Ultracentrifugation. Sedimentation equilibrium experiments were performed with the ProteomeLab XL-I (Beckman Coulter, Fullerton, CA) analytical ultracentrifuge. Purified Max protein samples, dissolved in 50 mM phosphate, pH 7.2, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, and 5% DMSO were loaded at concentrations of 0.5, 0.167 and 0.056 mg/ml in six-channel equilibrium cells and spun in An-50 Ti 8-place rotor at 30,000 rpm, 20°C for 24 h. Data were analyzed using HeteroAnalysis software (by J. L. Cole and J. W. Lary, University of Connecticut, Storrs, CT).
Focus Assays. Fertilized chicken eggs (White Leghorn) were obtained from Charles River Laboratories, Avian Products and Services (SPAFAS) (Preston, CT), and fertilized quail eggs were obtained from the Avian Science Research Facility at the University of California Davis (Davis, CA). Preparation and culture of primary chicken embryonic fibroblasts (CEF), quail embryonic fibroblasts (QEF), and focus assays with infectious retroviral vectors have been described previously (Berg et al., 2002). The oncogenic retroviruses used to transform CEF were 1) RCAS(A)-c-Myc (Petropoulos et al., 1996), 2) Prague strain A Rous sarcoma virus (Src) (Vogt, 1971), 3) avian sarcoma virus 17 (Jun) (Maki et al., 1987), and 4) RCAS(A)-myr-Δ72-c-p3k encoding a myristylated form of phosphatidylinositol 3-kinase p110α with a deletion of 72 N-terminal residues (Aoki et al., 2000). Cells were infected with retroviral vectors in the presence of compounds or DMSO and overlaid with agar medium the next day. Additional overlay agar containing compound or DMSO was added every second day. Foci were counted, and plates were stained with crystal violet after 1 to 3 weeks. Efficiencies of transformation are defined as the ratio of focus forming units per milliliter in the presence of compound divided by focus forming units per milliliter in the presence of DMSO alone.
Growth Curves. CEF or MCF7-35IM was seeded at 100,000 cells per 17-mm diameter well in 1 ml of media in 12-well tissue culture plates, in the presence of compound or DMSO. On certain days after seeding, cells were counted and replated at the original density. Growth was calculated by multiplying the -fold increase at each time point by the total number of cells. For QEF and Q8 cells, 4 × 104 cells/ml were seeded onto MP-6 plates and treated with 32, 16, 8, 4, 2, and 0 μM NSC13728, respectively. Cell numbers were counted from day 1 until day 5.
Reporter Assay. HEK293T cells were seeded into MP-24 tissue culture plates at 8 × 104 cells per well. The next day, the cultures were transfected using PolyFect (QIAGEN, Valencia, CA) with 100 ng of pGL2M4 firefly luciferase reporter driven by four copies of Myc binding site (E-box) and simian virus 40 promoter, 300 ng of pCMV3HuMyc construct expressing human Myc under the control of cytomegalovirus promoter in a pcDNA3 vector backbone, and 2 ng of pRLCMV, a Renilla reniformis luciferase construct, as internal control. pcDNA3 served as a negative control. Compounds were added to the culture 24 h later; then, after another 24 h of incubation, the cultures were lysed in 100 μl of passive lysis buffer (Promega) according to the manufacturer's protocol. Firefly luciferase activities and R. reniformis luciferase activities were measured by using the Dual-Luciferase reporter assay system (Promega) with a Berthold Biolumat model LB 9501. Firefly luciferase activities were normalized against R. reniformis luciferase activities. Assays were conducted in triplicate.
Northern Blots. Northern blots followed standard procedures. For Northern analyses of MCF7-35IM cells, all cells were incubated for 24 to 72 h as described under Results section, in the presence of 1 μM ICI 182,780 (Venditti et al., 2002). Cells were then given fresh media and fresh ICI 182,780 or 1 μg/ml doxycycline. RNA gels from CEF were probed with chicken cDNAs and RNA from MCF7-35IM with human cDNAs.
Western Blots. Western blotting followed standard procedures. The primary Myc antibody was rabbit anti-c-Myc (Cell Signaling Technology, Danvers, MA), used 1:1000 in 5% milk/Tris-buffered saline/Tween 20 overnight at 4°C.
Results
AutoDock “Blind Docking” VLS Identifies Lead Compounds
Virtual ligand screening of 1668 compounds from the NCI Diversity Set was performed against both the Max homodimer and the Myc-Max heterodimer. The compounds were clustered by the locations of their predicted binding sites, and results were sorted by predicted free energies. Because there was no prior small molecule binding information that could be used to narrow the docking search to a particular structural region, the search encompassed the entire bHLHLZ structure for each dimer. This search method, referred to as “blind docking” (Hetényi and van der Spoel, 2002), uses an unusually large search region for a small-molecule docking calculation (60 × 126 × 60 Å), particularly for a VLS calculation involving thousands of compounds. Because many potential binding sites were evaluated, the location of the predicted binding site, in addition to the predicted binding energy, was strongly considered when selecting compounds for experimental testing. It was unknown which binding sites would be most effective in stabilizing the homodimer and which would have the greatest impact in vivo, so it was desirable to test several of the different binding pockets predicted by AutoDock.
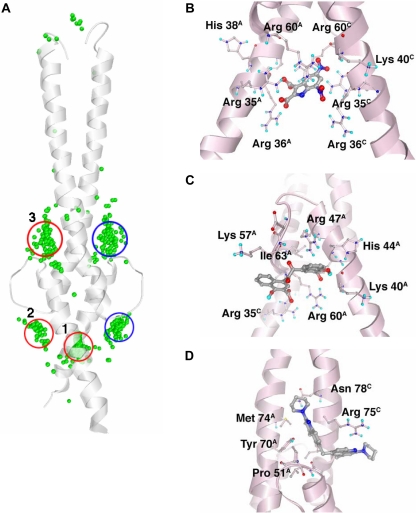
From a three-dimensional clustering of all docking solutions, 12 predicted binding pockets emerged. Of these, three main clusters contained 85% of the docked compounds and the lowest energy docking solutions from the entire chemical library (Fig. 1A, Table 1). The results from the Myc-Max VLS and from the Max-Max VLS were generally similar. Predicted binding energies of compounds docked to the Max homodimer were between -11.3 and -1.6 kcal/mol. Their distribution was approximately Gaussian, with a mean of -5.8 ± 1.4 kcal/mol (mean ± S.D.). The predicted binding energies of compounds docked to the Myc-Max heterodimer were between -10.4 and -1.3 kcal/mol, with a mean of -5.9 ± 1.2 kcal/mol. Forty compounds from the Max-Max VLS and 40 from the Myc-Max VLS were selected for experimental testing, representing the top docking solutions from the three major sites. These compounds varied in predicted binding energy from -11.3 to -8.4 kcal/mol. Because 12 compounds were common to both sets, 68 unique compounds were requested from the National Cancer Institute.
Fig. 1.
A, Max-Max binding sites predicted by AutoDock VLS. Shown is the Max-Max protein structure (ribbon) and the center-of-coordinates for the best docking solution for each of the 100 NCI Diversity Set compounds with the lowest predicted binding energy (green spheres). The locations of clusters 1, 2, and 3 (red) and 2b and 3b (blue) are circled. Clusters 2b and 3b are identical in the Max homodimer but are different in the Myc-Max heterodimer. B to D, the Max-Max homodimer is shown in pink ribbon with selected side chains colored by atom type and shown as ball-and-stick. The predicted bound structure of NSC131615 (B), representing cluster 1, NSC292215 (C), representing cluster 2, and NSC13728 (D), representing cluster 3, are shown.
Predicted Binding Sites Show Specific Chemical Preferences
Compounds in the three major clusters exhibit trends in their chemical properties consistent with the neighboring protein structure (Fig. 1A, Table 1). The binding sites for compounds in clusters 1, 2, and 3 are termed binding sites 1, 2, and 3, respectively. Compound structures are presented in Supplemental Fig. 1.
Binding Site 1. Cluster 1 compounds were predicted to bind almost exactly between the positively charged DNA-binding helices of the dimers. VLS results for the Max homodimer and the Myc-Max heterodimer were similar for cluster 1, consistent with the high degree of structural similarity between the Max homodimer and the Myc-Max heterodimer in this region. The cluster contained 456 compounds total, including those with the lowest predicted binding energies from the entire Diversity Set. Its high binding strength is attributed both to the strong electropositive potential of several basic residues in the binding site (Lys 40, Arg 35, Arg 60, Arg 36, and His 38 in the Max protein) and to a deep concave protein surface between the two basic helices (Fig. 1B). Appropriately, the general structure for lead compounds in this cluster is an abundance of negatively charged atoms or a high density of hydrogen bonding atoms; the average calculated octanol-water partition coefficient, logP, for the top 20 compounds in this cluster was -4.8.
Binding Site 2. Cluster 2 compounds interact with several positively charged residues from the protein basic regions (His 44, Arg 47, and Arg 60 in Max) and with neutral HLH region residues nearby (Fig. 1C); thus, binding site 2 neighbors binding site 1. Compounds in this cluster were predicted to bind residues of the basic helices and of the HLH structural motif. Cluster 2 contained only 90 compounds, yet it was the cluster with the second lowest binding energies, after cluster 1. Accordingly, the top binding compounds from this cluster are similar in charge to those from cluster 1 but are generally more hydrophobic; the average logP value for the top 20 compounds in this cluster is 1.4. Minor differences between the Max-Max and Myc-Max dimers exist in the HLH region of this binding site, such as the substitution of Phe 922 in Myc for His 44 in Max, and a loop backbone shift in Myc caused by an insertion at residue 933.
Binding Site 3. The binding site for cluster 3 is a shallow pocket located at the intersection of the leucine zipper and HLH regions. This cluster was the largest of the three, containing 863 compounds, yet compounds in cluster 3 were the weakest predicted binders among these top three clusters. The structure for cluster 3 compounds was the presence of one or more rigid, flat surfaces that fit within the relatively narrow pocket between leucine zipper residues Tyr 70 and Arg 75 and HLH loop residue Pro 51 (in Max) (Fig. 1D). Arg 75 is the only positively charged residue near the binding site. Consistent with this, the best binding compounds in this cluster lacked a strong negative charge, and the top 20 compounds in this cluster had an average logP of 4.7. Although hydrogen-bonding side chains neighbor the cluster 3 binding site, these are seldom used in the docking predictions by the cluster 3 compounds.
A major difference exists between the cluster 3 binding sites of the Myc-Max versus the Max-Max dimers. The Max-Max binding cavity used by the cluster 3 compounds is blocked in the Myc-Max structure by Max residues Arg 254 and Gln 261, so relatively few compounds are predicted to bind to this location with Myc-Max. Instead, the compounds that docked to site 3 in Max-Max are predicted to bind a variety of different sites around the Myc-Max HLH region. Of the three major cluster-binding sites, this site was therefore predicted to bind the compounds that are most specific to the Max homodimer or the Myc-Max heterodimer.
FRET Analyses Identified Specific Stabilizers of the Max Homodimer
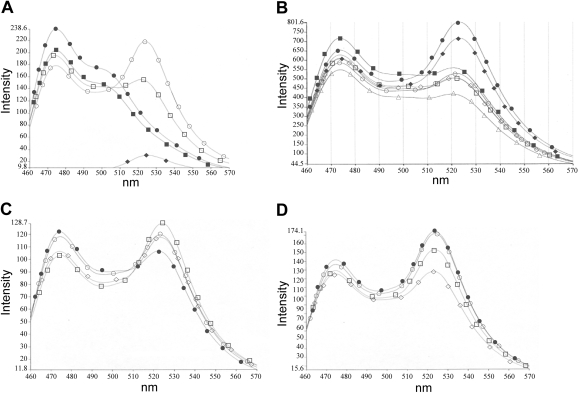
Construction and purification of MycCFP, MaxCFP, and MaxYFP fusion proteins have been described previously (Berg et al., 2002). Purified proteins were used to screen selected NCI compounds using FRET in single cuvette assays. In the absence of compound, the efficient dimerization of equimolar ratios of MycCFP with MaxYFP produced a strong FRET signal, whereas dimerization of MaxCFP with MaxYFP produced a weaker FRET signal (Fig. 2A). Hit molecules were defined as those that reproducibly stabilized Max homodimerization (increased FRET) or inhibited Myc-Max heterodimerization (decreased FRET) at the initial concentration of 25 μM. In a representative screen, compound NSC30188 stabilized the Max homodimer interaction, whereas four other compounds did not (Fig. 2B). At 25 μM, 17 of the 68 compounds (25%) produced at least some degree of Max-Max stabilization (not shown).
Fig. 2.
Stabilization of Max-Max homodimerization by NCI compounds is measured by FRET. Proteins are at 85 nM each. A, representative control FRET spectra. •, MycCFP; ▪, MaxCFP; ♦, MaxYFP; ○, MycCFP/MaxYFP; □, MaxCFP/MaxYFP. B, representative FRET spectra for compound screen. •, MycCFP/MaxYFP; ▪, MaxCFP/MaxYFP; ♦, MaxCFP/MaxYFP + NSC30188; ○, MaxCFP/MaxYFP + NSC73100; □, MaxCFP/MaxYFP + NSC38777; ⋄, MaxCFP/MaxYFP + NSC41309; ▵, MaxCFP/MaxYFP + NSC42258; proteins are at 85 nM; compounds are at 25 μM. C and D, titration of compound NSC13728 against the MaxCFP/MaxYFP (C) and MycCFP/MaxYFP (D) dimers. Compound concentrations: •, no compound; ○, 2.5 μM; □,5 μM; ⋄,10 μM.
Lead compounds were defined as molecules that specifically increased the Max homodimer FRET ratio by at least 10% when used at 10 μM or less. Those compounds were titrated in additional FRET assays against both the Max-Max and the Myc-Max dimers. Thirteen of the 68 compounds selected from the VLS results (approximately one in five) specifically stabilized Max homodimerization (Table 1). The concentration shown is that at which the compound was most effective in specifically stabilizing the Max homodimer or inhibiting the Myc-Max heterodimer (i.e., produced the greatest change in FRET ratio), and data are sorted accordingly.
FRET titrations for one dimer-specific compound are shown in Fig. 2, C and D. Compound NSC13728 specifically stabilizes the Max homodimer (Fig. 2C) while inhibiting the Myc-Max heterodimer (Fig. 2D).
FRET Data Correlate with Molecular Docking Results
As predicted, compounds docked in silico to the Max homodimer were more successful in stabilizing the homodimer in FRET analyses than those docked to the Myc-Max heterodimer. Sixty-six percent of the Max-Max stabilizers identified using VLS were specific to the Max homodimer. In contrast, none of the compounds identified in Myc-Max VLS specifically stabilized the Max homodimer. Of the 12 compounds common to both sets, three stabilized the Max homodimer; two of the three also stabilized Myc-Max. Thus, the Max-Max compounds had a hit rate twice that of Myc-Max, and the Max-Max VLS was much more effective than the Myc-Max VLS in finding compounds specific to Max-Max.
Hit rates for clusters 1, 2, and 3 were similar: 30, 23, and 25%, respectively. The stabilizers were evenly distributed over the sampled range of binding energies for clusters 1 and 2, but cluster 3 compounds showed a clear boundary for activity at a predicted binding energy of -8.45 kcal/mol. The 11 cluster 3 compounds with a binding energy below -8.45 kcal/mol had a hit rate of 46%, nearly double the overall hit rate. Only one stabilizer was found among the remaining 13 compounds of cluster 3. Hit rates for clusters 1 and 2 were relatively constant with respect to binding energy, and a similar drop-off in hit rate was not found among the sampled compounds. Compounds in clusters 1 and 2 also exhibited relatively poor specificity to either dimer, consistent with the structural similarities of Myc-Max and Max-Max dimers in this region. In contrast, cluster 3 compounds were much more specific. Eighty-five percent of the compounds predicted to bind to site 3 were specific for the Max homodimer, compared with 67% for site 2 and only 13% for site 1. Consistent with its specificity in FRET, compound NSC13728 docked in silico to cluster 3 (Table 1). Therefore, binding site 3 was the most effective docking site for identifying specific stabilizers and compound NSC13728 was chosen for further characterization.
Stabilization of the Max Homodimer Is Not a Function of DNA Binding
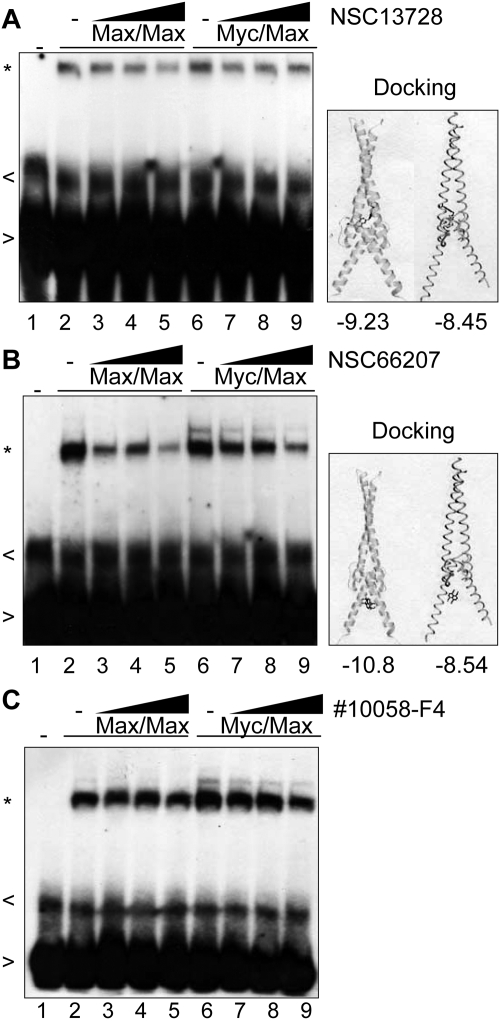
Max homodimer stabilizers could inhibit oncogenic transformation either by occupying the consensus E-box binding sites of the Myc network or by titrating the Max protein out of the network and thus making it unavailable for interaction with other partner proteins. EMSA analyses were performed to determine whether a stabilizer of Max homodimers would also stabilize the binding of homodimers to DNA. Assays used a 32P-labeled consensus Myc network binding site, the E-box. Compound NSC13728, which stabilized Max homodimerization in FRET, did not also stabilize DNA binding (Fig. 3A). Indeed, a slight decrease in DNA binding was observed for both Max homodimers and Myc-Max heterodimers. Consistent with other cluster 3 compounds, Autodock analyses predicted that this compound interacts stably with the HLH-leucine zipper dimerization domains of Myc and Max (shown alongside EMSA in Fig. 3). In contrast, a much greater decrease in DNA binding is observed when a cluster 1 compound (which docks in silico to the DNA binding region) is used in EMSA (Fig. 3B). An inhibitor of Myc-Max dimerization (10058-F4; Yin et al., 2003) also produces only a marginal decrease in DNA binding in EMSA analysis (Fig. 3C). The results suggest that DNA binding is significantly affected by the location of the docking site but not by stabilization of Max homodimers in solution.
Fig. 3.
Max-Max stabilization is not a function of DNA binding. MycCFP and MaxCFP were held constant at 85 nM each where indicated. Compounds were titrated against the Max-Max homodimer (lanes 3-5) and the Myc-Max heterodimer (lanes 7-9) using the compound concentration ranges that produced the greatest Max homodimer stabilization in FRET. A, NSC13728 was used at 2.5, 5, and 10 μM; NSC66207 was used at 2.5, 5, and 10 μM (B); and Calbiochem Myc inhibitor 10058-F4 was used at 7.5, 15, and 30 μM (C). Gel shift, indicated by (*), in the absence of compounds is shown in lanes 2 and 6. Free probe (>) and nonspecific bands (<) are marked. Molecular docking results are shown alongside EMSA, with predicted binding energies below.
In Vitro Binding Assays Demonstrate Interference by the Stabilizer with the Heterodimerization of Myc and Max and Enhancement of Max-Max Homodimerization
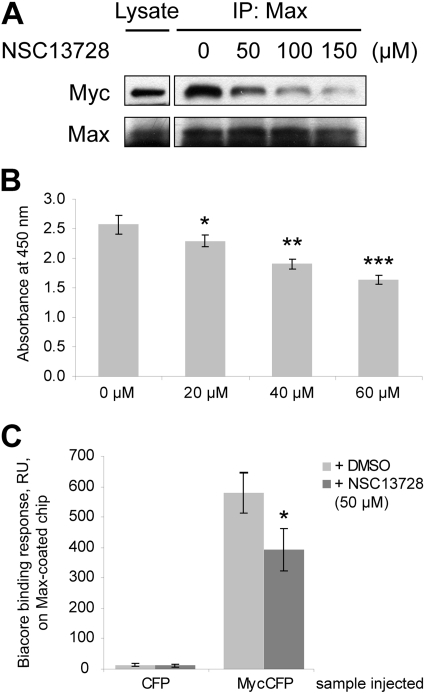
To test whether NSC13728 reduces the association of Myc with Max, coimmunoprecipitations were performed. For these experiments, we used the MCF7-35IM breast cancer cell line (Venditti et al., 2002). MCF7-35IM contains an inducible myc transgene as well as a regulatable endogenous myc gene. Addition of doxycycline to MCF7-35IM activates the exogenous myc transgene, leading to elevated levels of Myc protein. Induced MCF7-35IM cells were lysed, and protein lysates were incubated with NSC13728 at 0 (DMSO only), 50, 100, and 150 μM, respectively. Rabbit anti-Max polyclonal antibody was then added to pull down Max. After precipitation with agarose beads, bound proteins were separated in a 4-to-20% Tris-glycine SDS-PAGE gel, blotted, and probed with anti-Myc or anti-Max antibody. Figure 4A shows that NSC13728 reduces the association of Myc with Max in a dose-dependent manner.
Fig. 4.
Compound NSC13728 interferes with the heterodimerization of Myc-Max. A, NSC13728 reduced Myc association with Max in a dose-dependent manner in coimmunoprecipitation. MCF7-35IM cells were lysed after Myc expression was stimulated with doxycycline. Protein lysates were mixed with NSC13728 at 0 (DMSO only), 50, 100, and 150 μM, respectively, then rabbit anti-Max polyclonal antibody was added to pull down Max and bound Myc. Bound proteins were precipitated by Protein A/G PLUS-Agarose beads and separated in a 4-to-20% Tris-glycine SDS-PAGE gel. The blot was probed with anti-Myc or anti-Max antibody. B, NSC13728 reduced Myc association with Max in a dose-dependent manner in an ELISA assay. An ELISA plate was coated with Max, and the plate was blocked with nonfat milk. MycCFP was first incubated with NSC13728 at 0, 20, 40, and 60 μM, respectively. The mixture was added to the wells and incubated at 37°C for 1 h. After washing, anti-GFP-horseradish peroxidase conjugate was added to detect MycCFP. The substrate solution was added for color development, and the absorbance at 450 nm was read with a microplate reader. *, p = 0.064; **, p < 0.005; ***, p < 0.001 between DMSO-treated and NSC13728-treated samples. C, SPR measurements showed that the interaction between Myc and Max is inhibited by NSC13728. Max was directly immobilized onto a sensor chip. Analyte MycCFP or CFP control (100 nM) was injected over the Max-containing chip, and was allowed to dissociate in the Max buffer for 5 min. A positive binding is defined as the SPR response at the end of dissociation phase being equal or greater than 10 S.D. of response for the control, a buffer-only injection. *, p < 0.05 between DMSO-treated and NSC13728-treated samples.
The effect of NSC13728 on the heterodimerization of Myc and Max was further examined by ELISA assay. The purified bHLHLZ domain of Max was immobilized on an ELISA plate. MycCFP was mixed with NSC13728 at 0, 20, 40, and 60 μM, respectively, and then added to the plate. MycCFP bound to immobilized Max was detected colorimetrically by using a horseradish peroxidase-conjugated GFP antibody. The results again suggest that NSC13728 interferes with Myc-Max dimerization in a dose-dependent manner (Fig. 4B). Max preincubated with NSC13728 was also absorbed onto an ELISA plate. As expected, the stabilizer reduced binding of MycCFP to immobilized Max. However, this sequence of interactions gave less consistent results, probably because of the washing of the Max-compound mixture on the plates. Equal coating of Max on the plate was confirmed by using anti-Max antibody to directly detect Max in a parallel ELISA assay (data not shown).
Additional data on the action of the stabilizer were obtained with SPR. In this experiment, the purified bHLHLZ domain of Max was directly immobilized onto a sensor chip. The analyte MycCFP or CFP control was injected with or without NSC13728 over the chip coated with Max and was allowed to react in the buffer for 5 min. The CFP control yielded a weak SPR signal in either the presence or absence of NSC13728. In contrast, MycCFP injected with DMSO gave a strong SPR signal, indicative of an association of the immobilized Max and the injected MycCFP. When NSC13728 was coinjected with MycCFP, however, the signal was reduced approximately 30% (Fig. 4C). These data also support the conclusion that the stabilizer interferes with the interaction between Myc and Max.
Finally, analytical ultracentrifugation was employed to investigate the oligomeric state of Max in solution in the presence or absence of the stabilizer. The apparent molecular mass of Max in the absence of the compound was determined as 22,789 Da, a value between the molecular mass of monomer (13,134 Da) and dimer (26,268 Da), suggesting an equilibrium state between monomer and dimer (data not shown). In the presence of 13 μM stabilizer, the apparent molecular mass of Max was 26,245 Da, indicating a shift of the equilibrium to the dimeric state. To determine dimerization affinity, we have fitted the data to a monomer-dimer equilibrium model (HeteroAnalysis software), which yielded a Kd of 5.9 × 10-6 M in the absence of the compound, whereas in the presence of 13 μM compound, the Kd was 7 × 10-9 M. Thus, the compound strongly enhances dimerization of Max, possibly by binding to the dimer preferentially.
Compound NSC13728 Is Nontoxic and Does Not Change Growth Rates of CEFs
Growth rates of CEF were measured in the presence of compounds or DMSO alone. Compound NSC13728 did not alter the growth curves of CEF (Supplemental Fig. 2A). The Myc inhibitor 10058-F4, used for comparison, also did not change growth rates of CEF, although it decreases growth of nonavian cells (Yin et al., 2003). The data indicate that the compounds are nontoxic to CEF at the concentrations used and that the degree of Myc inhibition achieved by these compounds does not significantly alter the overall growth rates of CEF.
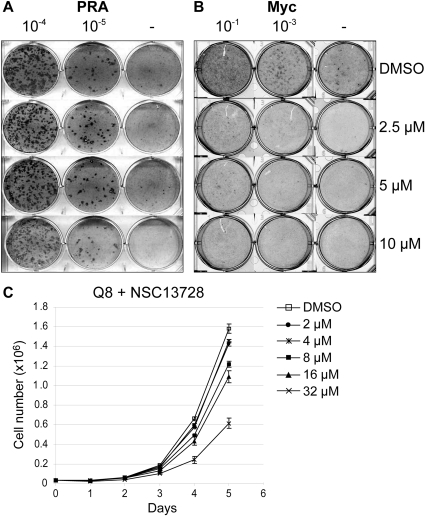
Compound NSC13728 Inhibits c-Myc-Mediated Oncogenic Transformation of CEF and the Growth of the Myc-Transformed QEF Clone Q8
To determine whether a Max-Max stabilizer could inhibit Myc-mediated oncogenic transformation, focus formation assay was performed with CEF. Cells were infected with retroviral vectors containing the myc, src, jun, or p3k oncogenes. A representative experiment shows that Src-mediated transformation is not inhibited by compound NSC13728 compared with DMSO (Fig. 5A). In contrast, transformation of CEF by Myc is inhibited approximately 1000-fold by NSC13728 (Fig. 5B). Efficiencies of transformation for multiple experiments are averaged and summarized in Table 2. The inhibition of Myc-mediated transformation by compound NSC13728 is specific, whereas transformation by Src, Jun, or phosphoinositide 3-kinase was not significantly affected by the compound. In cell culture, NSC13728 inhibited the Myc-induced formation of transformed cell foci with an IC50 of 3 μM.
Fig. 5.
Max homodimerization by compound NSC13728 leads to specific inhibition of Myc-mediated focus formation in CEF and growth inhibition in Q8 cells, an established QEF cell line transformed by the v-myc oncogene of avian acute leukemia virus MC29. Serial 10-fold dilutions of RCAS A avian retroviral vector containing the Prague A Src expression construct (A) or c-Myc (B) were used to infect CEF treated with DMSO or compound NSC13728 at the indicated concentrations. C, growth curves of Q8 cells. 4 × 104 cells/ml were seeded onto MP-6 plates and treated with 32, 16, 8, 4, 2, and 0 μM NSC13728, respectively. Cell numbers were counted on day 1 until day 5.
TABLE 2.
Summary of efficiencies of transformation
Efficiencies of transformation by Myc, Jun, P3K, or Src are shown for each compound at the indicated concentrations.
| Compound | Myc | Jun | P3K | Src |
|---|---|---|---|---|
| NSC13728 (2.5 μM) | 0.07 ± 0.04 | 0.90 ± 0.06 | 1.03 ± 0.19 | 1.14 ± 0.10 |
| NSC13728 (5 μM) | 0.01 ± 0.01 | 1.08 ± 0.07 | 0.90 ± 0.05 | 1.18 ± 0.20 |
| NSC13728 (10 μM) | 0.01 ± 0.00 | 0.57 ± 0.13 | 0.96 ± 0.20 | 0.78 ± 0.11 |
| 10058-F4 (7.5 μM) | 0.61 ± 0.11 | N.T. | N.T. | 0.95 ± 0.07 |
P3K, phosphoinositide 3-kinase; N.T., no transformation.
Growth curves were determined with the Q8, an established QEF cell line transformed by the v-myc oncogene of avian acute leukemia virus MC29 (Bister et al., 1977). Compared with nontransformed QEF, Q8 cells reach a much higher density at each time point (Supplemental Fig. 2B). Compound NSC13728 had no detectable inhibitory effect on control QEF at 10 μM (Supplemental Fig. 2B) but inhibited the growth of Q8 cells in a dose-dependent manner (Fig. 5C). These results suggest that NSC13728 also interferes with maintenance of Myc-induced transformation.
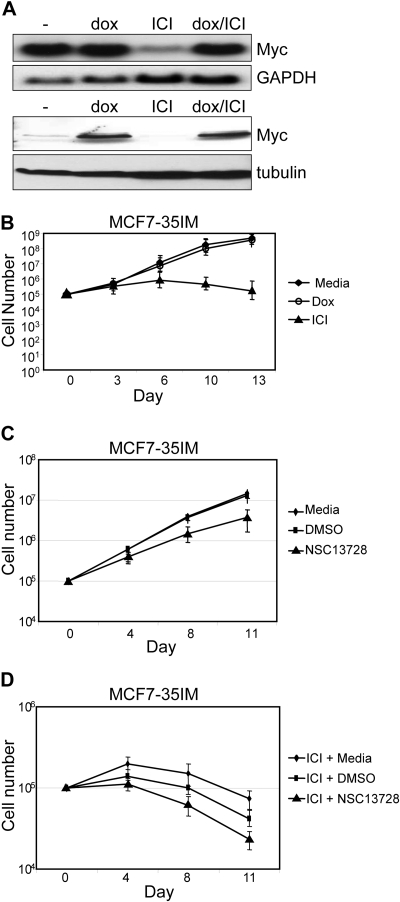
Compound NSC13728 Inhibits Growth and Complements Growth Reduction by ICI 182,780 in MCF7-35IM Cells
Growth curve experiments were also performed with the MCF7-35IM breast cancer cell line. Addition of doxycycline to MCF7-35IM activates the myc transgene carried by these cells, leading to an increase in total Myc. Addition of the estrogen antagonist ICI 182,780 almost completely inhibits endogenous Myc expression. When both doxycycline and ICI 182,780 are added to cells, exogenous Myc is expressed but endogenous Myc is down-regulated (Fig. 6A).
Fig. 6.
Max homodimerization stabilizers are nontoxic to and complement growth inhibition by Myc depletion in MCF7-35IM. A, Northern blot (top two panels) and Western blot (bottom two panels) of MCF7-35IM in the presence of 1 μg/ml doxycycline or 1 μM ICI 182,780, where indicated. B to D, growth curves of MCF7-35IM in the presence of doxycycline or ICI 182,780 (B), compound NSC13728 (C), or NSC13728 plus ICI 182,780 (D). Cells were plated at 105 cells per well of 12-well tissue culture plates and were grown to ∼80% confluence. At the indicated time points, cells were trypsinized, counted, and replated to the initial concentrations. To calculate growth, the -fold increase in cell number for each count was multiplied by the total number at the previous time point.
As shown previously, doxycycline does not significantly alter the growth rate of MCF7-35IM cells (Fig. 6B). In contrast, the reduction of Myc by ICI 182,780 leads to a significant decrease in growth rate of MCF7-35IM. Likewise, growth rates are also reduced in the presence of compound NSC13728, albeit not as drastically as with ICI 182,780 (Fig. 6C).
When compound NSC13728 is used in combination with ICI 182,780, an additive reduction in growth is observed (Fig. 6D). The fact that Myc transcription is not completely inhibited by ICI 182,780 (Fig. 6A) suggests that compound NSC13728 interferes with residual Myc activity in ICI 182,780-treated cells by stabilizing the Max homodimer. In addition, the fact that growth is reduced in the presence of NSC13728 versus DMSO at an early time point (4 days) suggests that NSC13728 may be active in stabilizing the Max homodimer before inhibition of Myc transcription by ICI 182,780.
Compound NSC13728 Inhibits Transcription of Myc Target Genes
We tested NSC13728 for its ability to inhibit Myc-induced transcriptional activity in HEK293T cells by reporter assay. A human Myc-expression construct, a firefly luciferase reporter driven by four copies of Myc binding site (E-box), and an R. reniformis luciferase internal control were cotransfected into HEK293T cells. pcDNA3 served as a negative control. Firefly luciferase activities were normalized against R. reniformis luciferase activities. The normalized reporter activity in NSC13728-treated cells were expressed as fractions of that in DMSO-treated cells, which was designated 1.00. As shown in Table 3, NSC13728 inhibited exogenous Myc-induced transcription in a dose-dependent manner.
TABLE 3.
Effect of NSC13728 on Myc transcriptional activity
HEK293T cells were transfected with three constructs: pGL2M4 firefly luciferase reporter driven by four copies of Myc binding site (E-box), pCMV3HuMyc construct expressing human Myc under the control of CMV promoter in a pcDNA3 vector backbone, and pRLCMV, a R. reniformis luciferase construct, as internal control. pcDNA3 served as a negative control. Compounds were added to the culture 24 h later; then, reporter activity was measured after another 24 h of incubation. Firefly luciferase activities were normalized against R. reniformis luciferase activities. The normalized reporter activity in DMSO-treated cells was designated 1.00. Data were expressed as mean ± S.D. (n = 3), followed by p value between DMSO-treated and compound-treated samples.
| Compound | Transcriptional Activity |
|---|---|
| DMSO | 1.00 |
| NSC13728 (2.5 μM) | 0.97 ± 0.07, P = 0.58 |
| NSC13728 (10 μM) | 0.85 ± 0.05, P = 0.051 |
| NSC13728 (20 μM) | 0.75 ± 0.06, P < 0.05 |
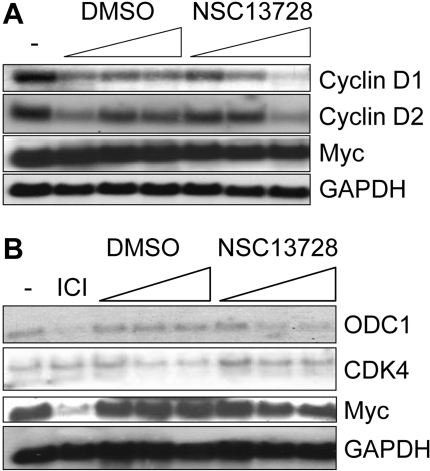
Transcription of genes responsive to Myc activity was further evaluated in the presence of compound NSC13728. Of the genes shown in Fig. 7, cyclin D2, ODC1, and CDK4 are directly bound in their respective promoter regions and up-regulated by Myc (Lee and Dang, 2006). Although cyclin D1 can be either up-regulated (Perez-Roger et al., 1999) or down-regulated (Philipp et al., 1994) by Myc in a cell type-dependent manner, its regulation and that of cyclin D2 have not previously been studied in CEF. We show a dose-dependent reduction in transcription of avian cyclins D1 and D2 in response to compound NSC13728 (Fig. 7A). Neither Myc nor GAPDH levels are affected by the compounds.
Fig. 7.
The Northern blot suggests that compound NSC13728 inhibits Myc target gene transcription in avian and mammalian cells. CEF (A) or MCF7-35IM (B) were incubated with increasing amounts of media alone (-), 1 μM ICI 182,780, DMSO alone, or compound NSC13728. Total DMSO volumes were constant at all three dilutions alone and with compounds. Compound NSC13728 was used at 2.5, 5, and 10 μM.
In the MCF7-35IM line, Myc and ODC1 are both reduced by ICI 182,780 as expected (Fig. 7B). A reduction of ODC1 is also observed in response to compound NSC13728, but the reduction is not as strong as with ICI 182,780. We were surprised that myc transcription may also be slightly reduced by the compound in this cell line. GAPDH is not affected. Although CDK4 is up-regulated by Myc in a variety of cell types (Lee and Dang, 2006), we show that it is not significantly reduced in response to ICI 182,780 in MCF7-35IM. In agreement with this observation, the effects of compound NSC13728 on transcription of this gene are marginal (Fig. 7B). The results demonstrate that in MCF7-35IM, the reduction in target gene transcription is correlated with depletion of Myc by ICI 182,780. Collectively, the data indicate that stabilization of the Max homodimer leads to an inhibition of Myc-mediated transcriptional activity.
Discussion
VLS drug discovery efforts generally target enzyme active sites, protein receptors and other “druggable” protein structures with deep, easily identifiable small molecule binding sites. In contrast, the dimerization interactions addressed here present few easily identified compound binding sites. In screening a large database of compounds over an entire dimerized bHLHLZ region, we were presented with the challenge that the calculations required significantly more computational power than typical docking calculations because of the larger search region. The benefit derived from this broad search was a distinct grouping of docked compounds according to predicted binding sites, in addition to predicted binding energy. This work represents the first time that virtual ligand screening of a large (60 × 126 × 60 Å) search region, also referred to as “blind docking,” has identified a small-molecule compound with significant biological function. The manuscript also provides an example for the utilization of small molecule modulators to stabilize, rather than inhibit, a protein-protein interaction.
The most obvious binding site for Myc network dimers is the basic helix region, which binds to DNA and presents a deep, concave surface for binding compounds. As predicted, the top scoring compounds from the AutoDock Diversity Set VLS (cluster 1 and 2 compounds) interacted with this region. Also as predicted, these compounds exhibited the poorest specificity for either Max homodimer or the Myc-Max heterodimer. As confirmed by EMSA, cluster 1 and 2 compounds have potential as inhibitors of Myc-Max DNA binding and transcriptional activation, but further studies are required to explore this possibility.
Cluster 3 compounds were the most successful specific Max-Max stabilizers and effectively inhibited Myc activity in cells. The generally neutral electrostatic potential and moderate number of hydrogen bond donors and acceptors of these compounds agree with the Lipinski “Rule of Five” specifying chemical properties for drug-like compounds (Lipinski et al., 2001). In addition, the hydrogen bonding properties of cluster 3 compounds could also be exploited to generate derivative libraries of more potent binders.
FRET analyses showed that stabilizers of the Max homodimer can also inhibit Myc-Max heterodimerization (e.g., NSC13728). This effect may result from direct inhibition of Myc-Max heterodimerization, or a reduction in levels of Max available for dimerization with Myc. These two mechanisms could also account for the observed inhibition of Myc-mediated oncogenic transformation and transcriptional modulation.
Compound NSC13728, the stabilizer of the Max homodimer, does not significantly enhance DNA binding of the Myc-Max heterodimer or of the Max homodimer, as determined by EMSA analysis. The small reduction in DNA binding observed in the presence of the compound requires further analysis. For the Myc-Max heterodimer, reduced DNA binding could result directly from a stabilization of the Max homodimer, leaving a fraction of the Myc protein without the partner essential for DNA binding. The absence of significant compound-induced changes in the in vitro DNA binding of Max-Max or Myc-Max supports the suggestion that interference with oncogenic transformation results from an effect of the compounds on the in vivo transcriptional activities of Myc.
In contrast to compound NSC13728, which is targeted to the intersection of leucine zipper and HLH, compounds of cluster 1 and 2 that are suggested to interact with the basic DNA-binding region of Myc or Max decrease DNA binding significantly. Such an effect would be predicted and could result from steric hindrance of the protein-DNA interaction. These observations suggest that DNA-binding is not the critical function that is affected in the inhibition of Myc-induced oncogenic cellular transformation. Compound NSC13728 also does not affect the growth of CEF at concentrations that strongly inhibit oncogenic transformation. Cell cycle progression at normal Myc levels may therefore be less sensitive to compound reduction of Myc function than transformation when Myc is overexpressed. The Max stabilizer does not reduce oncogenic transformation by the Src oncoprotein; this is consistent with evidence that the proliferative function of Src, but not Src-dependent transformation, depends on a basal level of Myc activity (Prathapam et al., 2006).
The FRET assay detects a relative stabilization of the Max homodimer by small-molecule compounds. In vitro binding assays, including coimmunoprecipitation, ELISA, and SPR analysis, demonstrated that the stabilizer interferes with the Myc-Max heterodimer formation in a dose-dependent manner. Analytical ultracentrifugation provided direct evidence that the stabilizer enhances the Max-Max homodimerization, resulting in a 1000-fold decrease of the dissociation constant. However, progression of the cell cycle in the presence of the compounds suggests that some residual levels of Myc-Max heterodimers persist in the cell. That level may be insufficient to initiate and maintain oncogenic transformation. Oncogenicity of the cellular Myc protein reflects a gain of function that depends on an increased level of Myc-Max heterodimers in the cell. It is conceivable that this quantitative gain of function is sensitive to a partial down-regulation that does not significantly affect basal activities of the Myc network.
Aberrantly high levels of Myc are present in many human cancers. Examples include Burkitt's lymphoma (characterized by a translocation of the c-myc gene), neuroblastomas carrying an amplified n-myc gene, and breast, pancreatic and lung cancers. The Myc network contains both positive and negative regulators of transcription. All regulators in the network require Max as a mandatory dimerization partner. The Myc-Max heterodimer represents the branch of the network that up-regulates the cell cycle. In cancers showing overexpression of Myc, stabilization of Max can be expected to selectively down-regulate Myc. In the present study, we show that stabilization of the Max homodimer can attenuate Myc-dependent transcription, cell cycle induction, and oncogenic transformation. The precise molecular mechanisms by which the stabilization of Max affects Myc function remain to be worked out. With modified screens, additional candidate compounds may be identified and may deserve scrutiny as useful and effective inhibitors of Myc.
Supplementary Material
Acknowledgments
We thank Dr. Andreas G. Bader for advice and materials and Lynn Ueno for expert technical assistance. This is manuscript number 18395 of The Scripps Research Institute.
This work was supported by the National Institutes of Health National Cancer Institute [Grant CA078045].
H.J., K.E.B., and A.E.B. contributed equally to this work.
ABBREVIATIONS: 10058-F4, (Z,E)-5-(4-ethylbenzylidine)-2-thioxothiazolidin-4-one; bHLHLZ, basic helix-loop-helix leucine zipper; CEF, chicken embryonic fibroblast; CFP, cyan fluorescence protein; DMSO, dimethyl sulfoxide; DTT, dithiothreitol; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assay; FRET, fluorescence resonance energy transfer; GFP, green fluorescence protein; HEK, human embryonic kidney; HLH, helix-loop-helix; ICI 182,780, fulvestrant; NCI, National Cancer Institute; NSC13728, 4-methyl-6-[(4-methyl-2-piperidin-1-ylquinolin-6-yl)methyl]-2-piperidin-1-ylquinoline; NSC292213, 4-[2-(3-carboxy-4-hydroxynaphthalen-1-yl)-2-oxoacetyl]-1-hydroxynaphthalene-2-carboxylic acid; NSC2979, 7,8,8a,9-tetrachloro-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,8,9,10,10a-decahydrophenanthrene-1-carboxylic acid; NSC299137, N-(9,10-dioxoanthracen-1-yl)-7-oxobenzo[e]perimidine-4-carboxamide; NSC30188, 4-[(3-carboxy-2-hydroxynaphthalen-1-yl)-methyl]-3-hydroxynaphthalene-2-carboxylic acid; NSC38777, 2-(2-nitrophenyl)butanedioic acid; NSC39863, 1,11a,13a-trimethyl-8-phenyl-2,3,3a,3b,4,5,5a,6,11,11a,11b,12,13,13a-tetradecahydro-1H-cyclopenta[5,6]naphtho[1,2-g]quinazolin-1-ol; NSC40837, 2-phosphonoethylphosphonic acid; NSC41309, furan-2,3,4,5-tetracarboxylic acid; NSC42258, 3-[[4-(1H-indol-3-ylmethyl)piperazin-1-yl]methyl]-1H-indole; NSC601364, 5-[3-pyridin-2-yl-6-(5-sulfofuran-2-yl)-1,2,4-triazin-5-yl]furan-2-sulfonic acid; NSC610938, 4-[(3-carboxy-4-oxonaphthalen-1-ylidene)-phenylmethyl]-1-hydroxynaphthalene-2-carboxylic acid; NSC66207, naphthalene-1,4,5,8-tetracarboxylic acid; NSC683770, (4R)-4-[(3R,5R, 8R,9S,10S,13R,14S,17R)-3-hydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid; NSC73100, 2-(9H-fluoren-2-ylcarbamoyl)terephthalic acid; NSC7616, achilleic acid; NSC93354, (8S,9S,10R,11S,13S,14S,17S)-11-hydroxy-10,13-dimethyl-17-(2-phenyl-1,3-thiazol-4-yl)-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]phenanthren-3-one; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; QEF, quail embryonic fibroblast; RCAS, replication-competent Avian sarcoma-leukosis virus long terminal repeat with a splice acceptor; SPR, surface plasmon resonance; Src, Rous sarcoma virus oncogene cellular homolog; VLS, virtual ligand screening; YFP, yellow fluorescent protein.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
References
- Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, and Chang HY (2006) Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet 38 421-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M, Schetter C, Himly M, Batista O, Chang HW, and Vogt PK (2000) The catalytic subunit of phosphoinositide 3-kinase: requirements for oncogenicity. J Biol Chem 275 6267-6275. [DOI] [PubMed] [Google Scholar]
- Berg T (2003) Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed 42 2462-2481. [DOI] [PubMed] [Google Scholar]
- Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, and Vogt PK (2002) Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci U S A 99 3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuscher AE, Olson AJ, and Goodsell DS (2005) Identifying protein binding sites and optimal ligands. Lett Drug Des Discov 2 483-489. [Google Scholar]
- Bister K, Hayman MJ, and Vogt PK (1977) Defectiveness of avian myelocytomatosis virus MC29: isolation of long-term nonproducer cultures and analysis of virus-specific polypeptide synthesis. Virology 82 431-448. [DOI] [PubMed] [Google Scholar]
- Blackwood EM and Eisenman RN. (1991) Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251 1211-1217. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Lüscher B, and Eisenman RN (1992) Myc and Max associate in vivo. Genes Dev 6 71-80. [DOI] [PubMed] [Google Scholar]
- Brooijmans N and Kuntz ID (2003) Molecular recognition and docking algorithms. Annu Rev Biophys Biomol Struct 32 335-373. [DOI] [PubMed] [Google Scholar]
- Cogliati T, Dunn BK, Bar-Ner M, Cultraro CM, and Segal S (1993) Transfected wild-type and mutant max regulate cell growth and differentiation of murine erythroleukemia cells. Oncogene 8 1263-1268. [PubMed] [Google Scholar]
- Dickerson TJ, Beuscher AE 4th, Rogers CJ, Hixon MS, Yamamoto N, Xu Y, Olson AJ, and Janda KD (2005) Discovery of acetylcholinesterase peripheral anionic site ligands through computational refinement of a directed library. Biochemistry 44 14845-14853. [DOI] [PubMed] [Google Scholar]
- Eilers M (1999) Control of cell proliferation by Myc family genes. Mol Cells 9 1-6. [PubMed] [Google Scholar]
- Fieber W, Schneider ML, Matt T, Kräutler B, Konrat R, and Bister K (2001) Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J Mol Biol 307 1395-1410. [DOI] [PubMed] [Google Scholar]
- Follis AV, Hammoudeh DI, Wang H, Prochownik EV, and Metallo SJ (2008) Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol 15 1149-1155. [DOI] [PubMed] [Google Scholar]
- Hetényi C and van der Spoel D (2002) Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci 11 1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L, Blackwood EM, and Eisenman RN (1992) Myc and Max proteins possess distinct transcriptional activities. Nature 359 426-429. [DOI] [PubMed] [Google Scholar]
- Lee LA and Dang CV (2006) Myc target transcriptomes. Curr Top Microbiol Immunol 302 145-167. [DOI] [PubMed] [Google Scholar]
- Li C, Xu L, Wolan DW, Wilson IA, and Olson AJ (2004) Virtual screening of human 5-aminoimidazole-4-carboxamide ribonucleotide transformylase against the NCI diversity set by use of AutoDock to identify novel nonfolate inhibitors. J Med Chem 47 6681-6690. [DOI] [PubMed] [Google Scholar]
- Lindeman GJ, Harris AW, Bath ML, Eisenman RN, and Adams JM (1995) Overexpressed max is not oncogenic and attenuates myc-induced lymphoproliferation and lymphomagenesis in transgenic mice. Oncogene 10 1013-1017. [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, and Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46 3-26. [DOI] [PubMed] [Google Scholar]
- Lüscher B (2001) Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 277 1. [DOI] [PubMed] [Google Scholar]
- Lutz W, Leon J, and Eilers M (2002) Contributions of Myc to tumorigenesis. Biochim Biophys Acta 1602 61-71. [DOI] [PubMed] [Google Scholar]
- Maki Y, Bos TJ, Davis C, Starbuck M, and Vogt PK (1987) Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A 84 2848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, and Cole MD (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94 363-374. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, and Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19 1639-1662. [Google Scholar]
- Nair SK and Burley SK (2003) X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112 193-205. [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, and Prochownik EV (1999) MYC oncogenes and human neoplastic disease. Oncogene 18 3004-3016. [DOI] [PubMed] [Google Scholar]
- Perez-Roger I, Kim SH, Griffiths B, Sewing A, and Land H (1999) Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1). EMBO J 18 5310-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos CJ, Givol I, and Hughes SH (1996) Comparative analysis of the structure and function of the chicken c-myc and v-myc genes: v-myc is a more potent inducer of cell proliferation and apoptosis than c-myc. Oncogene 12 2611-2621. [PubMed] [Google Scholar]
- Philipp A, Schneider A, Väsrik I, Finke K, Xiong Y, Beach D, Alitalo K, and Eilers M (1994) Repression of cyclin D1: a novel function of MYC. Mol Cell Biol 14 4032-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathapam T, Tegen S, Oskarsson T, Trumpp A, and Martin GS (2006) Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci U S A 103 2695-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JP, Beuscher AE 4th, Flajolet M, McAvoy T, Nairn AC, Olson AJ, and Greengard P (2006) Discovery of protein phosphatase 2C inhibitors by virtual screening. J Med Chem 49 1658-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, and Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45 2615-2623. [DOI] [PubMed] [Google Scholar]
- Venditti M, Iwasiow B, Orr FW, and Shiu RP (2002) C-myc gene expression alone is sufficient to confer resistance to antiestrogen in human breast cancer cells. Int J Cancer 99 35-42. [DOI] [PubMed] [Google Scholar]
- Vogt PK (1971) Spontaneous segregation of nontransforming viruses from cloned sarcoma viruses. Virology 46 939-946. [DOI] [PubMed] [Google Scholar]
- Wang R, Fu Y, and Lai L (1997) A new atom-additive method for calculating partition coefficients. J Chem Inf Comput Sci 37 615-621. [Google Scholar]
- Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, and Janda KD (2006) A credit-card library approach for disrupting protein-protein interactions. Bioorg Med Chem 14 2660-2673. [DOI] [PubMed] [Google Scholar]
- Yin X, Giap C, Lazo JS, and Prochownik EV (2003) Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22 6151-6159. [DOI] [PubMed] [Google Scholar]
- Yin X, Grove L, and Prochownik EV (1998) Lack of transcriptional repression by max homodimers. Oncogene 16 2629-2637. [DOI] [PubMed] [Google Scholar]
- Yuza Y, Kawakami M, Takagi K, Yamazaki Y, and Urashima M (1999) Max protein expression is associated with survival of children with lymphoblastic lymphoma. Pediatr Int 41 637-640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.