Abstract
TRAF2 and TRAF5 are adapter proteins involved in TNFα-induced activation of the JNK and NF-κB pathways. Currently, TNFα-induced NF-κB activation is believed to be impaired in TRAF2 and TRAF5 double knockout (T2/5 DKO) cells. Here, we report instead that T2/5 DKO cells exhibit high basal IKK activity and elevated expression of NF-κB-dependent genes in unstimulated conditions. Although TNFα-induced RIP1 ubiquitination is indeed impaired in T2/5 DKO cells, TNFα stimulation further increases IKK activity in these cells, resulting in significantly elevated expression of NF-κB target genes to a level higher than that in wild-type cells. Inhibition of NIK in T2/5 DKO cells attenuates basal IKK activity and restores robust TNFα-induced IKK activation to a level comparable to that seen in wild-type cells. This suggests that TNFα can activate IKK in the absence of TRAF2 and TRAF5 expression and RIP1 ubiquitination. In addition, both the basal and TNFα-induced expression of anti-apoptotic proteins are normal in T2/5 DKO cells, yet these DKO cells remain sensitive to TNFα-induced cell death, due to the impaired recruitment of anti-apoptotic proteins to the TNFR1 complex in the absence of TRAF2. Thus, our data demonstrate that TRAF2 negatively regulates basal IKK activity in resting cells, and inhibits TNFα-induced cell death by recruiting anti-apoptotic proteins to the TNFR1 complex rather than by activating the NF-κB pathway.
Keywords: TNFα, TRAF2, TRAF5, NF-κB, apoptosis
Introduction
TNF receptor (TNFR) associated factors (TRAFs) are characterized by the presence of a TRAF domain at the C-terminus. Except for TRAF1, other TRAFs contain N-terminal RING finger domain followed by five or seven zinc finger motifs 1; 2. Whereas the RING and zinc finger domains are essential for TRAFs to activate downstream signaling pathways, the TRAF domain is required for their interactions with relevant receptors and downstream effectors 2. TRAF2 is a prototypical member of the TRAF family and transduces signals from the TNFR superfamily members, leading to activation of the c-Jun N-terminal kinase (JNK) and the IκB kinase (IKK) pathways. JNK activates transcription factors of the AP-1 group (e.g. c-jun/ATF2) and IKK activates those of the NF-κB group. These transcription factors in turn induce the expression of genes involved in inflammation, the immune response, cell proliferation, cell differentiation, and the suppression of death receptor-induced apoptosis 3; 4.
IKK is a kinase complex that consists of two catalytic subunits (IKKα and IKKβ) and one regulatory subunit (IKKγ). TNFR family members activate NF-κB through a canonical and/or an alternative noncanonical pathways 5. The canonical NF-κB pathway depends on the presence of both IKKβ and IKKγ and is activated by most TNF superfamily members, resulting in the coordinate expression of multiple inflammatory and innate immune genes. The noncanonical pathway depends on NIK and IKKα and is activated by a subset of TNF family members, such as B cell-activating factor (BAFF), lymphotoxin α and β heterotrimers (LTα/β) and the CD40 ligand. This pathway triggers the expression of genes involved in adaptive immunity and lymphoid organogenesis 5.
Recently, an increasing number of studies has demonstrated that both TRAF2 and TRAF6 possess E3 ligase activity, and that they are capable of catalyzing the formation of non-canonical K63-linked polyubiquitin chains on themselves as well as on their substrates. These studies have also suggested that such K63-linked ubiquitination is essential for activation of the JNK and IKK pathways in response to TNFα and IL-1β stimulation 6; 7; 8. Ubc13/UEV1a is the only E2 ubiquitin conjugating enzyme complex that is currently known to bind to TRAF2 and TRAF6 and to catalyze K63-linked ubiquitination of these proteins 6. Inhibition of Ubc13 expression by siRNA, however, results in inhibition of TNFα-induced JNK activation, but not of IKK activation 9. A recent study also has demonstrated that conditional ablation of Ubc13 results in considerably impaired JNK activation in response to a variety of stimuli in B cells and mouse embryonic fibroblasts (MEFs), without affecting IKK activation under the same conditions. This suggests that either K63-linked polyubiquitination of TRAF2/TRAF6 is not essential for IKK activation, or IKK can also be activated by an alternative, ubiquitin-independent mechanism in response to cytokine stimulation 10.
TRAF2 knockout (T2 KO) MEFs are completely defective in TNFα-induced JNK activation, but only partially deficient in NF-κB activation 11. Interestingly, TRAF2-deficient macrophages overproduce TNFα and nitric oxide (NO) upon TNFα stimulation. In addition, whereas TRAF2 knockout mice die prematurely, mice double mutant for TRAF2 and either TNFα or TNFR1 survive for several months, suggesting that TRAF2 negatively regulates certain aspects of TNFR1 signaling, and thus the canonical NF-κB pathway 12. Also conditional knockout of TRAF2 in B-cells results in constitutive activation of the noncanonical NF-κB pathway, suggesting that TRAF2 also negatively regulates this pathway 13. Tada et al. have reported that, whereas TRAF5-null MEFs respond normally to TNFα-induced JNK and NF-κB activation, TRAF2 and TRAF5 double knockout (T2/5 DKO) MEFs exhibit an almost complete loss of TNFα-induced NF-κB activation 14. These data suggest that TRAF2 can regulate TNFR1 signaling in either a positive or negative direction, in a manner that likely varies according to cell type.
We identified a phosphorylation site at the N-terminal region of TRAF2 by a classical phosphopeptide mapping approach 15. While carrying out functional studies investigating the role of TRAF2 phosphorylation in TNFα-induced NF-κB activation in T2/5 DKO cells reconstituted with empty vector, wild-type (WT) or phosphomutant TRAF2, we repeatedly observed that control T2/5 DKO MEFs exhibit high basal IKK activity and elevated NF-κB target gene expression in the absence of stimulation with TNFα. Surprisingly, stimulation of these cells with TNFα, which merely activates the canonical NF-κB pathway through the TRAF2/5-RIP1-IKK cascade, further increased the expression of NF-κB target genes — to a level higher than that in TNFα-stimulated WT MEFs. To clarify these contradictory observations, we extensively analyzed the activation status of both the canonical and noncanonical NF-κB pathways in T2 KO and T2/5 DKO MEFs. Here we show that both the canonical and noncanonical NF-κB pathways are constitutively activated in these cells, and that the primary function of TRAF2 in TNFR1 signaling is to activate the JNK pathway, while inhibiting TNFα-induced cell death by recruiting antiapoptotic proteins to the TNFR1 complex.
Results
TRAF2 negatively regulates basal NF-κB activity
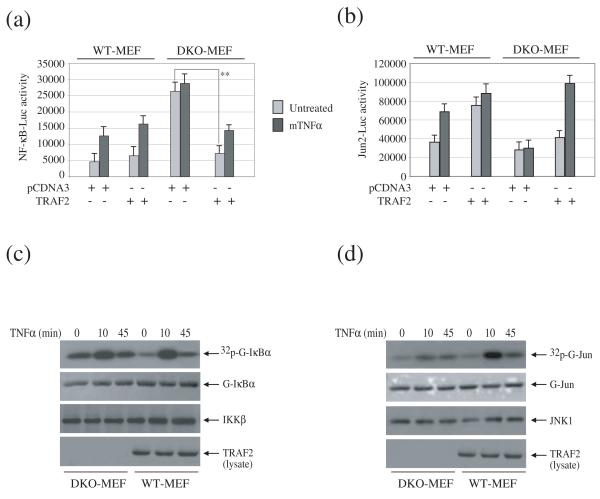
Numerous studies have shown that, in HeLa cells, transient TRAF2 expression induces NF-κB and c-Jun activation in the absence of TNFα stimulation 1; 4. As expected, expression of TRAF2 in HeLa cells increased both basal and TNFα-induced NF-κB and c-Jun activation (Fig. S1). Unexpectedly, we found that, in T2/5 DKO MEFs, transient expression of TRAF2 significantly reduced basal NF-κB activity, which was otherwise quite high compared to that in WT MEFs (Fig. 1a). In contrast, c-Jun activity — both the basal and induced — were lower in T2/5 DKO MEFs than in WT MEFs, and TRAF2 expression restored TNFα-induced c-Jun activation (Fig. 1b). These data suggest that TRAF2 negatively regulates basal NF-κB activity in unstimulated MEFs.
Fig. 1.
T2/5 DKO MEFs exhibit elevated basal IKK and NF-κB activities. (a, b) Wild-type (WT-MEF) and T2/5 DKO MEFs (DKO-MEF) were co-transfected with either NF-κB-Luc (a) or Jun2-Luc (b), plus pRL-TK and either pCDNA3 or TRAF2. 36 hrs after transfection, cells were left untreated or were treated with mouse TNFα (5 ng/ml) for 4 hrs, after which the NF-κB-Luc and Jun2-Luc activities were measured and normalized to pRL-TK activity. Data are presented as mean ± SD of the results from three independent experiments carried out in triplicate. (**) represents p< 0.01. (c, d) T2/5 DKO and WT MEF cells were left untreated or were treated with mTNFα (10ng/ml) as indicated. The IKK complex and JNK1 were then immunoprecipitated with anti-IKKγ and anti-JNK1 antibodies and subjected to in vitro kinase assays in which GST-IκBα1-55 and GST-jun1-87 served as substrates, respectively. The reaction mixtures were separated by SDS-PAGE, transferred onto nitrocellulose membranes and exposed to X-ray film for 4-6 hrs (32p-G-IκBα and 32p-G-jun). The same membranes were stained with Ponceau S (to detect G-IκBα and G-jun) and blotted with anti-IKKβ and anti-JNK1 antibodies, respectively.
TRAF2 suppress the basal activity of IKK complex
Processing of p100 to p52, a hallmark of activation of the noncanonical NF-κB pathway, takes place constitutively in both T2 KO and T2/5 DKO MEFs 16. Therefore, it is possible that the elevated basal NF-κB activity we detected in T2/5 DKO MEFs stems from constitutive activation of the noncanonical NF-κB pathway. The IKK immunokinase assay (using GST-IκBα fusion-protein as a substrate) is one of the most sensitive and reliable methods for assessing the activity of the IKK complex. To investigate the activity of the IKK complex and to rule out the possibility that an IKKα homodimer affects GST-IκBα phosphorylation in the kinase assay, we immunoprecipitated the IKK complex with anti-IKKγ antibody and extensively washed the IKK-bound G-protein beads with lysis buffer containing 350 mM NaCl. As shown in Fig. 1c, the IKK complex was constitutively activated in T2/5 DKO MEFs, and stimulation of these cells with TNFα led to a further, but weak, increase in IKK activity. In contrast, a JNK kinase assay revealed that, in T2/5 DKO MEFs, basal JNK activity was low prior to stimulation with TNFα and remained low even in the presence of TNFα (Fig. 1d). These data suggested that TRAF2 suppresses the basal activity of the IKK complex in resting cells, and confirmed that TRAF2 is required for TNFα-induced JNK activation.
TRAF2 and TRAF5 are not essential for TNFα-induced expression of certain NF-κB target genes
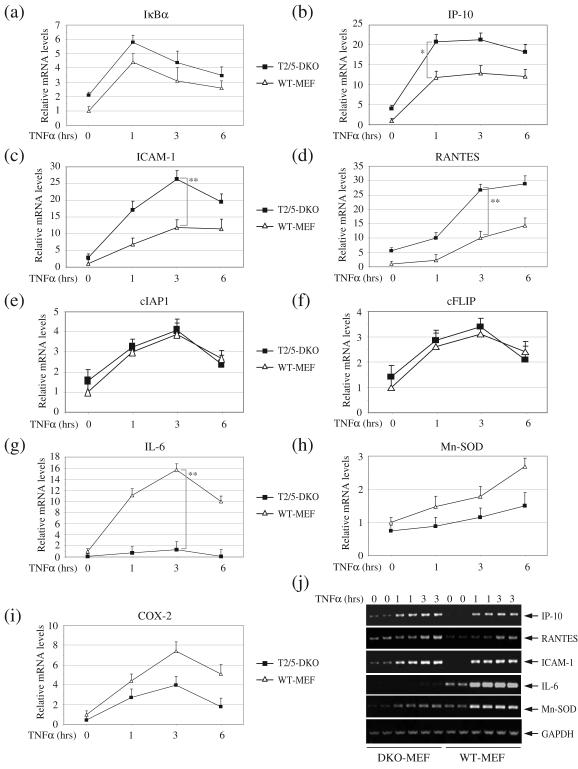
To further examine the activation status of the canonical NF-κB pathway in TRAF2/5 DKO cells, we analyzed the expression of nine well-known NF-κB target genes by quantitative real-time PCR. Consistent with the elevated basal NF-κB activity we had observed in these cells, the basal expression levels of IκBα, IP-10,RANTES and ICAM-1 were higher in T2/5 DKO MEFs than in WT MEFs (Fig. S2). Surprisingly, TNFα stimulation further increased the expression of these genes in T2/5 DKO MEFs, to levels higher than in WT MEFs (Fig. 2a-d). On the other hand, the basal and inducible expression of cIAP1 and cFLIP was comparable in both types of cells (Fig. 2e and 2f). In the cases of IL-6, Mn-SOD and COX-2, however, the basal expression levels were lower in the T2/5 DKO MEFs than in WT MEFs (Fig. 2g-i), and stimulation with TNFα failed to achieve the expression levels attained in WT MEFs; more specifically, the inducibility of IL-6 was almost completely impaired, and that of COX-2 and Mn-SOD was reduced (Fig. 2g-i). To confirm these results, we designed a second set of primers (Table S2) and performed conventional semi-quantitative RT-PCR analysis. As shown in Fig. 2j, the basal and inducible expression of IP-10, RANTES and ICAM-1 was indeed elevated in T2/5 DKO MEFs versus in WT MEFs, whereas in the case of IL-6, inducible expression was impaired. As TNFα merely activates the canonical NF-κB pathway, these data suggest that TRAF2 and TRAF5 are not essential for TNFα-induced expression of some NF-κB target genes.
Fig. 2.
In T2/5 DKO MEFs, TNFα induces the expression of certain NF-κB target genes. (a-i) WT and T2/5 DKO MEFs were left untreated or were treated with mTNFα (10ng/ml) for 1, 3 and 6 hrs. Expression levels of IκBα, IP-10, RANTES, ICAM-1, cIAP1, cFLIP, IL-6, Mn-SOD and COX-2 were then determined by real-time PCR. The relative expression level of each gene is presented as the ratio between the respective gene and the reference gene GAPDH, as mean ± SD from four independent experiments. (*) represents p < 0.05 and (**) p< 0.01. (j) WT and T2/5 DKO MEFs were left untreated or were treated with mTNFα (10ng/ml) as indicated, and the expression levels of IP-10, RANTES, ICAM-1, IL-6, COX-2 and GAPDH were determined by semiquantitative RT-PCR.
Restoration of TRAF2 expression leads to reduced basal IKK activity
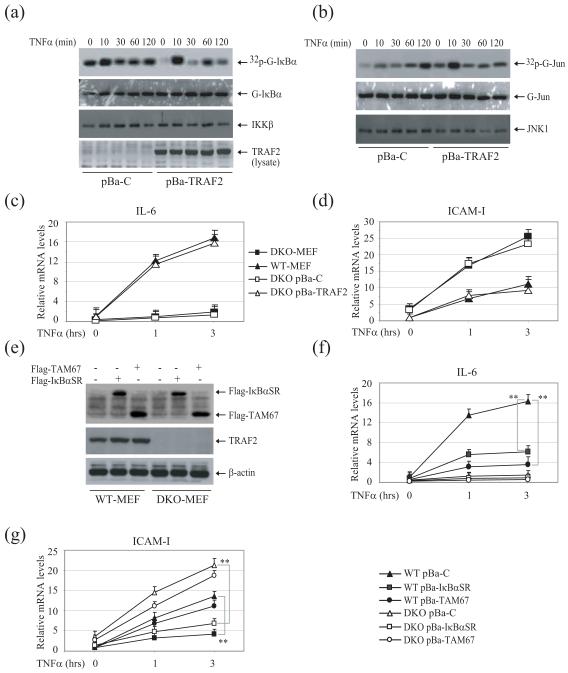
To rule out the possibility that the differences we observed were due to an immortalization effect, we established T2/5 DKO cell lines that stably express either empty vector or TRAF2 at a physiological level (Fig. S3). As expected, T2/5 DKO MEFs stably transfected with empty vector (pBa-C) exhibited high basal IKK activity, whereas their TRAF2-expressing counterparts (pBa-TRAF2) did not (Fig. 3a). In addition, pBa-TRAF2 cells exhibited immediate and robust JNK and IKK activation in response to TNFα stimulation (Fig. 3a and 3b). Consistent with a previous study 17, TNFα treatment did, however, induce JNK activation in pBa-C cells at later time point after stimulation (120 min). To further rule out the possibility that the elevated IKK activity in pBa-C cells was due to cell culture-associated artifacts, we independently established pBa-C and pBa-TRAF2 cell lines using T2/5 DKO MEFs that were at early passage numbers (less than 6) and late passage numbers (more than 30) and repeated the IKK immunokinase assay twice for each cell line, and obtained the same results as in the original experiments in both cases. Regardless of passage number, all pBa-C lines tested exhibited high basal IKK activity, and all pBa-TRAF2 lines exhibited reduced basal IKK activity (Fig. S4). Moreover, stable expression of TRAF2 in T2/5 DKO MEFs restored TNFα-induced IL-6 expression and suppressed the elevated basal and inducible expression of ICAM-I to levels comparable to those in WT MEFs (Fig. 3c and 3d). These data demonstrated that TRAF2 expression at a physiological level is essential for the inhibition of basal IKK activity in resting cells.
Fig. 3.
In T2/5 DKO MEFs, TRAF2 expression inhibits the basal IKK activity and restores TNFα-induced JNK activation and IL-6 expression. (a, b) T2/5 DKO MEFs reconstituted with empty vector (pBa-C) or TRAF2 (pBa-TRAF2) were untreated or treated with mTNFα (10ng/ml) as indicated. IKK and JNK kinase assays were then performed as in Fig. 1c and d. (c, d) WT, T2/5 DKO, pBa-C and pBa-TRAF2 MEFs were untreated or treated with mTNFα (10ng/ml) for 1 and 3 hrs, after which the expression of IL-6 and ICAM-I was determined by real-time PCR, as in Fig. 2c and 2g. (e) WT and DKO MEFs were stably transfected with Flag-IκBαSR or Flag-TAM67 by retroviral infection followed by puromycin selection. The expression of Flag-IκBαSR and Flag-TAM67 was then monitored by Western blotting. (f, g) WT and DKO MEFs stably transfected with Flag-IκBαSR or Flag-TAM67 were treated with TNFα for 1 and 3 hrs, after which the expression of IL-6 and ICAM-I was determined by real-time PCR.
Both c-Jun and NF-κB are required for TNFα-induced IL-6 expression in MEFs
TNFα-induced IL-6 expression is completely impaired in JNK1 and JNK2 DKO MEFs 18. To confirm that impaired TNFα-induced IL-6 expression in T2/5 DKO MEFs is due to impaired JNK/c-Jun activation in these cells, we stably expressed a dominant negative c-Jun (TAM67, having a truncated transactivation domain) or an IκBα super-repressor (IκBαSR, in which serine 32 and 36 were replaced with alanine) in WT and DKO MEFs (Fig. 3e). Interestingly, expression of either IκBαSR or TAM67 was found to be sufficient to inhibit significantly the TNFα-induced IL-6 expression in WT MEFs (Fig. 3f). On the other hand, expression of IκBαSR, but not TAM67, significantly inhibited TNFα-induced ICAM-I expression in both WT and DKO MEFs (Fig. 3g). In fact, TNFα-induced IL-6 expression is also almost completely impaired in p65 (a subunit of NF-κB) KO MEFs 19. This suggests that both c-Jun and NF-κB are required for TNFα-induced IL-6 expression in MEFs.
TNFα induces IκBα degradation and p65 nuclear translocation in T2/5 DKO MEFs
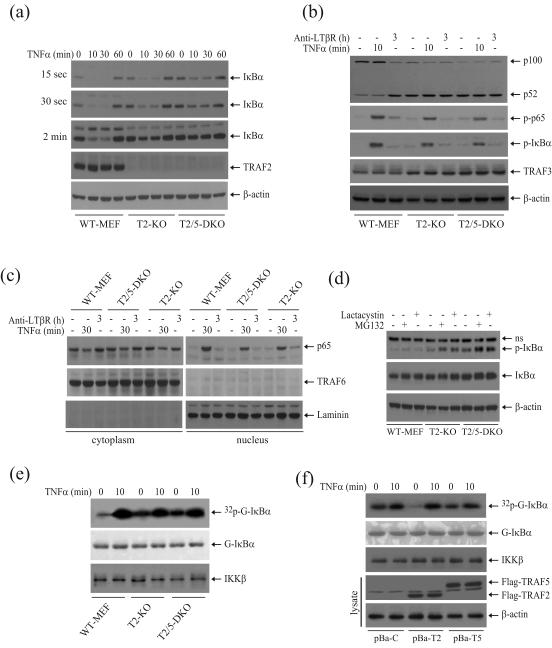
Currently, TNFα-induced NF-κB activation is believed to be impaired in TRAF2/5 DKO cells. This conclusion is based on an assessment of IκBα protein levels in T2/5 DKO MEFs following TNFα stimulation 14. To better assess the roles of TRAF2 and TRAF5 in TNFα-induced NF-κB activation, we examined IκBα degradation and p65 nuclear translocation in WT, T2 KO and T2/5 DKO MEFs by Western blotting. In WT MEFs, TNFα stimulation caused nearly complete degradation of IκBα within 30 min after stimulation, whereas it failed to do so in T2/5 DKO MEFs (Fig. 4a); these findings are consistent with a previous report 14. However, careful analysis of IκBα protein levels by shorter exposure of films revealed that in T2/5 DKO MEFs, TNFα-induced IκBα degradation is reduced rather than completely impaired (Fig. 4a, 15sec exposure). Surprisingly, TNFα-induced IκBα degradation was also attenuated in T2 KO MEFs. Consistent with this finding, TNFα stimulation triggered p65 translocation to the nucleus in both T2/5 DKO and T2 KO MEFs, at comparable levels (Fig. 4c). However, TNFα-induced p65 nuclear translocation in these cells was reduced in comparison to that in WT MEFs (Fig. 4c). These data suggested that TNFα does in fact induce the activation of the canonical NF-κB pathway in T2/5 DKO cells, but to a lesser than in WT MEFs.
Fig. 4.
In T2/5 DKO MEFs, TNFα induces IκBα degradation and p65 nuclear translocation. (a) WT, T2 KO and T2/5 DKO MEFs were left untreated or were treated with mTNFα (10ng/ml) as indicated, after which IκBα degradation was detected by Western blotting. 15sec, 30sec and 2min labels at left indicate the X-ray film exposure times. (b) WT, T2 KO and T2/5 DKO MEFs were untreated or treated with mTNFα (10ng/ml) for 10 min, or with anti-LTβR antibody (0.5μg/ml) for 3 hrs, after which p100 processing, p65 and IκBα phosphorylation and TRAF3 expression were monitored by Western blotting. (c) WT, T2 KO and T2/5 DKO MEFs were untreated or treated with mTNFα (10ng/ml) for 30 min, or with anti-LTβR antibody (0.5μg/ml) for 3 hrs, after which nuclear translocation of p65 was monitored by Western blotting. (d) WT, T2 KO and T2/5 DKO MEFs were untreated or treated with Lactacystin (10 μM) or MG132 (20 μM) for 90 min, after which IκBα phosphorylation was monitored by Western blotting. ns indicates nonspecific bands. (e) WT, T2 KO and T2/5 DKO MEFs were untreated or treated with mTNFα (10ng/ml) for 10 min, after which IKK activation was assessed by an in vitro kinase assay as in Fig. 1c. (f) T2/5 DKO MEFs stably expressing empty vector (pBa-C), TRAF2 (pBa-T2) and TRAF5 (pBa-T5) were untreated or treated with mTNFα (10ng/ml) for 10 min, after which IKK activation was assessed by an in vitro kinase assay as in Fig. 1c.
Both the canonical and noncanonical NF-κB pathways are constitutively activated in T2/5 DKO MEFs
To further assess the activation status of the canonical and noncanonical NF-κB pathways, we analyzed p100 processing and IκBα and p65 phosphorylation following the stimulation of cells with TNFα (inducer of the canonical NF-κB pathway) or agonistic anti-LTβR antibody (inducer of the noncanonical NF-κB pathway). As expected, in WT MEFs, anti-LTβR antibody but not TNFα induced p100 processing, whereas in T2 KO and T2/5 DKO MEFs, p100 was constitutively processed to p52 even in the absence of stimulation (Fig. 4b). In addition, a high level of processed p52 was found in the nuclear faction of T2/5 DKO but not in that of WT MEFs under unstimulated conditions (Fig. S5). Consistently, TNFα, but not the anti-LTβR antibody, induced the phosphorylation of IκBα and p65 in T2 KO and T2/5 DKO MEFs, albeit to a lesser degree than in WT MEFs (Fig. 4b). In light of a recent study showing that IκBα is constitutively phosphorylated and degraded in TRAF3 KO MEFs due to constitutive IKK activation 20, we wanted to test whether IκBα is constitutively phosphorylated and degraded in T2/5 DKO MEFs. To this end, we treated cells with the proteasome inhibitors lactacystin and MG132 for 90 minutes and then monitored IκBα phosphorylation by Western blotting. As shown in Fig. 4d, when the cells were treated with proteasome inhibitors, IκBα phosphorylation became clearly detectable in T2 KO and T2/5 DKO MEFs, but not in WT MEFs, suggesting that IκBα is constitutively phosphorylated and degraded in T2 KO and T2/5 DKO MEFs. These data suggest that both the canonical and noncanonical NF-κB pathways are constitutively activated in T2 KO and T2/5 DKO MEFs.
TRAF2 has a non-redundant role in suppression of basal IKK activity
In both T2 KO and T2/5 DKO MEFs, IκBα is constitutively phosphorylated and p100 is constitutively processed. This raises the question of what role TRAF5 plays in the regulation of basal and inducible IKK activity. Thus, we performed an IKK kinase assay assessing the difference in T2 KO and T2/5 DKO MEFs. Surprisingly, the IKK complex in T2 KO MEFs was also constitutively activated at a level comparable to that in T2/5 DKO MEFs (Fig. 4e). Consistent with this finding, stable expression of TRAF5 in T2/5 DKO MEFs failed to reduce the elevated basal IKK activity to a level like that seen in TRAF2-expressing cells (Fig. 4f). This suggests that TRAF2 plays a non-redundant role in the suppression of basal IKK activity in resting cells.
Expression of dominant negative (DN) NIK in T2/5 DKO MEFs partially restores TNFα-induced robust IKK activation
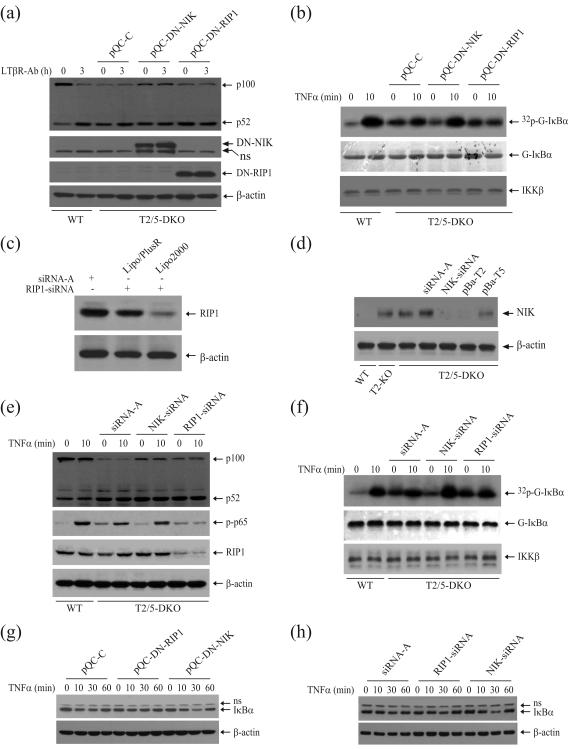
Whereas RIP1 is essential for activation of the canonical NF-κB pathway, it is NIK that activates the noncanonical NF-κB pathway 5. To examine which pathway accounts for the elevated activity of the IKK complex, we stably expressed DN-NIK and DN-RIP1 in T2/5 DKO MEFs, and then monitored p100 processing and IKK complex activity. As expected, the expression of DN-NIK partially suppressed p100 processing and basal IKK activity, whereas expression of DN-RIP1 inhibited only TNFα-induced (but not basal) IKK activation (Fig. 5a and 5b). Unexpectedly, the expression of DN-NIK led to an increase in TNFα-induced IKK activation that was comparable to that seen in WT MEFs (Fig. 5b). This suggests that NIK is responsible for the elevated basal IKK activity in T2/5 DKO MEFs, and that this elevated basal IKK activity masks TNFα-induced immediate and robust IKK activation in T2/5 DKO MEFs.
Fig. 5.
In T2/5 DKO MEFs, a reduction of NIK activity increases TNFα-induced IKK activation and IκBα degradation. (a) T2/5 DKO MEFs stably expressing empty vector (pQC-C), DN-NIK (pQC-DN-NIK) or DN-RIP1 (pQC-DN-RIP1) were untreated or treated with anti-LTβR antibody (0.5μg/ml) for 3 hrs, after which p100 processing and expression of DN-NIK and DN-RIP1 were monitored by Western blotting. (b) pQC-C, pQC-DN-NIK and pQC-DN-RIP1 cells were untreated or treated with mTNFα (10ng/ml) for 10 min, after which IKK activity was assessed by an in vitro kinase assay as in Fig. 1c. (c) T2/5 DKO MEFs were transfected with a scrabbled control siRNA-A or an siRNA specific to mouse RIP1 using either the Lipofectamine Plus-Reagent or Lipofectamine 2000. At 68 hrs after transfection, the expression level of RIP1 was assessed by Western blotting. (d) T2/5 DKO MEFs were transfected with an siRNA specific to mouse NIK using Lipofectamine 2000. At 68 hrs after transfection, the expression levels of NIK in siRNA transfected cells, as well as in the other cell lines indicated, were assessed by Western blotting. (e) T2/5 DKO MEFs were transfected with the indicated siRNA. At 68 hrs after transfection, p100 processing and p65 phosphorylation were monitored by Western blotting. (f) T2/5 DKO MEFs were transfected with siRNA as indicated. At 68 hrs after transfection, IKK activity was assessed by an in vitro kinase assay as in Fig. 1c. (g) pQC-C, pQC-DN-NIK and pQC-DN-RIP1 cells were untreated or treated with mTNFα (10ng/ml) as indicated, after which IκBα degradation was monitored by Western blotting. ns indicates nonspecific bands. (h) T2/5 DKO MEFs were transfected with the indicated siRNA using Lipofectamine 2000. At 68 hrs after transfection, the cells were untreated or treated with mTNFα (10ng/ml) as indicated, after which IκBα degradation was monitored by Western blotting.
Knockdown of NIK in T2/5 DKO MEFs restores TNFα-induced robust IKK activation
As expected, NIK is accumulated both in T2 KO and T2/5 DKO MEFs to a similar level, and stable expression of TRAF2, but not of TRAF5, suppressed NIK accumulation in T2/5 DKO MEFs (Fig. 5d). To confirm the role of NIK in constitutive activation of both NF-κB pathways in T2/5 DKO MEFs, we knocked down NIK and RIP1 by an siRNA-mediated approach (Fig. 5c and 5d). Consistent with the results of the DN-NIK and DN-RIP1 expression studies, knockdown of NIK in T2/5 DKO MEFs partially suppressed p100 processing and basal IKK activity, whereas knockdown of RIP1 decreased TNFα-induced, but not basal, IKK activation (Fig. 5e and 5f). Moreover, knockdown of NIK in T2/5 DKO MEFs almost completely restored TNFα-induced immediate and robust IKK activation (Fig. 5f and S6). These data support the notion that elevated basal IKK activity masks TNFα-induced robust IKK activation, and that TNFα-induced IKK activation is otherwise almost intact in T2/5 DKO MEFs.
Inhibition of NIK in T2/5 DKO MEFs restores TNFα-induced IκBα degradation
Given that IκBα itself is an NF-κB target gene, constitutive activation of IKK may induce constitutive phosphorylation, degradation and resynthesis of IκBα, thereby masking TNFα-induced robust and immediate degradation of IκBα. To test this hypothesis, we monitored IκBα degradation following the expression of DN-NIK and DN-RIP1 or the knockdown of NIK and RIP1 in T2/5 DKO MEFs. As shown in Fig. 5g, the expression of DN-NIK increased TNFα-induced IκBα degradation, whereas the expression of DN-RIP1 blocked this event. In line with this finding, the expression of DN-NIK in T2/5 DKO MEFs suppressed TNFα-induced ICAM-I expression to a level comparable to that in WT MEFs, whereas expression of DN-RIP1 inhibited its inducible expression (Fig. S7a). As expected, stable expression of DN-NIK and DN-RIP1 had no effect on TNFα-induced IL-6 expression in T2/5 DKO MEFs (Fig. S7b). Notably, siRNA-mediated knockdown of NIK, but not RIP1, increased TNFα-induced IκBα degradation (Fig. 5h). However, neither the expression of DN-NIK nor knockdown of endogenous NIK completely inhibited p100 processing or restored TNFα-induced IκBα degradation to the level seen in WT MEFs (Fig. S8, and data not shown). This is most likely because stably expressed DN-NIK is not sufficient to completely block endogenous NIK activity, and because the efficiency of siRNA transfection is relatively low in T2/5 DKO MEFs. Nevertheless, we consistently observed that expression of DN-NIK and knockdown of endogenous NIK suppressed basal IKK activity and increased TNFα-induced IKK activation and IκBα degradation. Collectively, these data suggest that elevated IKK activity and a high rate of constitutive IκBα degradation and resynthesis in T2/5 DKO MEFs mask TNFα-induced robust and immediate IKK activation and IκBα degradation. Otherwise, TNFα activation of the canonical NF-κB pathway is normal even in the absence of both TRAF2 and TRAF5, which explains why T2/5 DKO MEFs display normal, if not elevated, expression of NF-κB target genes in response to TNFα stimulation.
TRAF2, but not TRAF5, expression inhibits TNFα-induced cell death independent of NF-κB activation
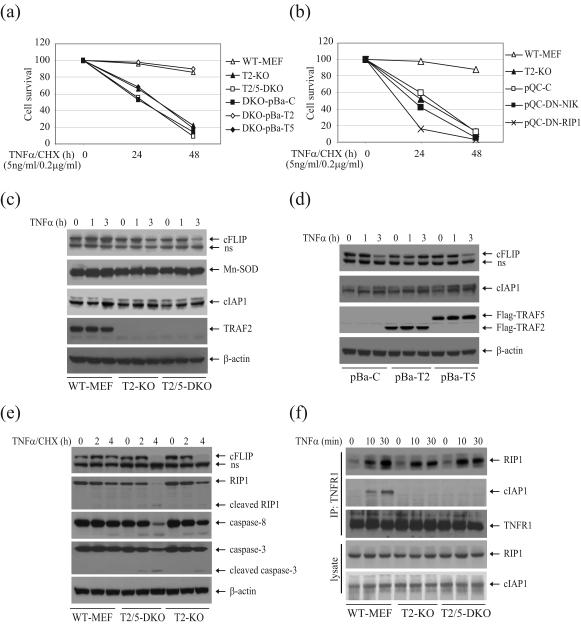
TNFα-induced cell death in TRAF2/5 DKO MEFs is believed to be due to impaired NF-κB activation 17. In fact, as we have demonstrated here, this is not the case. Interestingly, we noticed that T2/5 DKO MEFs naturally immortalized by a 3T3 protocol are sensitive to TNFα-induced cell death at early passages (Fig. S9a), but become less susceptible to TNFα-induced cell death after two-three months of in vitro culturing or following stable expression of puromycin-resistant empty vector (Fig. S9a and S9b). Nevertheless, both T2 KO and T2/5 DKO MEFs remained sensitive to cell death induced by TNFα (5 ng/ml) in the presence of very low levels of cycloheximide (CHX; 0.2 μg/ml), a condition that does not cause more than 10% cell death in WT MEFs (Fig. 6a and 6b). To examine the roles of TRAF2 and TRAF5 in TNFα-induced cell death under the same experimental conditions, we treated T2/5 DKO MEFs stably expressing TRAF2 or TRAF5 with TNFα (5 ng/ml) plus CHX (0.2 μg/ml). As shown in Fig. 6a, the expression of TRAF2, but not TRAF5, inhibited TNFα-induced cell death, suggesting that TRAF2 plays a primary role in the inhibition of TNFα-induced cell death. Although expression of DN-NIK suppressed basal IKK activity and increased inducible IKK activity in T2/5 DKO MEFs, it had no effect on TNFα-induced cell death in these cells (Fig. 6b). In contrast, expression of DN-RIP1 significantly sensitized T2/5 DKO MEFs to TNFα/CHX-induced cell death.
Fig. 6.
TRAF2 inhibits TNFα-induced cell death by indirectly inhibiting caspase-8 activation and cFLIP degradation. (a, b) The indicated cells were untreated or co-treated with mTNFα (5ng/ml) and CHX (0.2 μg/ml), and 24 and 48 hrs after treatment, the rate of cell death was assessed via the trypan blue exclusion assay. The data shown represent the average of three experiments performed in triplicate. (c) WT, T2 KO and T2/5 DKO MEFs were untreated or treated with mTNFα (10ng/ml) for 1 and 3 hrs, after which the expression levels of cFLIP, Mn-SOD, cIAP1 and TRAF2 were monitored by Western blotting. (d) pBa-C, pBa-T2 and pBa-T5 cells were left untreated or were treated with mTNFα (10ng/ml) for 1 and 3 hrs, after which the expression levels of cFLIP, cIAP1, TRAF2 and TRAF5 were monitored by Western blotting. (e) WT, T2 KO and T2/5 DKO MEFs were left untreated or were treated with mTNFα (5ng/ml) and CHX (0.2μg/ml) for 2 and 4 hrs, after which the expression and cleavage of cFLIP, RIP1, caspase-8 and caspase-3 were monitored by Western blotting. (f) WT, T2 KO and T2/5 DKO MEFs were left untreated or were treated with mTNFα (10ng/ml) for 10 and 30 min, after which the TNFR1 complexes were immunoprecipitated with anti-TNFR1 antibody. The recruitment of RIP1 and cIAP1 to the TNFR1 complexes was then monitored by Western blotting.
Mn-SOD, cIAP1 and cFLIP are among the anti-apoptotic proteins that inhibit TNFα-induced cell death by scavenging ROS or by blocking caspase activation 2. Therefore, we analyzed the expression of these anti-apoptotic proteins by Western blotting. As shown in Fig. 6c, the basal expression levels of Mn-SOD, cIAP1 and cFLIP are comparable among WT, T2 KO and T2/5 DKO MEFs. When each cell type was stimulated with TNFα for 1 or 3 hrs, cIAP1 and Mn-SOD protein levels were slightly increased in all cell types, whereas cFLIP protein levels were increased in WT-MEFs but reduced in both T2 KO and T2/5 DKO MEFs. Given that the cFLIP mRNA level increased upon TNFα stimulation in all these cells, the decrease in cFLIP protein levels in T2 KO and T2/5 DKO MEFs must be due to post-translational cleavage and degradation 21. To further examine the expression of these proteins in the same genetic background, we analyzed the expression of these proteins in T2/5 DKO MEFs that stably express empty vector (pBa-C), TRAF2 (pBa-T2) or TRAF5 (pBa-T5). As expected, the cIAP1 protein level was slightly increased in all cell types, whereas the cFLIP protein level was increased in pBa-T2 cells but decreased in pBa-C and pBa-T5 cells (Fig. 6d). These findings are consistent with the results that stable expression of TRAF2, but not of TRAF5, inhibits TNFα-induced cell death in T2/5 DKO MEFs (Fig. 6a). These data suggest that TRAF2, but not TRAF5, plays a primary role in the inhibition of TNFα-induced cell death, and that the anti-apoptotic role of TRAF2 is independent of NF-κB activation.
TRAF2 inhibits TNFα-induced cell death by recruiting anti-apoptotic proteins to the TNFR1 complex
Caspase-8 is essential for TNFα-induced cell death 4; 22. Therefore, we examined caspase-8 activation, as well as the cleavage of its well-known substrate RIP1, by Western blotting. Indeed, treatment of cells with TNFα in the presence of 0.2μg/ml CHX clearly induced caspase-8 activation and led to the subsequent cleavage of RIP1 in both T2 KO and TRAF2/5 DKO MEFs, but did not have a significant effect on this in WT MEFs (Fig. 6e). In addition, caspase-3 activation was also clearly detected in T2 KO and T2/5 DKO MEFs. NF-κB-dependent expression of TRAF2, cIAP1 and cIAP2 suppresses TNFα-induced caspase-8 activation, and TRAF2 and cIAP1 are components of the TNFR1 signaling complex 23; 24. Therefore, we further examined the recruitment of cIAP1 and cIAP2 to TNFR1 following TNFα stimulation. As shown in Fig. 6f, cIAP1 was recruited to TNFR1 in WT MEFs but not in T2 KO and T2/5 DKO MEFs. Consistently, stable expression of TRAF2, but not of TRAF5, in T2/5 DKO MEFs restored TNFα-induced cIAP1 recruitment to the TNFR1 complex (Fig. S10). We were not able to reproducibly detect endogenous cIAP2 expression in MEFs by Western blotting. Thus, it is not clear whether cIAP2 is also recruited to TNFR1 in a TRAF2-dependent manner. Nevertheless, these data suggest that TRAF2 inhibits caspase-8 activation by recruiting cIAP1 (and possibly also cIAP2) to the TNFR1 complex following TNFα stimulation.
TNFα-induced RIP1 ubiquitination is impaired in both T2 KO and T2/5 DKO MEFs
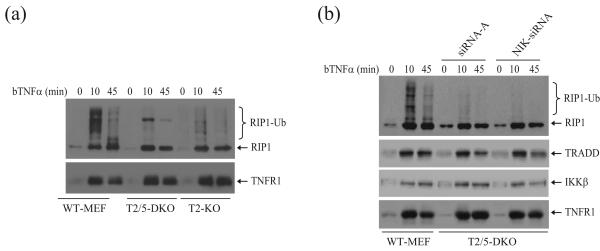
RIP1 is ubiquitinated immediately upon TNFα stimulation, and this ubiquitination is believed to be catalyzed by TRAF2 6; 7; 25. We found that TNFα-induced RIP1 ubiquitination is indeed reduced dramatically, but not impaired completely, in T2/ KO and T2/5 DKO MEFs (Fig. 7a). As NIK knockdown restored TNFα-induced IKK activation in T2/5 DKO MEFs, we next analyzed RIP1 ubiquitination in T2/5 DKO MEFs following NIK knockdown. As shown in Fig. 7b, the knockdown of NIK neither increased nor decreased TNFα-induced RIP1 ubiquitination in T2/5 DKO MEFs. TNFα-induced recruitment of TRADD and IKKβ to the TNFR1 complex also occurred equally in these cells regardless of the level of NIK protein. This suggests that either RIP1 ubiquitination has no role in TNFα-induced NF-κB activation, or that this pathway is redundant with another that does not require RIP1 ubiquitination.
Fig. 7.
TRAF2 is involved in TNFα-induced RIP1 ubiquitination in vivo. (a) WT, T2 KO and T2/5 DKO MEFs were left untreated or treated with bTNFα (100ng/ml) for 10 and 45 min, after which the TNFR1 complexes were pulled down with the aid of Dynabeads Streptavidin. RIP1 ubiquitination was then monitored by Western blotting using anti-RIP1 antibody. The same membrane was stripped and reprobed with anti-TNFR1 antibody. (b) T2/5 DKO MEFs were transfected with NIK-specific or control siRNA-A using Lipofectamine 2000. At 68 hours after transfection, cells were left untreated or treated with bTNFα (100ng/ml) for 10 and 45 min, after which the TNFR1 complexes were pulled down as above. The ubiquitination of RIP1 and the recruitment of TRADD and IKK were then monitored by Western blotting.
Discussion
Currently, it is widely accepted that TRAF2- and TRAF5-mediated K63-linked RIP1 ubiquitination is essential for TNFα-induced NF-κB activation 22. We report here that: i) TRAF2 suppresses basal IKK complex activity in resting cells by inhibiting NIK activity; ii) TNFα can also activate the NF-κB pathway in the absence of TRAF2 and TRAF5 expression and RIP1 ubiquitination; and iii) TRAF2 inhibits TNFα-induced cell death by recruiting cIAP1 to the TNFR1 complex rather than by activating the NF-κB pathway. In fact, our conclusions are supported directly or indirectly by the following published findings: i) TNFα-induced RIP1 ubiquitination is impaired in T2 KO MEFs 25, however, TNFα-induced NF-κB activation is not abrogated in TRAF2-deficient cells, although these cells are still sensitive to TNFα-induced cell death 11; 12; and ii) TRAF2 inhibits TNFα-induced cell death by recruiting cIAP1 to the TNFR1 complex 26. Recent gene knockout studies have revealed that in TRADD-deficient macrophages, RIP1 is still recruited to TNFR1 in response to TNFα stimulation, but is not efficiently ubiquitinated due to the lack of TRAF2 recruitment to TNFR1 in these cells. Surprisingly, such RIP1 recruitment to TNFR1 in TRADD-deficient macrophages is sufficient to activate IKK in the absence of K63-linked polyubiquitination 27; 28. Bertrand et al. have recently reported that cIAP1/2 constitutively target RIP1 for ubiquitination in cancer cells 29. It has now been well established that TRAF2 directly associates with and recruits cIAP1/2 to the TNFR1 complex following TNFα stimulation 22; 30; 31; 32. Thus, no matter whether TRAF2/5 or cIAP1/2 catalyze RIP1 ubiquitination, neither TRAF2/5 nor cIAP1/2 are present in the TNFR1 complex in T2/5 DKO MEFs. Therefore, the lack of RIP1 ubiquitination in T2/5 DKO MEFs following TNFα stimulation is not surprising. Collectively, all these published findings and our data presented here suggest that efficient RIP1 recruitment to TNFR1 is sufficient to activate IKK in the absence of TRAF2/5 expression and K63-linked RIP1 polyubiquitination.
Studies using a transient overexpression system have shown that TRAF2, TRAF5 and TRAF6 can positively regulate the canonical NF-κB pathway 1; 22. However, gene knockout studies have revealed that TRAF5 deficiency has no effect on TNFα-induced JNK and IKK activation, whereas TRAF2 deficiency abolishes TNFα-induced JNK, but not NF-κB, activation 11; 14. The conclusion that knockout of TRAF2 and TRAF5 abrogates TNFα-induced NF-κB activation was based on an analysis of IκBα protein level in these cells 14. In WT MEFs, TNFα-induced IκBα phosphorylation is robust and immediate, resulting in complete degradation of IκBα within 30 minutes. IκBα protein returns to a normal level within 60 min after TNFα stimulation, because the expression of IκBα itself is induced by NF-κB. If, however, the IKK complex is constitutively activated, IκBα will be constitutively phosphorylated, degraded and resynthesized, which will partially mask stimulation-induced immediate and complete IκBα degradation. Our data show that this is, in fact, what happens in T2/5 DKO MEFs upon TNFα stimulation. Therefore, the incomplete degradation of IκBα in T2/5 DKO MEFs following TNFα exposure is not caused by impaired IKK activation, but by the constitutive degradation and resynthesis of IκBα prior to stimulation. The IKK immunokinase assay we have used is one of the most sensitive and reliable methods for assessing the activation of the canonical NF-κB pathway, and it clearly demonstrates that basal IKK activity is elevated in T2 KO and T2/5 DKO MEFs, and that stable expression of TRAF2, but not of TRAF5, suppresses basal IKK activity in T2/5 DKO MEFs. In addition, we demonstrate the dependence of this elevated basal IKK activity on NIK, by showing that either the expression of DN-NIK or the knockdown of NIK in T2/5 DKO MEFs reduces the elevated basal IKK activity and increases TNFα-induced immediate activation of IKK (Fig. 5b and 5f). The fact that treatment of T2 KO and T2/5 DKO MEFs with proteasome inhibitors makes IκBα phosphorylation detectable in these cells, but that the same does not hold true in WT MEFs, lends further support to the notion that IKK is constitutively activated and IκBα is constitutively phosphorylated and degraded in T2 KO and T2/5 DKO MEFs (Fig. 4d). Moreover, our demonstration that the expression of DN-RIP1 in T2/5 DKO MEFs blocks TNFα-induced IκBα degradation and the expression of NF-κB target genes suggests that TNFα-induced expression of NF-κB target genes in T2/5 DKO MEFs depends on the RIP1-mediated canonical NF-κB pathway. Therefore, we conclude that TRAF2 and TRAF5 are not essential for TNFα-induced activation of the canonical NF-κB pathway.
Our analysis of NF-κB-dependent gene expression by real-time PCR revealed that cIAP1 and cFLIP are normally expressed in T2/5 DKO MEFs, whereas the basal and inducible expression of IκBα, IP-10, ICAM-I and RANTES were significantly increased in T2/5 DKO MEFs compared to that in WT MEFs. Expression of cIAP1 and cFLIP is induced primarily by the canonical NF-κB pathway, whereas the expression of IκBα, IP-10, ICAM-I and RANTES is regulated by both the canonical and non-canonical NF-κB pathways 22; 24; 33; 34. Thus, the two pathways synergistically induce the expression of IκBα, IP-10, ICAM-I and RANTES in T2/5 DKO MEFs in response to TNFα stimulation, thereby resulting in significantly elevated expression of these genes in T2/5 DKO MEFs. In contrast, IL-6 induction by TNFα was almost completely impaired in T2/5 DKO cells, and stable expression of TRAF2 fully restored TNFα-induced IL-6 expression (Fig. 3c). Likewise, TNFα-induced JNK activation was impaired in T2/5 DKO MEFs and restored by TRAF2 expression. These data suggest that TNFα-induced IL-6 expression is also controlled by c-Jun activity. At least it is clear that NF-κB alone is not sufficient to transactivate IL-6 expression in MEFs. The efficient induction of IL-6 depends on both c-Jun and NF-κB activities, as inhibition of either pathway almost completely abolish TNFα-induced IL-6 expression (Fig. 3f and 3g). This is supported by the fact that TNFα-induced IL-6 expression is impaired in both JNK1/2 DKO and p65 KO MEFs 18; 19. Many TNFα-inducible genes have both NF-κB and c-Jun binding sites 4. Therefore, efficient upregulation of some immediate genes, such as IL-6, in response to TNFα might be dependent on both NF-κB and c-Jun activities, while the inducible expression of other genes, such as ICAM-1, might be regulated primarily by NF-κB activity. Based on our data, we now believe that in TRAF2-deficient macrophages, the basal NF-κB activity is elevated, and that TNFα-induced NF-κB activation is not impaired. This explains why TRAF2-deficient macrophages overproduce TNFα and NO in response to TNFα stimulation 12.
A recent report has shown that in TRAF2-deficient B cells, CD40 fails to effectively activate the canonical NF-κB pathway 13. This conclusion was also based on an analysis of IκBα phosphorylation and degradation following the treatment of cells with anti-CD40 antibody. TNFR1 can recruit RIP1 independent of TRAF2 expression, and both TRAF2 and RIP1 can independently recruit IKK to the TNFR1 complex 4; 22. On the other hand, CD40 and TNFR2 directly associate with TRAF2, but not with RIP1. Therefore, it is possible that in the absence of TRAF2/5, TNFR1 is still able to recruit RIP1 and IKK and can thereby activate the canonical NF-κB pathway. However, in the absence of TRAF2, CD40 may not be able to recruit IKK and activate NF-κB.
Activation of the canonical NF-κB pathway is robust and transient, whereas activation of the noncanonical NF-κB pathway is slow and prolonged. The canonical NF-κB pathway is subject to strong negative feedback regulation, which involves the recruitment of TRAF1, A20 and cIAP1/2 to the TNFR1 complex, dephosphorylation of IKK at its T-loop, cleavage and degradation of p65 and rapid resynthesis of IκBα, and so on 5; 22. Therefore, it is possible that the negative regulators of the canonical NF-κB pathway are also activated constitutively, at least at some level, in T2/5 DKO MEFs. This would partially explain why p52, but not p65, accumulates in the nuclei of T2/5 DKO MEFs (Fig. S5), even though the IKK complex is constitutively activated in these cells. Notably, although the IKK complex is constitutively activated in T2/5 DKO MEFs, it is not activated maximally. Thus, TNFα stimulation results in a further increase in IKK activity, which leads to inducible expression of NF-κB-dependent genes, such as RANTES and ICAM-I. The constitutive activation of the IKK complex in T2/5 DKO MEFs is, at least partially, due to the accumulation and/or activation of NIK. Thus, NIK knockdown down-regulates the activities of both positive and negative regulators, thereby rendering the cells more responsive to TNFα stimulation.
In T2/5 DKO MEFs, TNFα triggers the cleavage and degradation of cFLIP, and ectopic expression of cFLIP inhibits TNFα-induced cell death 21, suggesting that cFLIP degradation is a critical event that leads to cell death in T2/5 DKO MEFs. cFLIP is an enzymatically inactive homologue of caspase-8, and is cleaved by caspase-8 35. Most recently, two studies have clearly demonstrated that small-molecule IAP antagonists induce cIAP1/2 degradation and TNFα-dependent apoptosis, as well as NF-κB activation 30; 31, suggesting that NF-κB activation is not sufficient to inhibit TNFα-induced cell death in the absence of cIAP1/2. As TRAF2 does not directly inhibit caspase-8 activity, activation of caspase-8 in T2/5 DKO MEFs is most likely due to the impaired recruitment of anti-apoptotic proteins such as cIAP1 to the TNFR1 complex. However, such caspase-8 activation is not sufficient to trigger apoptotic cell death, as the majority of T2/5 DKO MEFs undergo necrotic cell death upon TNFα treatment 17. cFLIP suppresses the prolonged phase of JNK activation and ROS accumulation induced by TNFα 21. cFLIP also inhibits TNFα-induced cell death by directly associating with and inhibiting the full activation of caspase-8 35. Therefore, in T2 KO and T2/5 DKO MEFs, caspase-8-mediated cleavage and proteasome-dependent degradation of cFLIP results in prolonged activation of JNK and the accumulation ROS, events that culminate in necrotic and apoptotic cell death. Notably, we repeatedly observed that although TNFα-induced cFLIP degradation was comparable in the T2 KO and T2/5 DKO MEFs, RIP1 cleavage and caspase-3 activation were more prominent in T2/5 DKO MEFs (Fig. 6e). Moreover, these DKO MEFs were more susceptible to TNFα-induced cell death than the T2 KO MEFs. This suggests that although TRAF5 does not recruit cIAP1 to TNFR1, it plays a certain role in inhibiting TNFα-induced cell death. However, the anti-apoptotic function of TRAF5 is marginal compared to that of TRAF2. IKKβ-/- and p65-/- cells are also sensitive to TNFα-induced cell death 22. Therefore, based on our data and on findings that have been published by others, we propose that two events are required to inhibit TNFα-induced cell death: i) NF-κB-dependent expression of anti-apoptotic proteins such as cIAP1/2 and cFLIP; and ii) TRAF2-mediated recruitment of anti-apoptotic proteins to the TNFR1 signaling complex. Ablation of either event sensitizes cells to TNFα-induced cell death.
Materials and Methods
Cell lines, plasmids and reagents
HeLa, WT MEF, T2 KO MEF and T2/5 DKO MEF cells were maintained in DMEM supplemented with 10% BCS and antibiotics. Antibodies (Abs) and reagents were purchased as follows: anti-TRAF2, anti-JNK, anti-IKKγ, anti-IKKβ, anti-cIAP1, anti-Mn-SOD and anti-NIK Abs from Santa Cruz Biotechnology (Santa Cruz, CA); anti-phospho-JNK Ab from Promega; anti-cFLIP Ab from Alexis (San Diego, CA); anti-IκBα, anti-caspae-8 and anti-caspase-2 Abs from Cell Signaling (Danvers, MA); mouse and human TNFα from Roche (Indianapolis, IN); anti-Flag Ab from Sigma Chemicals (St. Louis, MO); cocktail inhibitors of proteases and phosphatases from Pierce (Rockford, IL). siRNAs for mouse NIK, RIP1 and control siRNA-A (with scrambled sequences that will not lead to the specific degradation of any known cellular mRNA) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Constructs encoding 2xNF-kB-Luc and Jun2-Luc reporter genes and Flag-TRAF2 have been previously described 9. pCDNA3-TRAF5 and PEAK12-RIP1 plasmids were generously provided by Dr. Adrian Ting (Mount Sinai Medical Center, NY) and the dominant negative (DN) pCDNA3-NIK-KK429/430AA plasmid was provided by Dr. Gail Bishop (University of Iowa). The retroviral plasmids pBabe-Flag-TRAF2, pBabe-Flag-TRAF5, pQCXIH-DN-NIK and pQCXIH-DN-RIP1 (encoding the C-terminal 529-671aa region) were generated by subcloning the TRAF2, TRAF5, DN-NIK and RIP1-529-671 cDNAs into the pBabe-puro or pQCXIH-hygro plasmids.
Luciferase reporter gene assays
Cells cultured in 6-well plates were transfected with an NF-κB or c-Jun firefly luciferase reporter plasmid (NF-κB-Luc or Jun2-Luc; 0.2 μg) together with a Flag-TRAF2 (0.2 μg) and a Renilla luciferase reporter plasmid (pRL-TK; 0.01 μg) using Lipofectamine 2000 reagents. 36 hrs after transfection, test cells were treated with hTNFα (10 ng/ml) or mTNFα (5 ng/ml), and protein samples were prepared at 6 (HeLa) or 4 (MEFs) hrs after treatment. The firefly and Renilla luciferase activities were then measured by using the Dual-luciferase assay system according to the manufacturer’s instructions (Promega).
JNK and IKK immunokinase assays
MEF cells were treated with mTNFα (10 ng/ml) and protein samples were extracted using TNE lysis buffer (20 mM HEPES, pH 7.4, 350 mM NaCl, 1% Triton X-100, 1mM DTT, 1mM EDTA, 20% glycerol and a cocktail of protease and phosphatase inhibitors). Endogenous JNK1 or IKK complex was immunoprecipitated using anti-JNK1 or anti-IKKγ antibody, and then subjected to in vitro kinase assays, in which GST-Jun1-87 (for JNK) or GST-IκBα 1-55 (for IKK) served as substrate, as described previously 9.
Preparation of retroviral supernatants and infection of T2/5 DKO MEFs
293T cells at 60-70% confluent were co-transfected with 2 μg of pMD.OGP (encoding gag-pol), 2 μg of pMD.G (encoding vesicular stomatitis virus G protein) and 2 μg of pBabe-Flag-TRAF2, -TRAF5, pQCXIH-DN-NIK or pQCXIH-DN-RIP1 by the standard calcium phosphate precipitation method. 48 hrs after transfection, the viral supernatant was collected and filtered through a 0.45 μm filter. TRAF2 retroviral supernatant was diluted 1-, 2-, 3- and 4-fold with 10%FBS/DMEM and then immediately used for the infection of T2/5 DKO MEFs in the presence of 4 μg/ml polybrene for 6 hrs. 48 hrs after infection, cells were selected with puromycin (2.0μg/ml) for 6 days, and resistant cells were pooled together and frozen. Cells that stably express Flag-TRAF2 at the physiological level (Fig.S3; 3-fold) were used for the functional experiments, within one month of establishment. TRAF5 retroviral supernatant was diluted 3-fold before being used for the infection of T2/5 DKO MEFs. DN-NIK and DN-RIP1 retroviral supernatants were not diluted and were used immediately for the infection of T2/5 DKO MEFs.
Real time PCR
MEF cells were treated with mTNFα (10 ng/ml), and total RNA was prepared using the RNeasy Mini Kit (Qiagen). Five μg of total RNA was treated with RQ1 RNase-free DNase for 30 min at 37 °C, and then reverse transcribed using an oligo dT-primer. The resulting cDNA was subjected to quantitative real-time PCR using the Power SYBR Green AB Master Mix and an ABI Prism 7700 Sequence Detector (Applied Biosystems). Mouse GAPDH-specific primers were used to generate an internal control, and the average threshold cycle (CT) for samples in triplicate were used in the subsequent calculations. Relative expression levels of NF-κB target genes were calculated as the ratio with respect to the GAPDH expression level. The mean ± S.E. of four independent experiments was considered to be statistically significant at p < 0.05. Real-time PCR products were also separated on an agarose gel to confirm the presence of single bands (Fig. S11). All primer pair sets were designed to flank an intron (Table S1).
RIP1 ubiquitination
TNFR1 complex was immunoprecipitated using biotinylated mouse TNFα (bTNFα) in combination with Dynabeads-Streptavidin (Invitrogen). Recombinant mTNFα was biotinylated using Sulfo-NHS-LC-Biotin (Pierce) at 1 mg/ml for 1 hr according to the manufacturer’s instructions. Unincorporated biotin was removed from bTNFα by buffer exchange into PBS on PD-10 columns (Amersham). The biological activity of bTNFα was determined by its apoptosis-inducing capacity and found to be comparable to nonbiotinylated mTNFα. MEFs (4×100mm cells per treatment) were treated with bTNFα for 10 and 45 min, and then cells were washed twice with ice-coal PBS and lysed in TNE lysis buffer (containing 0.5 % CHAPS and 5 mM NEM) on ice for 30 min followed by centrifugation at 13,000 × g for 20 min at 4°C. The TNFR1 complex was then precipitated using 30μl of Dynabeads-Streptavidin at 4°C for 4 hours. Precipitates were washed four times with the same lysis buffer containing 2 mM NEM. RIP1 recruitment to TNFR1 and its ubiquitination were then monitored by Western blotting.
Supplementary Material
Acknowledgements
We thank Hiroyasu Nakano (Juntendo University School of Medicie, Japan) for providing us with TRAF2/5 DKO cells, Adrian Ting (Mount Sinai Medical Center, New York) for pMD.G, pMD.OGP and TRAF5 plasmids, Gail Bishop and Bruce Hostager (University of Iowa) for IκBαSR and NIK plasmid and helpful discussions. Support by NCI grant CA78419 (to HH) is gratefully acknowledged.
Abbreviations used
- cFLIP
cellular caspase-8 (FLICE)-like inhibitory protein
- CHX
cycloheximide
- IAP
inhibitor of apoptosis
- IκB
inhibitor of κB
- IKK
IκB kinase
- JNK
c-Jun N-terminal kinase
- MAPK
Mitogen-activated protein kinase
- NF-κB
nuclear factor κB
- RIP1
receptor interacting protein 1
- ROS
reactive oxygen species
- TNFα
tumor necrosis factor α
- TNFR
TNF receptor
- TRADD
TNFR associated death domain
- TRAF
TNFR associated factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors have conflicts of interest.
Reference
- 1.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–91. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 2.Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 2001;13:389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 3.Varfolomeev EE, Ashkenazi A. Tumor necrosis factor: an apoptosis JuNKie? Cell. 2004;116:491–7. doi: 10.1016/s0092-8674(04)00166-7. [DOI] [PubMed] [Google Scholar]
- 4.Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11:372–7. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 5.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–8. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–65. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 8.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. Embo J. 2004;23:322–32. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, Yamaoka S, Kawai T, Matsuura Y, Takeuchi O, Akira S. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7:962–70. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 11.Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, Ohashi P, Rothe M, Goeddel DV, Mak TW. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–25. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen LT, Duncan GS, Mirtsos C, Ng M, Speiser DE, Shahinian A, Marino MW, Mak TW, Ohashi PS, Yeh WC. TRAF2 deficiency results in hyperactivity of certain TNFR1 signals and impairment of CD40-mediated responses. Immunity. 1999;11:379–89. doi: 10.1016/s1074-7613(00)80113-2. [DOI] [PubMed] [Google Scholar]
- 13.Grech AP, Amesbury M, Chan T, Gardam S, Basten A, Brink R. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity. 2004;21:629–42. doi: 10.1016/j.immuni.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, Hashimoto H, Mak TW, Yagita H, Okumura K, Yeh WC, Nakano H. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem. 2001;276:36530–4. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 15.Ken Blackwell LZ, Thomas Gregory S., Sun Shujie, Nakano Hiroyasu, Habelhah Hasem. TRAF2 phosphorylation modulates TNFalpha-induced gene expression and cell resistance to apoptosis. Mol Cell Biol. 2008 doi: 10.1128/MCB.00699-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piao JH, Yoshida H, Yeh WC, Doi T, Xue X, Yagita H, Okumura K, Nakano H. TNF receptor-associated factor 2-dependent canonical pathway is crucial for the development of Peyer’s patches. J Immunol. 2007;178:2272–7. doi: 10.4049/jimmunol.178.4.2272. [DOI] [PubMed] [Google Scholar]
- 17.Sakon S, Xue X, Takekawa M, Sasazuki T, Okazaki T, Kojima Y, Piao JH, Yagita H, Okumura K, Doi T, Nakano H. NF-kappaB inhibits TNF-induced accumulation of ROS that mediate prolonged MAPK activation and necrotic cell death. Embo J. 2003;22:3898–909. doi: 10.1093/emboj/cdg379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura JJ, Kennedy NJ, Lamb JA, Flavell RA, Davis RJ. c-Jun NH(2)-terminal kinase is essential for the regulation of AP-1 by tumor necrosis factor. Mol Cell Biol. 2003;23:2871–82. doi: 10.1128/MCB.23.8.2871-2882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okazaki T, Sakon S, Sasazuki T, Sakurai H, Doi T, Yagita H, Okumura K, Nakano H. Phosphorylation of serine 276 is essential for p65 NF-kappaB subunit-dependent cellular responses. Biochem Biophys Res Commun. 2003;300:807–12. doi: 10.1016/s0006-291x(02)02932-7. [DOI] [PubMed] [Google Scholar]
- 20.Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci U S A. 2008;105:3503–8. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakajima A, Komazawa-Sakon S, Takekawa M, Sasazuki T, Yeh WC, Yagita H, Okumura K, Nakano H. An antiapoptotic protein, c-FLIPL, directly binds to MKK7 and inhibits the JNK pathway. Embo J. 2006;25:5549–59. doi: 10.1038/sj.emboj.7601423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Shu HB, Takeuchi M, Goeddel DV. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci U S A. 1996;93:13973–8. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–3. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 25.Lee TH, Shank J, Cusson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem. 2004;279:33185–91. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 26.Fotin-Mleczek M, Henkler F, Samel D, Reichwein M, Hausser A, Parmryd I, Scheurich P, Schmid JA, Wajant H. Apoptotic crosstalk of TNF receptors: TNF-R2-induces depletion of TRAF2 and IAP proteins and accelerates TNF-R1-dependent activation of caspase-8. J Cell Sci. 2002;115:2757–70. doi: 10.1242/jcs.115.13.2757. [DOI] [PubMed] [Google Scholar]
- 27.Pobezinskaya YL, Kim YS, Choksi S, Morgan MJ, Li T, Liu C, Liu Z. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol. 2008;9:1047–54. doi: 10.1038/ni.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G, Tschopp J, Pasparakis M. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9:1037–46. doi: 10.1038/ni.1638. [DOI] [PubMed] [Google Scholar]
- 29.Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–81. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 31.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. Embo J. 2003;22:5530–9. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 35.Budd RC, Yeh WC, Tschopp J. cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol. 2006;6:196–204. doi: 10.1038/nri1787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.