Abstract
The cellular immune system recognizes self epitopes in the context of MHC-I molecules. The immunological general view presumes that these self epitopes are just a background, both positively and negatively selecting T cells. We here estimate the number of epitopes in each human protein for many frequent HLA alleles, and a score representing over or under presentation of epitopes on these proteins. We further show that there is a clear selection for the presentation of specific self proteins types. Proteins presenting many epitopes include for example AIRE upregulated Tissue specific antigens, immune system receptors and proteins with a high expression level. On the other hand, proteins that may be considered less “useful” for the immune system, such as low expression level proteins, are under presented. We combine our epitope estimate with SNP measures to show that this selection can be directly observed through the fraction of non-synonymous SNPs (replacement fraction), which is significantly higher inside epitopes than outside
Introduction
The innate immune system uses germline-encoded receptors as a frontline response to most pathogens. These receptors are limited in their capacity to recognize pathogens that have rapidly evolved to avoid being detected by them. This limitation is balanced by the adaptive immune system that adapts itself to new pathogens using a large receptor repertoire, obtained through random V(D)J rearrangement [1, 2], randomization of the junction between these genes [3] and point mutations during B cells somatic hypermutations.
Theoretically, the randomly rearranged receptors of the adaptive immune system could respond equally to self and to non-self epitopes. However, cells reacting to self epitopes are believed to be deleted [4] or edited [5] during T cells' thymic education and the parallel education of B cells in the bone marrow [6]. Thus, according to the self/non-self selection theory, any T cell with a high affinity receptor for self epitopes should be removed from the T cell repertoire. This basic dogma has now been refined to include more complex tolerance mechanisms, such as Tregs and priming of dendritic cells [7, 8]. However, these more complex theories are mostly focused on CD4+ T cells. In the context of CD8+ T cells (CTL), the mechanisms of peripheral self reactive T cells regulation are still debated. If indeed self reactive CD8+ T cells are deleted, CD8+ T cell self epitopes should not be different than any other nonamer in the same gene, since they would not be recognized by CD8+ T cells (no CD8+ T cells present). Thus, the epitope number should be similar to the one expected from the statistical properties of the sequence. If this is indeed the case, all self proteins should have a similar number of epitopes (taking into account their length, and random fluctuations) and there should be no large differences in the number of epitopes between self protein groups. We here show that such large differences do exist.
CTLs recognize (through their T cell receptor (TCR)) antigens only in the context of MHC class I molecules. MHC molecules can bind 8-10 amino acid long peptides and present them to the cellular immune system [9]. The limited repertoire allows us to apply bioinformatics tools to the human genome and compute a part of the CTL epitope repertoire in detail. Using this repertoire, we compute the number of epitopes presented in different human proteins and show that specific proteins are selected to express a high number of epitopes, while others are selected to express fewer. We further show that the proteins selected to express a high number of epitopes are the one most “useful” for the immune system, leading to the conclusion that self epitopes are not ignored, but are actually used by the immune system. While the definition of “useful” is vague, we propose a few examples to show the elements selecting the number of epitopes presented in different proteins.
In general, CTL epitopes originate from short peptides cleaved by the proteasome [10] and transferred through TAP [11] that can associate non-covalently with the groove of the MHC-I molecules [9]. A cleaved nonamer is presented on a MHC-I molecule only if its affinity for the MHC molecule is high enough. This affinity is determined by the peptide's length and by its anchor and helping residues. In recent years the anchor residues for many human MHC-I alleles have been determined experimentally [12, 13]. We here apply MHC binding, TAP transfer and cleavage prediction algorithms to the human genome to compute the human CTL epitope repertoire. There are currently dozens of algorithms for the prediction of the different stages of epitope processing (e.g. NetMHC [14], IEDB [15], BIMAS [13], NetChop [16-19], ProPred and Pcleavage [16, 20] and many others). We have tested many of them, and chose the algorithms to use based on their sensitivity, specificity as well as the availability of the scoring matrices allowing us to perform a large scale genomic analysis.
Specifically, we predict, using bioinformatical tools in all the coding regions of each known human gene all the nonamers that are cleaved by the proteasome, that pass through the TAP channel and that are bound to (at least) one of the major HLA alleles and defined those as epitopes [21]. We then compute for each protein the number of epitopes presented per HLA allele (Figure 1). Note that not all epitopes pass the simple process described here. Some of the epitopes are octamers or decamers. Some of them use a TAP independent pathways or result from a cross-talk between the MHC-II and MHC-I presentation pathways. Not all epitopes are the direct result of proteasomal cleavage. Many occur for example through trimming of existing larger cleavage products. Still, the majority of epitopes are nonamers processed in the classical TAP dependent and proteasomal cleavage pathways. Furthermore, we are only interested in a comparison between different protein groups and in the absence of selection and specific biases, the number of such epitopes should be uniform among the different protein groups.
Figure 1.
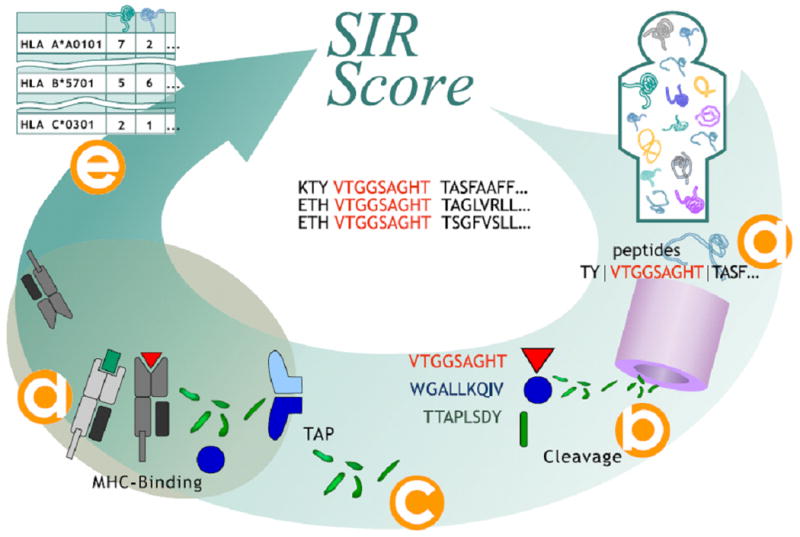
Algorithm for SIR score computation. Each Human protein is divided into all nonamers and the appropriate flanking regions (a). For each nonamer a cleavage score is computed (b). We compute for all nonamers with a positive cleavage score a TAP binding and choose only supra-threshold peptides (c). The MHC binding score of all TAP binding and cleaved nonamers is computed (d). Nonamers passing all these stages are defined as epitopes. We then compute the number of epitopes per protein per HLA allele (e).
Results
Epitope prediction
In order to produce an estimated library of the self-epitope repertoire, we scanned 33,677 known genes from the human genome (predicted by the ENSEMBL database) [22] and 31 different class I HLA molecules, most of them HLA-A and B (Table 1). Each gene was divided into all possible nonamers, and a score was computed for each nonamer, representing the probability that it should be cleaved in both C and N termini and not in its center [21, 23, 24]. For all high score peptides (i.e. peptides expected to be properly cleaved), the probability that they should pass TAP was computed [25], and for those successfully binding TAP, the probability to bind a given MHC-I molecule was estimated (Figure 1) [13]. Nonamers passing all stages above threshold were defined as predicted epitopes. The cleavage and MHC binding prediction algorithms were validated to have low error levels (Fig 2, Table 1).
Table 1.
List of HLA alleles, Allele frequencies normalized to 1, fraction of Falsely predicted epitope (FP) and fraction of measured epitope not predicted by the different algorithms. FP values were computed using 100,000 randome epitopes, while FN values were computed using measured epitopes. FN values of 0 were usually obtained when the number of observed epitopes was limited.
| Allele | Normalized Frequency | False Positives | False Negative |
|---|---|---|---|
| A_2402 | 1.29E-01 | 2.87E-02 | 1.82E-01 |
| A_0201 | 1.07E-01 | 6.16E-03 | 3.69E-01 |
| Cw_0702 | 9.66E-02 | 2.52E-02 | 1.16E-01 |
| Cw_0401 | 8.26E-02 | 1.99E-02 | 1.23E-01 |
| A_1101 | 7.52E-02 | 5.90E-03 | 6.77E-02 |
| B_4001 | 5.31E-02 | 1.89E-03 | 0.00E+00 |
| Cw_0602 | 5.09E-02 | 2.09E-02 | 0.00E+00 |
| A_0101 | 4.50E-02 | 3.38E-02 | 4.15E-01 |
| A_0301 | 3.69E-02 | 2.36E-02 | 2.32E-01 |
| B_0702 | 3.61E-02 | 1.64E-02 | 1.39E-01 |
| B_3501 | 3.24E-02 | 1.56E-02 | 5.65E-02 |
| B_5101 | 3.24E-02 | 7.85E-03 | 0.00E+00 |
| B_0801 | 2.95E-02 | 1.87E-02 | 3.76E-01 |
| B_5801 | 2.65E-02 | 3.36E-03 | 0.00E+00 |
| A_3101 | 2.43E-02 | 7.45E-03 | 2.18E-01 |
| B_4403 | 2.21E-02 | 8.77E-03 | 9.36E-02 |
| B_1501 | 2.06E-02 | 3.78E-02 | 3.70E-02 |
| A_6801 | 1.77E-02 | 7.68E-03 | 2.80E-01 |
| B_3901 | 1.77E-02 | 1.01E-02 | 0.00E+00 |
| B_0401 | 1.55E-02 | 5.95E-03 | 3.77E-01 |
| B_2705 | 1.11E-02 | 6.61E-03 | 1.55E-01 |
| A_0205 | 8.85E-03 | 1.18E-02 | 2.45E-01 |
| B_5201 | 8.85E-03 | 7.26E-03 | 3.30E-01 |
| B_3801 | 6.64E-03 | 1.04E-03 | 0.00E+00 |
| B_4006 | 5.16E-03 | 7.18E-03 | 0.00E+00 |
| B_3701 | 4.42E-03 | 1.57E-02 | 0.00E+00 |
| B_5102 | 2.21E-03 | 3.60E-03 | 5.56E-02 |
| B_2702 | 1.47E-03 | 3.50E-03 | 2.45E-02 |
| A_3302 | 1.00E-03 | 1.37E-03 | 0.00E+00 |
| B_40 | 1.00E-03 | 5.31E-03 | 4.95E-01 |
| B_5103 | 1.00E-03 | 9.98E-03 | 0.00E+00 |
Figure 2.
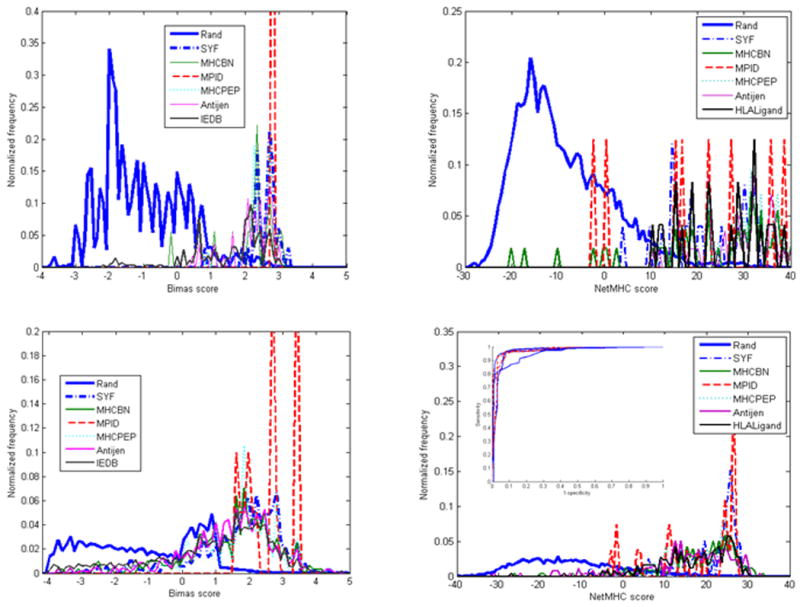
Typical validation results. The thick full blue line represents the distribution of random epitopes, while all other lines represent various epitope databases. Each sub-drawing represents a different HLA allele and different algorithms. The upper drawings are for B*5101 and the lower drawings are for A*0201. The left hand drawings are for the BIMAS algorithm and the right hand drawings are for NetMHC). For practically all alleles, the score of most random epitopes is low and the scores of epitopes from databases are high, although for some alleles (e.g. A*0201) a significant overlap exists. Note in the insert that the ROC curves are good for the algorithms tested and not very different among the algorithms tested.
SIR score
The Size of Immune Repertoire (SIR) score is defined as the ratio between the computed CTL epitope number (predicted epitopes) and the epitope number expected within the same number of random nonamers and a similar amino acid distribution (taking into account the correlation between the frequencies of neighboring amino acids).
This expected epitope fraction was computed by extracting the number of nonamers predicted to be epitope from a random 1.e6 amino acid sequence (999,992 overlapping nonamers). If such a sequence has 10,000 predicted epitopes on HLA B*2703 then the epitope fraction of B*2703 would be approximately 0.01 (10,000/999,992). If a given 408 amino acid long protein has 10 predicted HLA B*2703 epitopes then its SIR score for the same allele would be 10/(0.01*400)=2.5 (i.e. 10 computed epitopes divided by 4 expected epitopes).
The SIR score of a protein in a population is defined as the average SIR score over all available HLAs, weighted by the HLA frequency in the entire human population [26, 27]. The random sequence used to compute the random epitopes was matched to the human genome amino acid pair frequency. This matching was produced using a Markov model for the transition from one amino acid to the next trained on all human sequences.
The SIR score represents a large, albeit not full, subgroup of all epitopes. It misses for example, 8-mers and 10-mers as well as TAP independent epitopes. However, the ratio between expected and observed epitopes based on the same algorithms, should not be affected by the partiality of the list.
Viral Vs Human SIR score
The first observation resulting from the SIR score list is that, while the global average SIR score of human sequences is close to 1, the SIR score of a random sequence based on viral amino acid frequencies (based on the average of over 1,000 different fully sequenced most non-human viruses) is 19 % higher than its human counterpart (22 % higher for frequent HLA and 14 % higher for rare HLA) and the actual average SIR score of all viral sequences (not including viruses infecting humans) is 20 % higher than the one of human proteins. Even within the human sequences, significant differences can be observed. The SIR score of all human proteins for frequent HLA alleles is higher than 1, while for rare HLA alleles it is in general slightly lower than 1 (Figure 3) (r=0.47, p=0.02). This significant SIR score difference between frequent and rare alleles hints that in average, there is a weak selection on the MHC molecules to increase the number of presented human epitopes. This selection probably only occurs on the frequent alleles and not on the rare alleles. Note also that selection in the context of HLA can occur rapidly even for weak traits, through the selection in the population of people carrying a given HLA.
Figure 3.
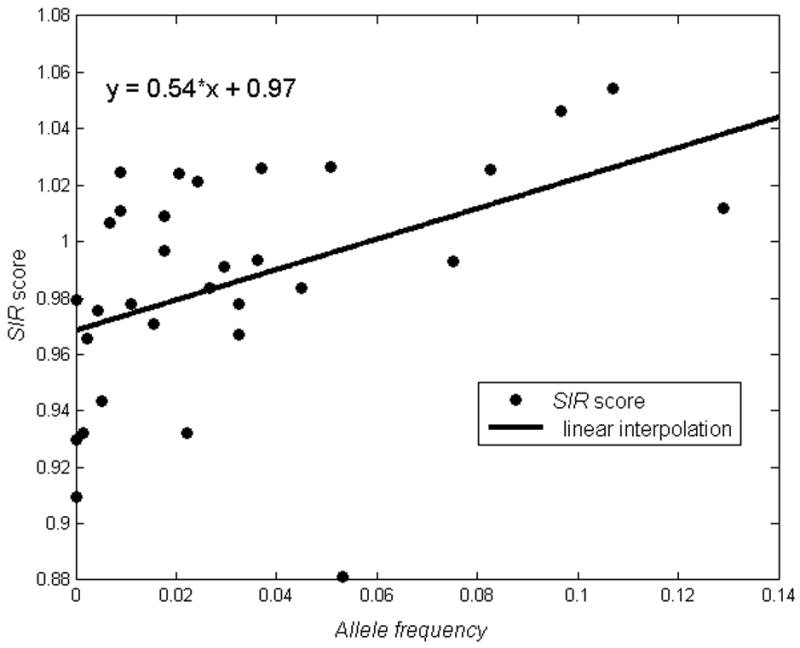
Correlation between HLA frequency (x axis) and SIR score (y axis). Each star represents a different HLA allele. Rare alleles have a low SIR score, while frequent alleles have a high one. The correlation between the allele frequency and the SIR score is significant (p=0.02). The full grey line represents a linear interpolation.
The very large difference between the number of viral and human epitopes presented shows a much more significant pressure to present viral epitopes. Considering the classical role of the immune system, this is not a surprising result. Moreover, focusing on the SIR scores of viruses, human viruses have a much lower SIR score than their non human counterparts [28]. From the evolution point of view this is also a reasonable result. The SIR score represents the relationship between the human immune system and its pathogens. The evasion of human viruses from the immune system is reflected by the lower SIR score of human viruses Vs non-human ones. This simply represents the fact the each specific human virus evolves faster than its human host.
Two possible evolutionary selection mechanisms can operate on epitope presentation on the MHC molecules, either the evolution of the MHC molecules, or the direct evolution of the epitopes. The first explanation is much simpler from an evolutionary point of view. It is easier to optimize the limited MHC locus than the entire genome. Furthermore, the MHC locus is the locus with the fastest evolution rate. It would be thus most natural to guess that the MHC locus has evolved and not the remaining parts of the genome. We here show that although the first explanation may still be partially valid, there are multiple evidences for specific selection of epitopes on proteins.
Selection of Epitopes in Human Protein Groups
We first test the specific selection of epitopes on a protein set most related to thymic education. A group of proteins directly known to be modulated for their presentation to the immune system are Tissue specific antigens (TSA) upregulated by AIRE in thymic medullary epithelial cells. These are tissue specific proteins expressed in various thymic cells [29-31]. AIRE itself is up-regulated in Medullary thymic epithelial cells (METC), and it induces the presentation of TSA in the thymus. An important goal of TSA presentation is thymic education of T cells [30]. TSAs are not known to have any other functional role in the thymus. The SIR score of TSA in METC as defined by Gotter et al [29] is significantly higher than all other human proteins (1.13 Vs 1, T test - p<1.e-50, Figure 4, last column). This result is consistent over most alleles. The over-presentation of epitopes on TSAs cannot be due to the properties of the MHC binding cleft or to the specific anchor residues, since TSAs in general have no known sequence similarity between themselves. A similar result is obtained on TSA regulated by AIRE in monocyte derived dendritic cells (MoDCs), as detailed by Sillanpaa et al [31] (Figure 4). The difference in SIR score between TSA and all other human proteins can either be explained as a selection of TSA antigens in general, or as the selection of the ones regulated by AIRE. The later explanation would make sense, given their role in immune regulation.
Figure 4.
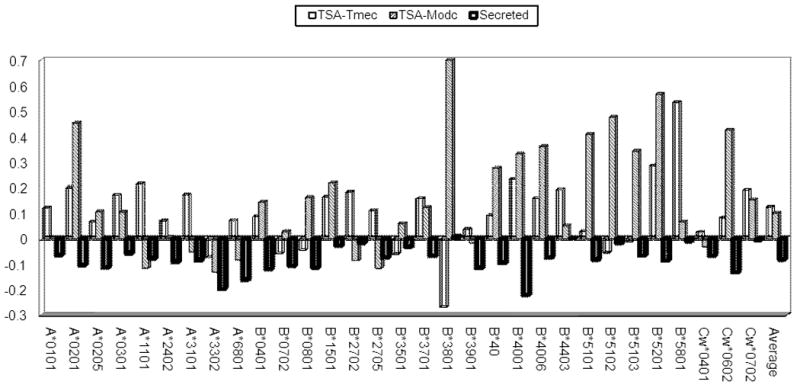
Difference between SIR score in specific protein groups and all human proteins. Three protein groups were used: TSA proteins in two cell types and secreted ones. One can see that the differences are actually common to most alleles. The results are presented for all alleles and for the weighted average over all alleles
A protein group expected to have a lesser impact on the cellular immune system is secreted proteins. Proteins can be presented to CTLs only if they are cleaved within the cytosol [10]. The resulting peptides can then be transferred to the ER and presented on MHC-I molecules. Secreted molecules have a higher probability than other proteins of being transferred to secretory compartments before their cleavage. Once in such compartments they are more likely to affect CD4+ T cells through the MHC-II pathway, and not CD8+ T cells through the MHC-I pathway [32]. These proteins thus do not present all their potential copy to MHC molecules, are thus slightly, yet systematically of a lesser use (on average) for the cellular immune system. These proteins are expected to present (on average) less epitopes. Assigning each protein its subcellular location in the GeneCards [33], we can measure the SIR score of all proteins classified as secreted. Indeed, the secreted proteins' average SIR score is significantly lower than the average SIR of all other proteins (0.93 Vs 1, p<1.e-40) (Figure 4). This result is again consistent over most HLA alleles, demonstrating that selection is systematic and not due to the binding pattern of a specific MHC molecule. These two examples show both positive and negative selection. Proteins selected for presentation to the immune system have a higher epitope number, while proteins with no significant importance for the immune system have a lower one. Note again that “importance” can be at many levels ranging from thymic selection to the activation of peripheral T cells. The SIR score differences can be either due to a different amino acid usage in different protein groups or to a direct selection for or against epitopes. In order to check the origin of the SIR score differences, we repeated the analysis, using as a background random proteins with an amino acid usage similar to either TSA or secreted proteins respectively. The new SIR score are much closer to 1 (1.06 for TSA and 0.96 for secreted proteins), but still statistically different from 1 for both groups (T-test, p<0.05). One can thus conclude that the selection for or against epitope can either be at the level of the amino acid MHC molecules tend to bind or at the specific level of epitopes within the proteins.
Correlation of SIR Score and RNA Expression Level
The total amount of epitopes presented from a protein is a combination of its transcription level (not necessarily its concentration, since a protein rapidly degraded has a low concentration, but can present many epitopes on MHC molecules) and the number of different potential epitopes in the protein (as measured by the SIR score).
A protein with a low RNA expression level produces a small total amount of epitopes. Even if these epitopes have a high affinity for the MHC molecule, the mere low expression level of the protein limits their amount. Such epitopes are thus less “important” for CTLs. Thus, as a first order approximation, a protein with a low RNA expression level is expected to be less “useful” for the immune system than one with a high RNA expression level, both at the level of thymic education and at the level of peripheral activation. If indeed specific proteins are selected for the presentation of epitopes, we expect the SIR score of a protein to be correlated with its RNA expression level. The correlation between the SIR score and the log average gene expression based on either GeneNote gene expression profiles or Electronic Northern Expression is indeed positive and significant (Pearson Correlation - r=0.076, p<1.e-14 and r=0.038, p<1.e-7 respectively). A similar correlation was obtained between the SIR score and the maximal expression level (among all organs tested). This positive correlation extends to the vast majority of organs and alleles (Figure 5). Namely, the RNA expression in most organs is positively correlated with the SIR score of most alleles (e.g. the RNA expression in the liver is significantly and positively correlated with the SIR score of most HLA alleles). Note that although the correlation itself is not very strong, it is highly significant. The mere presence of a correlation is very surprising when one considers the large number of factors affecting the RNA expression level, and that we only looked at the number of epitopes and not their characteristics. In addition, for MHC class I molecules, translation rather than transcription correlates with presentation. In the absence of such data, we compared the SIR score to the RNA expression level. One would suspect that removing all confounders and using protein translation rates, the correlation would be even more significant, although this needs further work.
Figure 5.
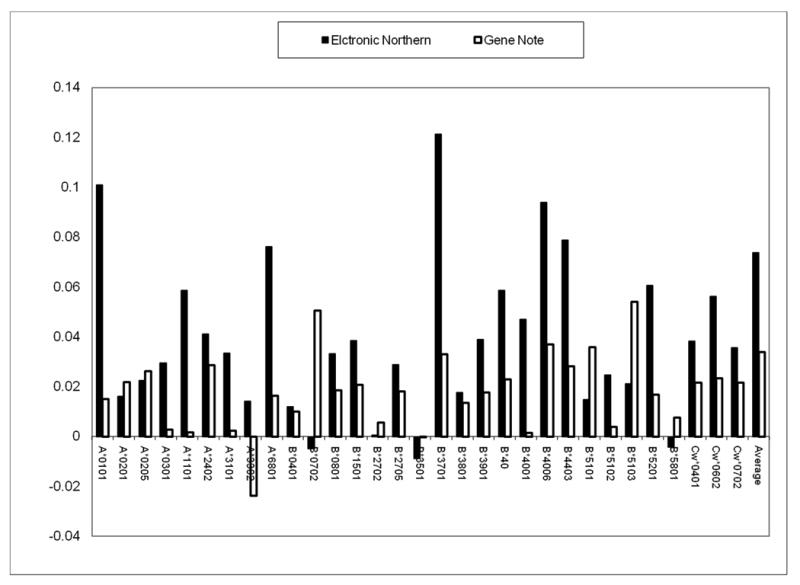
Correlation values between the SIR score and RNA average gene expression for the different alleles and for the weighted average over all alleles. The Y axis is the correlation coefficient with Electronic Northern or GeneNote human expression pattern profiles and the column is the allele. Practically all alleles have a significant positive correlation with the RNA expression level. No allele has a significant negative correlation.
The previous example cannot exclude the possibility that a third confounder raises both the epitope number and the protein expression in various situations. In order to directly test if epitopes are modulated, we looked for selection of point mutations in epitopes. We cannot directly measure mutations in the human genome, but we can measure the polymorphism they induce by looking at Single Nucleotide polymorphisms (SNPs).
SNPs within Epitopes
The correlation between epitopes and SNP is measured by dividing the 156,019 SNPs currently mapped in human genes in the Ensembl genome (33,677 genes) [22] into SNPs that are within an epitope and SNPs that are outside epitopes for the seven most highly expressed HLA alleles (A*0101, A*0201, A*1101, A*2402, B*4001, B*0702, Cw*0702 and Cw*0401). For example, if a protein has two epitopes for HLA A*0101, all SNPs outside these two epitopes and their first flanking residue are characterized as external and all the SNPs within at least one of the two epitopes are characterized as internal. Since we do not know the HLA haplotype of the population carrying each SNP, we only analyze the most frequent HLA alleles to maximize the probability that a SNP is within a person carrying these MHC alleles. The fraction of non- synonymous SNPs (replacement fraction), is 8% higher in average inside epitopes than outside epitopes (Chi square test, p<1.e-100) and is consistently higher in epitopes among all tested MHC alleles. Actually the fraction of mutations in epitopes is also slightly higher than outside epitopes (1.6 %, chi square, p<1.e-5), but this result is not consistent over alleles.
Proteins with High or Low SIR Score
To summarize, we observe a clear selection of the epitope number in proteins at the specific protein level, as well as clear evidence for the mechanism behind this selection. One can now compute which specific proteins are the most up or down regulated. We rank all proteins according to the chi square score of the SIR score. The resulting list (detailed in http://peptibase.cs.biu.ac.il:64080/SIR_51.html) shows that the high SIR protein list contains most inflammation response proteins as well as those related to virus detection. For example, all TLRs, but two (TLR1 and TLR6) are in the list of most highly presented proteins, as well as many transport proteins. Note that the sequence homology between TLRs is minimal. The number of epitopes expressed by TLRs is thus the result of selection for their function and not of specific sequences. A possible explanation for the selection of TLRs is that TLRs are upregulated, in the presence of an intra-cellular threat, and serve as an additional marker of infection for the immune system. On the other hand, most structural proteins, such as those related to bone structure, have a low SIR score. Thus, again one can see that the average number of self epitopes expressed per proteins is in some self proteins is adapted to the needs of the immune system. We only have shown the most obvious elements that can affect epitope selection. Yet the full list of factors that can make a protein “useful” for the immune system is very large, and requires a careful analysis to be defined more precisely.
Discusion
We have here shown that the cellular part of the adaptive immune system is not fully adaptive in the sense that it has evolved to detect the amino acid distribution of viruses Vs the one of self proteins. Even within human proteins, the epitope distribution is not random. Not all proteins express the same amount of epitopes. Some well defined proteins with some significance for the immune system, such as those highly over expressed in the thymus or inflammation related proteins and TLRs, express much more epitopes than others. The epitope repertoire obviously still contains a large random component. The adaptive immune system produces random receptors, but many of these receptors targets (epitopes on the MHC molecules) are found out to be selected. Interestingly, the selection of target proteins is not obtained only through the evolution of the immune system itself, but also through specific mutations in the target proteins, as is evidenced by the high fraction of replacement mutations inside epitopes. The common over/under-presentation of proteins groups is not due to a sequence similarity. We have compared the sequences of the different groups. These sequences contain no significant homology, specific amino acid usage or length distribution. This difference is thus probably purely immunological. The evolutionary driving force of epitope selection can occur either at the level of positive and negative selection in the thymus, or in the periphery. In the first case the goal of the repertoire modulations would be to better train the immune system. In the second case, the repertoire modulation could either affect the regulatory function of T cells, or their proposed “house-keeping” functions. CTL have indeed been proposed to have a regulatory function (parallel to the CD25+ CD4+ regulatory T cells) [34, 35].
The current analysis was focused on the CTL response. The CTL response is the simplest to analyze from a bioinformatics point of view. Most CTL epitopes are nonamers [35, 36] and the probability to pass each of the classical TAP dependent pathway processing stages can be well estimated by a linear function of the nonamer sequence. Finally, there are very large databases of experimentally measured epitopes allowing us to test the precision of the different algorithms. The error levels of the currently used algorithms are relatively low. However, the signal we are measuring still contains a significant amount of noise. Given the noise level, the presence of such a clear selection and non-synonymous SNP frequency hints that the observed selection would have been much stronger in the absence of the noise induced from the algorithms imprecision. An important caveat of this work is the assumption that there is no bias for the classical pathway in one group of protein or the other. However, there is currently no known group of proteins known to be processed specifically in one pathway or the other. If indeed such a bias existed it would show by itself that self proteins modulate the way they are expressed to the self immune system.
The limited number of CTL epitopes (approximately one per gene per HLA allele in average) makes this part of the immune system the most prone for selection. One cannot directly apply the conclusions from the effector part of the cellular immune system to the other parts of the adaptive immune system (CD4+ T cells and B cells). The question of how specific the other parts of the immune system are remains open.
Materials and Methods
Genomic data
Human protein sequences were used for this analysis. The human sequences were obtained from the Ensembl database [22]. All human predicted protein coding regions (exons) were used.
Gene classification
The list of the proteins upregulated by AIRE was extracted from published in vitro experiments of human cells [29, 31]. The secreted and membrane proteins were identified by the automatic extraction of the sub-cellular location of each protein from the information contained within GeneCards [33], and the subsequent categorization of proteins according to their location. All protein names were translated from their HUGO name (GeneCards) to the appropriate Ensembl entry in order to analyze their amino acid sequences. The conversion was made according to the GeneCards. In many cases a protein had several Ensembl transcripts. When this happened we used the Ensembl transcript containing the longest sequence.
Cleavage Score
Given a peptide and its two flanking regions FN-P1…..Pn-FC, the following score was defined:
FN and FC are the N and C termini flanking regions, while Pi are the residues within the peptide. A peptide with a high score, S, has a high probability of being produced, while a low score has a low creation probability. The appropriate values for S1-S5 were learned using a simulated annealing process [37]. Two learning sets were used. The positive learning set (containing 645 peptides) was composed of peptides observed to be produced in a variety of cleavage experiments. The negative learning set (containing 560 peptides) was composed of peptides observed to be either cleaved in their center or not cleaved at their extremities in the same cleavage experiments. Extra sub-units are added to the proteasome in the presence of interferon-γ, changing its cleavage properties. Since our main interest was in the production of peptides presented on MHC-I, we incorporated results from both the standard and so called “immunoproteasome” in our analysis. The algorithm was validated to have less than 16% false positives and 10% false negatives ([24] and http://peptibase.cs.biu.ac.il:64080/PepCleave_I/). This methodology takes into account N-Terminal Cleavage, which is not necessary for all peptide, but it prevents the bias of epitopes selected for analysis that may occur in epitope based cleavage algorithms.
TAP binding score
The probability that a peptide binds the TAP machinery is mainly a function of the residues at the first three N-term and the last C-term positions. Moreover it can be estimated through a linear combination of the binding energies of each residue. Multiple algorithms for TAP binding were checked. The score computed by Peters et al [25] gave the best differentiation between presented and random epitopes.
MHC binding motifs
Each gene was divided into all possible nonamers using a sliding window (e.g. a 300 amino acid protein was divided into 292 nonamers: position 1-9, position 2-10 and so on). For each nonamer, we computed the MHC binding energy of 31 different HLA class I molecules, most of them HLA-A and B. The affinity of a candidate peptide for each HLA allele was estimated using the binding coefficients predicted by Parker in the BIMAS software [13] (http://bimas.cit.nih.gov/.). These matrices estimate the contribution of each amino acid at each position to the total binding strength. Anchor residues have a high contribution, while neutral residues have a value of 1. The overall binding strength is the product of the binding strength of the amino acids at each of the nine positions. Although BIMAS is an old algorithm, it has many advantages over newer ones. Its main advantage is that most existing algorithms were learnt on published epitopes, limiting our possibility of performing an independent validation of their quality, while BIMAS was learnt using a regression on measured epitopes offrates [13]. Moreover, BIMAS provides the raw matrices for a large number of alleles and all the frequent ones. We have tested other algorithms, but the error level of much more modern algorithms, such as NetMHC were not significantly better than the one of BIMAS (Figure 2) and they seem to classify a much smaller number of epitopes, which given the other processing stages lead to no epitope for most proteins.
HLA frequency and weighted average
The number of currently known different HLA alleles ranges from hundreds to thousands for HLA A, B and C (A - 527, B -911, C − 283, http://www.ebi.ac.uk/imgt/hla/stats.html) [38]. Most individuals are heterozygous for HLA-A, HLA-B and HLA-C and express three different heterozygous MHC-I molecules. Different MHC-I molecules have different racial distributions [39]. The repertoire was computed for 31 alleles maximizing the population coverage (for the average human HLA distribution frequency). We used 9 HLA-A (60.8%), 19 HLA-B (44.2%), and 3 HLA-C (31.2%). In order to compute the properties of the repertoire in a given population, we used a weighted average of the SIR score on the available HLAs, based on the HLA frequency in the average human population. The average was done on HLA-A, B and C separately, and the weighted score over HLA-A, B and C was averaged with equal weight for A, B and C.
Validation
In order to validate the algorithms used, we checked the score of peptides present in 7 different databases: IEDB [40, 41], SYFPEITHI [36] - www.syfpeithi.de, MHCBN [42] -http://www.imtech.res.in/raghava/mhcbn/, MPID [43] -surya.bic.nus.edu.sg/mpid/, MHCPEP [35] - http://www3.oup.co.uk/nar/database/, AntiJen – http://www.jenner.ac.uk/AntiJen/ and HLALigand [27] -http://hlaligand.ouhsc.edu/LigandDB). Assuming that most peptides in the various databases are correct, we computed the threshold that would maximize the number of presented peptides from the positive databases, and minimize the number of peptides in a neutral set of 1,000,000 peptides using various amino acid distributions. We have checked for each HLA allele the level of type-I and type-II errors (e.g. Figure 2) and attempted to find a cutoff minimizing both. For most alleles we found cutoffs leading to low errors. A precise list of the false positive and false negative rates of the algorithms used can be found in http://peptibase.cs.biu.ac.il/peptibase/validation.htm.
SNPS
The Single Nucleotide Polymorphism (SNP) analysis was performed over 33,677 human genes from the 2005 version of the Ensembl. Given a HLA allele and an epitope within a gene, we divided the regions within and surrounding the epitope into four regions: A) The nucleotides flanking the C and N termini of the epitope (noted as positions 0, 10). B) The first and last nucleotides in the epitope (noted as positions 1, 9). C) The nucleotides in the center of the epitope (noted as positions 2 - 8). D) All nucleotides outside the epitope that are not directly flanking it.
Each SNP was assigned to one of these four groups according to its position. The regions size obviously varies between HLA alleles as each HLA has different epitopes. The Replacement fraction (R/(R+S) ratio) is defined as the fraction of non-synonymous mutations, where a synonymous mutation is a nucleotide mutation not leading to an amino acid change (e.g. CGG to CGC, which is at the amino acid level a R to R), and a non-synonymous mutation is a nucleotide mutation leading to an amino acid mutation (e.g. CCC to CAC, which is at the amino acid level a P to H). This fraction is directly related to the selection level operating on a gene. The R/R+S ratio of the SNPs was computed for each SNP and the R/R+S ratio in the center of the epitopes (position 2-8) was compared to the R/R+S ratio outside epitopes, in residues not flanking the epitope. We ignored at this stage the cleavage sites (positions 0, 1, 9 and 10), as they may be selected for reasons unrelated to MHC presentation, but including the cleavage sites does not significantly change the results.
RNA levels
The RNA expression levels of the different proteins used in the current analysis were extracted from the GeneNote normal tissue human expression profile [44-46]. For each gene in the GeneCards database, all the probe-sets representing a single gene were used. The RNA expression pattern used is the average expression over these probe-sets.
A second source of expression patterns was from electronic northern. The NCBI's Unigene dataset was mined for information about the number of unique clones per gene per tissue. Clones were assigned to particular tissues by applying data-mining heuristics to Unigene's library information file (Hs.lib.info). Electronic expression results were calculated by dividing the number of clones per gene by the number of clones per tissue. They were then normalized by multiplying by 1,000,000. We used the average expression over all tissues and the maximal expression supplied by the Genecards [33, 46].
Statistical analysis
The statistical measurement of selection was performed using various tests. When comparing two continuous variables, a Pearson correlation coefficient was used, and we tested the significance of this correlation. Such a test was used for the RNA expression level, and the correlation between the allele frequency and the SIR score. When comparing a few groups of proteins, a two tail unpaired T-test was used on all the proteins in the groups. This was used in the comparison of TSA, secreted proteins and all proteins. When comparing the expected and computed number of SNPs in epitopes or when comparing the number of viral epitopes compared to their expected number, a Chi-square test was used.
Acknowledgments
The work of Y.L, M.A, S.R, T.V, L.T and V.F was covered by NIH grant: 1 R01 AI61062-01. The work of T.V was also covered by a scholarship of Yeshaia Hororwitz foundation.
Bibliography
- 1.Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci U S A. 1976;73:3628–3632. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 3.Weigert M, Perry R, Kelley D, Hunkapiller T, Schilling J, Hood L. The joining of V and J gene segments creates antibody diversity. Nature. 1980;283:497–499. doi: 10.1038/283497a0. [DOI] [PubMed] [Google Scholar]
- 4.Nemazee D, Russell D, Arnold B, Haemmerling G, Allison J, Miller JF, Morahan G, Buerki K. Clonal deletion of autospecific B lymphocytes. Immunol Rev. 1991;122:117–132. doi: 10.1111/j.1600-065x.1991.tb00600.x. [DOI] [PubMed] [Google Scholar]
- 5.Prak EL, Weigert M. Light chain replacement: a new model for antibody gene rearrangement. J Exp Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller JF. Self-nonself discrimination and tolerance in T and B lymphocytes. Immunol Res. 1993;12:115–130. doi: 10.1007/BF02918299. [DOI] [PubMed] [Google Scholar]
- 7.Cope AP, Schulze-Koops H, Aringer M. The central role of T cells in rheumatoid arthritis. Clin Exp Rheumatol. 2007;25:S4–11. [PubMed] [Google Scholar]
- 8.Wilczynski JR, Kalinka J, Radwan M. The role of T-regulatory cells in pregnancy and cancer. Front Biosci. 2008;13:2275–2289. doi: 10.2741/2841. [DOI] [PubMed] [Google Scholar]
- 9.Williams A, Peh CA, Elliott T. The cell biology of MHC class I antigen presentation. Tissue Antigens. 2002;59:3–17. doi: 10.1034/j.1399-0039.2002.590103.x. [DOI] [PubMed] [Google Scholar]
- 10.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 11.Uebel S, Tampe R. Specificity of the proteasome and the TAP transporter. Curr Opin Immunol. 1999;11:203–208. doi: 10.1016/s0952-7915(99)80034-x. [DOI] [PubMed] [Google Scholar]
- 12.Rammensee HG, Bachmann J, S SS. MHC Ligands and Peptide Motifs. Austin Texas USA: 1997. [Google Scholar]
- 13.Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, Biddison WE, Coligan JE. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. 1992;149:3580–3587. [PubMed] [Google Scholar]
- 14.Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens. 2003;62:378–384. doi: 10.1034/j.1399-0039.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 15.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothe BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 16.Bhasin M, Raghava GP. Pcleavage: an SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences. Nucleic Acids Res. 2005;33:W202–207. doi: 10.1093/nar/gki587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxova P, Buus S, Brunak S, Kesmir C. Predicting proteasomal cleavage sites: a comparison of available methods. Int Immunol. 2003;15:781–787. doi: 10.1093/intimm/dxg084. [DOI] [PubMed] [Google Scholar]
- 18.Kesmir C, Nussbaum AK, Schild H, Detours V, Brunak S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 2002;15:287–296. doi: 10.1093/protein/15.4.287. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 20.Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 21.Louzoun Y, Vider T, Weigert M. T-cell epitope repertoire as predicted from human and viral genomes. Mol Immunol. 2006;43:559–569. doi: 10.1016/j.molimm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, Down T, Eyras E, Fernandez-Suarez XM, Gane P, Gibbins B, Gilbert J, Hammond M, Hotz HR, Iyer V, Jekosch K, Kahari A, Kasprzyk A, Keefe D, Keenan S, Lehvaslaiho H, McVicker G, Melsopp C, Meidl P, Mongin E, Pettett R, Potter S, Proctor G, Rae M, Searle S, Slater G, Smedley D, Smith J, Spooner W, Stabenau A, Stalker J, Storey R, Ureta-Vidal A, Woodwark KC, Cameron G, Durbin R, Cox A, Hubbard T, Clamp M, Andrews D, Cox T, Hotz H, Woodwark C, Egerton M, Scollay R, Shortman K, Schlosstein L, Terasaki PI, Bluestone R, Pearson CM. An overview of ensembl, Ensembl 2004. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louzoun Y, Vider T. Score for Proteasomal Peptide Production Probability. Immunology. 2004;1 [Google Scholar]
- 24.Ginodi I, Vider-Shalit T, Tsaban L, Louzoun Y. Precise score for the prediction of peptides cleaved by the proteasome. Bioinformatics. 2008;24:477–483. doi: 10.1093/bioinformatics/btm616. [DOI] [PubMed] [Google Scholar]
- 25.Peters B, Bulik S, Tampe R, Endert PMV, Holzhutter HG. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 26.Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 27.Sathiamurthy M, Hickman HD, Cavett JW, Zahoor A, Prilliman K, Metcalf S, Fernandez Vina M, Hildebrand WH. Population of the HLA ligand database. Tissue Antigens. 2003;61:12–19. doi: 10.1034/j.1399-0039.2003.610102.x. [DOI] [PubMed] [Google Scholar]
- 28.Vider-Shalit T, Fishbain V, Raffaeli S, Louzoun Y. Phase dependent immune evasion of Herpesviruses. J Virol. 2007 doi: 10.1128/JVI.02636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med. 2004;199:155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kont V, Laan M, Kisand K, Merits A, Scott HS, Peterson P. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol. 2007 doi: 10.1016/j.molimm.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sillanpaa N, Magureanu CG, Murumägi A, Reinikainen A, West A, Manninen A, Lahti M, Ranki A, Saksela K, Krohn K, Lahesmaa R, Peterson P. Autoimmune regulator induced changes in the gene expression profile of human monocyte-dendritic cell-lineage. Molecular Immunology. 2004;41:1185–1198. doi: 10.1016/j.molimm.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Rush C, Mitchell T, Garside P. Efficient priming of CD4+ and CD8+ T cells by DNA vaccination depends on appropriate targeting of sufficient levels of immunologically relevant antigen to appropriate processing pathways. J Immunol. 2002;169:4951–4960. doi: 10.4049/jimmunol.169.9.4951. [DOI] [PubMed] [Google Scholar]
- 33.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: encyclopedia for genes, proteins and diseases. Weizmann Institute of Science, Bioinformatics Unit and Genome Center; Rehovot, Israel: 1997. [Google Scholar]
- 34.Dhodapkar MV, Steinman RM. Antigen-bearing immature dendritic cells induce peptide-specific CD8(+) regulatory T cells in vivo in humans. Blood. 2002;100:174–177. doi: 10.1182/blood.v100.1.174. [DOI] [PubMed] [Google Scholar]
- 35.Brusic V, Rudy G, Harrison LC. MHCPEP, a database of MHC-binding peptides: update 1997. Nucleic Acids Res. 1998;26:368–371. doi: 10.1093/nar/26.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 37.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by Simulated Annealing. Science. 1983;220:671. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 38.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA Database - a sequence database for the human Major Histocompatibility Complex. Nucleic Acids Res. 2001;29:210–213. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh SGE, Parham P, Barber LD. The HLA Facts Book. Academic Press; London UK: 2000. [Google Scholar]
- 40.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger S, Stewart S, Surko P, Way S, Wilson S, Sette A. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sathiamurthy M, Peters B, Bui HH, Sidney J, Mokili J, Wilson SS, Fleri W, McGuinness DL, Bourne PE, Sette A. An ontology for immune epitopes: application to the design of a broad scope database of immune reactivities. Immunome Res. 2005;1:2. doi: 10.1186/1745-7580-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhasin M, Singh H, Raghava GP. MHCBN: a comprehensive database of MHC binding and non-binding peptides. Bioinformatics. 2003;19:665–666. doi: 10.1093/bioinformatics/btg055. [DOI] [PubMed] [Google Scholar]
- 43.Govindarajan KR, Kangueane P, Tan TW, Ranganathan S. MPID: MHC-Peptide Interaction Database for sequence-structure-function information on peptides binding to MHC molecules. Bioinformatics. 2003;19:309–310. doi: 10.1093/bioinformatics/19.2.309. [DOI] [PubMed] [Google Scholar]
- 44.Chalifa-Caspi V, Shmueli O, Benjamin-Rodrig H, Rosen N, Shmoish M, Yanai I, Ophir R, Kats P, Safran M, Lancet D. GeneAnnot: interfacing GeneCards with high-throughput gene expression compendia. Brief Bioinform. 2003;4:349–360. doi: 10.1093/bib/4.4.349. [DOI] [PubMed] [Google Scholar]
- 45.Shmueli O, Horn-Saban S, Chalifa-Caspi V, Shmoish M, Ophir R, Benjamin-Rodrig H, Safran M, Domany E, Lancet D. GeneNote: whole genome expression profiles in normal human tissues. C R Biol. 2003;326:1067–1072. doi: 10.1016/j.crvi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]


