Abstract
Cancer cells have diverse biological capabilities that are conferred by numerous genetic aberrations and epigenetic modifications. Today’s powerful technologies are enabling these changes to the genome to be catalogued in detail. Tomorrow is likely to bring a complete atlas of the reversible and irreversible alterations that occur in individual cancers. The challenge now is to work out which molecular abnormalities contribute to cancer and which are simply ‘noise’ at the genomic and epigenomic levels. Distinguishing between these will aid in understanding how the aberrations in a cancer cell collaborate to drive pathophysiology. Past successes in converting information from genomic discoveries into clinical tools provide valuable lessons to guide the translation of emerging insights from the genome into clinical end points that can affect the practice of cancer medicine.
A human ‘cancer genome’, or oncogenome, harbours numerous alterations at the level of the chromosomes, the chromatin (the fibres that constitute the chromosomes) and the nucleotides. These alterations include irreversible aberrations in the DNA sequence or structure and in the number of particular sequences, genes or chromosomes (that is, the copy number of the DNA). They also include potentially reversible changes, known as epigenetic modifications to the DNA and/or to the histone proteins, which are closely associated with the DNA in chromatin (Fig. 1). These reversible and irreversible changes can affect hundreds to thousands of genes and/or regulatory transcripts. Collectively, they result in the activation or inhibition of various biological events, thereby causing aspects of cancer pathophysiology, including angiogenesis, immune evasion, metastasis, and altered cell growth, death and metabolism1.
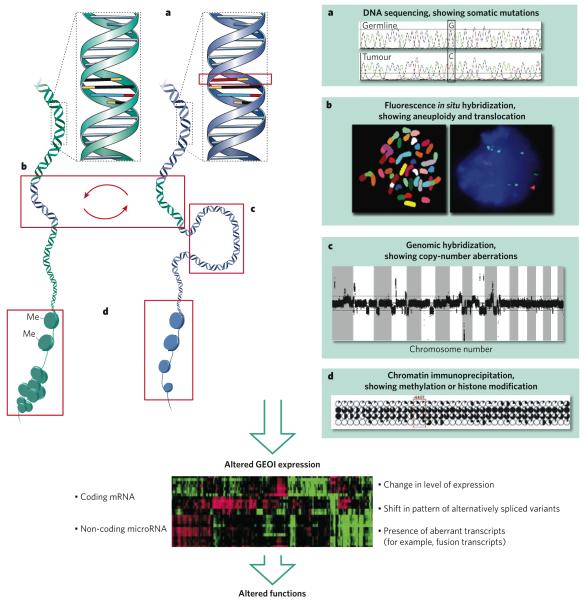
Figure 1. Various types of genomic and epigenomic aberration in cancers.
The main types of genomic and epigenomic aberration are illustrated together with examples of how they can be detected. a, Changes in DNA sequence, such as point mutations, can be assessed by DNA-sequencing techniques. b, Changes in genomic organization can be assessed by using fluorescence in situ hybridization. In the example shown, DNA segments are exchanged between the two (blue and green) DNA molecules. c, Changes in DNA copy number, such as those that result from amplification, can be assessed by using comparative genomic hybridization. d, Changes in DNA methylation and the resultant changes in chromatin structure can be assessed by using chromatin immunoprecipitation plus microarray analysis of immunoprecipitated DNA. Each of these types of change can alter the expression levels of genes or non-coding microRNAs (referred to here as genetic elements of interest, GEOIs), alter the splicing patterns of transcripts, or change gene function through mutation or through creating chimaeric genes. Many of these events can be as assessed by microarray analysis. These changes ultimately translate into altered functions, leading to a diseased state, such as cancer.
Mining the cancer genome and epigenome for aberrations that control these processes has become a major activity in cancer research, because it is widely understood that these aberrations provide clues to the mechanisms of disease pathogenesis. These studies can inform efforts to identify molecular events that can be targeted for therapy and to discover molecular biomarkers (biological indicators) that aid in early detection, diagnosis, prognosis (that is, prediction of clinical outcome) and the prediction of responses to therapies. Recognizing this, many national and international efforts, including The Cancer Genome Atlas pilot project by the US National Cancer Institute and the National Human Genome Research Institute2, have been initiated to accelerate the compilation of an atlas of alterations.
In recent years, cancer genomics — defined here as the study of the ensemble of DNA-associated abnormalities that allow and accompany cancer development — has exploded as a field, with studies facilitated by genome-wide, high-resolution, high-throughput platforms (Box 1). These technologies now yield informative, but dauntingly complex, multidimensional genomic data sets that describe in detail the myriad changes that occur within individual tumours and how these changes differ between individual tumours. Together with assays to detect these aberrations that are now used to stratify patients for treatment (discussed later), these data sets are now transforming the practice of cancer medicine, as is shown by the success of therapies that target distinct molecular events resulting from genomic aberrations. For example, patients with mutations in the gene encoding the epidermal growth-factor receptor (EGFR) can be treated with gefitinib or erlotinib3-5; those with the BCR-ABL translocation, with imatinib mesylate6; and those with amplification (that is, increased copy number) of the oncogene ERBB2 (also known as HER2 or NEU), with trastuzumab or lapatinib7. In parallel, assays for mutations in germline DNA can identify individuals who are at high risk of developing cancer. For example, mutations in TP53 (which encodes the tumour-suppressor protein p53) are associated with Li-Fraumeni syndrome8; mutations in BRCA1 or BRCA2 indicate an increased risk of breast and ovarian cancer9-11; mutations in genes whose products are involved in DNA-mismatch repair (such as MLH1, MSH2 or MSH6) are associated with hereditary non-polyposis colorectal cancer12; and mutations in CDKN2A (which encodes a tumour-suppressor protein known as INK4A (or p16), which is involved in regulating the cell cycle) indicate an increased risk of familial atypical multiple mole melanoma-pancreatic cancer13.
Box 1|Techniques for analysing the cancer genome.
Comprehensive analyses of the genome of various types of cancer cell — in terms of DNA copy number, DNA sequence, DNA organization, gene expression and epigenomic modification — are underway worldwide. A rapidly evolving suite of technological solutions is allowing cancer genomes to be characterized with remarkable resolution and accuracy. Several of the techniques used to analyse the various aberrations and modifications are summarized here.
Copy-number aberrations
Changes in the copy number of genetic regions or chromosomes across the entire genome of a cancer cell can be mapped onto a representation of the normal genome by using comparative genomic hybridization. This technique readily allows the genes involved in copy-number aberrations to be identified31. Modern analysis platforms for comparative genomic hybridization map copy-number changes onto DNA sequences arranged in microarrays77 and allow these changes to be assessed quantitatively (including for individual alleles in some platforms) with sub-gene resolution. Even at this resolution, aberrations can be missed, especially when using platforms that are gene-oriented. Emerging next-generation technologies that efficiently sequence small genome fragments that have been collected randomly from tumour-cell genomes will complement such DNA-microarray-based strategies for analysing copy number. These work by sequencing tens of millions of short DNA fragments and then summing the number of fragments in equal-sized bins distributed along the genome. The relative number of DNA fragments in each bin is an estimate of the relative copy number at that genomic location. The resolution of this approach can be made arbitrarily high by sequencing to an increasing depth.
Structural aberrations
Structural changes can involve segmental deletions or insertions, and translocations or more complex rearrangements (for example, those occurring during gene amplification or copy-number change). These changes can be uncovered by using cytogenetic techniques such as banding analysis or fluorescence in situ hybridization or by using DNA-sequence-based strategies such as end-sequence profiling. End-sequence profiling is an adaptation of whole-genome shotgun sequencing that allows structural aberrations to be detected23. DNA from a tumour is cloned into a large-insert vector, and the ends of the resultant clones are sequenced and then mapped onto the normal human DNA sequence. Paired ends that map farther apart than the maximum size tolerated by the cloning vector indicate the presence of a structural aberration. This approach has the advantage that clones containing aberrant DNA from gene fusions can be sequenced to identify the exact DNA sequence at the breakpoint. But it has the disadvantage that millions of tumour DNA clones must be maintained. Alternatively, cloning strategies known as paired-end sequencing, which retain only the ends of the cloned DNA fragment, can be used78. These paired ends are then sequenced to identify structural aberrations (as described above). This strategy is efficient but does not yield the DNA sequence across the breakpoints.
DNA-sequence abnormalities
Recent efforts in large-scale DNA-sequence analysis have identified several hundred candidate genes that might have functional roles in various human cancers40,41. Some occur at a relatively high frequency, but most are present in only a few per cent of tumours. Results from the extensive sequencing and mutation-validation efforts that are now underway will be necessary to establish the prevalence of, and clinicopathological associations for, these genetic elements of interest (GEOIs). Both established and next-generation sequencing technologies will be brought to bear on this issue. There are several current techniques for DNA sequencing, including sequencing by hybridization, dideoxy sequencing and cyclic array sequencing. Sequencing by hybridization79 is a DNA-array-based strategy in which mutations are detected based on the intensity of hybridization of sample DNA to microarrays comprising short oligonucleotide probes that are designed to be perfectly complementary to the reference sequence plus oligonucleotide probes that differ by one base at each ‘substitution position’ in the genome to be tested. This approach is well suited to resequencing. Dideoxy sequencing80 is the current standard method for detecting mutations. It is typically applied to PCR products that result from the amplification of sample DNA by using primers that flank regions of interest, and it generates collections of DNA fragments in which each fragment terminates with a base-specific fluorescent label. The fragments are then separated according to size by using capillary electrophoresis, and the terminating base is identified by fluorescence emission analysis. Sequence ‘reads’ are generally about 750 bases. In most cases, dideoxy sequencing will not detect mutations that are present in less than about 20% of the cells represented in the PCR-amplified population. Mutations that have been discovered so far are summarized in the Catalogue of Somatic Mutations In Cancer (http://www.sanger.ac.uk/genetics/CGP/cosmic). The efficiency of sequencing can be increased by using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry to measure the masses of DNA fragments generated by primer extension with dideoxy termination. However, sequence reads are typically less than 100 bases with this read-out method. Cyclic array sequencing allows millions to billions of DNA fragments to be sequenced in parallel by arranging these fragments on a sequencing substrate and using a cyclic enzymatic process to interrogate the sequence of all fragments in parallel81,82. Current read lengths range from about 30 bases to 300 bases, and the number of reads per analysis ranges from 0.3 million to 30 million. Cyclic array sequencing techniques facilitate the detection of rare mutations. Recent affinity-enrichment techniques allow subsets of the genome to be enriched before sequencing (for example, all known exons), thereby decreasing the cost of targeted sequencing83.
Epigenomic analysis
It is clear that epigenomic modifications are major contributors to the formation and progression of tumours, especially during the early stages of tumour development. Several techniques for the genome-wide assessment of DNA methylation and chromatin structure have now been established, and others are emerging; these techniques help to elucidate the role of epigenomic modifications in the cancer genome. The five established techniques are restriction-landmark genomic scanning, microarray-based epigenomic analysis, reduced representation bisulphate sequencing, methylation-specific digital karyotyping, and chromatin immunoprecipitation plus microarray analysis. First, restriction-landmark genomic scanning84 using methylation-sensitive enzymes was the first method developed as a genome-wide screen for methylation of CpG islands. This technique, which involves two restriction digests followed by electrophoretic separation, allows methylation to be analysed in up to 4,000 loci85,86. Second, microarray-based epigenomic analysis methods87 involve hybridization of tumour and reference DNA samples to DNA microarrays. These microarrays comprise oligonucleotides derived from CpG-island sequences (which are generated by digestion of CpG islands with methylation-sensitive restriction enzymes that cleave preferentially within the islands). Comparing the signal intensities from the tumour and reference samples provides a profile of sequences that are methylated in the tumour but not in the references (or vice versa). Third, reduced representation bisulphate sequencing88 is a genome-wide shotgun sequencing approach in which the tumour and reference DNA samples are treated with sodium bisulphate to convert cytosine to uracil while leaving 5-methylcytosine unconverted, then digested with a methylation-specific enzyme and sequenced. Comparison of CpG sequences in the tumour and reference genomes then reveals bisulphate-induced changes. This method is well suited to next-generation single-molecule sequencing strategies. Fourth, methylation-specific digital karyotyping89 is a modified technique for DNA copy-number profiling90. Sequencing is carried out to accurately count tags to compare CpG sequences in tumour and reference samples, thereby allowing quantitative measurement of methylation events. Fifth, chromatin immunoprecipitation plus microarray analysis (ChIP on chip)91 involves an initial immunoprecipitation step, thereby enriching DNA sequences associated with histone modifications (for example, methylation or acetylation of histone H3) for which specific antibodies are available. Immunoprecipitated DNA sequences are then analysed by using DNA-microarray-based methods or single-molecule DNA-sequencing strategies.
These examples have demonstrated the promise of cancer genomics, stimulated rapid advances in genomic technologies and computational science, and galvanized an entire generation of multidisciplinary scientists to identify the next set of key therapeutic targets and disease biomarkers for cancer. Although there has been tremendous success in the rapid accumulation of genomic data, most of these enormous data sets have not yet been translated into meaningful clinical end points. In the past, the translation of each genomic aberration into improved management of patients has taken at least a decade and sometimes billions of dollars. Given this situation, it is important to understand the barriers that prevent more rapid and less costly conversion of genomic information into useful diagnostic tests and effective therapeutic agents. Is statistical significance in the absence of mechanistic insight sufficient to harness the full potential of these complex genomic data sets in a cost-efficient and effective way? Or is some degree of understanding of the molecular biological function required for efficient translation? The BCR-ABL, ERBB2 and EGFR examples seem to support the view that coupling insights into the genome with pathobiological findings holds the greatest promise for making an impact in the clinic. In this article, we review lessons from past genomic discoveries that have been translated successfully into the clinic and describe strategies (including integrative analyses and model systems) that have been useful for the identification of genetic elements of interest (GEOIs). We conclude with a discussion of the challenges that are faced and potential ways to move forwards in this field.
Lessons from the past
There are several pioneering examples of genomic aberrations being discovered in cancer cells and the findings being successfully translated into therapeutic agents and tests for cancer risk, prognosis or response to therapy, with considerable effects on the practice of cancer medicine. Although many of these successes predated the current genome-wide, high-throughput technologies — indeed, some resulted from decades of painstaking work — they nevertheless presage the translation of information from the cancer genome into clinical tools. These translational efforts can be considered in terms of the type of genomic aberration studied — translocations, gene amplification, mutations and germline susceptibility — and the examples described in this section might help to guide and accelerate translation of the genomic aberrations now being discovered.
Translocations
The first genomic aberration found to be associated consistently with a human malignancy (that is, recurrent) was the Philadelphia chromosome, discovered by Peter Nowell and David Hungerford in 1960 (discussed in ref. 14). In the ensuing decades, cytogenetic and molecular studies showed this to be a translocation between chromosomes 9 and 22, resulting in a fusion product, BCR-ABL. As a result of this fusion, the activity of the non-receptor tyrosine kinase ABL is dysregulated in patients with chronic myeloid leukaemia or with some forms of acute lymphoblastic leukaemia. More than 30 years after the discovery of the Philadelphia chromosome, a small-molecule inhibitor of ABL, imatinib mesylate, was developed as an effective therapeutic agent against the effects of the BCR-ABL translocation in patients with chronic myeloid leukaemia6. However, despite marked initial responses, this targeted therapy does not lead to a lasting cure, because resistant cancer cells emerge15. Genomic analyses of the resistant cells showed that point mutations were acquired (and sometimes amplified) that abrogated the inhibitory effects of the drug. This result guided the development of new small-molecule inhibitors to counter this resistance mechanism, culminating in the recent approval of nilotinib and dasatinib by the US Food and Drug Administration (FDA)16. These findings suggest that the development of anticancer drugs will occur by an iterative process in which genomic analyses are first used to guide the development of targeted therapies and associated predictive biomarkers, and then genomic studies of resistant cancer cells aid in the development of second-generation and third-generation inhibitors to counter the mechanisms of resistance that have arisen against the first-generation inhibitors. Banking tumour tissues from patients who are sensitive or resistant to drugs will be essential to support these studies. In many cases, this will require biopsy of metastatic lesions, a process that is not regularly carried out in clinical trials. Another lesson from the imatinib mesylate story is that genomic analyses can guide the use of small-molecule inhibitors that are effective against several targets. Imatinib mesylate, for example, also inhibits the receptor tyrosine kinase c-KIT. Following genomic analyses of gastrointestinal stromal tumours (GIST sarcomas)17 and mucosal melanomas18, which showed that both cancers harbour c-KIT mutations, imatinib mesylate has been used successfully to treat patients with GIST sarcomas or mucosal melanomas17-19.
Since the pioneering discovery of the Philadelphia chromosome, numerous recurrent translocations that cause cancer have been discovered in human leukaemias and lymphomas by using molecular cytogenetic analyses20. However, finding causal translocations in solid tumours has been difficult, possibly reflecting the complex genomic profiles and heterogeneous nature of these malignancies. With the current ability to analyse the genome, together with the sophisticated analytical approaches available and the ever increasing amounts of genomic information, recurrent structural aberrations are now being discovered in solid tumours and might be more prevalent than previously thought. A notable discovery is the high frequency of translocations between TMPRSS2 (which is upregulated in response to androgenic hormones) and the ETS-family genes ERG, ETV1 and ETV4 (which encode transcription factors) in human prostate cancer. Using a new integrative analytical methodology called cancer outlier profile analysis, which identifies associations between genomic and transcriptional abnormalities, Arul Chinnaiyan and colleagues21 identified a family of common translocations that brings ETS-family genes under the control of TMPRSS2, in effect placing the expression of these genes under androgen-mediated regulation. Molecular assays for fusion events are now being developed and evaluated for use as early detection markers for prostate cancers22. It is hoped that applying similar computational approaches to emerging multidimensional data sets will allow the detection of other causal structural aberrations in solid tumours. And this is only the beginning. Next-generation sequencing technologies that allow the entire genomes of tumour cells to be sequenced will be particularly valuable for discovering fusion genes and other structural rearrange ments. The promise of this approach is illustrated by the remarkable structural complexity found in cancer genomes by using end-sequence profiling23, genomic-region sequencing24 or genome-wide parallel paired-end sequencing25 (Box 1).
Gene amplification
Another prominent success story involves the now well-established ERBB2 oncogene. ERBB2, which is homologous to mouse Erbb and the gene encoding tumour antigen p185, was initially identified as a transforming oncogene in NIH/3T3 cells26 and was also found to be amplified in human breast-cancer cell lines27-29. Shortly after these findings, ERBB2 amplification was found in ∼30% of primary breast-cancer tumours, and this amplification was associated with a short survival time and short time to relapse30. On the basis of these observations, trastuzumab (a monoclonal antibody specific for the extracellular domain of ERBB2) was developed to treat breast tumours that had ERBB2 amplification7. Clinical introduction of trastuzumab was guided by molecular assays for ERBB2 amplification31 or overexpression. More recently, molecular diagnostic assays that assess ERBB2 amplification or overexpression have guided clinical use of the small molecule lapatinib, which targets ERBB2 and EGFR32.
Mutations
Since the completion of the Human Genome Project, several important discoveries in genomics have come from the systematic resequencing of genes, gene families or genes in pathways that are relevant to cancer. One of the first, and perhaps most celebrated, successes from such large-scale resequencing projects was the discovery that BRAF, which encodes a serine/threonine kinase, frequently contains activating somatic mutations: in 60% of malignant melanomas, in 10% of colorectal cancers and in a smaller percentage of other cancers33. This discovery has driven many programmes aimed at developing BRAF inhibitors, and several drugs are now in clinical trials. Other notable discoveries from large-scale resequencing efforts include frequent mutations in PIK3CA34 (which encodes the catalytic subunit of phosphatidylinositol-3-OH kinase) and AKT1 (ref. 35) (which encodes a serine/threonine kinase) in many cancer types, as well as in ERBB2 and EGFR in non-small-cell lung cancer36,37. In addition to gender, ethnicity, smoking history and the histopathological subtype of the cancer, it was found that the mutation status of EGFR predicts responses to treatment with the EGFR inhibitors gefitinib or erlotinib in patients with advanced non-small-cell lung cancer3-5. Testing for mutations in EGFR before decisions are made about treatment with EGFR inhibitors is becoming routine37. The ability to determine EGFR genotype retrospectively by using banked tumour tissues with matched germline DNA from the ongoing clinical trials was crucial for allowing the stratification of responders and for showing efficacy38. These studies highlight the importance of uniformly collecting pretreatment and post-treatment tumour specimens with matched normal controls from clinical trials.
Germline susceptibility
In addition to its impact on somatic genetics studies, genomics is revolutionizing the search for germline genes that confer susceptibility to cancer and for polymorphisms that are responsible for inherited predisposition to disease, including cancers. One of the early successes in this area was the discovery that inactivating mutations in BRCA1 are associated with familial breast cancer9,10. Genetic screening for germline mutations in BRCA1 — and now in a second cancer-susceptibility gene, BRCA2 (ref. 11) — is being rolled out worldwide to identify patients who are at a high risk of developing early-onset breast and ovarian cancer. Moreover, the knowledge that BRCA1 is required for error-free repair of DNA double-strand breaks led to the development of inhibitors of poly(ADP-ribose) polymerase 1 (PARP1, an enzyme involved in the recognition of DNA single-strand breaks)39. These and subsequent studies showed that discovery of inactivating germline mutations associated with increased susceptibility to cancer can be guided by analyses of loss of heterozygosity or reduction in DNA copy number and/or DNA methylation in the tumours that eventually develop. Applying current (and future) genomic technologies in coordinated germline and tumour studies should considerably accelerate the discovery of susceptibility genes of this class, thereby increasing our ability to identify high-risk individuals who can then be managed using aggressive surveillance and prevention strategies. Identifying susceptibility genes by this method will require the coordinated collection of tumour specimens, together with germline DNA, in large-cohort genetic susceptibility studies.
Making sense of the cancer genome
Empowered by the improved ability to survey the cancer genome with increasing accuracy and resolution, numerous studies have been carried out or initiated with the hope of discovering the next EGFR, ERBB2 or BRAF. Instead, these analyses are uncovering hundreds of recurrent genomic or genetic alterations that affect thousands of GEOIs — including annotated genes, non-coding microRNAs and other conserved elements — that might contribute to the pathophysiology of human cancers. The nature and strength of each GEOI, the certainty of its contribution to cancer, and therefore its translational importance, varies substantially. Some GEOIs will be strong, causal ‘drivers’ of important cancer hallmarks1. Others will be weaker but important ‘contributors’ to the development of cancer pathophysiology. And many will be genomic ‘noise’ (or ‘passengers’): that is, elements that are biologically ‘neutral’ and have been accumulated by chance during the cancer’s lifespan. Distinguishing the drivers and contributors from the passengers is a central challenge in genomic research. This is made more difficult by the diversity of GEOI function and the likelihood that GEOI function might depend on the tumour type (or subtype), as well as on the tumour microenvironment.
The assignment of GEOIs as drivers is compelling in the case of high-frequency events: for example, the amplification of regions that contain EGFR in glioblastomas (in 45% of tumours) or ERBB2 in breast cancer (in 20% of tumours); deletions of regions that contain CDKN2A or the tumour-suppressor gene PTEN (in many solid tumours); or mutations in TP53, RAS, BRAF or PIK3CA in a wide range of solid tumours (see the Cancer Gene Census, http://www.sanger.ac.uk/genetics/CGP/Census). Such assignments rest on the weight of functional evidence built up over decades, a luxury not afforded for GEOIs that are being found and will be found by using modern high-throughput genomic technologies. Furthermore, these prominent ‘gene mountains’ seem to be few and far between relative to the numerous ‘hills and valleys’ stretching broadly over large regions of the cancer genome40,41. Which of these GEOIs are involved in the crucial paths to malignancy? And what are their relative contributions? These are challenging questions without simple answers, but progress can be made by integrating data from multiple systems and then searching for common patterns (Fig. 2): that is, searching for GEOIs that are recurrently dysregulated by multiple mechanisms in several biological systems. In this section, we discuss several approaches that have been used successfully to find drivers and contributors — the needles in the haystack of cancer genome data — including integrative analyses of multidimensional data, interspecies comparative genomics and analyses of human cancer cell-line systems.
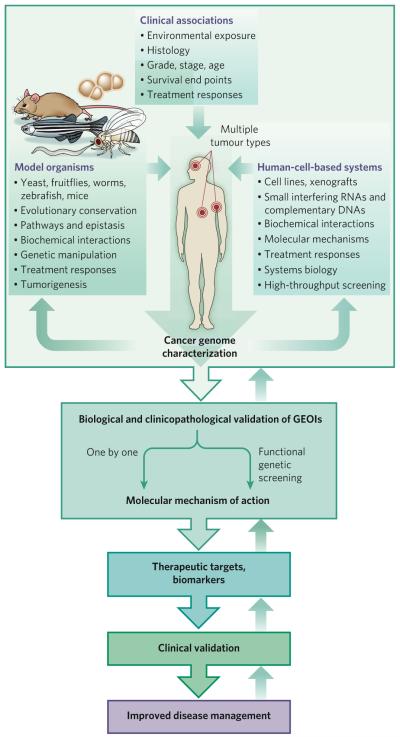
Figure 2. Integration of complex multidimensional genomic data with insights from other model systems.
The identification of cancer drivers or contributors from multidimensional genomic data (such as that shown in Fig. 1) from a particular human tumour type can be facilitated by integration with similar data from other tumour types (for example, by searching for GEOIs that are common to cancers of different lineages). Incorporating clinical information into the analysis of this genomic data helps to narrow the focus to clinically relevant GEOIs. This genomic knowledge can be further filtered by integrating it with insights obtained from studies in model systems. These systems can include non-human model organisms (such as mice, zebrafish, nematodes, fruitflies and yeast), which can be studied to identify evolutionarily conserved GEOIs, to define pathways that GEOIs influence and to elucidate the roles of GEOIs in normal development. Cell-line model systems also can be useful, particularly for functional genetic screening or monitoring responses to drugs. This type of integrative analysis, which extends beyond the cancer genome, is an informative way to identify GEOIs that are likely to be drivers or contributors. After such GEOIs have been identified, they need to undergo stringent biological and clinicopathological validation (Box 2), a labour-intensive process that can be accelerated by carrying out functional screening with a library of GEOIs rather than by assessing one GEOI at a time. For successful translation into the clinic — that is, development of a therapeutic agent that targets the GEOI or a biomarker for the GEOI — a basic understanding of the molecular mechanism of action of the GEOI is helpful, particularly in terms of the specific cellular and genetic context in which it maintains the tumour. Such a biomarker or therapeutic agent will then need to be clinically validated before it can be adopted for routine clinical practice. At each step of this process, the results can be fed back to inform and refine the analyses and to help improve the validation platforms.
Integrative analyses of multidimensional data
The cancer genome can be dysregulated through multiple mechanisms. These include modifications to the DNA and the histones, changes in the DNA structure and copy number, and mutations in the coding and non-coding sequences. These changes can lead to alterations in transcription, translation, post-translational modification and, ultimately, gene and protein function. Technological advances that allow the cancer genome to be examined in multiple ‘omic’ dimensions are helping to focus the search for drivers and contributors, by uncovering GEOIs that tend to be dysregulated by several mechanisms. A classic example is the tumour-suppressor protein INK4A (encoded by CDKN2A), which can be inactivated in three ways: through the homozygous deletion of 9p21 or the region of 9p21 that contains CDKN2A; through the epigenetic silencing of gene expression (by promoter methylation); or through point mutations that cripple the function of INK4A42. Similarly, the PIK3CA oncogene can be activated through amplification and overexpression43 and/or through activating mutations34. Such dysregulation through multiple mechanisms is clearly illustrated when examining well-known oncogenes in a typical signalling pathway (Fig. 3). In other words, if a genetic element is important, then the cancer will find a way to dysregulate it by any means possible. For this reason, the targeted resequencing of genes located in regions of amplification has borne fruit, such as identifying the c-KIT oncogene as a therapeutic target for mucosal and acral melanomas18. Thus, data showing that a GEOI can be dysregulated in several complementary ways in cancer, through the integ ration of more than one dimension of genomic information, provide strong evidence that a GEOI is likely to be pathogenetic. The current large-scale cancer genome projects that are carrying out genome-wide characterization in a coordinated and comprehensive manner will be the most powerful at leveraging such multidimensional data for integrative analyses. In addition, integration across tumour types can be highly informative, because it is clear that the mechanisms of dysregulation of many oncogenes, including MYC, EGFR, AKT1, RAS, TP53, PTEN and CDKN2A, vary according to tumour type. For example, genes, such as MYC, that are activated by translocation in leukaemias can be activated by amplification in solid tumours. The convergence of genomic data that implicate a particular GEOI across tumour types can help to rapidly prioritize GEOIs that are likely to have broad importance. As a by-product, it is probable that the power of genomic biomarkers to determine prognosis or predict responses to therapies will increase substantially if assays are developed to assess the cumulative effect of all mechanisms of dysregulation, including effects on protein structure and abundance.
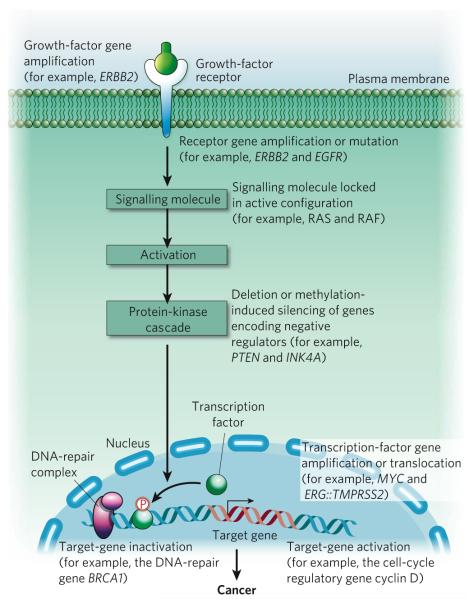
Figure 3. Disruption of intracellular signalling by alterations in the cancer genome.
A simplified signalling pathway is depicted to highlight known examples of bona fide oncogenes that are subjected to dysregulation by various mechanisms. It is clear that a signalling pathway can be disrupted at multiple points, and a variety of genomic and epigenomic alterations can contribute to this, ultimately leading to cancer.
Interspecies comparative cancer genomics
Another approach to uncovering drivers and contributors is to use evolutionary conservation as a guide. This can be a powerful way to find oncogenes, because genes that are involved in pathways that are dysregulated in cancers — such as receptor-tyrosine-kinase signalling, cell-cycle regulation and apoptosis — are strongly conserved across species44,45. This comparative approach was enormously helpful in refining the draft of the human genome sequence. With respect to cancer, it has been established that oncogenes from one species can induce the malignant transformation of cells from different species, despite poor sequence conservation (for example, the Drosophila spp. homologue of MYC, diminutive, can transform rodent cells46). Recent large-scale, cross-species comparisons have established that mouse and human tumours sustain orthologous genomic events in diverse tumour types47-49. This finding supports the view that genomic alterations conserved across species are more likely to represent crucial events in tumorigenesis and that using evolutionary conservation as a filter can provide a powerful solution to the central problem of noise in genomic data sets.
Early studies of cancer across species involved histopathological diagnoses, but such cross-species comparisons now include genetic and genomic analyses to show, for example, that genetically engineered mice can be used to model genetic aspects of human cancer. That mouse models are valid for studying human cancer is exemplified by cross-species conservation of gene-expression patterns that result in activation of the gene encoding Ki-RAS in lung cancers50, as well as conservation of somatic mutations in the gene encoding NOTCH1 in mouse and human T-cell acute lymphoblastic leukaemia51. These findings were followed by studies providing proof of the concept that comparing genomic profiles of mouse and human tumours allows previously unidentified oncogenes to be uncovered47,48. In one of these studies, by Minjung Kim et al.47, the ability to manipulate stages of mouse tumour development in vivo — from regression to recurrence to escape — was used to force the selection of aberrations conferring metastatic capability on tumours. Genome-wide copy-number profiles of these ‘escaper’ tumours revealed focal amplification in regions syntenic (that is, on a chromosomal region of common evolutionary ancestry) to human 6p24-25, a region that sustains copy-number gain in 36% of metastatic melanomas but not in primary melanomas52. Although 6p gain is highly recurrent, indicative of potential pathogenetic and/or prognostic importance in human tumours, the large region of amplification in human tumours makes the identification of drivers and contributors difficult or even impossible. Given the focal nature of the event in mice, cross-species comparison was able to narrow down the area of interest to an 850-kilobase region encompassing only eight annotated genes, with NEDD9 (which encodes an adaptor protein) as a putative driver. With that information as a guide, further functional and clinico-pathological studies documented the metastasis-promoting activities of NEDD9 and uncovered its molecular mechanism of action (interaction with focal adhesion kinase). Likewise, when looking at recurrent copy-number aberrations in tumours with ERBB2 amplification, comparisons between human breast tumours and a transgenic mouse model (in which an oncogenic form of Erbb2 called NeuNT was expressed under the control of the endogenous Erbb2 promoter) implicated the genes encoding GRB7 and 14-3-3-σ as contributors to the ERBB2-mediated oncogenic process53.
Although syntenic aberrations have been observed between mouse and human tumours, it is important to note that the genomes of most mouse tumours accumulate far fewer aberrations than do solid tumours in humans. For example, in oncogene-driven mouse models of cancer, tumours often have few or no copy-number aberrations, and the infrequent (and typically simple) copy-number aberrations that are present presumably occur only under strong selective pressures. This simplicity facilitates the identification of drivers and contributors targeted by such copy-number aberrations, as exemplified by the studies of Kim et al.47 (discussed earlier) and Lars Zender et al.48. The disadvantage, however, is that this method does not lend itself to widespread use of cross-species comparison.
On the basis of observations that DNA-breakage events induced by telomere dysfunction can drive regional amplifications and deletions and that laboratory mice do not experience telomere-based crisis, Ronald DePinho and colleagues knocked out the gene encoding the RNA component of the telomerase holoenzyme from the mouse germ line in an effort to humanize the mouse genome. The resultant telomerase-deficient mice experienced progressive shortening of telomeres with each successive generation of mice, eventually leading to telomerebased crisis54. Tumours from these animals indeed showed high levels of instability, harbouring numerous non-reciprocal translocations and complex copy-number aberrations55-57. A genome-wide comparison of such genome-unstable mouse tumours with several human cancers of diverse origins showed non-random overlaps between the copy-number aberrations. This finding proves that mouse and human tumours experience common biological processes that are driven by orthologous genetic events49.
Attesting to the potential of such cross-species comparisons in oncogene discovery, the focused resequencing of GEOIs within syntenic deletions uncovered a high frequency of mutations in FBXW7 and PTEN in human T-cell acute lymphoblastic leukaemia49. Mutations in PTEN were also shown to modify responses to NOTCH1 inhibitors in the clinic58. These studies support the idea that cross-species synteny is both a measure of validation, by virtue of evolutionary conservation and use of different genetic mechanisms (that is, a GEOI can be dysregulated by different mechanisms, such as mutation and copy number), and a guide for discriminating drivers and contributors from passengers.
Another way in which mice are valuable for comparative genomic studies is in the identification of susceptibility loci. Extending the concepts used to identify BRCA1, it might be expected that mutations or polymorphisms that contribute to cancer susceptibility are subjected to positive selection during the ‘evolution’ of the cancer genome. Thus, these mutations or polymorphism might be found by allele-specific analysis of copy number and gene expression in defined model systems. For example, using genomic strategies, Allan Balmain and colleagues59,60 identified that polymorphic variants of AURKA (also known as STK15), which encodes an aurora kinase, are associated with an increased risk of developing cancer at several sites in humans. These studies began by analysing the position of quantitative trait loci that control susceptibility to skintumour formation in mice from interspecific crosses (Mus musculus × Mus spretus). One of these loci, Skts13, was orthologous to a region that is frequently increased in copy number in human cancers of the breast, colon and ovary; this region, 20q13, contains the gene encoding AURKA. Analyses of the expression of the mouse orthologue of AURKA, Stk6, showed an allele-specific difference in the mouse interspecific crosses, and copy-number analyses of two alleles AURKA 91A and AURKA 91T showed that AURKA 91A is preferentially amplified in human colon tumours. A subsequent meta-analysis of the association between the alleles AURKA 91T and AURKA 91A and the risk of developing cancer of the colon, breast, prostate, skin, lung and oesophagus showed an increased risk in both homozygotes and heterozygotes. These results confirmed that the AURKA 91A is a low-penetrance cancer-susceptibility allele that increases the risk of developing many cancer types. This integrative analysis of quantitative cancer traits in mice, allele-specific copy-number change and expression, and susceptibility to cancer in large population-based studies serves as a model for the definitive identification of the (probably large number of) low-penetrance, high-prevalence polymorphisms that influence cancer risk.
Finally, model systems, including mouse models, are well suited to forward genetic screens, in which researchers can ‘listen’ and let the cancer cells ‘tell’ which events are required or preferred on the path towards full malignant transformation. For example, retroviral insertional mutagenesis in mice has yielded recurrent and common insertion sites at loci containing genes such as Ras, Myc, Notch1, Flt3, c-Kit and Tp53 (ref. 61), attesting to the power of this method to identify oncogenes when the results are integrated with existing and emerging human cancer genome data.
Cell-line model systems
Much of our understanding of tumour cell biology, including aspects of gene regulation and signalling, has come from studies of tumour cells in culture. The roughly 50,000 publications describing uses of the HeLa cell line and the 20,000 publications describing uses of the NIH/3T3 cell line attest to this fact. That said, established tumour cell lines grown on plastic dishes, in three-dimensional cultures or in immunocompromised mice cannot fully recapitulate all the biological aspects of tumours that are growing in the complex human microenvironment. Nor can any model fully represent the responses of the various human tumours to therapy — in part because of differences in the biological environment and in part because the models do not capture the range of biological, genomic and epigenomic diversity found in human tumours. Therefore, it is expected that each model system has strengths and weaknesses. Mice are one such system. As we have described, and as is discussed in greater detail elsewhere62, the value of mouse models is unequivocal. As long as researchers are aware of the limitations of any one model, then the information that such a system offers can be used. Integrating data from several models will help to build a true picture of cancer.
So, what can be learned about genomic aberrations by studying cell-line models? And why are these models important? To put it simply, cell lines are essential for the functional and biological validation of GEOIs (Box 2). Almost without exception, the functional validation of a GEOI and establishment of its molecular basis of action begins with various cell-line model systems, including established tumour cell lines (which are versatile and easy to manipulate). These systems allow the possible roles of GEOIs in the pathophysiology of cancer to be tested. For example, the driver or contributor role of a GEOI found in a region of recurrent amplification might be studied by assessing the consequences of enforced expression of the GEOI in cell lines in which it is expressed at a normal level. Likewise, the role of a GEOI in a region of recurrent deletion might be assessed by decreasing its expression by using RNA inter ference (RNAi)-mediated knockdown in a cell line in which it is expressed at a normal level. Cell lines derived from tumours in which GEOIs are dysregulated by genomic or epigenomic aberrations are valuable ‘experiments of nature’ that also provide information about GEOI function, for example through assessing the biological consequences of restoring dysregulated GEOI expression to levels that are closer to normal.
Box 2 | Validation of GEOIs.
The points below outline the basic approach to validating GEOIs that have been identified in the cancer genome. GEOIs need to be validated in terms of their biological activity and their clinicopathological association, and each validation should be confirmed by using several assays. It is important to note that it is the cumulative weight of evidence — as assessed by several of the assays outlined below but not any single assay — that determines whether a GEOI contributes to, or drives, cancer.
Biological validation
The types of assay for biological validation are listed.
Model systems for manipulation of GEOIs
Established mouse or human tumour cell lines with detailed genomic characterization
Relatively naive, non-transformed primary cell cultures established from normal tissues
Genetically engineered animal models for in vivo studies
Candidate GEOI manipulation
Loss of function, by RNAi-mediated knockdown of the GEOI or by pharmacological inhibition using available drugs
Gain of function, by expression of cDNA containing the GEOI or by pharmacological activation using available drugs
Functional assays for biological activity
Cell proliferation and/or apoptosis, and migration and invasion in two-dimensional or three-dimensional culture models
Anchorage independence in vitro
Migration, invasion and in vivo lung seeding through tail-vein injection
Tumorigenicity of subcutaneous or orthotopic xenotransplants
GEOI-specific assays
Biochemical or intracellular signalling activities
Clinicopathological validation
The properties of GEOIs that are likely to drive or contribute to cancer are listed, together with ways to search for these properties.
Evidence of dysregulation at the DNA level through various mechanisms
Prevalence
Assay genetic and/or epigenetic events in a large number of tumours across a broad range of cancer types
Evidence of altered expression
Measure level of expression
Search for altered splicing variants
Search for novel transcripts of translocated regions
Assess changes in proteins
Correlation with clinical parameters
Cancer subtype
Survival duration or response to treatment
A major obstacle to the accurate interpretation of functional data derived from established tumour cell lines is the lack of clarity about the complements of genetic alterations that these cell lines carry. It has become clear that the genotype of the system — be it a cell line, a model or even a patient — can dictate the behaviour of tumour cells and can alter their response to a manipulation such as RNAi-mediated knockdown or pharmacological inhibition. As is the case for the original tumours from which they were derived, no two tumour cell lines are alike. Moreover, there is the legitimate concern that genomic aberrations will be gained or lost during extended passages in culture. Therefore, it is important that cell-line models — whether grown on plastic, in three-dimensional culture or in xenografts (that is, a grafted into a different species) — are subjected to the same level of comprehensive genomic characterization as human tumour specimens. In this way, the interpretation of functional studies can be guided by the knowledge of the similarities and differences between the cell lines and tumours that they are intended to model. It is also important that any cell-line system used for functional studies of the cancer genome comprises multiple independent cell lines that are molecularly diverse. If there is sufficient diversity, analyses of such cell-line collections minimize the risk that the elucidated function of an aberration will be idiosyncratic to a particular cell line.
As is the case for model organisms, forward genetic screening using a tumour cell-line model (particularly given recent advances in RNAi technology) can be used to identify cancer-relevant genes. Such in vitro screens are limited by the kinds of phenotypes amenable to high-throughput screens in culture (such as viability and growth assays). Nonetheless, recent studies that combine high-throughput RNAi-based screening with in vitro genomic profiling of primary human tumour specimens have led to the identification of the transcription factor REST as a tumour-suppressor protein in colon cancer63, IKBKE (which encodes a signalling molecule) as an oncogene in breast cancers64, and PIK3CA mutations as important determinants of resistance to treatment with trastuzumab65.
Cell lines are also important models for assessing drug sensitivity and resistance in the quest to identify biomarkers that can guide early-phase clinical-trial studies; to identify drugs that might be effective in cancer subtypes that are resistant to the drug(s) used in the current standard of care; and to identify effective drug combinations. Although still in its infancy, an increasing number of studies support the concept that molecular biomarkers for predicting drug responses can be uncovered by analysing how molecularly characterized tumour cell lines respond to particular chemotherapeutic agents (which target molecular mechanisms that are intrinsic to the tumour cells)66-70. As a corollary, these analyses also identify drugs with a high specificity for subsets of tumour cells defined by certain molecular characteristics. Examples are in vitro analyses that predict the known sensitivities to trastuzumab71 and lapatinib68 of tumours in which ERBB2 has been amplified, sensitivity to gefitinib of tumours harbouring EGFR mutations 3-5, resistance to gefitinib conferred by an acquired mutation in EGFR72, and resistance to imatinib mesylate in tumours with mutated or amplified BCR-ABL73.
Using cell-line model systems that include large numbers of independent, established tumour cell lines of broad molecular and cellular diversity, together with comprehensive genomic characterization, can be and will be tremendously effective for translating genomic insights into clinical end points. But these systems could be further improved by developing co-culture or three-dimensional culture conditions that more closely model in vivo microenvironments, as well as by developing strategies to establish primary or short-term cultures that minimize the ‘culture shock’ associated with adapting to plastics.
A molecular understanding
The identification of driver or contributor GEOIs, especially the weaker or less prevalent ones, can be greatly accelerated by integrative analyses of multidimensional data and by comparisons with data from multiple model systems or species (Fig. 2). But identification of a GEOI is insufficient for its translation into a clinical end point. Cancer is a complex and heterogeneous collection of disease entities that are defined by clinical, histopathological and genetic parameters. Given this disease heterogeneity, even if a strong correlation between a GEOI and cancer is found in the laboratory in a test validation set (for example, a collection of genomic data, behaviour in a model system or even responses in a clinical trial), this correlation, no matter how significant, might not apply to every patient or trial subject. Without a definition of the genomic and biological context under which a GEOI exerts its cancer-associated activities, the full diagnostic and prognostic and therapeutic value of these genomic insights will not be realized.
Consider the example of EGFR mutations in non-small-cell lung cancer and glioblastoma multiforme (GBM). Mutational activation of EGFR in non-small-cell lung cancer is present in a subpopulation of patients who are highly responsive to targeted inhibition of EGFR. The proportion of patients with non-small-cell lung cancer who have an activating mutation in EGFR is small (about 10% in studies carried out in the United States and somewhat higher in Asian populations)37. Thus, the response of these patients to gefitinib, which inhibits the tyrosine-kinase activity of EGFR, would not have emerged in the absence of genetic stratification of this clinically distinct population. Conversely, amplification of a mutant form of EGFR known as EGFRvIII is prevalent in GBM (in about 45% of primary GBM cases)74, yet EGFR-specific tyrosine-kinase inhibitors have strikingly little clinical effect. A positive, albeit transient, clinical response has been detected in subsets of patients in whom EGFR is amplified or mutated but PTEN is intact 75, indicating that this key molecule downstream of EGFR in the signalling pathway can modify the biological response of the tumour (Fig. 3). However, these positive responses do not last, despite documented pharmacological extinction (that is, inactivation) of mutated or amplified EGFR. In this case, the proteomic profiling of receptor-tyrosine-kinase activation patterns in solid tumours, including GBM and lung cancer, has provided a rational explanation for the patterns of clinical responses. Specifically, Jayne Stommel et al.76 showed that established GBM cell lines, GBM xenotransplants and GBM primary tumour specimens from patients contain several coactivated receptor tyrosine kinases and that inhibition of EGFR alone can lead to its replacement with other coactivated receptor tyrosine kinases in the phosphatidylinositol-3-OH-kinase (PI(3)K) signalling complex, thus maintaining downstream signalling and cell survival. Signalling downstream of PI(3)K was extinguished only when multiple receptor tyrosine kinases were targeted by RNAi or by a combination receptor-tyrosine-kinase inhibitor76. Thus, the integration of genomic and proteomic insights with the molecular dissection of the signalling complex now provides a more accurate blueprint for the rational deployment of receptor-tyrosine-kinase inhibitors for treating GBM, tumours of the lung and other solid tumours.
Establishing the molecular basis of action of a GEOI in a specific tumour-biological context is perhaps the most difficult step in cancer genomics. Compounding the challenges of lengthy and laborious functional and clinicopathological validation (Box 2) is the biological phenomenon of false negatives. False negatives can arise in many ways; for example, when the cancer-associated biological activities of a GEOI (such as interaction with the host stroma) are not captured by standard cell-based assays; when a GEOI has a relevant role but only in a particular cellular or genetic context that is not recreated in the validation assay; and when a GEOI contributes only part of the overall activity conferred by a genomic event (so that the activity of a single GEOI is negligible in the absence of this cooperating partner or partners). Therefore, validation must not rely on just a single type of assay that involves a single manipulation.
Gain-of-function and loss-of-function manipulations for multiple tumour phenotypes using multiple cell lines should be carried out to search for the context in which biological activity can be uncovered. This process can be aided by knowledge obtained from other analyses, such as information about the biology of the tumour, the gene family of the GEOI, the pathways that the GEOI product is involved in, and insights from integrative analyses that nominated the GEOI. For example, if a GEOI identified by integrative genomic analyses is prioritized further on the basis of its known role in neural stem-cell homeostasis, then the next step would be to assess how manipulation of the GEOI affects the renewal, maintenance and differentiation of neural stem cells, in addition to carrying out the more generic assays of anchorage independence or cell proliferation (Box 2). Similarly, if a GEOI is identified in a subset of tumours with a particular genotype (such as with activated RAS or a mutation in EGFR), then its biological importance needs to be assayed in the appropriate context. This process has been demonstrated in two recent studies47,48. Kim et al.47 showed that NEDD9 had gain-of-function pro-invasion activities only in cells in which BRAF or RAS was concomitantly activated, an experimental design that was informed by the characteristics of the metastatic escapers harbouring NEDD9 amplification. Zender et al.48 showed that the inhibitor of apoptosis IAP1 (also known as BIRC2) and the transcription factor YAP had oncogenic activities in Tp53+/- hepatoblasts with Myc activation but not in those with Akt1 or Ras activation. This finding is consistent with the presence of an amplicon in the chromosomal region 9qA1 (which contains the genes encoding IAP1 and YAP) in this mouse model of hepatocellular carcinoma. In the study by Zender et al.48, both IAP1 and YAP were shown to be targets of 9qA1 amplification, showing that a single genomic aberration can dysregulate more than one gene that contributes to the pathophysiology of the cancer. The chances of missing important GEOIs in a region of recurrent aberration can be reduced by using efficient functional genomic assays to assess the consequences of changing the expression levels of all GEOIs associated with the aberration. For example, genetic screens can be carried out with low-complexity libraries representing GEOIs resident in a particular genomic event (which is especially useful for regions that are large and gene-rich), allowing the identification of cooperating contributors (which together confer the biological advantage selected for in the cancer cells). This functional genomic approach will be important for sorting out which of the less impressive ‘hills and valleys’ are biologically important.
Similarly challenging is the issue of biological false positives. For example, an RNAi-mediated loss-of-function assay is a powerful way to determine whether the expression of a GEOI is required in a cell for a specific tumorigenic phenotype (such as cell survival, anchorage independence or invasion). However, given the innumerable genetic and epigenetic alterations that are present in established tumour cells (and, consequently, the altered signalling between pathways and networks), the observed phenotype might be an artefact. In this case, finding a complementary gain-of-function activity can help to increase the evidence in support of a particular GEOI being a true driver or contributor to cancer. In addition, the type of functional activity also conveys a different level of confidence; for example, anchorage-independent growth in soft agar is a more stringent assay than increased proliferation in fully supplemented culture medium.
Biological false positives can also emerge as a direct consequence of the artificial nature of the assays used. Consider the possibility that overexpression of a GEOI confers a strong anchorage-independent phenotype; this effect might, however, result from the supraphysiological level of expression in vitro. Conversely, knockdown of a GEOI might result in cell death because its expression is required for the survival of all cells not just cancerous ones. To this end, clinicopathological validation through analysis of the DNA, messenger RNA and protein levels in normal samples and tumour samples arranged in microarrays can provide support for cancer relevance, by demonstrating the prevalence of genomic aberrations or dysregulated GEOI expression in large independent cohorts of specific tumour types. This can be particularly informative if the tumour cohorts are annotated with the clinical outcome because such a survey will not only add to the evidence but also provide invaluable insight into possible clinical contexts for therapeutic development. Ultimately, it is the cumulative weight of evidence based on the strength of particular functional activities, the magnitude of clinicopathological data and the importance of mechanistic clues that provides the confidence to assign a GEOI as a cancer-relevant driver or contributor rather than a mere passenger.
Conclusion
Cancer is the phenotypic end point of numerous genomic and/or epigenomic alterations that have accumulated within cells, and of the interactions of such altered cells with the stromal components in a unique host microenvironment. Some of the major challenges in translating the knowledge gained from cancer genomics into clinical practice stem from the fact that many cancer-associated changes in the genome are noise, as well as from the incomplete understanding of the biological functions of many of the genetic elements that are present in recurrent genomic alterations. Compounding these issues is the unfortunate reality that cancer is a highly complex, nimble and versatile disease.
We argue here that making sense of this complexity can be greatly facilitated by integrating genomic and biological insights from model systems with clinical knowledge of the disease. Translation can further be accelerated by rigorous biological validation and mechanistic exploration in preclinical settings to better define the clinical context(s) in which a genetic element (or components of the pathways or networks that is involved in) is an effective point of intervention for therapy. At the same time, we need to consider that the current understanding of what makes a strong driver, a cooperating contributor or, for that matter, a genomic passenger is limited at best and might be incorrect. Therefore, this must be an iterative learning process in which the results of downstream biological validation and mechanistic studies — and even of clinical experiences from which inhibitors or biomarkers are developed and used — can and must inform the integrative analyses and the validation approaches. This effort will be facilitated by the development or assembly of model systems that are characterized to the same degree as primary tumours and that can be used to quickly test hypotheses suggested by ‘omic’ analyses of tumours.
For the efficient translation of cancer genome information into the clinic, studies must go beyond statistical analyses of large genomic data sets. This process will require the amalgamation of expertise and insights from cancer biology, cancer genetics, cancer modelling and systems biology, as well as clinical experiences. We suggest that this integrative process will be facilitated by establishing international centres or cooperatives that organize the information obtained from diverse genomic, biological and clinical studies in ways that guide functional analyses and optimize the translation of the cancer genome into effective biomarkers or therapeutics.
Acknowledgements
We thank R. DePinho, A. Futreal, P. Mischel, A. Kimmelman, K.-K. Wong, W. Hahn and K. Polyak for discussions and critical reading of the manuscript. This work was supported in part by the US Department of Energy, the Office of Science, the Office of Biological and Environmental Research, the National Institutes of Health and the National Cancer Institute.
Footnotes
The authors declare no competing financial interests.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci. Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 3.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101.References 3-5 show that a subset of patients with lung cancer have EGFR mutations and are responsive to an EGFR-specific tyrosine kinase inhibitor, a finding based on prospective analyses of retrospective data.
- 6.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401.This paper provides the first proof of concept of targeted therapy: chronic myeloid leukaemia harbouring BCR-ABL was shown to be sensitive to treatment with a BCR-ABL-specific tyrosine-kinase inhibitor, imatinib mesylate.
- 7.Pegram M, Slamon D. Biological rationale for HER2/neu (c-erbB2) as a target for monoclonal antibody therapy. Semin. Oncol. 2000;27:13–19. [PubMed] [Google Scholar]
- 8.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 9.Futreal PA, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 10.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 11.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 12.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J. Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 13.Gruis NA, et al. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nature Genet. 1995;10:351–3. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- 14.Nowell PC. Discovery of the Philadelphia chromosome: a personal perspective. J. Clin. Invest. 2007;117:2033–2035. doi: 10.1172/JCI31771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nardi V, Azam M, Daley GQ. Mechanisms and implications of imatinib resistance mutations in BCR-ABL. Curr. Opin. Hematol. 2004;11:35–43. doi: 10.1097/00062752-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Quintas-Cardama A, Kantarjian H, Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nature Rev. Drug Discov. 2007;6:834–848. doi: 10.1038/nrd2324. [DOI] [PubMed] [Google Scholar]
- 17.Demetri GD. Targeting c-kit mutations in solid tumors: scientific rationale and novel therapeutic options. Semin. Oncol. 2001;28:19–26. [PubMed] [Google Scholar]
- 18.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 19.Hodi F, et al. A major response to Imatinib mesylate in KIT mutated melanoma. J. Clin. Orthod. doi: 10.1200/JCO.2007.14.0707. in the press. [DOI] [PubMed] [Google Scholar]
- 20.Rowley JD. The role of chromosome translocations in leukemogenesis. Semin. Hematol. 1999;36:59–72. [PubMed] [Google Scholar]
- 21.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 22.Bradford TJ, Tomlins SA, Wang X, Chinnaiyan AM. Molecular markers of prostate cancer. Urol. Oncol. 2006;24:538–551. doi: 10.1016/j.urolonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Volik S, et al. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc. Natl Acad. Sci. USA. 2003;100:7696–7701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. doi: 10.1038/ng.128. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schechter AL, et al. The neu oncogene: an erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- 27.King CR, Kraus MH, Aaronson SA. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science. 1985;229:974–976. doi: 10.1126/science.2992089. [DOI] [PubMed] [Google Scholar]
- 28.Semba K, Kamata N, Toyoshima K, Yamamoto T. A v-erbB-related protooncogene, c-erbB-2, is distinct from the c-erbB-1/epidermal growth factor-receptor gene and is amplified in a human salivary gland adenocarcinoma. Proc. Natl Acad. Sci. USA. 1985;82:6497–6501. doi: 10.1073/pnas.82.19.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coussens L, et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science. 1985;230:1132–1139. doi: 10.1126/science.2999974. [DOI] [PubMed] [Google Scholar]
- 30.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106.This study correlated ERBB2 amplification with outcome for individuals with breast cancer.
- 31.Kallioniemi A, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 32.Cameron D, et al. A phase III randomized comparison of lapatinib plus capecitabine versus capecitabine alone in women with advanced breast cancer that has progressed on trastuzumab: updated efficacy and biomarker analyses. Breast Cancer Res. Treat. doi: 10.1007/s10549-007-9885-0. doi:10.1007/s10549-007-9885-0 (in the press) [DOI] [PubMed] [Google Scholar]
- 33.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 35.Carpten JD, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 36.Stephens P, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 37.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nature Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 38.Blackhall F, Ranson M, Thatcher N. Where next for gefitinib in patients with lung cancer? Lancet Oncol. 2006;7:499–507. doi: 10.1016/S1470-2045(06)70725-2. [DOI] [PubMed] [Google Scholar]
- 39.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 40.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720.References 40 and 41 report on large-scale sequencing studies aimed at identifying somatic mutations in human cancers.
- 42.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat. Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 43.Shayesteh L, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nature Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 44.Horvitz HR, Shaham S, Hengartner MO. The genetics of programmed cell death in the nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 1994;59:377–385. doi: 10.1101/sqb.1994.059.01.042. [DOI] [PubMed] [Google Scholar]
- 45.Nurse P, Masui Y, Hartwell L. Understanding the cell cycle. Nature Med. 1998;4:1103–1106. doi: 10.1038/2594. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber-Agus N, et al. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc. Natl Acad. Sci. USA. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030.References 47 and 48 show the power of cross-species integration of cancer genome data for oncogene discovery.
- 49.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886.This paper compares the genomes of mouse tumour cells with genetically engineered chromosomal instability to the genomes of various human cancers and shows that there is a significant non-random number of syntenic events, proving that mouse and human cells can experience common biological processes driven by orthologous genetic events during transformation.
- 50.Sweet-Cordero A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nature Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 51.O’Neil J, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 53.Hodgson JG, et al. Copy number aberrations in mouse breast tumors reveal loci and genes important in tumorigenic receptor tyrosine kinase signaling. Cancer Res. 2005;65:9695–9704. doi: 10.1158/0008-5472.CAN-05-0755. [DOI] [PubMed] [Google Scholar]
- 54.Artandi SE, DePinho RA. Mice without telomerase: what can they teach us about human cancer? Nature Med. 2000;6:852–855. doi: 10.1038/78595. [DOI] [PubMed] [Google Scholar]
- 55.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 56.Maser RS, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Hagan RC, et al. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 58.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ewart-Toland A, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nature Genet. 2003;34:403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- 60.Ewart-Toland A, et al. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- 61.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 62.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nature Rev. Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 63.Westbrook TF, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 64.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052.References 63 and 64 integrate hits from forward genetic screening, using RNAi, with genomic profiles of human cancers to find previously unidentified oncogenes.
- 65.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumasb resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 66.Staunton JE, et al. Chemosensitivity prediction by transcriptional profiling. Proc. Natl Acad. Sci. USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 68.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib ( GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 69.Hieronymus H, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 70.Wei G, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008.This study shows that the cancer genomes of a panel of human cancer cell lines reflect the genomic diversity of human cancers.
- 72.Wong KK. HKI-272 in non small cell lung cancer. Clin. Cancer Res. 2007;13:s4593–s4596. doi: 10.1158/1078-0432.CCR-07-0369. [DOI] [PubMed] [Google Scholar]
- 73.Scappini B, et al. Changes associated with the development of resistance to imatinib (STI571) in two leukemia cell lines expressing p210 Bcr/Abl protein. Cancer. 2004;100:1459–1471. doi: 10.1002/cncr.20131. [DOI] [PubMed] [Google Scholar]
- 74.Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 75.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 76.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 77.Greshock J, et al. A comparison of DNA copy number profiling platforms. Cancer Res. 2007;67:10173–10180. doi: 10.1158/0008-5472.CAN-07-2102. [DOI] [PubMed] [Google Scholar]
- 78.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drmanac R, et al. DNA sequence determination by hybridization: a strategy for efficient large-scale sequencing. Science. 1993;260:1649–1652. doi: 10.1126/science.8503011. [DOI] [PubMed] [Google Scholar]
- 80.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 81.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 83.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nature Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 84.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 85.Dai Z, et al. An AscI boundary library for the studies of genetic and epigenetic alterations in CpG islands. Genome Res. 2002;12:1591–1598. doi: 10.1101/gr.197402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Plass C, et al. An arrayed human not I-EcoRV boundary library as a tool for RLGS spot analysis. DNA Res. 1997;4:253–255. doi: 10.1093/dnares/4.3.253. [DOI] [PubMed] [Google Scholar]
- 87.van Steensel B, Henikoff S. Epigenomic profiling using microarrays. Biotechniques. 2003;35:346–350. 352–354, 356–357. doi: 10.2144/03352rv01. [DOI] [PubMed] [Google Scholar]
- 88.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu M, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nature Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 90.Leary RJ, Cummins J, Wang TL, Velculescu VE. Digital karyotyping. Nature Protoc. 2007;2:1973–1986. doi: 10.1038/nprot.2007.276. [DOI] [PubMed] [Google Scholar]
- 91.Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front. Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]