Abstract
SUMMARY
The Aurora-A kinase gene is amplified in a subset of human tumors and in radiation-induced lymphomas from p53 heterozygous mice. Normal tissues from p53−/− mice have increased Aurora-A protein levels, but lymphomas from these mice exhibit heterozygous deletions of Aurora-A and/or reduced protein expression. A similar correlation between low p53 levels and Aurora-A gene deletions and expression is found in human breast cancer cell lines. In vitro studies using mouse embryo fibroblasts demonstrate that inhibition of Aurora-A can have either positive or negative effects on cell growth as a function of p53 status. These data have implications for the design of approaches to targeted cancer therapy involving the crosstalk between Aurora-A kinase and p53 pathways.
SIGNIFICANCE
Inhibitors of the Aurora-A kinase have been developed by several pharmaceutical companies as potential cancer therapeutics, based on the known oncogenic activities of this gene in induction of genetic instability and cell transformation. The data presented here demonstrate that inhibition of Aurora-A can in fact provide a growth advantage to cells that have suffered prior loss of p53. In addition, inhibition of Aurora-A may lead to premature loss of wild-type p53 function. We conclude that there is an important functional interaction in vivo between p53 and Aurora kinase, and that this network should be taken into account in the design of clinical trials of these agents for cancer therapy.
INTRODUCTION
Recent successes in the development of “targeted” therapeutic drugs such as the BCR-ABL kinase inhibitor Gleevec (Druker, 2004) and the EGFR inhibitors Iressa and Tarceva (Paez et al., 2004; Lynch et al., 2004) have stimulated interest in the extension of these approaches to other cancer targets, in particular other members of the kinase family. One of the challenges that will have to be faced during development of these approaches is acquisition of drug resistance by treated tumor cells, either through additional mutations in the target gene (Pao et al., 2005; Kobayashi et al., 2005; Sawyers, 2005) or by rewiring of signaling pathways that allows escape from the effects of target inhibition. Analyses of signaling in bacteria, yeast, and mammalian cells have demonstrated the existence of complex networks that can be rapidly rewired in response to external stimuli (Almaas et al., 2004; Wuchty et al., 2003; Luscombe et al., 2004). These complex networks offer many possibilities for tumor cells to circumvent the negative effects of targeted inhibitors without necessarily mutating the target gene itself. Increased knowledge of crosstalk between signaling pathways can therefore help in the design of therapeutic strategies, as well as in the selection of patients to be entered into clinical trials.
The Aurora-A kinase gene has attracted a great deal of attention as a potential therapeutic target because of its identification as an oncogene (Bischoff et al., 1998). Common germline polymorphisms in this gene have also been shown to confer increased risk of development of a number of tumor types (Ewart-Toland et al., 2003, 2005). Aurora-A kinase has been implicated in the control of chromosome segregation during mitosis and has been found frequently amplified in many human cancers (Bischoff et al., 1998; Sen et al., 1997; Katayama et al., 2003; Meraldi et al. 2004). Elevated expression of Aurora-A was also reported to correlate with genomic instability and clinically aggressive disease (Zhou et al., 1998; Tanaka et al., 1999). Aurora-A kinase is involved at multiple levels in interactions with the p53 pathway, suggesting that these proteins form part of an integrated functional network. Aurora-A interferes with p53 suppressor function by at least two mechanisms: first, in vitro studies have shown that Aurora-A kinase phosphorylates p53 at Ser315, facilitating MDM2-mediated degradation of p53 in cancer cell lines (Katayama et al., 2004); second, Aurora-A also phosphorylates p53 at Ser215 and inactivates its transcriptional activity (Liu et al., 2004). On the other hand, p53 interacts with Aurora-A to suppress its oncogenic activity in a transactivation-independent manner (Chen et al., 2002). Taken together, these data suggest that deregulation of the functional balance between Aurora-A and p53 might trigger checkpoint abnormalities, chromosome instability, and carcinogenesis. However, the in vivo functional relationship between these pathways in tumor development has not been comprehensively investigated.
We have used a genetic approach to study the reciprocal interactions between Aurora-A kinase and p53 during development of radiation-induced mouse lymphomas. Wild-type p53 protein is induced after exposure to γ-radiation and is essential for an efficient response to DNA damage and repair of induced lesions (for review, see Fei and El-Deiry, 2003). Germline deficiency of p53 has been reported to lead to increased chromosomal abnormalities (Cross et al., 1995; Fukasawa et al., 1996) and susceptibility to development of a spectrum of tumors, the most frequent being lymphoma (Donehower et al., 1992). Tumorigenesis in p53+/− mice can be accelerated by exposure to a single dose of γ radiation (Kemp et al., 1994). Analysis of genetic instability using microsatellite imbalance as well as whole-genome comparative genomic hybridization (CGH) arrays demonstrated, in addition to extreme genetic instability in an important subset of these tumors, the existence of p53-dependent genetic alterations at several loci (Mao et al., 2004, 2005). We have now applied CGH array analysis to tumors derived from p53 null mice and show that the latter have, remarkably, rather stable genomes compared to tumors from equivalent p53 heterozygous mice. Among the loci that clearly differed between tumors from heterozygous and null mice was the Aurora-A kinase locus on distal mouse chromosome 2. This locus was found to be frequently gained or amplified in tumors from p53+/− mice, but showed deletions in a substantial proportion of tumors from the p53−/− mice. These results demonstrate the existence of a complex reciprocal relationship between Aurora-A and p53 in vivo, by which inhibition of Aurora-A may act positively or negatively during tumor development in a p53-dependent manner.
RESULTS
Genetic Signatures in Lymphomas from p53+/− and p53−/− Mice
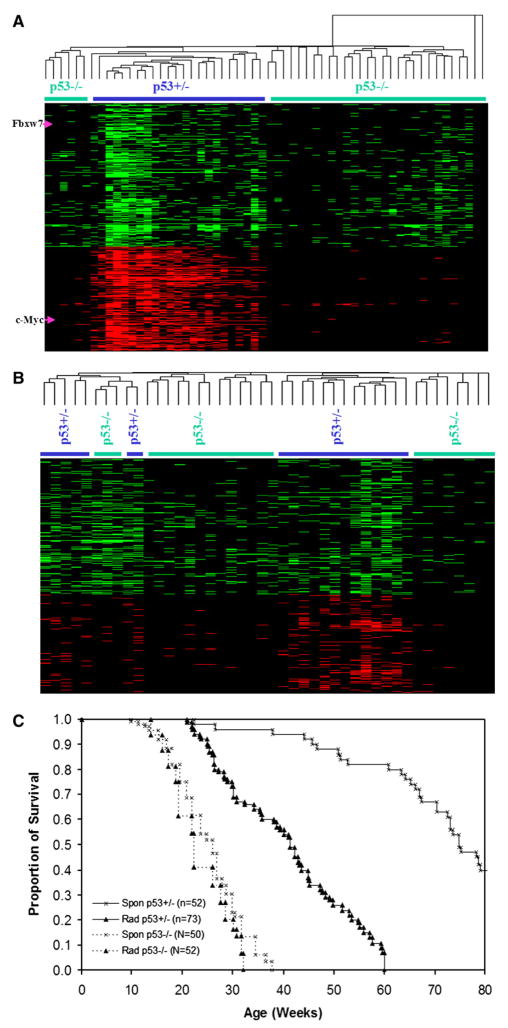
We carried out whole-genome bacterial artificial chromosome (BAC) CGH array analysis to compare the patterns of genomic instability in radiation-induced tumors from p53+/− and p53−/− mice. In an attempt to recognize global patterns of genetic alterations in these tumors, we carried out unsupervised cluster analysis of the whole-genome BAC profiles generated from these tumors (Figure 1A). For this purpose, the genome was divided into bins of variable size based on the gain/loss frequency of all samples, and tumors showing gene copy number losses within a particular bin were denoted in green, while those regions showing gains were represented in red (Figure 1). Unsupervised cluster analysis showed that, on average, there were many more genetic changes in tumors from irradiated p53+/− mice than in those from p53−/− mice. Detailed inspection of these patterns identified a large number of chromosomal changes that were specific to tumors from mice with at least one functional p53 allele (Figure 1A). For example, gain of the c-Myc locus and loss of Fbxw7 (Mao et al., 2004) were found only in tumors from p53+/− mice. These results obtained from genome-wide BAC CGH array analysis were consistent with data obtained by microsatellite analysis of allelic imbalances in tumors, which also demonstrated the relative stability of tumors from mice with complete germline deletions of p53 (Mao et al., 2004).
Figure 1. Unsupervised Cluster Analysis of Genetic Changes in Thymic Lympho-mas from p53+/− Compared to p53−/− Mice.
p53 status of the mice is denoted by the colored bars.
(A) Comparison of genetic changes in radiation-induced thymic lymphomas from p53+/− and p53−/− mice. The locations of the Fbwx7 and c-Myc loci are denoted by arrows.
(B) Comparison of genetic changes in spontaneous thymic lymphomas from p53+/− and p53−/− mice.
(C) Lack of effect of radiation in determination of tumor latency in p53−/− mice. The graph shows the proportion of surviving mice at different time points after birth. There is no statistically significant difference between mice in both groups (tested by Kaplan-Meier method using SPSS package). p53+/− mice are very sensitive to radiation, and a single dose of 4 Gy decreases their average lifespan by about 30 weeks.
The cluster analyses were carried out using Cluster V3.0 (http://rana.stanford.edu/software).
We next compared the spectrum of changes in spontaneously arising, as opposed to radiation-induced, tumors from both p53+/− and p53−/− mice (Figure 1B). Overall, the spontaneous tumors derived from p53+/− mice, although showing less heterogeneity and instability than in the corresponding tumors that arose after radiation exposure (Figure 1A and Mao et al., 2005), had higher levels of gene copy number gains and losses than equivalent tumors from the p53 null animals (Figure 1B). Tumors from p53−/− mice tended to cluster together, as did those from p53+/− mice, with a few exceptions (Figure 1B). We conclude that the difference in genetic instability between these tumors is an intrinsic consequence of p53 status and is not simply due to a difference in response to radiation.
In contrast to the dramatic increase in genetic instability evident in tumors from irradiated p53+/− mice, irradiation of p53−/− mice at the same dose level did not lead to any substantial change in genomic instability patterns (Figure S1; see the Supplemental Data available with this article online). This is in agreement with reports indicating that 4 Gy γ radiation of p53−/− mice at the age of 5 weeks did not significantly influence tumor latency, while similar treatment of p53+/− mice dramatically decreased tumor latency (Figure 1C; Kemp et al., 1994). The reasons for the apparent immunity to the effects of radiation in cells devoid of p53 are unclear, especially in view of the existence of p53-independent checkpoints that can lead to apoptosis (Frenkel et al., 1999).
p53-Dependent Gene Copy Gains or Losses at the Aurora-A Locus
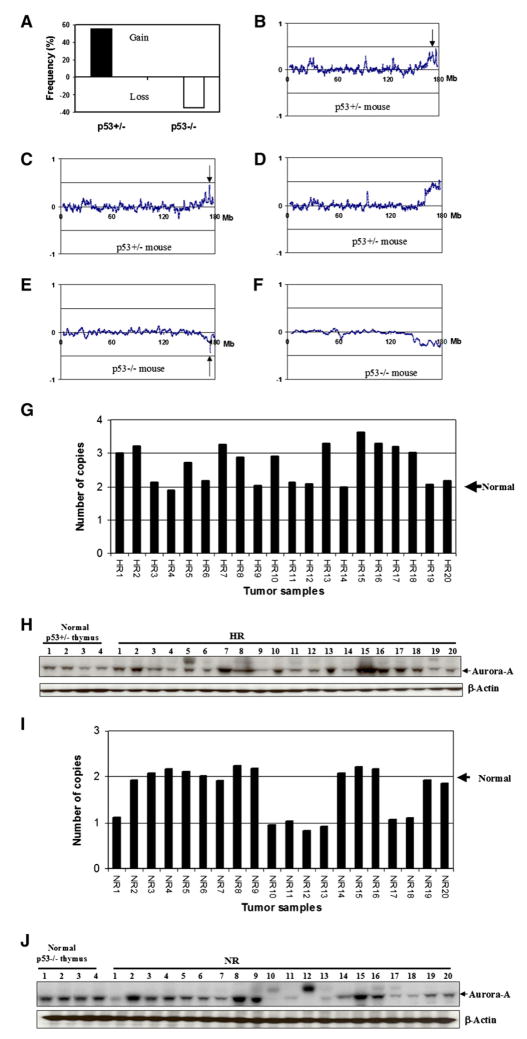
The Aurora-A gene is frequently gained or amplified in tumors from a wide variety of human tissues, including the colon, lung, pancreas, and breast (Sen et al., 1997; Bischoff et al., 1998; Katayama et al., 2003). In agreement with these results, mouse lymphomas from p53+/− mice showed, in over 55% cases, gains on distal chromosome 2 in the region containing Aurora-A (Figure 2A). Detailed dissection of the amplicon on chromosome 2 in p53+/− tumors showed that there is a complex pattern of amplification (Figures 2B–2D), an observation which mimics the situation seen in several human cancers (Albertson et al., 2000). One of these regions includes the Aurora-A gene. In some cases, the BAC containing the Aurora-A gene (BAC ID RP23-414K7) was the single most highly amplified sequence in the region (e.g., Figure 2C). To verify these genetic imbalances we carried out quantitative Taq-Man analysis using Aurora-A-specific probes which confirmed the data found by the BAC CGH array studies (Figure 2G). Approximately half of the samples had at least three copies of the gene, while the remainder appeared to be diploid at this locus. In complete contrast to the situation seen in tumors from p53+/− mice, similar tumors from the p53−/− animals not only showed no examples of amplification or gain, but in fact gene deletions were observed in 35% of the lymphomas (Figure 2A). In some cases, the deletions were quite specific, involving a minimal region of only 200 kb containing the Aurora-A gene (e.g., Figure 2E). Seven of 20 tumors from p53 null mice analyzed by TaqMan quantitation of Aurora-A gene levels showed heterozygous deletions, having only the equivalent of one copy (Figure 2I). These data, taken together, indicate that Aurora-A can be a target for either amplification or deletion, dependent on p53 status.
Figure 2. Gene Copy Number Changes at the Aurora-A Locus in Tumors.
(A) Frequency of somatic gain/loss of Aurora-A in lymphomas as a function of p53 status.
(B–D) Representative BAC array profiles of chromosome 2 in tumors from p53+/− mice. The arrows indicate the position of the Aurora-A gene, which is frequently gained in tumors from p53+/− mice.
(E and F) Loss of the Aurora-A gene in tumors from p53−/− mice.
(G) Detection of Aurora-A genetic alterations in radiation-induced tumors from p53 heterozygous mice (HR) by TaqMan assay.
(H) Detection of Aurora-A protein levels in radiation-induced tumors from p53 heterozygous mice (HR) by western blotting. The first four lanes show levels of Aurora-A in normal thymus from unirradiated p53+/− mice. Tumors in lanes labeled HR 1–20 correspond to those labeled HR1–HR20 in (G).
(I) Detection of Aurora-A genetic alterations in radiation-induced tumors from p53 null mice (NR) by TaqMan assay.
(J) Detection of Aurora-A protein levels in radiation-induced tumors from p53 null mice (NR) by western blotting. The first four lanes show levels of Aurora-A in normal thymus from unirradiated p53−/− mice. These are consistently higher than in corresponding p53+/− mice (H). Tumors in lanes labeled NR 1–20 correspond to those labeled NR1–NR20 in (I).
Correlation of Aurora-A Protein Levels and Gene Copy Number
The effects of the genetic imbalances at the Aurora-A locus on protein levels in the tumors analyzed by CGH were investigated by western blotting of tumor extracts. The results shown in Figures 2H and 2J demonstrate that there is a strong correlation between gene copy numbers as determined by quantitative TaqMan analysis and protein levels both in p53+/− and p53−/− mouse tumors. The numbers under Figures 2H and 2J correspond to the tumor numbers in Figures 2G and 2I, respectively. Tumors from p53+/− mice that have increased Aurora-A gene copy number have relatively high protein levels (compare Figures 2G and 2H), whereas those with deletions have, on average, lower protein levels than p53−/− mouse tumors with two copies of the gene (compare Figures 2I and 2J). Some tumors from p53−/− mice that did not show genetic loss of Aurora-A nevertheless exhibited low levels of protein (e.g., tumor numbers NR14, NR19, and NR20 in Figure 2J), suggesting that different mechanisms can lead to downregulation of Aurora-A protein levels in p53 null tumors.
One consistent observation was that normal thymus tissue from p53−/− mice had higher protein levels of Aurora-A than the equivalent tissue from p53+/− mice (compare Figure 2H, lanes 1–4, with Figure 2J, lanes 1–4). Similar observations were made with other tissues, such as spleen, from the same animals (data not shown). Interestingly, although tumors from p53 heterozygous and null mice show diametrically opposed genetic alterations leading to gains or losses at the Aurora-A locus, the protein levels, overall, lie within a similar range (compare Figures 2H and 2J). These data suggest that there may be an optimal level of Aurora-A protein that is compatible with rapid cell growth as required for tumor progression. While this level is generally attained in most tumors containing wild-type p53 by gene copy number gains, tumors that develop from cells with no functional p53, and consequently higher starting levels of endogenous Aurora-A protein (Figure 2J), reduce the amount of Aurora-A protein to “acceptable” levels by mechanisms that commonly involve gene deletion. The data suggest that in rapidly dividing cells, levels of Aurora-A have to be within a specific window compatible with ordered progression of mitosis. Cells that are p53 wild-type or null achieve this goal in very different ways. The data in Figures 2H and 2J also show however that some tumors, both from p53 heterozygous and null mice (e.g., HR9, NR10, and NR12), have only very low levels of Aurora-A and presumably have compensated for this loss by upregulating alternative mechanisms, the nature of which remain unclear. In order to exclude the possibility that activating mutations in Aurora-A might influence the patterns seen, the complete coding region of Aurora-A was sequenced in a series of 40 tumors from p53+/− or p53−/− mice. This study did not reveal the presence of any coding sequence alterations that might influence Aurora-A activity.
Reduced Levels of AURORA-A in a Subset of Human Breast Cancers
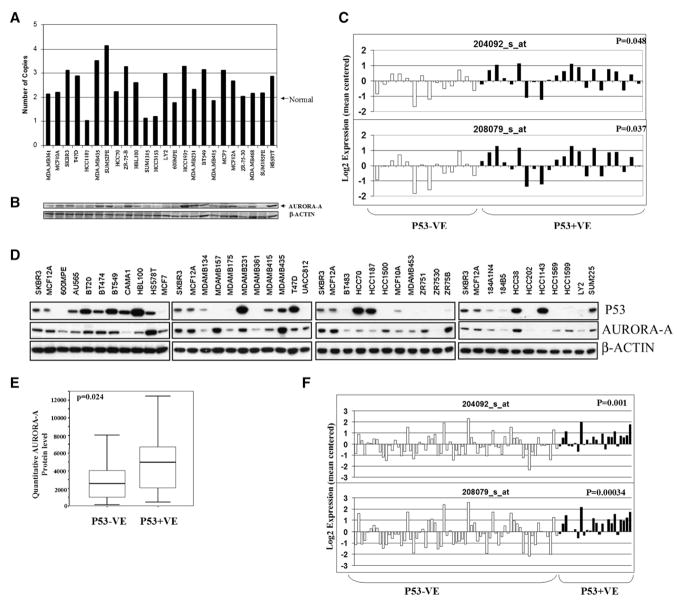
In order to determine whether any evidence for deletion or reduced expression of Aurora-A can be found in human cancers, we measured gene copy number of AURORA-A in a series of breast cancer cell lines by quantitative Taq-Man analysis using an AURORA-A-specific probe. As shown in Figure 3A, about 50% of breast cancer cell lines had increased copy number of the AURORA-A gene, in agreement with what has been previously reported for breast tumors (Sen et al., 1997). However, three cell lines showed the loss of one copy of the Aurora-A gene, similar to the situation seen in tumors from p53−/− mice. All three tumors showing reduced copy number also had low levels of AURORA-A protein, as did some tumors with normal gene copy number (e.g., HCC70, 600MPE, ZR-75-30, and SUM185PE; Figure 3B). We conclude that some human breast tumors exhibit reduced gene copy number and protein levels of Aurora-A, similar to the lymphomas from p53−/− mice. Clearly, these human tumors cannot have developed from p53−/− normal cells, but it is possible that mutations leading to loss of p53 function occurred relatively early in the tumorigenesis process, exerting selective pressure for loss rather than gain of Aurora-A. The complete coding region of AURORA-A was sequenced in all breast cancer lines shown in Figure 3A. As was also observed for the mouse tumors, no mutations were detected that might influence the conclusions from these experiments.
Figure 3. Alteration of Aurora-A Gene Copy Number and Expression in Human Breast Cancer Cell Lines and Primary Tumors.
(A) Detection of Aurora-A gene copy number changes in human breast cancer cell lines by TaqMan assay using Aurora-A-specific probes. The names of the cell lines used are shown below the TaqMan data.
(B) Detection of Aurora-A protein levels in same set of human breast cancer cell lines by western blotting. The order of the cell lines used from left to right is the same as in (A).
(C) Comparison of Aurora-A RNA levels between p53+VE (on the right, dark bars) and p53−VE (on the left, open bars) human breast cancer cell lines. Aurora-A RNA levels were derived from Affymetrix array data using two separate probes (204092_s_at and 208709_s_at). p53 protein level was determined by western blotting. p53+VE denotes tumors with detectable p53 protein by western blotting, whereas p53−VE denotes tumors with no p53 protein. A low Aurora-A RNA level was significantly correlated with a low p53 protein level.
(D) Protein levels of p53 and AURORA-A in human breast cancer cell lines determined by western blotting.
(E) Comparison of quantitated Aurora-A protein level between p53+VE and p53−VE human breast cancer cell lines. Low Aurora-A protein level was significantly correlated with low p53 protein level. The thick line represents the median, whereas thin lines are ranges, i.e., minimum and maximum number. The box shows 95% confident intervals of median.
(F) Comparison of Aurora-A RNA levels between p53+VE and p53−VE human primary breast tumors. Aurora-A RNA levels were derived from Affymetrix microarray data. P53 protein levels were determined by immunobiochemical staining. For details, see Experimental Procedures.
Correlation between P53 and AURORA-A Expression in Breast Cancers
Since it has been shown that genetic changes at the Aurora-A locus in mouse lymphomas were p53 dependent, we examined the relationship between the levels of P53 and AURORA-A in human breast cancer cell lines by Affymetrix microarray analysis and western blotting. Genome-wide expression array analysis using the Affymetrix platform has been carried out on a large panel of human breast cancer cell lines (Neve et al., 2006). Inspection of these array data showed that there was a statistically significant correlation between the RNA levels of AURORA-A and protein levels of p53 (Figure 3C). Tumor cell lines were separated into two groups based on the presence or absence of p53 detectable by western blotting. The association between p53 protein status and Aurora-A RNA levels was statistically significant using two independent probe sets for Aurora-A (p = 0.037 and p = 0.048; Figure 3C). We also found a significant association between AURORA-A and P53 at the protein level. Western blotting using AURORA-A-specific antibodies demonstrated a significant correlation between RNA expression and protein levels (for probe 204092_s_at: R = 0.72, p < 0.001; for probe 208079_s_at: R = 0.67, p < 0.001). The data showed that p53-positive tumors, as defined in the Experimental Procedures, had on average higher levels of Aurora-A than tumors with low levels of p53 (p = 0.024; Figures 3D and 3E). Finally, we searched for further confirmation of these observations in an independent set of Affymetrix RNA expression array data on primary breast cancers (Chin et al., 2006). Although western blots of these tumors for p53 were not available, there was a highly significant association between tumors designated as p53 positive or negative by immunohistochemistry and RNA levels of AURORA-A (Figure 3F). In spite of the complexity of genetic changes in human tumors, as opposed to the controlled situation investigated in the mouse, we conclude that levels of p53 and AURORA-A are significantly correlated in human breast cancer cell lines and primary tumors.
Aurora-A Downregulation in p53 Heterozygous and Null MEFs
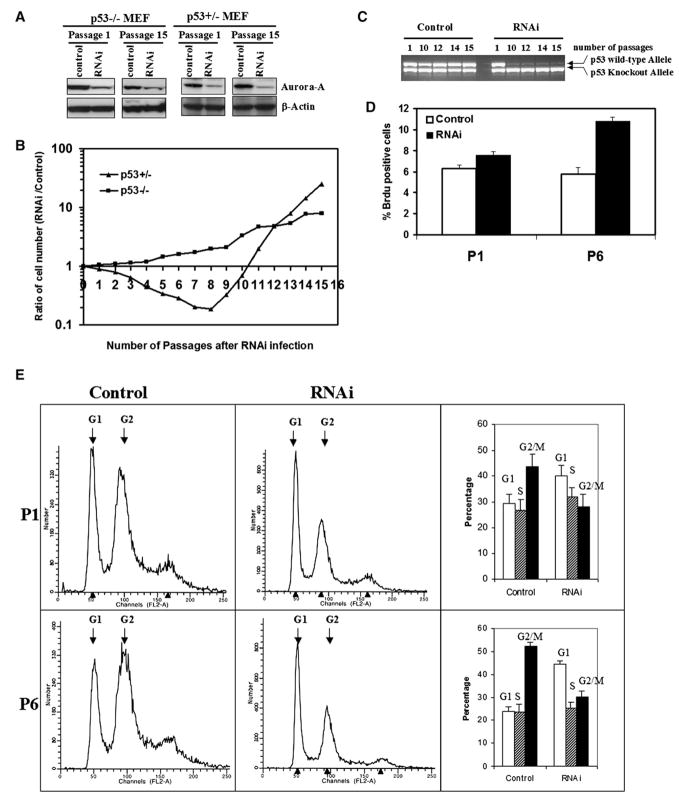
The opposite behavior of the Aurora-A locus in tumors from p53+/− and p53−/− mice suggested that Aurora-A may have either positive or negative effects on cell growth as a function of p53 status. We designed a small interfering RNA (RNAi) against Aurora-A and generated stable transfectants in p53+/− and p53−/− MEFs to examine the effects of Aurora-A downregulation on cell proliferation and apoptosis. The RNAi effectively reduced Aurora-A protein expression in MEFs (Figure 4A), but had mimimal effects on cell morphology (Figure S2). Downregulation of Aurora-A in p53+/− MEFs initially led to a reduction in cell proliferation in comparison with controls (Figure 4B, triangle symbols; Figure S3, lower panel). This reduction in relative growth continued over eight passages in the continuous presence of the RNAi, at which point the transfected cells entered a phase of rapid growth that clearly exceeded the growth rate of control cultures (Figure 4B, triangle symbols; Figure S3, lower panel). Subsequent analysis of the p53 status of these cells showed that this transition was tightly linked to the loss of the wild-type p53 allele (Figure 4C). Prolonged culture of p53+/− MEFs under normal conditions ultimately leads to loss of the remaining p53 allele, but usually after about 25 passages (Harvey et al., 1993). In contrast, downregulation of Aurora-A by RNAi transfection leads to acceleration of this loss to somewhere between passages 5 and 10, suggesting that inhibition of Aurora-A function selects for complete loss of p53 function. We conclude that downregulation of Aurora-A using RNAi may stimulate a p53 checkpoint leading to selection for complete loss of the remaining gene copy.
Figure 4. Aurora-A Downregulation by RNAi in p53 Heterozygous and Null MEFs.
Two controls for the experiment were pSUPER vector without RNAi or with scrambled RNAi (see Experimental Procedures), both of which gave the same results.
(A) Aurora-A protein levels are downregulated by RNAi in p53−/− MEFs after selection with 2 μg/ml puromycin.
(B) Downregulation of Aurora-A in p53+/− MEFS produces a clear growth disadvantage at early passages, but growth advantage at late passages, compared with their untransfected counterparts. p53 null MEFs transfected with the same RNAi construct showed similar behavior independently of the presence of Aurora-A RNAi at early passages, and a clear growth advantage at late passages.
(C) Loss of the remaining p53 allele in p53+/− MEFs treated with Aurora-A RNAi. p53+/− cells in the presence of scrambled (control) RNAi maintain the normal p53 allele until around passage 25, but p53+/− cells treated with Aurora-A RNAi lose the wild-type p53 allele between passages 5 and 10.
(D) Percentage of BrdU-positive cells in p53−/− cultures treated with Aurora-A RNAi (right column) compared to controls (left column). Experiments were carried out with cells at passages 1 and 6. Inhibition of Aurora-A leads to a net increase in the number of proliferating cells. This experiment was repeated three times and the results are statistically significant (*p < 0.01). Error bars stand for mean ± SD of three different experiments.
(E) FACS profiles of p53−/− cells in the presence or absence of Aurora-A RNAi. The same cell populations used for the analyses shown in (D) were analyzed by flow cytometry. The proportion of cells in the G2/M phases of the cell cycle was substantially reduced, suggesting, when taken together with the BrdU incorporation assay shown in (D), that inhibition of Aurora-A releases a G2/M checkpoint and stimulates cell cycle progression. Error bars stand for mean ± SD of three different experiments.
In contrast to the situation seen with p53+/− MEFs, downregulation of Aurora-A in p53−/− MEFs did not cause any obvious decrease in cell growth or proliferation but in fact induced a slightly higher growth rate than in the corresponding control p53−/− cells transfected with the empty vector or random RNAi constructs (Figure 4B, square symbols; Figure S3, upper panel). The difference between RNAi-treated cells and controls appeared to be due to stimulation of growth rather than increased apoptosis, shown by increased BrdU incorporation in treated compared to control cell populations (p < 0.01) (Figure 4D). Comparison of numbers of apoptotic cells by Annexin V staining did not reveal any significant differences between treated and control cells, indicating that decreased cell death was not the explanation for the increase in cell number (Figure S4). Further analysis of FACS profiles of the treated and untreated cells showed that those expressing Aurora-A RNAi had a substantially lower proportion of cells in the G2/M phases of the cell cycle (Figure 4E). These data suggested that the reduction in Aurora-A protein levels in the p53 null cells by treatment with RNAi may serve to relieve a block at the G2/M stage of the cell cycle, allowing more rapid progression through mitosis.
Downregulation of Aurora-A Partially Rescues Genomic Instability in p53 Null MEFs
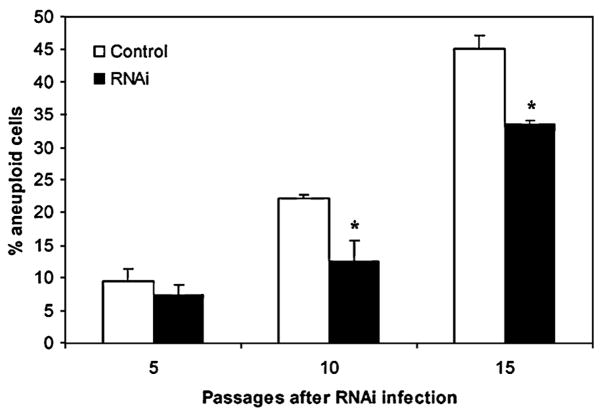
Loss of the p53 tumor suppressor gene is known to result in genomic instability (Cross et al., 1995; Fukasawa et al., 1996; Wahl et al., 1997). Although p53 function has been extensively investigated in the context of the DNA damage response checkpoint, the mechanisms underlying genomic instability in p53−/− cells have not been well established. Recently it has been demonstrated that in the context of p53 deficiency there is an increase in the number of tetraploid cells, and that these are more likely than diploid cells toundergo transformation (Fujiwara et al., 2005). We carried out detailed FACS analysis of MEFs from p53−/− mice before and after treatment with Aurora-A RNAi. The results demonstrated that the increased aneuploidy seen in p53 null MEFs (Figure 4E, left panel) was significantly decreased after RNAi-mediated downregulation of Aurora-A at several different passage levels (Figure 5). These data, taken together with the observations of increased G2/M phase cells and high Aurora levels in p53 null cells (Figure 4E), suggest that increased Aurora levels are a major contributing factor to the elevated instability and aneuploidy in p53 null fibroblasts. This deregulation of mitosis however comes at the expense of relatively retarded growth, and both aneuploidy development and growth defects are at least partially alleviated by inhibition of Aurora-A.
Figure 5. Downregulation of Aurora-A Partially Rescues Genomic Instability in p53 Null MEFs.
p53 null MEFs grown in the presence or absence of Aurora-A RNAi were harvested at the passage levels shown and subjected to FACS analysis of cell cycle profiles. The percentage of aneuploid cells (de-fined as cells with >2N DNA content) increases with passage level of both treated and untreated cultures, but the degree of aneuploidy was significantly reduced by expression of the Aurora-A RNAi (*p < 0.01). Error bars stand for mean ± SD of three different experiments.
DISCUSSION
Control of mitosis is critical for the ordered regulation of cell division, and aberrant expression of various components of the molecular circuitry responsible for this control is an important contributing factor to neoplasia. Studies of the mitotic cycle in Drosophila embryos have identified many of the critical players in this process and have revealed the complexity of the interactions that ensure accurate execution of the entry into and exit from mitosis (O’Farrell et al., 1989; Glover 1991). The Aurora A and B kinases interact with and phosphorylate a number of proteins involved in mitotic spindle assembly, and consequently the concentrations of these proteins have to be maintained within specific limits: either over- or underexpression leads to chromosome missegregation and aneuploidy (Glover et al., 1995; Meraldi et al. 2004). The consequences of aneuploidy development in normal cells are growth arrest or cell death, but in tumors this process is thought to be a major contributor to the neoplastic phenotype (Michor et al., 2005; Rajagopalan and Lengauer, 2004). Deregulation of mitotic control can take place in tumors by amplification and/or overexpression of Aurora-A kinase, but can also be caused by deregulation of other members of the Aurora family or their interacting proteins such as Mad2L1 (Hernando et al., 2004; Meraldi et al., 2004).
The p53 gene has been shown to be involved in control of genetic stability, and loss of even a single copy of this gene in the mouse can result in karyotypic instability and the appearance of abnormal centrosomes and mitotic figures (Bouffler et al., 1995; Fukasawa et al., 1996). Complete loss of p53 recently has been shown to lead to tetraploidy and, subsequently, to development of malignant aneuploid tumor cells (Fujiwara et al., 2005). Whole-genome CGH array analysis, however, demonstrates that tumors from p53 null mice exhibit less instability than corresponding tumors from p53+/− mice, in spite of the fact that the latter have lost the remaining wild-type p53 allele and are functionally p53 null. We interpret these data to mean that the timing of p53 loss is a major determinant of the level of induced genetic instability. The presence of a functional p53 protein presumably stimulates downstream targets in response to radiation exposure, or to other forms of stress, and the resultant selective pressures lead to deletions or other genomic rearrangements that circumvent the induced checkpoints. In the complete absence of functional p53 at the earliest stages of tumor development, fewer checkpoints are activated and there are consequently less requirements for gene copy number gains or losses leading to their inactivation.
In line with the involvement of both Aurora and p53 in mitotic control, several laboratories have identified functional interactions between these two proteins in cell culture model systems (Katayama et al., 2003; Liu et al., 2004). In a wide a variety of human tumors, and in mouse tumors that arise in mice with wild-type p53 function, the gene encoding Aurora-A is frequently amplified and associated with aneuploidy development (Zhou et al., 1998). In the present study, we have demonstrated that prior loss of p53, as in mice carrying nonfunctional p53 alleles (Donehower et al., 1992), leads to a rewiring of this interaction. Complete loss of p53 leads to upregulation of Aurora-A through reduced expression of the p53-dependent tumor suppressor gene Fbxw7, which controls Aurora-A at the protein level (Mao et al., 2004; Perez-Losada et al., 2005; Fujii et al., 2006). This mechanism may contribute to the well-documented chromosome abnormalities, in particular the tetraploidization, seen in p53 null cells. Notably, development of tetraploidy is stimulated by overexpression of Aurora-A, and this precedes the detection of centrosome abnormalities in mouse cells (Meraldi et al., 2002). This interpretation is further supported by the observation shown in Figure 5 that downregulation of Aurora-A in p53 null fibroblasts can partially reduce the level of aneuploidy, while simultaneously allowing more rapid cell growth.
With the onset of lymphoma development in vivo in p53 null mice, these high Aurora levels may be incompatible with ordered progression through mitosis, particularly if additional components of the mitotic apparatus are also deregulated by genetic or epigenetic events. As a consequence, in a substantial proportion of tumors, levels of Aurora that are compatible with rapid cell growth are restored by deletion, or in some cases by downregulation by other mechanisms. In this context, Aurora-A is not a tumor suppressor gene in the classical sense but acts as a rheostat in control of mitosis.
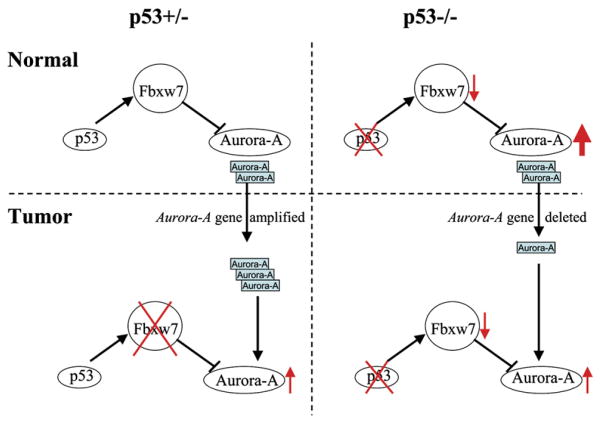
The relationship between p53 status and Aurora-A levels is likely to be much more complex in human tumors than in the mouse, as p53 function can be abrogated by deletions or truncation mutations leading to loss of function, or by alternative point mutations that introduce dominant-negative or gain-of-function mutations (Soussi and Lozano, 2005; Vousden and Prives, 2005). In addition, the relative timing of genetic events involving Aurora-A and p53 in human tumors is unclear. Nevertheless, we observed a significant correlation between the levels of p53 and Aurora-A both in breast cancer cell lines and in primary human tumors. As in the mouse studies, we see examples of tumor cells with single copy losses of the Aurora-A gene and correspondingly low protein levels. One possible explanation is that in a subset of human tumors, p53 mutations leading to loss of function occur prior to amplification or activation of Aurora-A, such that Aurora-A deletions are required during further progression, as seen in the mouse models. This explanation is compatible with the model shown in Figure 6, by which the normal feedback loop between p53 and Aurora-A levels is disrupted in tumor cells. However, the temporal series of events is very difficult to establish from human tumor analysis, and this mechanism, although compatible with the mouse data, remains unproven.
Figure 6. Proposed Model for the Disruption of the Feedback Loop between Aurora-A and p53 in Mouse Lymphoma-genesis.
In normal cells with wild-type p53 function, Aurora-A levels are controlled at least in part by the Fbxw7 ubiquitin ligase (left upper panel) (Mao et al., 2004; Fujii et al., 2006). Loss of p53 in normal thymus leads to downregulation of Fbxw7 expression and consequent upregulation of Aurora-A at the protein level (right upper panel). In transformed cells from p53+/− mice, Aurora-A levels are increased by gene copy number gains or by loss of Fbxw7 (or a combination of both, since Fbxw7 also affects other targets). Subsequent loss of p53 in this case presumably does not change Aurora-A levels, because of disruption of this feedback loop. In p53−/− normal cells, Aurora-A levels are already very high, such that in some cases, additional events are required to reduce the protein level to within an acceptable window. This is accomplished in some cases, both in mouse and human tumors, by single copy gene deletions. The range of levels of Aurora-A seen in tumors from p53+/− and p53−/− mice is quite similar, but the mechanisms that result in these levels can be different depending on p53 status.
The observation of opposite consequences of Aurora-A inhibition in cells with or without functional p53 has important implications for the development of cancer therapeutics aimed at inhibition of this kinase. Our data suggest that pharmacological inhibition of Aurora-A may in some cases, depending on individual tumor stage and p53 status, lead to decreased aneuploidy but increased growth, or alternatively to complete loss of wild-type p53 activity. A recent study of progression of breast ductal carcinoma in situ (DCIS) demonstrated high expression of Aurora-A at the preinvasive stage, but decreased expression associated with development of adjacent invasive lesions in the same patients (Hoque et al., 2003). In the present manuscript, we have identified a subset of human breast cancers with genetic loss of Aurora-A and low levels of protein. While it seems likely that small-molecule inhibitors of these mitotic kinases will be an important addition to the armory of agents that may be used for cancer therapy, our data underline the importance of individualized assessment of the genetic status of Aurora family members and p53 in human cancers before embarking on extensive clinical trials of these agents. Further studies of the complex networks of interactions between these and other important cancer signaling hubs (Vogelstein et al., 2000) will be required to identify the specific combinations of drugs that will be necessary for successful therapy of malignant disease.
EXPERIMENTAL PROCEDURES
Tumor Induction and Preparation of DNA
Five-week-old p53+/− and p53−/− 129/Sv mice of both sexes were exposed to a single dose of 4 Gy whole-body radiation and observed daily until moribund, then sacrificed and autopsied. We performed all animal experiments in compliance with UCSF Laboratory Animal Resource Center (LARC)-approved protocols.
Thymic lymphomas and normal tails were cut, minced in small pieces, and placed in a microfuge tube containing 0.5 ml lysis buffer (100 mM Tris·HCl [pH 8.5], 5 mM EDTA, 0.2% SDS, 200 mM NaCl). Proteinase K was added at a final concentration of 100 mg/ml and incubated at 55°C overnight. The lysate was extracted twice with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v) (GIBCO-BRL). DNA in the lysate was precipitated by addition of an equal volume of isopropanol. The DNA precipitates were dissolved in TE buffer (10 mM Tris·HCl [pH 8.0] and 1 mM EDTA).
Genome-Wide BAC Microarray
Detailed methods are described in Mao et al. (2005). Normal genomic DNA from mouse strain C57BL/6J was used as the control, and Cot-1 DNA was used for blocking repetitive sequences in BAC clones and genomic probes. The Cy5:Cy3 ratios were plotted together along individual chromosomes. For each mouse tumor sample, two experiments were done with reversal of the dye labels to remove any ratio artifact.
TaqMan Assay
We designed TaqMan primers and probes using the Primer Express Oligo Design Software v1.0 (Applied Biosystems, Foster City, CA). Probes were FAM probes designed specifically for TaqMan (Applied Biosystems). All primer sets were used to perform amplifications in triplicate on the ABI 7700 instrument (Applied Biosystems). Reactions (50 μl) were performed in 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 1.6 M primer, 0.4 M probe, 12.5 ng DNA. Cycling parameters were as follows: 95°C for 12 min × 1 cycle (95°C for 20 s, 60°C for 20 s, 72°C for 60 s) × 40 cycles. Copy number is determined from the PCR cycle number (CT) at which DNAs reach a threshold amount of fluorescence above background. To normalize for differences in the amount of total input DNA, amplification at a reference locus was performed once per plate in triplicate for each individual DNA. The CT values for each set of triplicates were averaged. The Ct of the pooled reference was subtracted from the CT for each locus to obtain the ΔCT [ΔCT = CT (locus) − CT (reference)]. ΔCt values were determined for locus in tumor samples and a set of six normal genomic DNAs. The average of the six ΔCT values [ΔCT(normal)] measured from the normal DNAs was calculated once for each locus in this study and used in the subsequent calculations for all experiments performed on a single ABI 7700. ΔΔCT = ΔCt (tumor) − Average ΔCT (normal). Number of copies = 2**(ΔΔCT).
RNAi Generation, Transfection, and MEF Growth Determination
MEFs were prepared from 13.5-day-old embryos from p53 wild-type, heterozygous, and null mice (129/Sv background). All experiments were performed with MEFs prepared from embryos from at least two different litters. The genotype of the MEFs was confirmed by PCR-based analysis of DNA.
MEFs were infected with high-titer retroviral stocks produced by transient transfection of 293T ecotropic Phoenix cells. After infection with the pSUPER retrovirus (OligoEngine) allowing the expression of RNAi molecules, MEFs were selected with 1–2 μg/ml of puromycin in the culture medium. The oligonucleotide for Aurora-A RNAi is AACTGTGTCTCCAGGCCTG. Two controls for the experiment were pSUPER vector without RNAi or with scrambled RNAi: GGAAGCCAAGCCAAATGGC. The same results were obtained from both controls.
For growth (proliferation) curve determinations, cells were seeded into three 100 mm tissue culture plates at 3 × 105 cells per plate in DMEM (high glucose) supplemented with 10% FBS and penicillin-streptomycin. Cell numbers were determined every 3 days by Coulter counter (Beckman Coulter). Accumulative cell numbers were calculated at each passage. Experiments were repeated at least three times.
Western Blotting
Total protein extracts were prepared from tumor MEFs with RIPA lysis buffer. For western blots, 50 μg of protein extract per lane was electrophoresed, transferred to PVDF membranes (Millipore), and incubated with Aurora-A antibody (BD Transduction Laboratories); as a control, the same membranes were stripped and immunoblotted again with anti-β-actin antibody (AC-15, Sigma). The membranes were washed and treated with rat anti-specie IgG κ-chain secondary antibody conjugated to horseradish peroxidase (Amersham Pharmacia). The antigen-antibody reactions were visualized by using an enhanced chemiluminescence assay (ECL; Amersham Pharmacia).
Immunochemical Techniques and Immunoblot Quantification
Immunoblot analyses were performed using 20 μg cleared cell lysates. This material was electrophoretically resolved on denaturing sodium doedecyl sulfate (SDS)-polyacrylamide gels (4%–12%), transferred to polyvinylidene difluoride membranes (PVDF, Millipore), and probed with specific antisera using standard techniques. Bound antibodies on immunoblots were detected by either chemiluminescent (ECL; Pierce) or infrared imaging (Li-Cor, Odyssey). Images were recorded as TIFF files for quantitation (see below). Antibodies: Aurora kinase (mouse monoclonal IAK1, BD Biosciences), p53 (Ab-6, Oncogene).
Protein Quantification
Protein levels were measured by quantifying emitted chemiluminescence or infrared radiation recorded from labeled antibodies using Scion Image (http://www.scioncorp.com/) or Odyssey software (http://www.licor.com/) (R.M.N. et al., unpublished data). For each protein, the blots were made for 4 sets of 11 cell lines, each set including the same pair (SKBR3 and MCF12A) to permit intensity normalization across sets. A basic multiplicative normalization was carried out by fitting a linear mixed effects model to log intensity values and adjusting within each set to equalize the log intensities of the pair of reference cell lines across the sets.
Detection of Cell Cycle Changes and Aneuploid Cells by FACS Analysis
Cells were trypsinized, washed with PBS + 1% FBS, fixed with cold 70% ethanol, treated with propidium iodide (10 μg/ml) and ribonuclease A (100 μg/ml), and subjected to cell cycle analysis using FACS Calibur (Becton Dickinson). Percentage of aneuploid cells was calculated with ModFit LT cell-cycle analysis software (Verity Software House, Topsham, ME).
Detection of Apoptosis by Annexin V
The annexin V-FITC apoptosis detection kit was purchased from BD Biosciences PharMingen. All procedures were carried out according to the manufacturer’s instructions.
Detection of BrdU Incorporation in DNA-Synthesizing Cells
When growing cells in a tissue culture flask reached 70% confluence, BrdU was added at a final concentration of 10 μM. After 1 hr incubation, cells were collected and processed according to the manufacturer’s instructions (BD Biosciences). Levels of BrDU incorporation were analyzed by flow cytometry.
Supplementary Material
Acknowledgments
These studies were initially supported by the Commission of the European Communities and the Cancer Research Campaign (UK), and subsequently by NCI grant U01 CA84244 and by the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-03ER63630 to A.B. The UCSF Cancer Center Genome Core was essential for the sequencing and study design of TaqMan assay. Special thanks go to the staff of the CRUK Beatson Institute and UCSF Comprehensive Cancer Center animal house for animal husbandry.
Footnotes
Supplemental Data
The Supplemental Data include Supplemental Experimental Procedures, six supplemental figures, and microarray data and can be found with this article online at http://www.cancercell.org/cgi/content/full/11/2/161/DC1/.
References
- Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffler SD, Kemp CJ, Balmain A, Cox R. Spontaneous and ionizing radiation-induced chromosomal abnormalities in p53-deficient mice. Cancer Res. 1995;55:3883–3889. [PubMed] [Google Scholar]
- Chen SS, Chang PC, Cheng YW, Tang FM, Lin YS. Suppression of the STK15 oncogenic activity requires a transactivation-independent p53 function. EMBO J. 2002;21:4491–4499. doi: 10.1093/emboj/cdf409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman P, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, MacCarthy-Morrogh L, Ponder BA, Nagase H, Burn J, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34:403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- Ewart-Toland A, Dai Q, Gao YT, Nagase H, Dunlop MG, Farrington SM, Barnetson RA, Anton-Culver H, Peel D, Ziogas A, et al. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- Fei P, El-Deiry WS. P53 and radiation responses. Oncogene. 2003;22:5774–5783. doi: 10.1038/sj.onc.1206677. [DOI] [PubMed] [Google Scholar]
- Frenkel J, Sherman D, Fein A, Schwartz D, Almog N, Kapon A, Goldfinger N, Rotter V. Accentuated apoptosis in normally developing p53 knockout mouse embryos following genotoxic stress. Oncogene. 1999;18:2901–2907. doi: 10.1038/sj.onc.1202518. [DOI] [PubMed] [Google Scholar]
- Fujii Y, Yada M, Nishiyama M, Kamura T, Takahashi H, Tsunematsu R, Susaki E, Nakagawa T, Matsumoto A, Nakayama KI. Fbxw7 contributes to tumor suppression by targeting multiple proteins for ubiquitin-dependent degradation. Cancer Sci. 2006;97:729–736. doi: 10.1111/j.1349-7006.2006.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Glover DM. Mitosis in the Drosophila embryo—in and out of control. Trends Genet. 1991;7:125–132. doi: 10.1016/0168-9525(91)90457-2. [DOI] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Hoque A, Carter J, Xia W, Hung MC, Sahin AA, Sen S, Lippman SM. Loss of aurora A/STK15/BTAK overexpression correlates with transition of in situ to invasive ductal carcinoma of the breast. Cancer Epidemiol Biomarkers Prev. 2003;12:1518–1522. [PubMed] [Google Scholar]
- Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, Cheng JQ. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Mao JH, Li J, Jiang T, Li Q, Wu D, Perez-Losada J, Delrosario R, Peterson L, Balmain A, Cai WW. Genomic instability in radiation-induced mouse lymphoma from p53 heterozygous mice. Oncogene. 2005;24:7924–7934. doi: 10.1038/sj.onc.1208926. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol. 2005;15:43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH, Edgar BA, Lakich D, Lehner CF. Directing cell division during development. Science. 1989;246:635–640. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Losada J, Mao JH, Balmain A. Control of genomic instability and epithelial tumor development by the p53-Fbxw7/Cdc4pathway. Cancer Res. 2005;65:6488–6492. doi: 10.1158/0008-5472.CAN-05-1294. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. Calculated resistance in cancer. Nat Med. 2005;11:824–825. doi: 10.1038/nm0805-824. [DOI] [PubMed] [Google Scholar]
- Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005;331:834–842. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kimura M, Matsunaga K, Fukada D, Mori H, Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 1999;59:2041–2044. [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. P53 and prognosis: new insights and further complexity. Cell. 2005;120:7–10. doi: 10.1016/j.cell.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Wahl GM, Linke SP, Paulson TG, Huang LC. Maintaining genetic stability through TP53 mediated checkpoint control. Cancer Surv. 1997;29:183–219. [PubMed] [Google Scholar]
- Wuchty S, Oltvai ZN, Barabasi AL. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nat Genet. 2003;35:176–179. doi: 10.1038/ng1242. [DOI] [PubMed] [Google Scholar]
- Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.