Abstract
Free energy calculations are fundamental to obtaining accurate theoretical estimates of many important biological phenomena including hydration energies, protein-ligand binding affinities and energetics of conformational changes. Unlike traditional free energy perturbation and thermodynamic integration methods, λ-dynamics treats the conventional "λ" as a dynamic variable in free energy simulations and simultaneously evaluates thermodynamic properties for multiple states in a single simulation. In the present paper, we provide an overview of the theory of λ-dynamics, including the use of biasing and restraining potentials to facilitate conformational sampling. We review how λ-dynamics has been used to rapidly and reliably compute relative hydration free energies and binding affinities for series of ligands, to accurately identify crystallographically observed binding modes starting from incorrect orientations, and to model the effects of mutations upon protein stability. Finally, we suggest how λ-dynamics may be extended to facilitate modeling efforts in structure-based drug design.
Keywords: free energy, protein-ligand, sampling, drug design
1 Introduction
Free energy simulation methods are fundamental to understanding the thermodynamic properties of many biologically important systems and phenomena1,2. These methods have been employed to estimate hydration free energies of ions3,4 and small molecules5,6, protein-ligand binding affinities7, and protein stability8. Since potency often correlates well with the binding affinity of a drug to its targeted receptor, theoretical methods which can reliably estimate binding free energies can facilitate the design or optimization of new therapeutics7,9,10. Rigorous free energies methods, while being more expensive and not amenable to high-throughput screens of large libraries of drug candidates, may be very effective in providing better estimates of binding affinities and provide more reliable information for guiding drug design programs.
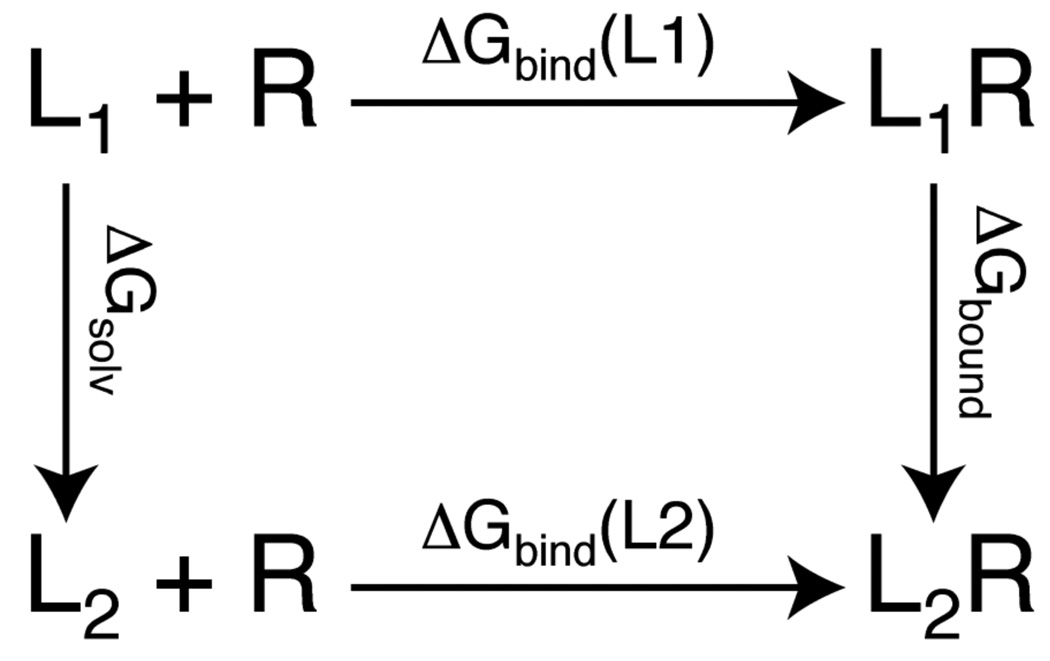
Using the thermodynamic cycle illustrated in Figure 1, the relative binding affinities of two ligands can be described by the difference in the free energies associated with the chemical transformation of one ligand into the other in the bound and solvent environments respectively, i.e.
| (1) |
In traditional free energy perturbation (FEP)11 and thermodynamic integration (TI)12, the free energy changes associated with these vertical “arms” of the thermodynamic cycle are evaluated by alchemically morphing one ligand into the other. In these methods, multiple simulations are performed at discrete points along the transformation pathway between the end-states. For these simulations, the Hamiltonian governing the dynamics of the system is defined as:
| (2) |
where Vo and V1 are the potential energy contributions associated with the two endpoints and the “λ” parameter indicates the distance along the transformational pathway. Using FEP, the free energy change is computed from the ensemble averages generated at discrete λ values:
| (3) |
where β=(kBT)−1. To ensure adequate sampling within each of these simulations, the free energy change barrier between λ and λ+δλ should be less than ~2 kcal/mol1,13.
Figure 1.
Thermodynamic cycle for computing the relative binding free energies of two ligands (L1 and L2) to a common receptor (R).
For drug design purposes, the computational demands of these traditional free energy calculations are significant as simulations at multiple λ values must be performed for each pair of ligands that are investigated. By contrast, the λ-dynamics simulation method which was developed in the Brooks’ lab is an appreciably more efficient method for computing relative hydration or binding free energies for a series of molecules14. In λ-dynamics, which was inspired by the work of Liu and Berne15 and Tidor16, the λ parameter is treated as a dynamic variable with a fictitious mass and is propagated along with the atomic coordinates throughout the course of a simulation. In this way, it is possible to obtain free energy estimates for a full “arm” of the thermodynamic cycle within one simulation. Furthermore, λ-dynamics is no longer limited to a comparative analysis of two ligands, but rather can simultaneously evaluate multiple ligands in a given simulation. In this case, each ligand is assigned a value of λ with the constraint that for N ligands. In the context of structure-based drug design, these λ-dynamics simulations are akin to competitive binding experiments in which all the ligands compete for a common receptor on the basis of their relative free energies using multiple copy simultaneous search approaches17.
This review focuses on λ-dynamics simulation methods in which the λ parameters scale the potential energy contributions of the corresponding ligands and where λ is treated as a dynamic variable whose value is modified at each time step. First, we present the general theory and the main features of λ-dynamics simulations. Second, we will highlight the chemical applications that have been explored using λ-dynamics simulation methods and finally, we will provide a prospective about how λ-dynamics can be used increasingly in structure-based drug design.
2 λ-dynamics theory and methods
2.1 λ-dynamics theory
The λ-dynamics methodology has been described in detail in Kong and Brooks14 and Guo et al.18. Here, we provide a brief overview of the key equations governing the λ-dynamics simulations and analyses in the context of computing relative binding affinities for multiple ligands. Unlike traditional FEP or TI calculations, λ is treated as a dynamic particle with fictious mass mλ. The dynamics of the system is generated from the extended Hamiltonian:
| (4) |
where the first two terms represent the kinetic energies of the atomic coordinates and λ variables respectively. The hybrid potential energy function for a protein and a total of L ligands is constructed as:
| (5) |
where X and xi are the coordinates of the environment and ligand i respectively and λi2 and Fi are the coupling parameter and biasing potential, respectively, that are associated with ligand i. The use of λi2 ensures that the individual λ values are positive and the restraint ensures that the λ values are in the range of [0,1] throughout the simulation trajectory. If all potential energy terms are scaled by λ then significant geometric distortions of the ligand are prevalent at very small values of λ. To retain near-equilibrium conformations of the ligands, it is suggested that the intramolecular bond and angle energy terms should not be scaled.
The difference in free energy between any two ligands i and j, with biasing potentials Fi and Fj respectively, can then be determined from a given simulation by:
| (6) |
where corresponds to the probability that the hybrid system is in a state that is dominated by ligand i. In practice, the ratio of the probability of states dominated by λi2 and λj2 are obtained from the relative amount of time that λi2 and λj2 are greater than a given cutoff value during a given simulation. In general, cutoff values between 0.8 and 0.9 are employed; however, the sensitivity of the computed relative binding affinities to the cutoff value will depend on the smoothness of the free energy landscape with respect to λ. To obtain accurate estimates of the relative free energies, each ligand must regularly sample the dominant state. Ligands with free energies that are within 2–3 kcal/mol of each other will adequately sample all λi=1 states. In the following section, the use of biasing potentials will be discussed that enable ligands whose free energy differences are greater than 3 kcal/mol to successfully compete with each other for the dominant state.
Bitetti-Putzer et al. introduced an alternate method for obtaining relative free energies from a single λ-dynamics simulations19. In this generalized ensemble thermodynamic integration (GETI) method, snapshots from a λ-dynamics trajectory are sorted into bins according to their λi value and then the traditional TI equation:
| (7) |
can be used to compute the change in free energy associated with each individual bin. With thorough sampling of both ligand states, Bitetti-Putzer et al. demonstrated that the free energy estimate converged significantly more quickly using GETI than via standard TI methods and was relatively insensitive to the adjustable parameters of this method, that is, the choice of the width and number of bins. While this method enables relative free energies to be obtained from a single simulation, unlike the probability ratio method in Eq 6, it is restricted to sampling two ligands simultaneously.
2.2 Biasing potentials
The biasing potentials, Fi, that were introduced in Eq 5 have two purposes. First, Fi can be used as a reference free energy. For example, if the biasing potentials {F} are 0 for all ligands then Eq 5 is the free energy associated with one arm of the thermodynamic cycle in Figure 1. Alternatively, the set of Fi values could be the free energies associated with the corresponding ligand transformations in the solvent environment, i.e. one-half of the thermodynamic cycle; by using these reference free energies in the protein-ligand simulations, Eq 5 simply becomes the relative free energy for the full thermodynamic cycle. This latter strategy has been successfully applied for rapidly screening and ranking ligands directly from the probabilities of each ligand being in the λ=1 state in the bound simulations when their {F} are assigned from the corresponding solvent simulations.
Second, Fi can serve as biasing potentials to focus the sampling in a particular region of phase space. Since accurate estimates of the relative free energies depend upon adequate sampling of all λi2=1 (or, in practice, λi2>0.8) within a given simulation, biasing potentials can be chosen to reduce the barrier height between different states along the reaction coordinates.
Guo et al. proposed an iterative strategy, which is similar in spirit to the entropy sampling method of Lee20 and Hao and Scheraga21, to optimize the biasing potentials for a given set of ligands18. In this strategy, the estimated free energies of each ligand from a simulation are selected as the biasing potentials for the corresponding ligands in the subsequent simulation. Data from multiple simulation trajectories may be combined using the weighted histogram analysis method (WHAM)22,23 to obtain more accurate estimates of the free energy. The probability histogram from R simulations with L ligands is given by:
| (8) |
where Nk is the number of snapshots with {λ2} in the kth simulation and nm is the total number of snapshots taken in the mth simulation and
| (9) |
The estimated free energy relative to the reference free energy {F} after the Rth simulation is
| (10) |
By using Eq 8– Eq 10 in an iterative fashion, in each successive simulation, the updated biasing potentials should render the ligands increasingly competitive with one another for the dominant state and, thus, the convergence of the free energies should improve. However, in the presence of large barriers between the endpoints, these biases of Fi, which are linearly scaled by λi, may not sufficiently flatten the free energy surface. In these situations, biasing potentials that depend on λ may be utilized to provide better control over simulation efficiency and sampling space. In the following section, the use of biasing potentials for restraining the conformational space that a given ligand samples is described.
2.3 Restraining potentials for multiple topology representations
In performing these simulations in which ligands are alchemically transformed from one to another either a non-physical hybrid ligand must be constructed or each ligand must be explicitly represented. In the former construction, or single hybrid representation, atoms that are common to the ligand series are represented only once as a common core and are treated as “environment” atoms in the Hamiltonian. The portions of the ligands which vary are each represented by individual non-interacting moieties that are attached to the common core. This approach helps to reduce the degrees of freedom within the system and focuses the sampling on the specific chemical variations on the ligand core. This is an efficient strategy when the ligands adopt similar binding modes within the receptor. However, if a series of ligands adopts several distinct conformations within the binding pocket, it may be difficult to adequately sample all conformations within a given simulation and obtain reliable estimates of the relative binding affinities.
By contrast, in multiple topology representations, multiple complete ligands are represented explicitly in a simulation though they do not interact with one another directly. In this representation, the ligands sample independent conformations within the binding pocket. While a variety of binding modes can be well-sampled throughout the λ-dynamics simulations, adding restraining potentials to the hybrid potential in Eq 5 is recommended to ensure that ligands that are only weakly coupled to the environment (i.e. when their respective λ values are low) do not wander outside the vicinity of the binding pocket. This restraining potential depends on λi and the atomic coordinates of ligand i as well as the average coordinates of the environment atoms, Xo, such that the hybrid potential energy is described by:
| (11) |
The restraining potential ensures that the ligands remain in low-energy regions of conformational space and is defined as:
| (12) |
where α determines how to scale the biasing potential when λi2 approaches 0 and thus the extent of the conformational space about the initial binding orientation that may be sampled. Small values of α give rise to weak restraining potentials and enlarge the sampling space for the ligand; by contrast, large values of α limit the sampling space even when λi2 is near 0. Studies have shown that an α value of 0.3 provides a good balance of the probabilities and restraining of the ligands24. By contrast, α > 5 tends to result in large populations of states with intermediate λ values and, with small values of α, the restraining potential is not sufficiently strong to keep the ligand within the binding pocket. This restraining potential has the added advantage of preventing high-energy states that cause instability in the integration algorithm when small increases in λi when λi approaches 0 can cause significant spikes in the energy.
By including this restraining potential, the relative binding free energy is estimated by:
| (13) |
where the second and third terms represent the average restraining potentials for ligands i and j when their respective λ values are 0 (or in practice, when λi<0.05 and λj<0.05). In this case, the environment atoms are assumed to move more slowly than the ligand atoms and so the time invariant coordinates Xo can be approximated by the instantaneous coordinates X(t). Thus, the restraining potential Ri(Xo, xi) has been replaced by Ri(X(t),xi). Using this approximation, the system does not strictly obey Newton’s equations of motion since the restraining potential is only “seen” by the ligands with low λ values. However, the dominant ligand and environment atoms do retain Newtonian motion in the λ-dynamics simulations. Eq 13 also assumes that the entropy terms related to the restraining potential cancel due to the similarity of the ligands. It has been shown that the second and third terms in Eq 13 converge more rapidly than the probabilities computed from the trajectories, especially for weak binders25 and that the specific value of α influences the sampling efficiency but not in principle the final free energy estimates24.
When R simulations are generated with the mth simulation using a set of biasing potentials , the probability at the desired biasing potentials (i.e. the solvation free energies of the ligands) may be estimated by:
| (14) |
where
| (15) |
The relative binding free energies can then be estimated by:
| (16) |
where the second and third terms correspond to the restraining potentials of ligand i and j and are summed over the states in which the respective λ2 values are zero.
2.4 λ-dynamics implementation in CHARMM
λ-dynamics has been incorporated into the CHARMM macromolecular modeling software package26 within the BLOCK module. In this module, the system is partitioned into the environment (including any atoms which are invariant among the ligands) and the individual ligands (or atoms that constitute the unique portions of the ligands). Initial λ values, mλ and F are specified for each partition of the system and additional restraining potentials can be applied to a specific partition or can be used to couple two partitions. Once the system partitions and biasing and restraining potentials are defined, molecular dynamics simulations can be used to sample atomic coordinates and λ values.
In Tidor’s original studies using simulated annealing techniques16,27 and in the chemical-MC/MD method of Pitera and Kollman28, λ was treated as a dynamic variable that was varied via a Monte Carlo scheme after every molecular dynamics step in which the atomic coordinates were propagated. While Tidor sampled intermediate λ values throughout the simulations, Pitera and Kollman only sampled the end-states, i.e. where at each timestep one ligand was “real” (i.e. λ=1) and all others were “ghosts” (i.e. λ=0). In these latter simulations, jumping between λ values of 0 and 1 did not hinder the effective sampling of each ligands in the systems that were investigated. However, the authors confess that other systems may not be amenable to sampling in this manner and that intermediate λ values might be required. In developing λ-dynamics simulations in our group, appropriately biased λ-dynamics propagated by molecular dynamics simulations yielded improved transition frequencies over those propagated by trial moves based on Monte Carlo methods (unpublished data).
3 λ-dynamics applications
3.1 Rapid screening and accurate calculations for multiple ligands
One of the key advantages of λ-dynamics over traditional FEP and TI methods arises from its ability to assess the thermodynamic properties of multiple chemical moieties in a single simulation. These chemical moieties can include individual complete ligands or multiple substituents attached to a common ligand core. Using Eq 6, relative free energies of all ligand pairs may be assessed using probabilities obtained from the same trajectory. In addition to the computational gains afforded by λ-dynamics by sampling all λ values and multiple chemical moieties simultaneously, studies using λ-dynamics have demonstrated its ability to rapidly and reliably differentiate between good and poor binders (whether the “binding site” is a traditional protein environment in binding free energies or an analogous shell of solvent molecules in hydration free energies). When the overall binding affinity among the species differs by less than 2 kcal/mol, they all compete reasonably well for binding, and λ≈1 is well represented for each species. In general, any species whose binding affinity differs by more than 3 kcal/mol from the most favorable binder can be easily screened out within tens of picoseconds of simulation time because it cannot adequately compete, i.e., it never reaches the λ≈1 state. For most drug design applications, it is sufficient to identify the poor binders and then to obtain more precise estimates of the relative free energies among the good binders by lengthening the simulations and, if need be, by incorporating additional biasing potentials. The following examples provide concrete illustrations of how λ-dynamics simulations are effective at rapid screening and obtaining accurate free energy estimates.
Kong and Brooks, in their initial paper, investigated the hydration free energies of four small organic molecules14. Within short simulations (100ps) in explicit solvent, the compound with the poorest hydration free energy could be identified unambiguously. However, due to the large energy barrier between two of the substituents, the system was trapped in different local minima depending on the initial assigned λ values and the accurate ordering of the remaining compounds required the addition of biasing potentials. Once the biasing potentials were applied, the computed relative hydration free energies using Eq 6 for pairs of compounds from the subsequent simulation agreed very well with other published theoretical calculations and experimental data.
Similar results were achieved by Guo et al. for hydration free energies for a series of larger benzamidine derivatives18,29. By using the gas phase free energies as the reference free energy values {F}, a single λ-dynamics trajectory of only 30ps provided the correct relative ranking of the four compounds. Different combinations of initial λ values yielded similar results suggesting that the free energy landscape for this system was relatively smooth. By studying a pair of benzamidine derivatives and using the iterative technique described in section 2.2, Guo et al. confirmed that the λ-dynamics results were in excellent agreement with FEP calculations and achieved a comparable degree of precision with two times greater efficiency.
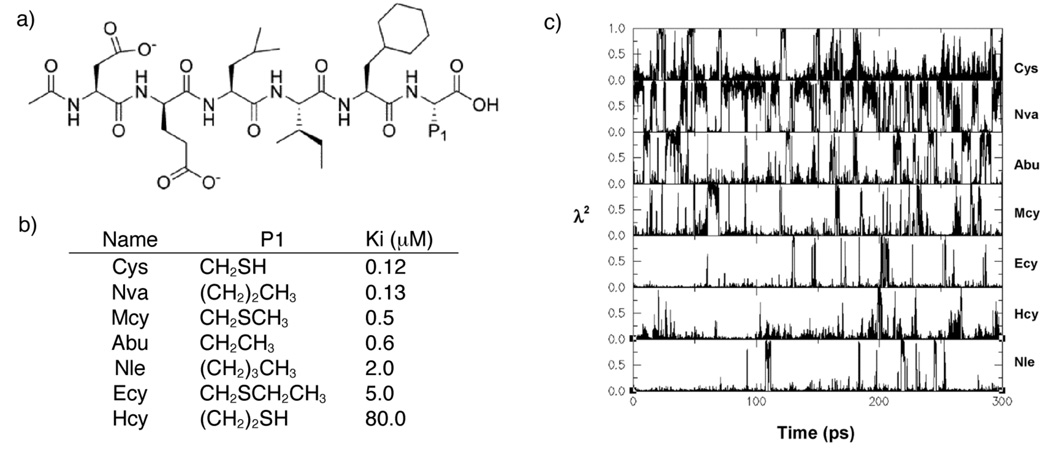
Researchers at Schering-Plough used λ-dynamics to compute the relative binding affinities for seven 6-mer peptide inhibitors of hepatitis C virus (HCV) protease30. A hybrid inhibitor was constructed in which five residues formed the common core and the sixth residue contained seven discrete sidechains. Within a single λ-dynamics trajectory, these seven variants of the inhibitor could be classified as strong or weak binders. Figure 2 illustrates how λi=1 was sampled sufficiently often for the strong binders that their relative binding affinities could be reliably computed. Additional simulations which contained the weak binders and only one of the strong binders as a reference molecule enabled all relative binding free energies to be computed accurately. These theoretical estimates were within 0.6 kcal/mol of the experimentally determined value for five of the 7 ligands and all agreed with experiment within 2.0 kcal/mol. In performing a series of FEP simulations for several pairs of these 6-mer peptide inhibitors, similar results were obtained but at five times the computational expense of the corresponding λ-dynamics simulations.
Figure 2.
a) Schematic of the HCV protease inhibitors. b) Chemical identities and binding data for the HCV protease inhibitors. c) Reprinted by permission from Guo et al30. Example of the λ-dynamics trajectory of seven inhibitors during a 300 ps simulation. The names of the inhibitors are shown on the right.
3.2 Exploring alternate binding modes with λ-dynamics simulations
Multiple topology λ-dynamics with the restraining potential described in section 2.3 has been used to investigate different ligand orientations within a single simulation and to compute relative free energies for series of ligands that have different binding modes within a common protein target. Banba and Brooks studied alternate binding modes of a series of heterocycles within an artificial cavity in cytochrome c peroxidase25. Each ligand was explicitly represented and, without the restraining potential, six ligands became trapped in local minima associated with their respective incorrect initial ligand orientations and rendered the simulations unstable due to high-energy states sampled by the unbound ligands. By applying restraining potentials (Eq 12), within 300 ps the best binder of the ligand series was readily identified. These trends were further validated by Damodaran et al. in which a series of monosubstituted benzene derivatives were bound to β-cyclodextrin24.
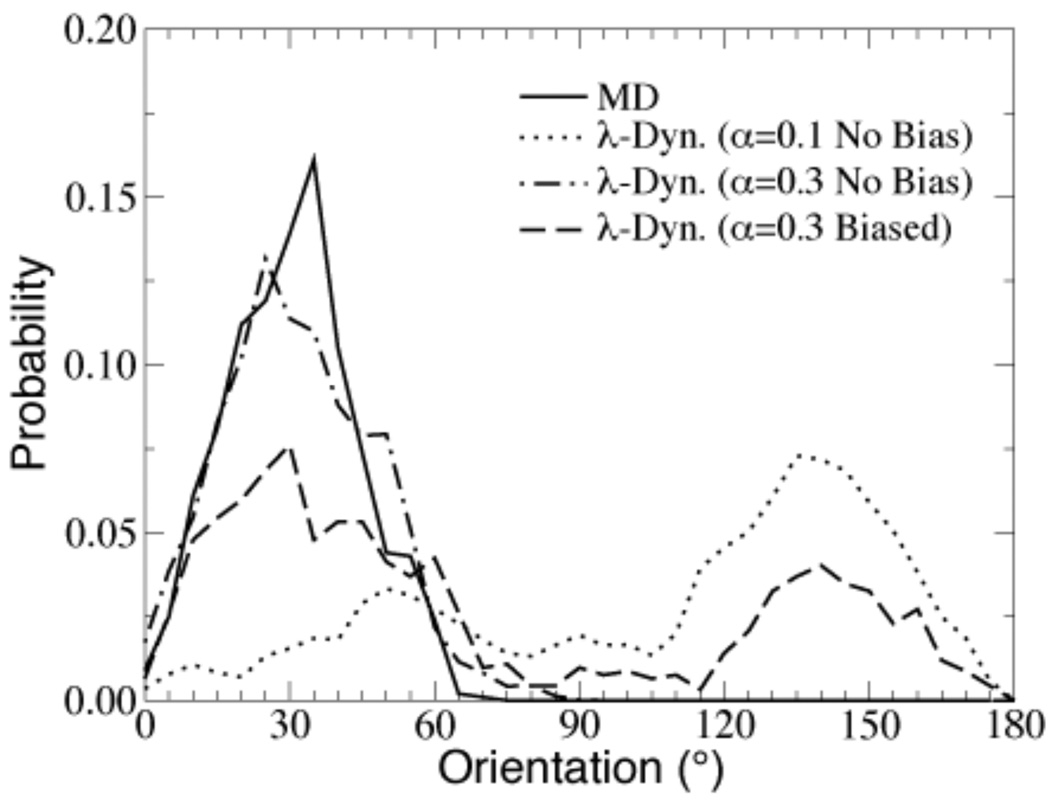
Conventional molecular dynamics simulations will tend to restrict the sampling of ligands to their initial orientation within a given binding pocket. By contrast, with appropriately selected biases and restraining potentials λ-dynamics simulations allow ligands to sample alternative binding modes31. For example, Figure 3 depicts the orientations that toluene samples within the β-cyclodextrin host cavity throughout a λ-dynamics as well as a conventional molecular dynamics trajectory24. In addition, λ-dynamics was able to identify the crystallographic binding mode of five-membered ring heterocycles bound to the cytochrome c peroxidase cavity regardless of the initial binding orientation and determine the relative free energy contributions of different binding modes based on their relative populations of dominant states in the course of a single simulation trajectory25. Conformational sampling via MD simulations was restricted to the ligand’s initial orientation due to the large energy barriers between the alternate binding orientations. Traditional FEP and TI simulations also tend to be restricted to the initial binding orientation since simulations are performed for values of λ that are large enough that they can not sufficiently lower the potential energy barrier between alternative binding orientations. Thus, λ-dynamics is a robust free energy method even when the binding mode of a potential new therapeutic is unknown. In fact, it has been suggested that the value of α and Fi in Eq 11 and Eq 12 can be tuned to sample alternate binding orientations31.
Figure 3.
Reprinted by permission from Damodaran et al. (2001). Distributions in the host cavity of the toluene ligand from MD and λ-dynamics trajectories.
3.3 Modeling protein mutations
Pitera and Kollman used chemical-MC/MD to compute the relative free energies of a series of single amino acid sidechains32. For these simulations, they determined the relative hydration free energies of blocked alanine, serine and valine residues in good agreement with TI calculations. In addition, they modeled the effect that mutations at a critical site in T4 lysozyme exert on the stability of the protein. For most of the mutants, the relative free energies of folding obtained from the chemical-MC/MD calculations (i.e. by using the relative populations of the dominant sidechains that were observed in simulations of both the folded protein and a free tripeptide mimicking the unfolded chain) were within 1 kcal/mol of the values obtained from experiments and from traditional TI calculations.
In this work, Pitera and Kollman highlight the importance of including all relevant sidechain rotamer states of the residues under investigation as well as the need for long simulation times (i.e. >5ns) to reach convergence. They experienced difficulties in sampling between sidechains that differed significantly in volume (e.g. arginine and glycine) and in charge (e.g. glutamic acid and lysine) and suggest that chemical-MC/MD will be most effective in comparing natural or non-natural sidechains that are similar in size. λ-dynamics as it is implemented in CHARMM where fractional values of λ can be obtained (as opposed to the discrete “real, λ=1” and “ghost, λ=0” assignments associated with chemical-MC/MD; see section 2.4) should enable more frequent transitions among the end-states and facilitate the sampling even among sidechains that differ significantly from one another. This is currently under investigation in our group in the context of modeling protein stability as well as discerning the influence of binding site mutations on ligand binding affinities.
3.4 Variations of λ-dynamics
In the past decade, λ-dynamics simulation methods have stimulated the development of other theoretical approaches in which the λ parameter scales the potential energy and dynamically varies throughout the course of a simulation.
Constant pHMD (CPHMD) was also developed in the Brooks’ lab and may be considered an application of λ-dynamics in which fictious λ particles are used to propagate titration degrees of freedom. The end-states in this approach represent the deprotonated (λ=0) and protonated (λ=1) states33. This work was extended into two dimensions to model simultaneous titration at two competing sites, such as the two histidine protonation sites and two oxygen atoms in carboxyl groups34,35. CPHMD has been used to study a variety of applications including pH-dependent helix folding36, folding and aggregation propensities in amyloid peptides37, identification of folding intermediates that involve ionizable sidechains38, and the elucidation of acid-base catalytic mechanisms39.
Recently, Abrams et al. incorporated a dynamic λ parameter into their adiabatic free energy dynamics (AFED) for generating free energy profiles along reaction paths40. In this λ-AFED method, by choosing switching functions that generate a high barrier between the endpoints and by carefully assigning the temperature and mass of the “λ”, λ can be effectively adiabatically decoupled from the rest of the system and thus the probability distribution in λ directly leads to the free energy profiles of interest. This method was successfully used to compute hydration free energies of methane and methanol in explicit water. These results are in agreement both with experimental values and published FEP and TI calculations yet were obtained with significantly less computational expense than the traditional free energy methods.
In another strategy, Bitetti-Putzer et al. employ λ as a self-regulating sampling variable to efficiently traverse high-energy barriers and to thoroughly explore low-energy basins41. By using multiple copies of a subset of the system and intermittently varying their associated λ values a copy in a high-energy region will adopt a small λ value and will sample more broadly until it finds a low-energy region at which time its λ value will increase and the copy will begin to sample more finely. Bitteti-Putzer et al. implemented λ-dynamics in which atomic coordinates and λ values were propagated via Monte Carlo sampling. In their study of the entry of an indole fragment into the nonpancreatic secretory phospholipase A2 binding pocket, λ MC moves for the multiple copies of the indole fragments were proposed much less frequently than those for the atomic coordinates and were limited to a maximum move of 0.2. This method was shown to be more effective than multiple copy simulation search methods (i.e. where equal fixed λ values are assigned for each indole fragment) for crossing the free energy barrier associated with diffusion of the ligand from the surface of the protein into the binding pocket and for identifying the correct low-energy position and orientation of the bound indole fragment.
4 Advancing λ-dynamics in structure-based drug design
4.1 Developing multi-site λ-dynamics
Except for CPHMD, λ-dynamics applications to date have focused on simulating differences at single sites in a given system. These differences have ranged from chemical moieties attached to a specific substituent site on a common core compound14,18,29 to complete, explicitly represented ligands24,25,31 to amino acids sidechains within peptides30 and larger proteins32. λ-dynamics simulation methods are currently being extended to model differences at multiple distinct sites simultaneously in a given system. In this way approach, chemical modifications at multiple substituent sites on a common core may be investigated within a single λ-dynamics simulation. Structure-activity relationship (SAR) studies that form the backbone of most drug optimization strategies often investigate the effect of altering chemical moieties at different positions on a lead compound. However, not all combinations of moieties are exhaustively synthesized to determine their experimental binding affinities with respect to a given protein target. Multi-site λ-dynamics would mimic these SAR experiments yet would effectively evaluate all combinations of substituents to identify the drug candidate which would have the optimal binding affinity. In these simulations, each substituent site is identified with a specific λ parameter and the sum of the λ values at each site is restrained to 1.
To obtain reliable relative free energies, each combination of λi2=1 across the substituent sites would need to be adequately sampled. Thus, increasing the number of substituent sites under investigation may require more aggressive sampling methods. In practice, the most favorable substituent at each site should be readily identified if the effects of the substitutions at different sites on the common core are primarily additive in nature. In these simulations, the moieties at each site that are associated with the optimal binder would be the dominant states that were sampled with the highest probability. More complicated relationships among sites can be envisioned where a drug may experience favorable interactions with the binding pocket residues due to the presence of a fragment type at either one substituent site or another, but not both sites simultaneously. For example, for substituent sites that are located in close proximity to one another, the drug may orient itself such that the favorable fragment can adopt the necessary interactions with the binding pocket. Alternatively, spatially separated substituents might influence binding in mutually exclusive ways. In a binding pocket of relatively fixed dimensions, the common core may be able to successfully accommodate a bulky fragment at either “end” of the core, but not at both “ends” simultaneously. In our group, we will be exploring the ability of λ-dynamics to address both of these contexts.
4.2 Predicting effects of drug resistance
In addition to investigating multiple substituent sites on a common core to model a series of ligands, variations in the binding pockets themselves may also be explored. By developing a hybrid model of the binding pocket for a protein target, the effects of anticipated mutations upon a ligand’s binding ability may also be assessed. This may be especially important in the case of developing drug molecules for virus proteins, for example, whose ready ability to mutate leads to substantial drug resistance and reduced long-term efficacy of pharmaceuticals42–44. In practice, ligands that demonstrate favorable binding affinities to the wild-type protein may be exposed to alternate binding pockets via simulation methods. λ-dynamics simulations can be performed in which a ligand effectively “sees” and responds to multiple sidechains at highly-mutatable residues that line the binding pocket. In this case, the hybrid molecule now consists of a protein target in which, at multiple distinct sites, different amino acid sidechains are attached to a common α-carbon. Sidechains at each site do not “see” each other and their respective interactions with the environment are scaled by λ; the mutant sidechains would “see” those from other sites in proportion to the product of their λ values. The relative binding affinity between the ligand and the wild-type and mutant proteins indicates the ability of the ligand to retain its efficacy in the presence of clinically important mutations. With multi-site λ-dynamics, variations in both the binding pocket as well as the ligand core itself may be assessed simultaneously. From this multi-dimensional hybrid system, multiple ligands can be investigated simultaneously and ligands that do not bind well to all clinically-relevant proteins are effectively screened out.
5 Conclusions
The λ-dynamics free energy methodology has been developed to obtain accurate estimates of relative hydration free energies and binding affinities. This approach is based on the idea that multiple ligands compete for a common receptor on the basis of their relative free energies and that multiple ligands are modeled concurrently in a common receptor environment using a multiple copy simultaneous search strategy. In λ-dynamics simulations, the λ parameter is treated as a dynamic variable and is propagated along with the atomic coordinates and velocities throughout the course of a simulation; in this way, only one simulation is required to sample all λ values between the ligand end-states (i.e. λ=0 and λ=1). Furthermore, multiple ligands can be assessed simultaneously where the potential energy term associated with each ligand is scaled according to its λ value.
To date, studies of hydration free energies and binding affinities of small molecules have been investigated using λ-dynamics simulations. It is clear from these studies, that λ-dynamics simulations can rapidly screen out poor binders among a series of compounds. When the free energy surface is relatively smooth the λ=1 states for the remaining compounds will be well sampled which enable accurate calculations of the relative free energies. When the free energy surface is more rugged, biasing potentials can be readily employed to overcome large energy barriers between the end-points.
λ-dynamics simulations are also able to sample more extensive conformational space and are less likely to get trapped in local energy minima than traditional molecular dynamics simulations methods. For a series of ligands which have alternate binding conformations, λ-dynamics simulations have been successful at identifying binding modes that are observed in crystal structures even when the simulations are initialized from incorrect orientations.
Results from λ-dynamics simulations agree well with other theoretical methods, like FEP, and with experimental results where available; although, agreement between the theoretical estimates and experimental data will also depend heavily on the quality of the force field parameters that are used in the simulations. By enabling the range of λ values to be sampled within a single simulation trajectory and by simultaneously modeling on the order of 5 to 10 ligands, the computational expense of free energy calculations relative to traditional free energy perturbation and thermodynamic integration methods decreases dramatically.
λ-dynamics has been implemented in CHARMM and efforts are ongoing to extend the scope of λ-dynamics in structure-based drug design, specifically to allow multiple sites of variations to be targeted within a single simulation. Multi-dimensional hybrid ligands will allow relative binding affinities of a significantly larger number of unique ligands to be evaluated. Using multi-site λ-dynamics for hybrid binding pockets, ligands can be screened against clinically-important binding pockets. λ-dynamics simulations are not intended to be a panacea for all stages of drug optimization, but can be used in conjunction with other methods as an effective and efficient method to refine docked poses, systematically screen variations of a lead compound or obtain reliable free energy estimates for promising ligands45. Finally, λ-dynamics has also stimulated the development of free energy methods that enable the efficient exploration of important biological phenomena such as pH-dependent conformational changes and mechanisms via constant pH molecular dynamics33–35 and barrier crossings in protein-ligand complexes by using λ as a self-regulating sampling variable41.
Acknowledgement
Support for this work from the NIH (GM037554) is acknowledged.
References
- 1.Beveridge DL, Dicapua FM. Annu Rev Biophys Biophys Chem. 1989;18:431–492. doi: 10.1146/annurev.bb.18.060189.002243. [DOI] [PubMed] [Google Scholar]
- 2.Foloppe N, Hubbard R. Curr Med Chem. 2006;13(29):3583–3608. doi: 10.2174/092986706779026165. [DOI] [PubMed] [Google Scholar]
- 3.Kelly CP, Cramer CJ, Truhlar DG. J Phys Chem B. 2006;110(32):16066–16081. doi: 10.1021/jp063552y. [DOI] [PubMed] [Google Scholar]
- 4.Ashbaugh HS, Asthagiri D. J Chem Phys. 2008;129(20):204501. doi: 10.1063/1.3013865. [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen WL, Ravimohan C. J Chem Phys. 1985;83(6):3050–3054. [Google Scholar]
- 6.Fleischman SH, Brooks CL. J Chem Phys. 1987;87(5):3029–3037. [Google Scholar]
- 7.Gilson MK, Zhou H. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 8.Brooks CL., III Acc Chem Res. 2002;35(6):447–454. doi: 10.1021/ar0100172. [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen WL. Science. 2004;303(5665):1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 10.Jorgensen WL, Ruiz-Caro J, Tirado-Rives J, Basavapathruni A, Anderson KS, Hamilton AD. Bioorg Med Chem Lett. 2006;16(3):663–667. doi: 10.1016/j.bmcl.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 11.Zwanzig RW. J Chem Phys. 1954;22(8):1420–1426. [Google Scholar]
- 12.Straatsma TP, Berendsen HJC. J Chem Phys. 1988;89(9):5876–5886. [Google Scholar]
- 13.Straatsma TP, Berendsen HJC, Postma JPM. J Chem Phys. 1986;85(11):6720–6727. [Google Scholar]
- 14.Kong X, Brooks CL., III J Chem Phys. 1996;105(6):2414–2423. [Google Scholar]
- 15.Liu ZH, Berne BJ. J Chem Phys. 1993;99(8):6071–6077. [Google Scholar]
- 16.Tidor B. J Phys Chem. 1993;97(5):1069–1073. [Google Scholar]
- 17.Caflisch A, Miranker A, Karplus M. J Med Chem. 1993;36(15):2142–2167. doi: 10.1021/jm00067a013. [DOI] [PubMed] [Google Scholar]
- 18.Guo Z, Brooks CL, III, Kong X. J Phys Chem B. 1998;102:2032–2036. [Google Scholar]
- 19.Bitetti-Putzer R, Yang W, Karplus M. Chem Phys Lett. 2003;377(5–6):633–641. [Google Scholar]
- 20.Lee J. Phys Rev Lett. 1993;71(2):211–214. doi: 10.1103/PhysRevLett.71.211. [DOI] [PubMed] [Google Scholar]
- 21.Hao MH, Scheraga HA. J Phys Chem. 1994;98(18):4940–4948. [Google Scholar]
- 22.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13(8):1011–1021. [Google Scholar]
- 23.Boczko EM, Brooks CL., III J Phys Chem. 1993;97(17):4509–4513. [Google Scholar]
- 24.Damodaran KV, Banba S, Brooks CL., III J Phys Chem B. 2001;105(38):9316–9322. [Google Scholar]
- 25.Banba S, Brooks CL., III J Chem Phys. 2000;113(8):3423–3433. [Google Scholar]
- 26.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 27.Jarque C, Tidor B. J Phys Chem B. 1997;101(45):9362–9374. [Google Scholar]
- 28.Pitera J, Kollman PA. J Am Chem Soc. 1998;120(30):7557–7567. [Google Scholar]
- 29.Guo Z, Brooks CL., III J Am Chem Soc. 1998;120:1920–1921. [Google Scholar]
- 30.Guo ZY, Durkin J, Fischmann T, Ingram R, Prongay A, Zhang RM, Madison V. J Med Chem. 2003;46(25):5360–5364. doi: 10.1021/jm030040o. [DOI] [PubMed] [Google Scholar]
- 31.Banba S, Guo Z, Brooks CL., III J Phys Chem B. 2000;104:6903–6910. [Google Scholar]
- 32.Pitera JW, Kollman PA. Proteins. 2000;41(3):385–397. [PubMed] [Google Scholar]
- 33.Lee MS, Salsbury FR, Brooks CL. Proteins. 2004;56(4):738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 34.Khandogin J, Brooks CL., III Biophys J. 2005;89(1):141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khandogin J, Brooks CL., III Biochemistry. 2006;45(31):9363–9373. doi: 10.1021/bi060706r. [DOI] [PubMed] [Google Scholar]
- 36.Khandogin J, Chen J, Brooks CL., III Proc Natl Acad Sci U S A. 2006;103(49):18546–18550. doi: 10.1073/pnas.0605216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khandogin J, Brooks CL., III Proc Natl Acad Sci U S A. 2007;104(43):16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khandogin J, Raleigh DP, Brooks CL., III J Am Chem Soc. 2007;129(11):3056–3057. doi: 10.1021/ja0688880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian J, Khandogin J, West AH, Cook PF. Biochemistry. 2008;47(26):6851–6858. doi: 10.1021/bi800087k. [DOI] [PubMed] [Google Scholar]
- 40.Abrams JB, Rosso L, Tuckerman ME. J Chem Phys. 2006;125(7):074115. doi: 10.1063/1.2232082. [DOI] [PubMed] [Google Scholar]
- 41.Bitetti-Putzer R, Dinner AR, Yang W, Karplus M. J Chem Phys. 2006;124(17):174901. doi: 10.1063/1.2171194. [DOI] [PubMed] [Google Scholar]
- 42.Rousseau MN, Vergne L, Montes B, Peeters M, Reynes J, Delaporte E, Segondy M. Journal Of Acquired Immune Deficiency Syndromes. 2001;26(1):36–43. doi: 10.1097/00126334-200101010-00005. [DOI] [PubMed] [Google Scholar]
- 43.Salomon H, Wainberg MA, Brenner B, Quan YD, Rouleau D, Cote P, LeBlanc R, Lefebvre E, Spira B, Tsoukas C, Sekaly RP, Conway B, Mayers D, Routy JP. Aids. 2000;14(2):F17–F23. doi: 10.1097/00002030-200001280-00003. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien WA. Clin Infect Dis. 2000;30:S185–S192. doi: 10.1086/313858. [DOI] [PubMed] [Google Scholar]
- 45.Eriksson MAL, Pitera J, Kollman PA. J Med Chem. 1999;42(5):868–881. doi: 10.1021/jm980277y. [DOI] [PubMed] [Google Scholar]