SUMMARY
The duplex formed between the branch site (BS) of a spliceosomal intron and its cognate sequence in U2 snRNA is important for spliceosome assembly and the first catalytic step of splicing. We describe the development of an orthogonal BS-U2 system in S. cerevisiae in which spliceosomes containing a grossly substituted second copy U2 snRNA mediate the in vivo splicing of a single reporter transcript carrying a cognate substitution. Systematic use of this approach to investigate requirements for branching catalysis reveals considerable flexibility in the sequence of the BS-U2 duplex and its positioning relative to the catalytic centre. Branching efficiency depends on the identity of the branch nucleotide, its position within the BS-U2 duplex, and its distance from U2/U6 helix Ia. These results provide previously unavailable insights into substrate selection during spliceosomal branching catalysis; additionally, this system provides a foundation and tool for future mechanistic splicing research.
INTRODUCTION
Pre-mRNA splicing can be considered as three broad phases, each comprising multiple steps: these phases are spliceosome assembly, splicing catalysis, and spliceosome disassembly/mRNA release. Although the majority of work in the splicing field is focused on assembly, there is growing consensus that transitions during the catalytic phase can limit the rate and fidelity of splicing and act as potential points of regulation (Park et al., 2004; Pleiss et al., 2007; Smith et al., 2008).
The nature of the two chemical reactions of splicing (Padgett et al., 1984) and a model for the mechanism by which fidelity is maintained (Burgess and Guthrie, 1993) were proposed twenty-five and fifteen years ago, respectively, yet, despite more recent advances (reviewed in Smith et al., 2008) many details of splicing catalysis remain enigmatic. The relative neglect of the catalytic phase is in part due to the complex assembly phase that precedes it, as many defects will manifest themselves during assembly and thus preclude the investigation of catalysis; this highlights the need for new experimental systems for the study of the catalytic phase of splicing. The duplex between the BS and the highly conserved cognate region of U2 snRNA (Parker et al., 1987; Wu and Manley, 1989; Zhuang and Weiner, 1989), shown schematically in Fig. 1a, exemplifies a structure believed to be important in multiple aspects of the splicing reaction, whose investigation has been hindered by the limitations of current experimental models.
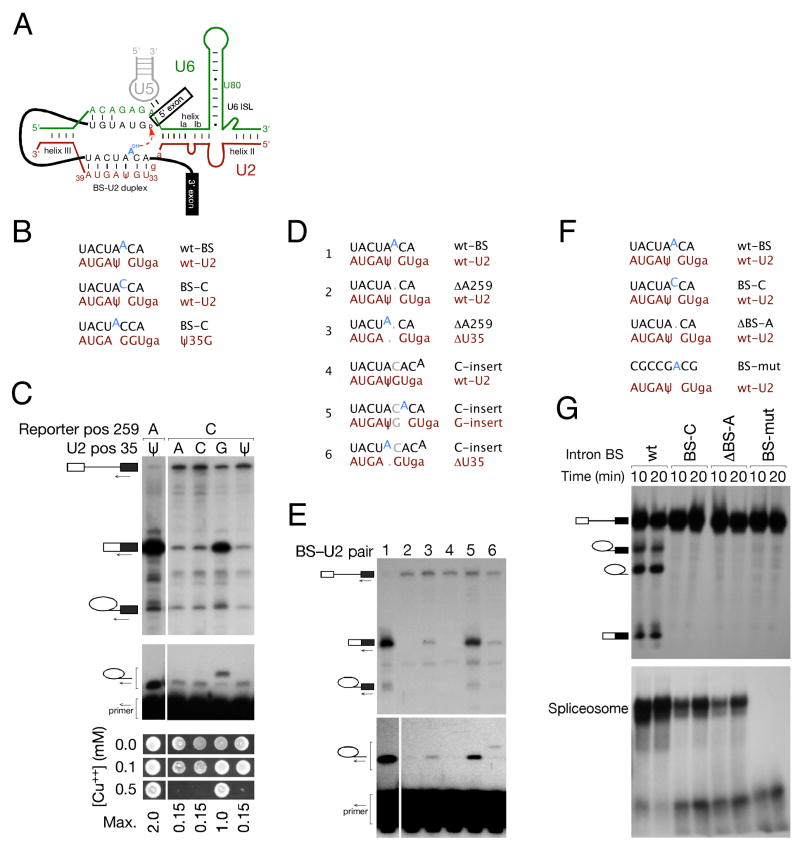
Figure 1. Bulging of the branch site nucleophile is required for splicing in S. cerevisiae, but the position of the bulge within the BS-U2 snRNA duplex is flexible.
(A) Schematic of RNA-RNA interactions that contribute to the first step of splicing. The pre-mRNA is shown in black, with the branch site (BS) adenosine highlighted in blue: U2 snRNA is shown in red, U5 in grey and U6 in green. U2 positions A31 and G32 are shown in lowercase throughout for positional reference.
(B) Schematic of the BS-U2 pairing regions of wt and BS-C mutant reporters, and wt and Ψ35G U2 snRNAs used in (C).
(C) An intron BS-C mutation can be suppressed by U2 Ψ35G but not by other mutations at this position: upper panel – splicing efficiency monitored by primer extension with a 3′ exon primer on S. cerevisiae total RNA from strains carrying the indicated U2 snRNA and reporter constructs; middle panel – primer extension with an intronic primer immediately upstream of BS indicates that the Ψ35G suppressor promotes the use of a non-canonical adenosine as the branch nucleophile; lower panel – copper growth assay to verify that the mRNA observed in the upper panel is functional.
(D) Schematic of reporter and U2 constructs used in (E).
(E) Primer extensions with 3′ exon (upper panel) and BS-proximal intronic (lower panel) primers on total RNA from the indicated strains indicate that branch nucleotide bulging is required for splicing, but the position of the bulge within the BS-U2 duplex is not fixed.
(F) Schematic of reporter and U2 constructs used in (G).
(G) In vitro splicing (upper panel) and spliceosome assembly (lower panel) of the S. cerevisiae UBC4 intron carrying the indicated BS mutations, as assayed by denaturing and native PAGE, respectively. BS-C and ΔBS-A mutations allow reduced spliceosome assembly but no branching, indicating the importance of the bulged nucleotide for spliceosomal catalysis.
During spliceosome assembly, the BS is transiently bound by SF1/BBP before being transferred to U2 snRNA (Berglund et al., 1997). The stability of the resulting BS-U2 duplex is proofread by kinetic competition between duplex formation and ATP hydrolysis by Prp5 (Xu and Query, 2007), with sufficiently stable BS-U2 interaction allowing progression into the productive splicing pathway. Mutations that weaken the interaction between the BS and U2 therefore strongly inhibit spliceosome assembly.
In the first step of splicing, the 2′ hydroxyl group of a conserved adenosine at the branch site (BS) attacks the 5′ splice site (5′SS), and the BS-U2 duplex has been proposed to play a role at this stage. The conserved 5′-GΨAGUA-3′ (where Ψ is pseudouridine) motif in U2 allows the second consecutive A in the conserved 5′-UACUAAC-3′ branch site (the canonical BS nucleophile is highlighted) to bulge from the helix (Fig. 1a). Substitution of the first adenosine in the AA pair with arabinosyl adenosine favours bulging of this unreactive nucleotide rather than the adjacent and normally favoured adenosine, and inhibits first step catalysis in HeLa extract (Query et al., 1994). In addition, NMR studies on model duplexes suggest that the Ψ at position 35 in U2 snRNA (S. cerevisiae numbering used throughout) favours extrusion of the BS-A from the helix (Newby and Greenbaum, 2002). Deletion of Pus7, the pseudouridine synthase responsible for this modification in S. cerevisiae, confers sickness (presumably due to a splicing defect) on cells with mildly mutated U2 snRNA (Yang et al., 2005). Finally, abrogation of an analogous bulged adenosine in domain 6 of a group II self-splicing intron virtually abolishes first step catalysis by branching, with little to no negative impact on the hydrolytic first step pathway (Chu et al., 1998). Together, these studies suggest that bulging of the BS nucleophile via interaction with U2 is important for branching catalysis by the spliceosome.
The ability of the higher eukaryotic spliceosome to catalyse this reaction using either adenosine in -UAAC as a nucleophile (Query et al., 1994) and poor conservation of both the BS and 5′SS in higher eukaryotes (reviewed in Burge et al., 1999) suggest flexibility with respect to the selection and activation of nucleophile and electrophile. Yeast, by contrast, have strong splice site consensus sequences and appear to have strict positional requirements for the BS nucleophile: mutation of the BS-A to a pyrimidine, instead of activating the upstream adenosine, results in inefficient splicing from the mutated nucleotide (Jacquier and Rosbash, 1986). However, the observation that the BS-binding region of U2 snRNA can be used as a 5′SS for both steps of splicing in vivo (Smith et al., 2007) suggests active site flexibility and implies that the BS-U2 interaction might be more dynamic than previously thought.
Here, we demonstrate the importance of nucleophile bulging for splicing catalysis. From the systematic movement of a bulge motif through the BS-U2 duplex in vivo in S. cerevisiae, we conclude that adenosines bulged from at least three positions within the BS-U2 duplex are competent to act as the first-step nucleophile. Mutations in the BS-binding region of U2 have unpredictable effects on splicing and cell growth. An orthogonal system is therefore desirable in which a constant second copy U2 snRNA, capable of splicing a reporter transcript but not of interacting with endogenous introns, can be used for the in vivo investigation of nucleophile positioning. We report the development and characterisation of such a BS-U2 system. We show unambiguously that three bulge positions can participate in first step catalysis, and confirm the importance of nucleotide identity at the branch site. We also show that the branch site and 5′SS are positioned independently for first step catalysis, and that the favoured position of the branch nucleophile is dependent on distance from the U2 component of U2/U6 helix Ia.
RESULTS
Bulging is required for splicing, but the position of the bulge within the BS-U2 duplex is flexible
Our continued interest in the branch site-U2 snRNA duplex was spurred both by the observation of trans-splicing to U2 in S. cerevisiae (Smith et al., 2007) and by the results of a screen for U2-based suppressors of branch site mutations. Because U2 is essential and sensitive to mutation in its BS-binding region, this mutagenesis screen and all subsequent experiments involving mutant U2 snRNAs were performed using a strain carrying both a wild-type (wt) copy of U2 and a second gene encoding the mutant. One U2 allele capable of suppressing the splicing defect of BS-C (5′-UACUACC-3′) was identified as Ψ35G (5′-GGAGUA-3′). Other mutations at this position failed to suppress BS-C (Fig. 1b&c), and other BS mutations could be suppressed by analogous mutations of position 35 to the Watson-Crick base-pairing partner of the mutated BS nucleotide (data not shown). This suggested that the splicing defect due to BS mutation was overcome via bulging and activation of the preceding adenosine – a conclusion confirmed by primer extension analysed at single nucleotide resolution (Fig. 1c, middle panel). The ACT1-CUP1 reporter encodes a metallothionein whose expression correlates over a large dynamic range with resistance to copper in the growth medium (Lesser and Guthrie, 1993). All primer extension data in this and subsequent experiments were confirmed by copper growth assays (Fig. 1c, lower panel, and data not shown); this verifies that mRNA is released from the spliceosome, exported to the cytoplasm and translated.
The use of a positionally non-canonical BS nucleotide does not normally occur in S. cerevisiae; our data suggested both that bulging of the BS was likely required for catalysis as well as assembly, and that S. cerevisiae could be induced to display the same flexibility as higher eukaryotes with respect to branch nucleophile positioning. We introduced pairs of mutations into the BS and a second copy U2 snRNA such that the bulge was flattened by insertion or deletion of a nucleotide in the BS sequence, and restored by corresponding deletion or insertion in the second copy U2 (Fig. 1d). As expected for an essential bulge, introns that generated flat duplexes failed to support splicing (Fig. 1e, lanes 2&4), and restoration of the bulge via either insertion or deletion in U2 snRNA restored splicing activity, albeit with reduced efficiency relative to the wt BS-U2 pair (Fig. 1e, lanes 3, 5&6). To distinguish between the assembly and catalytic phases, we assayed spliceosome assembly and splicing progression in vitro using S. cerevisiae whole cell extract and the efficiently spliced UBC4 transcript (John Abelson, personal communication) carrying a variety of mutations at and around the BS. Analysis of splicing reactions in native and denaturing polyacrylamide gels indicated that deletion of the branch nucleophile to produce a flat BS-U2 duplex allowed spliceosome assembly at a level comparable to that shown by an intron carrying a BS-C mutation: neither of these mutant introns, however, showed detectable splicing catalysis, in contrast to the efficient splicing of the wt UBC4 intron (Fig. 1f&g). As expected, an intron with a severely mutated BS region failed to elicit detectable spliceosome assembly (Fig. 1f&g). Further analysis of assembled spliceosomes, using low concentrations of EDTA to separate assembly intermediates (Cheng and Abelson, 1987), indicated that the bulge-less transcript progressed at least to the stage of a U2/5/6 spliceosome – i.e. that U1 and U4 snRNAs can be lost to generate a normal assembly intermediate (Fig. S1a), and that a similar array of splicing complexes can form on wild-type and bulge-less UBC4 introns in the absence of Prp2, the ATPase whose action immediately precedes the catalytic phase of splicing (Fig. S1b&c). We can therefore conclude that the absence of a bulged branch nucleotide confers a post-assembly defect in addition to impairing spliceosome assembly, although the resolution of current assays does not allow us to conclude that the branch nucleophile is bulged at the time of catalysis per se.
High-resolution primer extension of in vivo splicing indicated that the expected branch nucleophile was used for each BS-U2 pair that showed detectable splicing (Fig. 1e, lower panel – the background band at the wt position is due to extension on endogenous actin), indicating that changing the length of the BS-U2 duplex, and correspondingly moving the bulged adenosine towards or away from its helix III-proximal end (Fig. 1a&d), does not abolish the ability of this adenosine to act as the nucleophile for the first step of splicing in this context.
Systematic movement of a CAC motif through the BS-U2 duplex indicates that three bulge positions can participate in catalysis
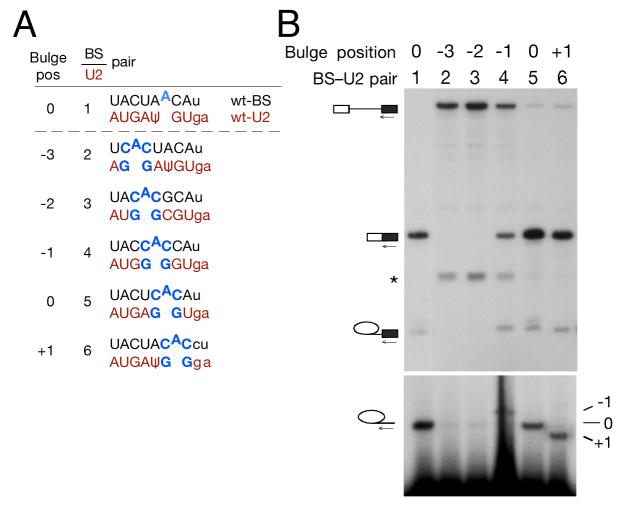
To further investigate the apparent flexibility of nucleophile location within the BS-U2 duplex, we generated a series of BS-U2 pairs in which a 5′-CAC-3′ motif was systematically placed at all positions in the BS-U2 duplex opposite a 5′-GG-3′ motif in the second copy U2 to produce a bulge whose nature remained constant while its position within the helix was varied (Fig. 2a). Primer extension analysis of total RNA isolated from strains carrying these BS-U2 pairs indicated that a bulged adenosine placed at any of three positions in the duplex could participate in first step splicing catalysis (Fig. 2b). If the canonical BS position is defined as 0, these positions are −1 (immediately upstream of the BS in the pre-mRNA), 0 and +1 (immediately downstream) (Fig. 2b, lanes 4–6). Bulged adenosines further up- or downstream showed no reactivity (Fig. 2b, lanes 2–3 and data not shown).
Figure 2. Systematic variation of bulge position indicates that at least three positions within the BS-U2 duplex are competent for first step catalysis.
(A) Schematic of reporter and U2 constructs used in (B).
(B) Primer extension with a 3′ exon primer indicates that −1, 0 and +1 BS bulge positions allow splicing catalysis. In this and subsequent figures, a band resulting from degradation of pre-mRNA is marked by an asterisk.
Grossly substituted BS-U2 pairs function in splicing: development of an orthogonal BS-U2 system
The BS-binding region of U2 snRNA is universally conserved in crown group eukaryotes, although U2s from highly divergent eukaryotes (e.g. kinetoplastids) have different sequences (Fig. 3a): minor changes both in this sequence and throughout the highly conserved 5′ end of U2 are generally lethal as a sole copy and can impair cell growth as a second copy (Parker et al., 1987). These negative effects, together with potentially variable expression levels of mutant U2s, preclude direct, quantitative comparison of branching efficiency from different BS-U2 duplex positions because a different U2 is required for each reporter gene. We sought to generate BS-U2 pairs to allow the fair comparison of different branch positions and nucleophiles within the context of a constant U2 sequence. These second copy U2 snRNAs were designed to be orthogonal to wt U2 – that is, they would interact with the reporter transcript but not with endogenous introns, while the reporter transcript in turn would interact only with its cognate U2.
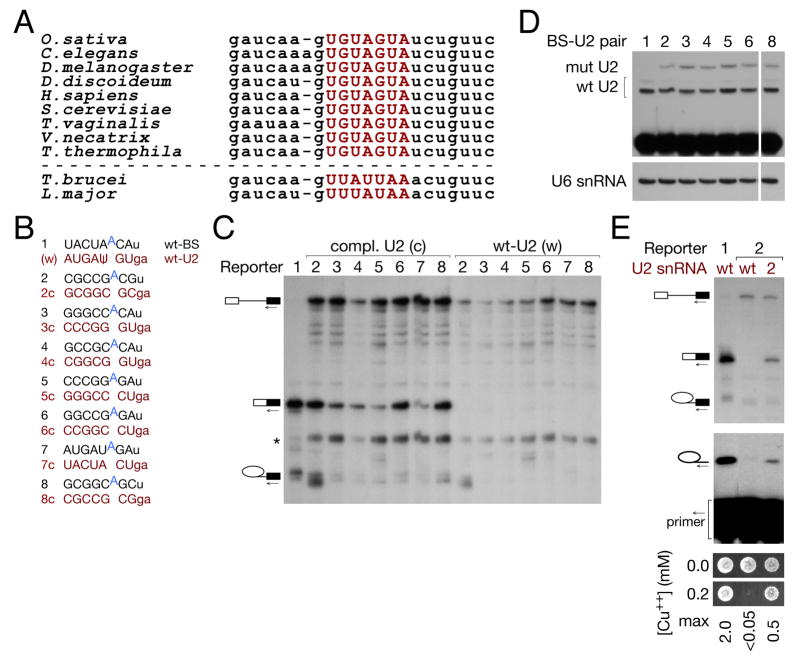
Figure 3. A wide variety of grossly substituted BS-U2 pairs are functional for splicing, and are orthogonal to the endogenous splicing machinery.
(A) Alignment of U2 snRNA sequences from (S. cerevisiae) positions 26 to 49 from diverse organisms: the BS-binding region, highlighted in red, is 100% conserved in crown group eukaryotes.
(B) Schematic of wt and grossly substituted reporter and U2 constructs used in (C).
(C) Primer extension with a 3′ exon primer indicates that grossly substituted branch sites allow splicing catalysis in the presence of their cognate U2 (c) [numbers refer to (B)], but not wt U2 (w).
(D) Primer extension analysis with ddTTP replacing dTTP to assay the expression of U2 snRNAs with wild-type and GC-substituted (orthogonal) BS-binding regions.
(E) Primer extension on total RNA from the indicated strains with a BS-proximal intronic primer (middle panel) and copper growth (lower panel) indicate that the expected branch nucleotide is used and that the mRNA resulting from splicing is functional, for representative grossly substituted BS-U2 duplex 2 from (B).
We generated several U2 snRNAs in which the entire BS-binding region was substituted, together with corresponding reporter transcripts for each (a selection is shown schematically in Fig. 3b). The U2s tested included ones in which all nucleotides were mutated to C or G, with purine/pyrimidine identity preserved (Fig. 3b, #2) or transverted (#8) at each position. Virtually all grossly substituted BS-U2 pairs could participate in both steps of splicing, although the efficiency of the first step was variable (Fig. 3c, lanes 2–8(c), and data not shown). As expected, the substituted reporter failed to splice in the absence of its corresponding U2 (Fig. 3c, lanes 2–8(w), and data not shown). For all BS-U2 pairs, we verified by high-resolution primer extension that the anticipated branch nucleophile was used, and confirmed splicing efficiency by copper growth (Fig. 3e and data not shown). Thus, despite its extreme conservation, the BS-U2 duplex is remarkably tolerant to substitution.
Using a primer extension assay in which the primer abutted the BS-binding sequence in U2, and ddTTP replaced dTTP, we were able to distinguish between, and thus assay the levels of, wt and most orthogonal U2 snRNAs (Fig. 3d); we presume these to reflect the level of U2 snRNP, as we expect unassembled U2 snRNA to be rapidly degraded. Orthogonal U2 expression, while variable, shows no correlation with splicing efficiency (Fig. 3c&d, quantitation in Fig. S2c), suggesting that U2 snRNA expression does not normally limit splicing in our system. The first step defect indicated by the accumulation of pre-mRNA (Fig. 3c) may instead reflect suboptimal packing of substituted BS-U2 duplexes with other spliceosome components. This is discussed more fully below in the context of Fig. 5.
Strains carrying certain mutant second copy U2 alleles exhibited slow growth (Fig. S2d&e). We do not know the basis of this growth defect, as it does not correlate with reduced or excessive expression of either wt or mutant U2 snRNA (Fig. S2b&e and data not shown). Deleterious effects arising from the generation of orthogonal BS-containing introns are also unlikely: we identified candidate pre-mRNAs in which new introns may be generated by our orthogonal U2s, but were unable to detect products of such new splicing events by RT-PCR (data not shown).
Establishment of nucleophile requirements for the first step of splicing
The orthogonal BS-U2 system minimises potential interaction between the mutant BS reporter and wt U2 snRNA, allowing systematic investigation of the effect of branch nucleotide identity, context and positioning in a constant U2 background. Our previous analysis (Fig. 2) had suggested that bulges from at least three positions within the BS-U2 duplex were competent for first step catalysis. Using reporters in which all BS flanking sequence was mutated to G/C with purine/pyrimidine identity either maintained or transverted (UACUAACA to CGCCGACG or GCGGCAGC, respectively – the zero position branch nucleotide is highlighted), we placed a bulged adenosine at all positions from −4 to +1 (Fig. 4a&c) and assayed splicing by primer extension and copper growth (Fig. 4b&d and data not shown). We observed relatively efficient branching from the 0 or −1 positions in both constructs (Fig. 4b&d), and additionally from the +1 position in the transverted duplex, but not from bulged nucleotides further upstream than the −1 position (Fig. 4d and data not shown). This confirms the positional flexibility of branching in the context of a constant U2 snRNA background – bulges at the −1 and +1 positions are expected to be separated by ~8 Å and 90° (Berglund et al., 2001).
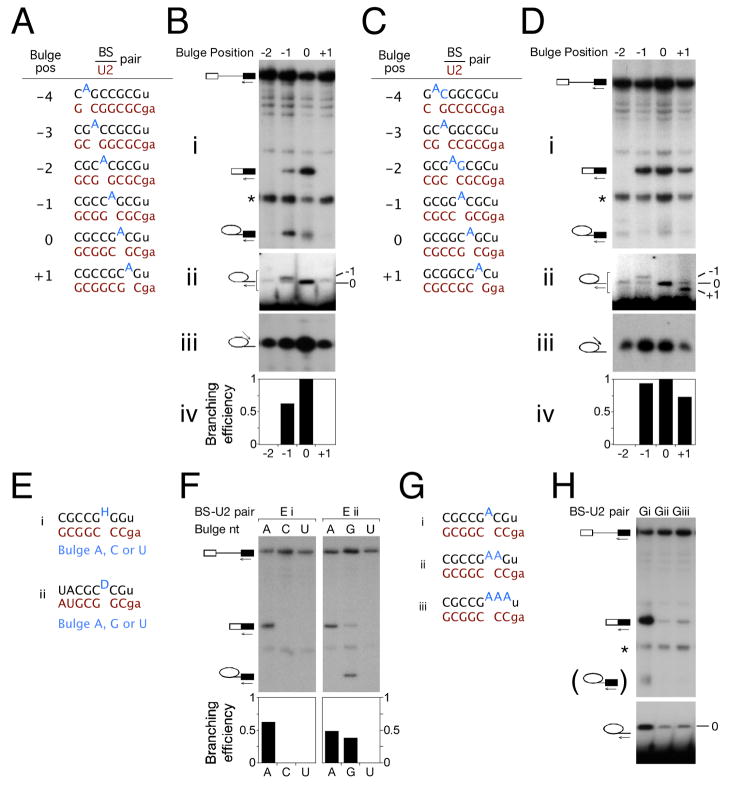
Figure 4. Positional flexibility and preferred identity of the branch nucleophile in orthogonal BS-U2 systems.
(A) Schematic of reporter and U2 constructs used in (B).
(B) Primer extension on total RNA from the indicated strains with 3′ exon (panel i), BS-proximal intronic (panel ii), and 5′SS-proximal intronic (panel iii) primers indicates that two bulge positions in this duplex are catalytically competent, that the expected branch nucleotide is used in each case, and that the location of the branch nucleotide in the BS-U2 duplex does not affect the position of 5′SS cleavage. Panel iv, quantitation of the 3′ exon primer extension in (i).
(C) Schematic of reporter and U2 constructs used in (D).
(D) Primer extension on total RNA from the indicated strains primers as in (B) indicates that three bulge positions in this duplex are catalytically competent, and, as in (B), that the branch nucleophile can be moved without changing the position of 5′SS cleavage.
(E) Schematic of reporter and U2 constructs used in (F).
(F) Primer extension using a 3′ exon primer indicates an intrinsic nucleotide preference at the branch site, with A>G>>C~U.
(G) Schematic of reporter and U2 constructs used in (H).
(H) Primer extension indicates that branching occurs from the upstream-most nucleotide in a run of consecutive single-stranded adenosines.
We believe the disparity between the two constructs to be related to base pair identity flanking the bulge. Base pairing at both flanking positions strongly favours splicing (Fig. S3a), and nucleotide identity within the pair impacts splicing efficiency (Fig. S3b). Consistent with flanking base pair identity underlying the observed difference in the use of the +1 position in the pseudo wild-type CG (CGCCGCAG) and transverted GC (GCGGCGAC) duplexes (both sequences shown with a highlighted +1 adenosine), splicing is more efficient for branch nucleotides preceded by a purine and followed by a pyrimidine than the converse (Fig. S3b). Thus, one would expect the +1 position to be relatively favoured in the context of the transverted GC duplex, and relatively disfavoured in the pseudo wild-type CG context. Moving the branch site A in the context of some orthogonal BS-U2 duplexes imposes a strong block on the second step of splicing (Fig. 4b and data not shown): we do not know the mechanistic basis for this defect.
Splicing, both in yeast (Vijayraghavan et al., 1986) and in higher eukaryotes (Hornig et al., 1986), is most efficient with an adenosine branch nucleophile: guanosine is mildly suboptimal and either pyrimidine nucleotide even more so. The effects of BS mutation, however, have been studied in a small number of genes, and in an otherwise wt BS-U2 context: alternative pairings for such duplexes can be envisaged in which an ‘incorrect’ nucleotide is bulged – i.e. the observed splicing defect may arise due to bulging nucleotides suboptimal in terms of identity or of position. We produced duplexes with only one pairing register, and a BS nucleotide flanked by a pair of either guanosine or cytosine nucleotides (Fig. 4e), to facilitate the predictable bulging of A/C/U or A/G/U, respectively. The splicing profile of these reporters mirrored that of BS mutants in a wt context, with guanosine supporting a fairly efficient first step and pyrimidines showing little to no branching (Fig. 4f). These data support an inherent preference for adenosine at the branch position, as previously suggested by work showing recognition of multiple groups on the adenosine nucleotide during spliceosome assembly and catalysis (Query et al., 1996).
A remaining question was the behaviour of branch sites with multiple consecutive bulged nucleotides. We constructed BS-U2 pairs containing one, two or three consecutive unpaired adenosines (Fig. 4g). The presence of additional unpaired adenosines impaired splicing of the reporter (Fig. 4h, upper panel), presumably due to the lack of base pairing downstream of the branch site adenosine (cf. Fig. S3). Splicing did, however, occur to detectable levels and high-resolution primer extension indicated that the upstream-most adenosine was invariably used as the branch nucleophile (Fig. 4h). These results recall data regarding branch site selection in the minor spliceosome (McConnell et al., 2002), and suggest a model whereby the most upstream unpaired adenosine in the BS-U2 or BS-U12 duplex is specified as the branch nucleophile.
Grossly substituted BS-U2 duplexes predominantly impair spliceosome assembly: inappropriately bulged nucleotides limit catalysis
The BS-U2 duplex, as a highly conserved element containing a first-step substrate, likely forms several tertiary interactions in the spliceosome core; our constructs, although stably base-paired, may be defective in some such interactions. Although virtually all grossly substituted BS-U2 duplexes supported splicing, first-step efficiency was invariably diminished relative to wt BS-U2 (Fig. 3c). Spliceosome assembly precedes branching catalysis, and pre-mRNA could accumulate due to a defect in either or both of these steps.
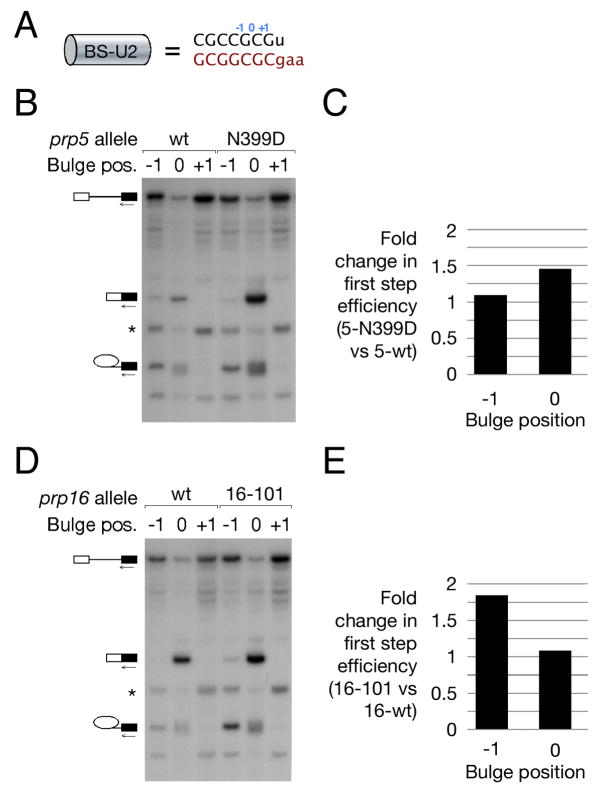
To clarify the nature of the defect shown by our constructs, we investigated their splicing in the context of mutant alleles of the spliceosomal ATPases Prp5 and Prp16. Prp5 has been proposed to monitor the stability of BS-U2 interaction during spliceosome assembly via kinetic competition between its ATPase activity and BS-U2 pairing – failure to establish stable interaction prior to ATP hydrolysis by Prp5 leads to pre-mRNA discard (Xu and Query, 2007). Mutant prp5 alleles, by relaxing the stability requirement for the nascent complex, thus facilitate spliceosome assembly on introns with suboptimal BS-U2 pairing and presumably also those in which a stable duplex is imperfectly packed. Prp16 enhances splicing fidelity by acting in competition with splicing catalysis – failure to complete first step catalysis prior to ATP hydrolysis by Prp16 leads to pre-mRNA discard (Burgess and Guthrie, 1993). A suboptimally packed BS-U2 duplex could inhibit first step catalysis by globally destabilising the first step conformation of the spliceosome.
For each ATPase we assayed the splicing efficiency, in a wt and a mutant strain, of substrates with a CGCCGAC branch site with an adenosine at either the −1, 0 (highlighted here), or +1 positions (Fig. 5a). The prp5 N399D allele (Xu and Query, 2007) stimulated splicing from the 0 position ~1.5-fold, yet had little effect on the splicing of a reporter with a −1 position adenosine (Fig. 5b&c). By contrast, the prp16-101 allele (Burgess and Guthrie, 1993) stimulated the splicing of the −1 position reporter 1.8-fold but had little effect on the 0 position reporter (Fig. 5d&e). Taken together, these observations suggest distinct limiting steps in the splicing pathway for the −1 and 0 position reporters. In this context, spliceosome assembly is impaired by duplex substitution: the mutant prp5 allele suppresses this assembly defect. In a substrate with an optimal 0 position bulge, enhanced assembly leads to enhanced splicing. With the nucleophile bulged from the −1 position, however, increased assembly does little to stimulate splicing because the overall efficiency of the reaction is limited at a later stage: in this case, the slowed exit from the first step conformation afforded by prp16 mutation strongly stimulates branching, suggesting that the position of the bulged nucleophile within the BS-U2 duplex is important for first step catalysis by the spliceosome.
Figure 5. Grossly substituted BS-U2 duplexes predominantly impair spliceosome assembly: inappropriately bulged nucleotides predominantly impair splicing catalysis.
(A) Schematic of the BS-U2 duplex used in this figure, with the positions of bulged adenosines indicated.
(B) Primer extension using a 3′ exon primer on total RNA from strains carrying reporters with adenosines bulged from the indicated positions in a grossly substituted BS-U2 duplex and either wt or mutant Prp5.
(C) Graph indicating the fold change in branching efficiency [calculated as (M+LI)/(P+M+LI)] at the −1 and 0 bulge positions as a result of Prp5 mutation.
(D, E) As (B, C), but comparing wt and mutant Prp16.
The preferred nucleophile position depends on distance from the U2 portion of U2/U6 helix Ia, and does not determine 5′SS selection
The analysis described above demonstrates the flexibility of branch site activation within a constantly positioned BS-U2 duplex, without addressing how movement of the duplex itself may affect branch site selection. U2 snRNA is involved in several intra- and intermolecular structures within the catalytic spliceosome (Burge et al., 1999). We therefore sought, again using a constant BS sequence from which an adenosine could be bulged at multiple positions, to investigate the impact on preferred bulge position of changing the distance between BS-U2 and these spliceosomal structures. We were relatively unconstrained in our choice of mutations, and made a large number of insertion and deletion mutants throughout the 5′ end of U2, including into either the stem or loop of the 5′ stem loop, up- and downstream of helix I, and upstream of helix III (Fig. S4). Several of these mutant U2s failed to show stable expression, presumably due to an inability to assemble into snRNPs (data not shown); all expressed U2s with insertions/deletions anywhere upstream of helix I showed at worst a mild splicing defect with no impact on branch nucleophile selection (data not shown). Of most interest were insertions predicted to alter the distance between helices Ia & III and the BS-U2 duplex, which lies between the U2 components of each (Fig. 1a), as this network of interactions may help to juxtapose key catalytic components and splicing substrates.
An all-CG branch site with the (0 position branch) sequence CGCCGAC shows branching from the −1 and 0 positions in the presence of its cognate U2 snRNA (Fig. 6a). Insertion of two nucleotides into U2 upstream of helix III, with accompanying deletion of two nucleotides downstream of helix Ia, predicted to shift the BS-U2 duplex towards helix Ia and away from -III, altered this splicing profile such that the −2 position was now highly active for branching and the 0 and −1 positions less favoured than in the original context (Fig. 6b). An analogous 1-nt insertion/deletion pair also shifted the observed branching preference towards upstream bulges (data not shown). Conversely, deleting one nucleotide upstream of helix III and inserting one downstream of helix Ia, predicted to shift BS-U2 away from helix Ia and towards -III, disfavoured the use of the −1 position while maintaining robust use of the 0 position BS (Fig. 6c). Collectively, these data suggest that branch position relative to either or both of these helices impacts the bulge positions within the BS-U2 duplex that are competent for first step catalysis.
Figure 6. The preferred position of the BS nucleophile within the BS-U2 duplex is partially determined by distance from the U2 component of U2/U6 helix Ia.
For each panel: (i) Simplified schematic of the spliceosome catalytic centre with helices Ia and III indicated; BS adenosines in blue are sized proportionally to their ability to participate in splicing catalysis based on quantitation (v) of the 3′ exon primer extension in (ii). Branch site and 5′SS use are assayed by primer extension in (iii) and (iv), respectively: the position of the branch nucleotide both within the BS-U2 duplex and relative to the U2 component of helix Ia does not impact the position of 5′SS cleavage. All panels use an all-CG BS-U2 duplex, indicated below the figure, in which purine-pyrimidine identity is maintained at each position relative to wild-type.
(A) Profile of −3 to +1 bulged reporters with CG-substituted but otherwise wt U2.
(B) Deletion of two nucleotides in U2 downstream of helix Ia, and insertion of two nucleotides upstream of helix III, predicted to move the BS-U2 duplex towards helix Ia and away from helix III, activates upstream bulge positions for catalysis.
(C) Insertion of one nucleotide in U2 downstream of helix Ia, and deletion of one nucleotide upstream of helix III, predicted to move the BS-U2 duplex away from helix Ia and towards helix III, represses the use of upstream bulge positions for catalysis.
(D) Deletion of two nucleotides in U2 downstream of helix Ia recapitulates the effect seen in (B), activating upstream bulge positions for catalysis.
(E) Deletion of one nucleotide in U2 upstream of helix III does not strongly impact the catalytic potential of different bulge positions.
To investigate the relative importance of BS distance from helices Ia and III, we produced constructs with single-site deletions either downstream of helix Ia or upstream of helix III. Deletion of two nucleotides downstream of helix Ia again activated branching from the −2 position (Fig. 6d), recapitulating the result of such a deletion in the context of an accompanying helix III-proximal insertion (cf. Fig. 6b&d). By contrast, a 1-nt deletion upstream of helix III produced a splicing profile similar to that of the original orthogonal U2 (cf. Fig. 6e&a). These data show that the distance from the branch nucleophile to the U2 component of helix Ia is an important determinant of nucleophile specification for catalysis. We note that this distance requirement may be relative either to helix Ia or to a rigid higher-order structure containing this helix. Despite the activation of upstream bulge positions for catalysis by helix Ia-proximal deletion in U2, branching remains most efficient from the zero position (Fig. 6b&d). We hypothesise that this position within the duplex remains the optimal one, and that the distance between the bulge and helix Ia is an (at least partially) independent factor in nucleophile determination.
To confirm the above analysis, we recapitulated the results in the context of the transverted GC duplex. Again, reducing the distance between the BS-U2 duplex and helix Ia led to relative activation of upstream bulge positions for first step splicing catalysis (Fig. S5 a&b). We note that the distribution of upstream branch use differs between the two constructs; as previously discussed, we believe that flanking base pair identity underlies this observed difference.
Although either or both of the branch nucleophile and the entire duplex from which it was bulged were moved in the above analysis (Fig. 4&6), and high-resolution primer extension confirmed the potential to use various different branch sites, the 5′SS remained unchanged and no activation of cryptic sites was observed in any of this work (Fig. 4b&d, panel iii, and panel iv of Fig. 6). If 5′SS positioning were dependent on BS positioning, or if the structures that contained the 5′SS and BS, rather than the reactive groups themselves, were positioned relative to one another, movement of the branch site nucleophile and/or BS-U2 duplex would be expected to result in an analogous movement of the 5′SS. Therefore, the observation that moving the BS or the duplex containing it fails to impact 5′SS selection suggests that the position of the BS nucleophile does not determine the position of the 5′SS electrophile for the first step of pre-mRNA splicing.
DISCUSSION
In vivo systems incontrovertibly provide the most valid environment for the study of biological processes. Unfortunately, however, in vivo mutational analyses are restricted by a requirement for cell viability. Orthogonal systems, in which a nonessential second copy enzyme exclusively targets its cognate substrate (which is in turn not a substrate for the wild-type cellular enzyme) offer the possibility of combining the power of the in vivo setting with the mutational freedom of in vitro analysis. Essential, highly conserved base pairing interactions, when not complicated by competing pairings, are ideally suited to the generation of such systems: an approach based on the replacement of the Shine-Dalgarno interaction has been used to produce orthogonal ribosomes in E. coli (Rackham and Chin, 2005). Here we describe a system for the study of pre-mRNA splicing, based on the analogously conserved interaction between the intron branch site and its cognate sequence in U2 snRNA. We use a variety of BS-U2 pairs to investigate the flexibility of nucleophile selection for the first step of splicing in yeast. Our results establish conclusively that bulging of the nucleophile is required during the catalytic phase of splicing, confirm the importance of branch nucleophile identity and location, and suggest a model whereby the first unpaired nucleotide in the BS-U2 duplex is specified as the nucleophile. Additionally, our data indicate a high degree of flexibility in the active site of the yeast spliceosome, and strongly argue that 5′SS and branch site specification within the catalytic centre are independent. Finally, they extend the evidence for similarity between the spliceosome and group II self-splicing introns to include a common mechanism of first step nucleophile specification and positioning.
Flexibility of first step nucleophile specification
The yeast spliceosome has stringent substrate requirements for assembly and catalysis; although nucleophile bulging is required, outside of this constraint, there is flexibility with respect to branch nucleophile selection and positioning for splicing catalysis. Because the spliceosome is robust, several deleterious changes may be required to generate a visible effect on splicing efficiency in vivo, particularly if these changes predominantly affect steps that are not close to being rate-limiting. In the generation of an orthogonal BS-U2 system tailored for the investigation of nucleophile specification we show that, although BS-U2 pairing is required for splicing, the sequence of the BS and its cognate region of U2 are much less important. Complete replacement of the BS-U2 duplex results in both spliceosome assembly and first step catalysis being partially limiting; this allows a systematic analysis of the determinants of branch nucleophile use. We identify three major determinants of branching efficiency: the identity of the branch nucleotide (and to a lesser extent the identity of flanking base pairs), the position of the bulged branch nucleotide within the BS-U2 duplex and, independently, its position relative to the U2 component of U2/U6 helix Ia.
In a case in which multiple unpaired adenosines are present with little potential for downstream pairing to U2, branching occurs at the first unpaired A (Fig. 4h). This suggests that, although appropriate positioning of a bulge relative to the catalytic core provides the strongest determinant for efficient branching, flexibility at the edge of a helix can also contribute to nucleophile selection. The distribution of mapped branch positions in the U12 spliceosome (McConnell et al., 2002) obeys similar rules, with the strong features being a well-defined BS-U12 duplex containing an unpaired A nucleophile positioned similarly to that in the BS-U2 duplex.
The branch site adenosine of a pre-mRNA can be bulged from almost any Watson-Crick duplex, as indicated by the observation that virtually all BS-U2 pairs we tested were functional. The extreme conservation of the BS-binding region of U2 (the GUA sequence opposite the bulged branch nucleophile is conserved between U2 and U12) is therefore not explained by a dependence of spliceosome function on a specific sequence. The splicing defect of highly substituted BS-U2 pairs with an optimally positioned bulge appears to be due predominantly to impaired spliceosome assembly, as suggested by partial suppression by prp5 alleles, and to a lesser impact on splicing catalysis (Fig. 5). In contrast, BS-U2 pairs with a sub-optimally positioned bulge are catalytically defective, indicated by strong improvement by ATPase-defective prp16 alleles. Thus, assembly is primarily affected by alteration of the BS-U2 duplex, whereas catalysis is sensitive to altered positioning of the nucleophile.
Independent positioning of 5′SS and branch site
If higher-order structures containing the branch nucleophile and 5′SS electrophile for the first step of splicing were fixed relative to one another, moving the branch nucleophile should alter the position of 5′SS cleavage. We observe, however, that moving the branch nucleophile relative to the BS-U2 duplex or to helix Ia does not alter 5′SS selection, indicating that this is not governed by BS selection. This observation is in agreement with the ability of higher eukaryotic spliceosomes to cleave the same 5′SS at the same position using more than one possible branch nucleotide (Query et al., 1994), and highlights the commonality of substrate selection in the S. cerevisiae and metazoan spliceosomes.
The position of branching within the BS-U2 duplex can impact the second step of splicing. The mechanistic basis of the defect sometimes observed with non-zero branch positions and/or helix Ia-proximal insertions and deletions (e.g. Fig. 6) is unclear, especially as branching from non-zero positions (e.g. Fig. 4d) and insertions into U2 (e.g. Fig. 1d) do not in themselves invariably lead to second step defects. Crosslinks have been observed between the 3′SS and helix I-proximal nucleotides in U2 (Newman et al., 1995), and insertions around the U2 component of helix Ia block the second step in vitro (McGrail et al., 2006 and references therein). Thus, despite the apparent lack of requirement for BS-U2 base pairing for exon ligation by the spliceosome (Smith et al., 2007), U2 must play a role in this step. The formation of the branch structure likely eliminates the positional and rotational flexibility of the BS-U2 duplex; this in turn would influence the repositioning of the branch structure required for entry into the second step. A BS-U2 duplex containing an aberrant branch may be sterically or torsionally suboptimal for repositioning if this is required, or the putative disruption of the BS-U2 duplex may be inhibited. Although we do not address it in this work, the role of U2 in the second catalytic step merits further attention, and our orthogonal system should prove to be a valuable tool for such analysis.
Evolutionary considerations
The spliceosome and group II self-splicing introns share a number of features that suggest a common evolutionary ancestor. Support for significant overlap of catalytic mechanism between the two systems has recently been provided by the crystal structure of a spliced group II intron: in this structure, motifs that are conserved between group II introns and U6 snRNA, in which they are close to helix Ia, coordinate magnesium ions (Toor et al., 2008). Our data extend this catalytic similarity by suggesting a conserved mechanism of branch nucleophile selection and positioning. A branch site adenosine incapable of bulging cannot efficiently participate in first step branching catalysis in either system (Fig. 1; Chu et al., 1998), and the identity of flanking base pairs influences branching efficiency (Fig. S3; cf. Fig. 6&S5; Chu et al., 2001). In addition, the use of cryptic branch sites in group II introns can be observed upon alteration of the distance between domains 5 and 6, the equivalents of the U6 ISL and BS-U2 duplex, respectively (Chu et al., 2001). In our BS-U2 system, the use of upstream bulges is favoured by deletions predicted to decrease the distance between the U6 ISL and the BS-U2 duplex, and disfavoured by insertions predicted to increase it (Fig. 6). Thus, our data support a spatially similar organisation of conserved elements in group II introns and the spliceosome.
We have built on previous genetic analyses of the spliceosome, employing compensatory base pairs and second copy mutants; these approaches have identified and validated many RNA-RNA interactions in both the yeast and human spliceosomes (e.g. Datta and Weiner, 1991; Madhani and Guthrie, 1992; Shukla and Padgett, 2002; Sun and Manley, 1995). However, our system is distinguished by its conception to address a specific mechanistic question regarding splicing catalysis, rather than to identify/validate an intermolecular interaction. In addition, our results show the acceptability of gross sequence substitutions within an extremely conserved element in the core of the spliceosome. We note that the minor spliceosome may represent a natural example of an orthogonal BS-U2 system, specific for a subset of introns. This system relies on several dedicated interactions (e.g., U11-5′SS, U12-BS, U12-U6atac), suggesting that analogous extensions to our system should be feasible. We expect that other highly conserved interacting partners will be similarly amenable to substitution; this will allow the generation and in vivo analysis of spliceosomes with more and/or different components separate from the endogenous splicing machinery. Further such spliceosomes, both derived from that described here and entirely novel, will be invaluable in the continuing analysis of the catalytic phase of splicing.
EXPERIMENTAL PROCEDURES
Strains and reporter plasmids
Details of S. cerevisiae strains used in this analysis can be found in the supplemental methods section.
Primer extension and copper growth
Primer extensions were carried out as described (Siatecka et al., 1999). Extension products were separated in 7%, 10%, 20% or 25% polyacrylamide/8M urea gels and visualized by autoradiography. Sequences of primers used can be found in the supplemental methods section. Branching efficiency was calculated from YAC6 primer extensions as (M+LI)/(M+LI+P), where M, LI and P represent the signals due to mRNA, lariat intermediate and pre-mRNA, respectively. Copper growth assays were performed as described (Lesser and Guthrie, 1993).
Extract preparation and in vitro splicing
Yeast whole cell extracts were prepared and in vitro splicing carried out as described (Cheng and Abelson, 1987) using 1 fmol of body-labelled transcript. Aliquots of in vitro splicing reactions were removed at 10 and 20 min time points and either separated in native 3.5% acrylamide/agarose gels or stopped by phenol/chloroform extraction, ethanol precipitated, and separated in 6% acrylamide/8M urea gels.
Acknowledgments
We thank Bibhusita Pani and other members of the Konarska and Query labs for help and advice, and we thank John Abelson for communicating results prior to publication. This work was supported by NIH grants GM49044 to M.M.K., GM57829 to C.C.Q, and by a Cancer Center Support (core) grant from the NCI to the Albert Einstein College of Medicine. C.C.Q. is a scholar of the Irma T. Hirschl Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Rosbash M, Schultz SC. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. RNA. 2001;7:682–691. doi: 10.1017/s1355838201002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- Cheng SC, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Chu VT, Liu Q, Podar M, Perlman PS, Pyle AM. More than one way to splice an RNA: branching without a bulge and splicing without branching in group II introns. RNA. 1998;4:1186–1202. doi: 10.1017/s1355838298980724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Adamidi C, Liu Q, Perlman PS, Pyle AM. Control of branch-site choice by a group II intron. EMBO J. 2001;20:6866–6876. doi: 10.1093/emboj/20.23.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta B, Weiner AM. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991;352:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- Hornig H, Aebi M, Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986;324:589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Jacquier A, Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A. 1986;83:5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in S. cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- McConnell TS, Cho SJ, Frilander MJ, Steitz JA. Branchpoint selection in the splicing of U12-dependent introns in vitro. RNA. 2002;8:579–586. doi: 10.1017/s1355838202028029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail JC, Tatum EM, O’Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9:958–965. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, Sharp PA. Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Moore MJ, Sharp PA. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- Query CC, Strobel SA, Sharp PA. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell. 2002;9:1145–1150. doi: 10.1016/s1097-2765(02)00505-1. [DOI] [PubMed] [Google Scholar]
- Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing. Mol Cell. 2007;26:883–890. doi: 10.1016/j.molcel.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Manley JL. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in S. cerevisiae. J Biol Chem. 2005;280:6655–6662. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]