Initiation of the cell division cycle in yeast is controlled by two distinct kinases that coordinately regulate the interaction of Whi5, a repressor of initiation, with histone deacetylases.
Abstract
START-dependent transcription in Saccharomyces cerevisiae is regulated by two transcription factors SBF and MBF, whose activity is controlled by the binding of the repressor Whi5. Phosphorylation and removal of Whi5 by the cyclin-dependent kinase (CDK) Cln3-Cdc28 alleviates the Whi5-dependent repression on SBF and MBF, initiating entry into a new cell cycle. This Whi5-SBF/MBF transcriptional circuit is analogous to the regulatory pathway in mammalian cells that features the E2F family of G1 transcription factors and the retinoblastoma tumor suppressor protein (Rb). Here we describe genetic and biochemical evidence for the involvement of another CDK, Pcl-Pho85, in regulating G1 transcription, via phosphorylation and inhibition of Whi5. We show that a strain deleted for both PHO85 and CLN3 has a slow growth phenotype, a G1 delay, and is severely compromised for SBF-dependent reporter gene expression, yet all of these defects are alleviated by deletion of WHI5. Our biochemical and genetic tests suggest Whi5 mediates repression in part through interaction with two histone deacetylases (HDACs), Hos3 and Rpd3. In a manner analogous to cyclin D/CDK4/6, which phosphorylates Rb in mammalian cells disrupting its association with HDACs, phosphorylation by the early G1 CDKs Cln3-Cdc28 and Pcl9-Pho85 inhibits association of Whi5 with the HDACs. Contributions from multiple CDKs may provide the precision and accuracy necessary to activate G1 transcription when both internal and external cues are optimal.
Author Summary
Eukaryotic cells grow and divide by progressing through carefully orchestrated stages of the cell cycle characterized by stage-specific patterns of gene expression, DNA replication, and scission. How stage-specific gene expression is coordinated with cell cycle progression is only partially understood. The phase known as G1 marks the initiation of the cell cycle (called START in yeast) and involves the coordinated expression of more than 200 genes regulated by two transcription factors, SBF and MBF. The activity of SBF and MBF is restrained by binding of the repressor protein Whi5 to the two transcription factors early in G1 phase. Phosphorylation of Whi5 by G1-specific forms of the cyclin-dependent kinase (CDK) Cdc28 promotes dissociation of Whi5 from SBF and its export from the nucleus; this, in turn, releases SBF to activate G1-specific transcription. This G1 transcriptional circuit is analogous to that defined in mammals by the E2F family of transcription factors and the retinoblastoma (Rb) tumor suppressor protein. Rb further contributes to the repression of G1-specific transcription in mammals by recruiting histone deacetylases (HDACs), which are chromatin remodeling complexes that regulate promoter accessibility. Here, we show that regulation of G1-specific transcription in yeast also involves repressor-mediated recruitment of HDACs. We demonstrate that repression by Whi5 is modulated by both Cln-Cdc28 and a second G1-specific CDK, Pcl-Pho85, and further show that both kinases regulate the interaction of Whi5 with HDACs. We propose that regulation of the repressor by more than one G1-specific CDK ensures definitive inactivation of Whi5, a critical event for appropriate cell cycle initiation.
Introduction
Cyclin-dependent protein kinases (CDKs) act as molecular machines that drive cell division, and cell cycle progression is dependent on oscillation between CDK active and inactive states. In S. cerevisiae, the CDK Cdc28 associates with nine different cyclin subunits to promote and coordinate a complex network of events necessary for smooth cell cycle transitions [1]. Irreversible commitment to a new round of cell division occurs toward the end of G1 phase in a process called Start in yeast. The analogous regulatory event is called the restriction point in mammalian cells [2],[3]. In yeast, three G1 cyclins, Cln1, Cln2, and Cln3, associate with Cdc28 to initiate events required for progression through Start. Passage through Start catalyzes a defined molecular program that initiates DNA replication, budding, spindle maturation, and chromosome segregation [3].
One key feature of Start in yeast, and G1 progression in other eukaryotic cells, is the induction of a transcriptional program involving over 200 genes, including those encoding the G1 (CLN1, CLN2, PCL1, and PCL2) and B-type cyclins (CLB5 and CLB6) [4],[5]. G1/S phase-specific transcription depends on two heterodimeric transcription factors called SBF (Swi4,6 cell cycle box binding factor) and MBF (MluI binding factor). These complexes share a common regulatory subunit, Swi6, which is tethered to DNA via its binding partners, encoded by SWI4 in SBF and MBP1 in MBF [5]. At the well-studied HO locus, binding of the zinc-finger transcription factor Swi5 is followed by recruitment of the Swi/Snf chromatin remodeling complex and the SAGA histone acetyltransferase complex [6]–[8]. These events set the stage for SBF binding and recruitment of the SRB/mediator complex [6]. Importantly, subsequent recruitment of PolII and transcription initiation is dependent on CDK activity [9]. Although any one of the three G1 cyclins is sufficient to drive Start, genetic studies indicate a key role for Cln3-Cdc28 in activating SBF and MBF. At the same time Cln1 and Cln2 are required for the proper execution of other Start-related events such as budding and DNA synthesis. Cells lacking CLN3 are large and severely delayed for onset of G1/S transcription, while ectopic induction of CLN3 in small G1 cells activates transcription and accelerates passage through Start [10].
Start does not occur until cells have passed a critical cell size threshold, a barrier modulated by nutrient conditions, among other regulatory inputs [11]. A systematic analysis of cell size profiles for the entire set of yeast deletion mutants uncovered many new regulators of Start including Whi5 and implicated it as an inhibitor of G1/S-specific transcription [12],[13]. Whi5 occupies specific promoters early in G1 phase when CDK activity is low. However, Cdc28-dependent phosphorylation of both Whi5 and SBF/MBF late in G1 phase results in disengagement from SBF and nuclear export of Whi5 consequently leading to activation of SBF- and MBF-dependent transcription [12],[13].
Whi5 is proposed to function in a manner analogous to the well-characterized Rb family proteins in metazoans. E2F, the functional analog of SBF/MBF, regulates G1-specific gene expression required for passage through the restriction point [14]. E2F activity is restricted to late G1 phase because of inhibition by the retinoblastoma protein (Rb). Rb associates with E2F to restrain its activity until late G1, at which point stepwise phosphorylation of Rb by two CDKs, cyclin D-Cdk4/6 and cyclin E-Cdk2, causes the dissociation of Rb from E2F [15]. This process appears to be regulated by a positive feedback loop in which Rb phosphorylation by cyclinE-Cdk2 leads to further dissociation of Rb from promoters and enhancement of G1-transcription. At the molecular level, Rb interacts with both E2F and chromatin remodeling complexes such as histone deacetylases (HDACs) [16]–[18]. Rb appears to repress transcription through at least three distinct mechanisms: (1) Rb can bind directly to the activation domain of E2F thereby blocking its activity [19]; (2) recruitment of Rb can block the assembly of the pre-initiation complex thus inhibiting the activity of adjacent transcription factors [20] and; (3) Rb can recruit remodelers such as HDAC1 and BRG1 to modify chromatin structure. BRG1 is one of the human Swi/Snf adenosine triphosphatases (ATPases) that remodel nucleosomes by utilizing ATP to weaken the interactions between DNA and histones [16],[17]. The specific roles of different CDKs in regulating E2F-Rb function, however, remain unclear.
Another yeast CDK Pho85 was originally discovered as a regulator of phosphate metabolism, but has since been shown to play numerous roles in the regulation of cell division and other processes [21]–[23]. Ten genes encoding Pho85 cyclins (Pcls) have been identified and they appear to dictate substrate and functional specificity of Pho85 [24]–[26]. Expression of three Pcls, PCL1, PCL2, and PCL9, is restricted to G1 phase of the cell cycle [25]. Specifically, PCL9 expression peaks early in G1, whereas maximal expression of PCL1 and PCL2 is observed at Start and is dependent largely on SBF [27]–[29]. Although Pho85 is not essential for viability, it is required for cell cycle progression in the absence of the Cdc28 cyclins CLN1 and CLN2 [29], and its absence leads to catastrophic morphogenic changes that culminate in a G2 arrest [30]. Consistent with this observation, inactivation of both Cdc28 and Pho85 CDKs specifically inhibits expression of G1-regulated genes involved in polarized growth [31].
As noted above, transcriptional repression by Rb has been linked to its interaction with histone modification complexes, in particular HDACs. Recent work highlights the importance of post-translational modifications of histone and nucleosome positioning in regulating gene expression [32],[33]. Histone acetylation neutralizes the positive charge generated by lysine-rich regions present in the N-terminal tails of histones, thereby disrupting nucleosome structure and increasing promoter accessibility [34]. As a result, many transcription activators have been shown to interact with histone acetyltransferases, whereas transcriptional repressors often associate with HDACs to promote nucleosome formation to occlude transcription factor binding [35],[36]. Histone deacetylation in S. cerevisae is mediated by a family of HDACs including Rpd3, Hda1, Hda2, Hos1, Hos2, and Hos3 [37]. Similar to their mammalian counterparts, some yeast HDACs are recruited to promoters by sequence-specific regulatory factors to repress gene expression. For example, the Rpd3 deacetylase complex is recruited to the INO1 promoter by the DNA binding protein Ume6 [35],[38]–[40]. This recruitment results in local histone deacetylation and repression of INO1 gene expression [41]. Hda1 is another example of this type of regulation, and is recruited to its target promoters by the repressor Tup1 [42].
In this study, we provide detailed mechanistic insights into Whi5-dependent regulation of G1-specific transcription and cell cycle progression. Specifically, we identify Whi5, to our knowledge, as the first demonstrated physiological substrate for the G1-specific Pcl9-Pho85 CDK and provide genetic and biochemical evidence supporting a direct role for Pho85 at Start. Furthermore, we show that in a manner similar to Rb in mammalian cells, Whi5-mediated repression involves the HDACs Rpd3 and Hos3. Dual phosphorylation of Whi5 by Cdc28 and Pho85 inhibits Whi5 activity in at least two ways. Both kinases appear to regulate interaction of Whi5 with different HDACs, whereas Cdc28 is also involved in disrupting Whi5 association with SBF and promoting its nuclear export [12],[13]. G1-specific CDKs thus are specialized to regulate different aspects of the same critical cell cycle event—inhibition of Whi5—resulting in definitive inactivation of the Whi5 repressor.
Results
A Synthetic Dosage Lethality Screen Identifies Whi5 as a Putative Substrate for the CDK Pho85
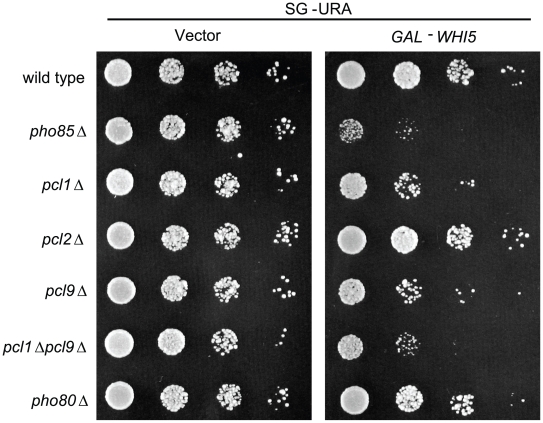
Synthetic dosage lethality (SDL) is a genetic assay that is based on the rationale that increasing levels of a protein may have no effect on the growth of an otherwise wild-type (wt) strain but may cause a measurable phenotype—such as lethality—in a mutant strain with reduced activity of an interacting protein [43],[44]. Previous studies suggest that SDL can be used effectively to identify novel enzyme targets and a genome-wide SDL screen in cells lacking Pho85 identified known targets of the CDK [23]. In addition to known substrates, several putative Pho85 targets were also identified, including the G1-specific transcription repressor Whi5 [24]. To further explore the role of Pho85 in G1 phase-specific transcription we examined the WHI5-PHO85 SDL interaction in greater detail. As noted previously, Pho85 activity and substrate specificity depends on its interaction with cyclin subunits known as Pcls [25]. To implicate specific Pcl-Pho85 complexes in modulating Whi5 function we examined the effects of WHI5 overexpression in cells lacking different Pcls (Figure 1). Similar to effects observed in cln3Δ and cln1Δ cln2Δ mutants [13], overexpression of WHI5 resulted in growth inhibition of pcl1Δ and pcl9Δ deletion strains and this growth defect was exacerbated in a pcl1Δ pcl9Δ double mutant (Figure 1). Unlike pcl1Δ or pcl9Δ mutants, strains lacking PCL2 or PHO80 cyclins were not adversely affected by increased WHI5 dosage suggesting that the WHI5-PHO85 genetic interaction is dependent on the PCL1,2 cyclin subfamily and more specifically on PCL1 and PCL9 (Figure 1). This observation is consistent with the fact that Pcl1 and Pcl9 (but not Pcl2) are the two G1-specific cyclins that localize to the nucleus [30],[45]. The growth phenotype seen in the plating assay was confirmed by measuring growth rates in liquid culture (unpublished data). On the basis of these results, Pcl1/9-Pho85 may contribute to Whi5 regulation in a manner similar to Cln3-Cdc28.
Figure 1. WHI5 overexpression is toxic to strains compromised for Pho85 CDK activity.
Isogenic wt (BY263), pho85Δ (BY391), pcl1Δ (BY628), pcl2Δ (BY451), pcl9Δ (BY694), pcl1Δ pcl9Δ (BY760), and pho80Δ (BY490) strains bearing either GAL1-WHI5 (pBA1973) or empty vector control (pEG-H) were spotted in serial 10-fold dilutions on galactose media and incubated for 72 h at 30°C.
Whi5 Is a Substrate for Pcl9-Pho85 Phosphorylation
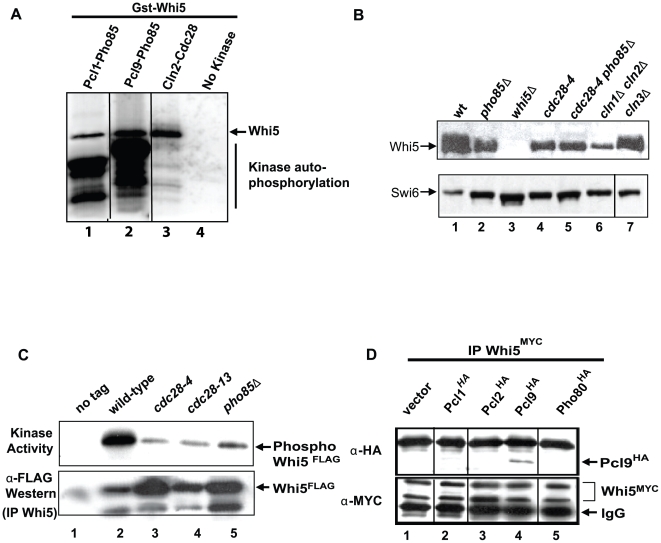
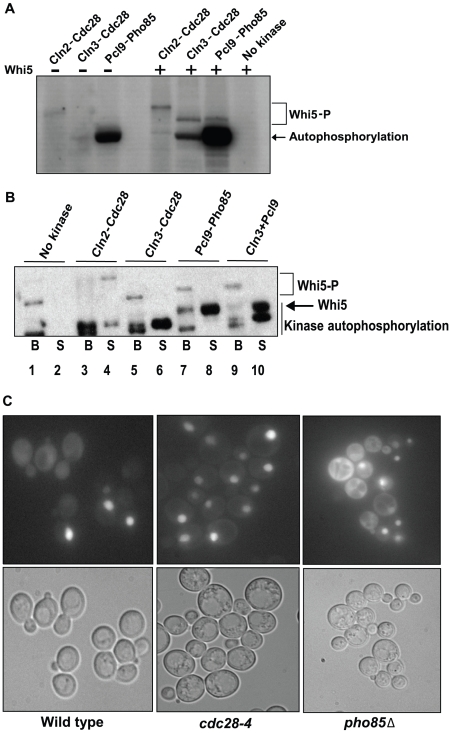
The genetic interactions described above suggest Whi5 may be a direct target of Pho85. Evidence supporting this hypothesis is provided by protein microarray assays where Whi5 is phosphorylated in vitro by Pcl1-Pho85 [46]. We characterized the Whi5-Pho85 interaction biochemically by performing in vitro kinase assays using recombinant Pcl-Pho85 CDK complexes and purified Whi5 as substrate (Figure 2A). Incorporation of [32P] into Whi5 was not detected in the absence of CDKs (Figure 2A, lane 4). However, Whi5 phosphorylation was observed in the presence of Pcl1- and Pcl9-Pho85 (Figure 2A, lanes 1,2) and when compared to Cln2-Cdc28 kinase activity, Pho85 and Cdc28 phosphorylated Whi5 at similar levels in vitro (Figure 2A, lanes 1–3).
Figure 2. Whi5 is a substrate for Pcl9-Pho85 CDK-dependent phosphorylation.
(A) In vitro phosphorylation of Whi5 by Pho85 kinase. Purified Whi5GST and γ-32P-ATP were incubated alone (lane 4) or in the presence of recombinant Pcl1-Pho85 (lane 1), Pcl9-Pho85 (lane 2), or Cln2-Cdc28 (lane 3) kinases. Phosphorylated Whi5 protein was resolved by SDS-PAGE and autoradiography. (B) Slower-migrating forms of Whi5 are dependent on Cdc28 and Pho85. Cell extracts were prepared from wt (BY2507, lane 1), pho85Δ (BY4152, lane 2), cdc28-4 (BY4153, lane 4), cdc28-4 pho85Δ (BY4154, lane 5), cln1Δ cln2Δ (BY4289, lane 6), and cln3Δ (BY4288, lane 7) strains expressing WHI5MYC along with a whi5Δ control strain (BY4454, lane 3). Cells were grown at 30°C (semipermissive temperature for cdc28-4 strains) to log phase (Optical Density [OD] = 0.6) before harvesting. cdc28-4 cells were placed at 37°C for 2 h to inactivate Cdc28 before harvesting. Whi5MYC mobility was assessed by immunoblotting. (C) Whi5 associates with Pho85-dependent kinase activity. Wt (BY263) (lane 2), cdc28-4 (BY465) (lane 3), cdc28-13 (BY462) (lane 4), or pho85Δ (BY867) (lane 5) strains bearing a GAL-WHI5FLAG plasmid (pMT3586) or control vector control (pMT3164) were grown at 30°C (semipermissive temperature for cdc28-4 and cdc28-13 strains) in galactose media for 3 h. Whi5 complexes were recovered on anti-FLAG resin, incubated in kinase buffer with γ-32P-ATP at 30°C and resolved by SDS-PAGE. Capture of Whi5 protein was detected with anti-FLAG antibody. (D) Whi5 interacts with the Pcl, Pcl9. Anti-MYC immune precipitates of WHI5MYC strain lysates (BY2507) bearing either PCL1HA (pBA1820, lane 2), PCL2HA (pBA1821, lane 3), PCL9HA (pBA1822, lane 4), PHO80HA (pBA1823, lane 5), or a vector control (pBA330v, lane 1) were probed with 9E10 anti-MYC and 12CA5 anti-HA antibodies. The black lines in (A, B, and D) indicate empty lanes that were removed from the original blot.
Previous studies revealed multiple Whi5 slow-migrating isoforms that correlate with its phosphorylation state [12],[47]. We examined the effect of various cyclin or CDK mutants on Whi5 mobility (Figure 2B). Because of genetic redundancy of Pcl cyclins [27], we were unable to reproducibly detect changes in Whi5 phosphoforms in cyclin mutant strains. Therefore, a Pho85 mutant was used to asses the phosphorylation status of Whi5. Consistent with previous findings [12],[13], slow migrating Whi5 isoforms present in asynchronous wt extracts (Figure 2B, lane 1) were modestly reduced in cells lacking CLN3 (Figure 2B, lane 7) and completely absent in a cln1Δ cln2Δ double mutant (Figure 2B, lane 6), confirming that Whi5 phosphorylation depends on Cln-Cdc28 kinase complexes. Consistent with our SDL results and in vitro kinase assays, we observed a significant reduction in Whi5 mobility in extracts from a pho85 mutant strain (Figure 2B, lane 2). Thus, similar to Cdc28, phosphorylation of Whi5 also depends on Pho85 in vivo.
To determine if Whi5 physically associates with Pho85 in yeast, we first assayed Whi5FLAG immune complexes for kinase activity. A robust autophosphorylation activity was recovered from Whi5FLAG immunoprecipitates derived from wt cell extracts when radiolabeled ATP was added to the immunoprecipitated sample (Figure 2C, lane 2). This activity was partially dependent on both CDC28 and PHO85 (Figure 2C, lanes 3–5). We also confirmed a physical interaction between Whi5 and Pcls using a co-immunoprecipitation assay (Figure 2D). Immunoprecipitation of Whi5MYC from epitope-tagged cyclin extracts revealed a specific association between Pcl9 and Whi5 (Figure 2D, lane 4). We failed to reproducibly detect a physical interaction between Whi5 and Pcl1 (Figure 2D, lane 2) suggesting that Pcl9-Pho85 is the primary Whi5 CDK. Taken together, the phosphorylation and co-immunoprecipitation assays strongly suggest that, in addition to Cdc28, Pho85 also phosphorylates Whi5. Furthermore these results identify Whi5 as the first reported substrate for Pcl9-Pho85, one of two Pcls whose activity is restricted to early G1 phase.
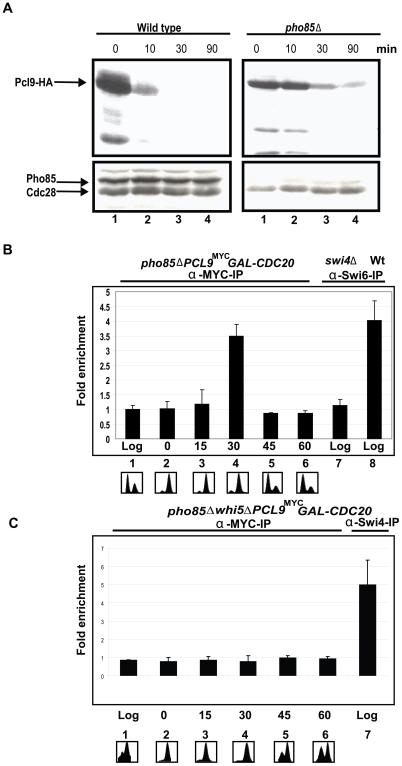
Whi5 associates indirectly with G1 phase-regulated promoters through interaction with SBF and MBF. Interactions with these transcription factors and subsequent promoter binding are disrupted by CDK-dependent phosphorylation [12],[13]. Because Whi5 appears to be a Pho85 substrate, we assessed the occupancy of SBF promoters by Pcl9. To date, cyclins have not been detected at yeast promoters. Pcl9 is normally an unstable short-lived protein [27]; however, similar to other cyclins, Pcl9 turnover appears to be catalyzed in part by its cognate CDK, Pho85 (Figure 3A) [48]. Therefore, to test Pcl9 promoter localization in a more sensitive genetic background, we performed ChIP (Chromatin immunoprecipitation) experiments in a pho85Δ strain (Figure 3B). The highest levels of CLN2 promoter DNA were detected in Pcl9MYC immune complexes 30 min following release from a metaphase-anaphase arrest (Figure 3B). The Pcl9-chromatin association was no longer detectable 45 min after GAL-CDC20 induction indicating that the interaction is short-lived and transient as predicted for a regulator of Start. The association was Whi5-dependent since Pcl9 was not detected at the CLN2 promoter in a strain lacking Whi5 (Figure 3C). The localization of Pcl9 to CLN2, a G1 promoter, is consistent with a direct role for Pcl9-Pho85 in regulating G1 transcription.
Figure 3. Pcl9 localizes to G1-specific promoters in a cell cycle-dependent manner.
(A) Pho85 regulates Pcl9 protein stability. Wt (BY263) and pho85Δ strains (BY391) harboring a GAL1-PCL9HA plasmid (pBA2112) were grown to exponential phase in galactose media (lane 1). PCL9 expression was repressed by addition of glucose to final concentration of 2% and cells were harvested 10 (lane 2), 30 (lane 3), and 90 (lane 4) min after addition of glucose. Pcl9 abundance was assessed by immunoblotting using 12CA5 anti-HA antibodies. (B) Pcl9 localizes to SBF-dependent promoters. An exponentially growing GAL1-CDC20 pho85ΔPCL9MYCstrain (BY4148, lane 1) was arrested at M/G1 phase in glucose-containing medium (lane 2). Cultures were harvested 15 (lane 3), 30 (lane 4), 45 (lane 5), and 60 (lane 6) min after release from CDC20-induced arrest in galactose medium. Cell cycle progression was monitored by FACS analysis. Anti-MYC and anti-Swi6 ChIPs from the indicated strains were analyzed for CLN2 promoter sequences by quantitative RT-PCR. (C) In a strain lacking Whi5, GAL1-CDC20 pho85Δ whi5ΔPCL9MYC, Pcl9 no longer localizes to the CLN2 promoter. Anti-Swi4 ChIPs are shown as a positive control.
Pcl9-Pho85 Regulates Whi5 Function via Phosphorylation
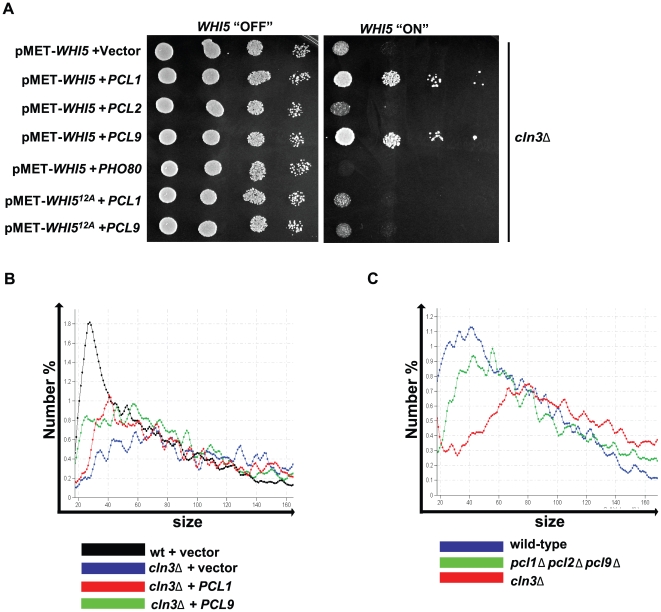
As mentioned above, cln3Δ mutants arrest in G1 phase as large unbudded cells in response to increased WHI5 dosage, indicating that Whi5 is a dose-dependent regulator of Start. Therefore, if Pho85 and Cdc28 function analogously to inhibit Whi5 activity, we predict that elevated Pho85 kinase activity would antagonize the toxic effects of WHI5 overexpression and suppress the growth defects observed in a cln3Δ mutant. To test this prediction, high copy plasmids expressing PCL1, PCL2, PCL9, or PHO80 were introduced into a cln3Δ strain expressing WHI5 from a conditional MET25 promoter (Figure 4A). Plasmid-based expression of Pcls and Whi5 was confirmed by immunoblotting (Figure S1). Induction of WHI5 expression in a cln3Δ mutant resulted in cell death whereas overexpression of PCL1 or PCL9 partially suppressed this toxicity and restored growth (Figure 4A). Consistent with results from SDL analyses (Figure 1), this suppression was specific to PCL1 and PCL9 since neither PCL2 nor PHO80 were able to function effectively in the assay (Figure 4A). Furthermore, PCL1/9-mediated suppression was dependent on phosphorylation since growth of a cln3Δ mutant expressing a nonphosphorylatable form of WHI5 (Whi512A) [13] could not be restored (Figure 4A). These genetic results corroborate the biochemical evidence that Pcl-Pho85 regulates Whi5 activity through phosphorylation.
Figure 4. PHO85 affects growth and cell size defects associated with cln3Δ.
(A) Ectopic PCL1 and PCL9 expression alleviates WHI5 toxicity in a cln3Δ strain. A cln3Δ strain (BY653) bearing a methionine-repressible WHI5GST (pBA1975) or WHI512A-GST low-copy plasmid (pBA2249) along with an additional vector control (pBA330v), PCL1HA (pBA1820), PCL2HA (pBA1821, PCL9HA, pBA1822), or PHO80HA (pBA1823) construct were spotted in serial 10-fold dilutions on media supplemented with or lacking methionine (WHI5 “OFF”, WHI5 “ON,” respectively) and incubated for 72 h at 30°C. (B) PCL1 and PCL9 cyclins modulate cell size. Cell size distributions were analyzed for wt (BY263) and cln3Δ strains (BY653) bearing vector control (pBA330v), PCL9HA (pBA1822), or PCL1HA (pBA1820) plasmids. The median cell volume based on three replicates was: 42.33 fl±1.13 (wt+vector control); 71.78 fl±1.43 (cln3Δ+vector control); 55.67 fl±1.66 (cln3Δ+PCL1); 54.25 fl±1.21 (cln3Δ+PCL9). (C) Cells lacking PHO85 G1 cyclins exhibit an enlarged cell size. Cell size distributions were analyzed for wt (BY263), pcl1Δ pcl2Δ pcl9Δ (BY764), and cln3Δ strains (BY653). The median cell volume based on three replicates was: 46.73 fl±0.63 (wt); 53.96 fl±0.75 (pcl1Δ pcl2Δ pcl9Δ); 72.72 fl±1.22 (cln3Δ).
Given its effect on WHI5 overexpression, we next examined PCL effects on other CLN3-associated phenotypes. CLN3 is required to activate G1-specific transcription once cells have achieved a critical size [49]–[51]. A cln3Δ mutant exhibits a large cell size phenotype because of its inability to inhibit Whi5 and activate Start-specific transcription [12],[13]. Ectopic expression of PCL1 or PCL9 reduced cln3Δ cell size to an intermediate level between that of wt and cln3Δ cells (Figure 4B). Conversely, deletion of PCL9, PCL1, and the partially redundant cyclin PCL2 resulted in a cell size increase (Figure 4C). These results suggest that Pcl-Pho85 and Cln3-Cdc28 share a common role in cell cycle progression to regulate Whi5 activity and promote passage through Start.
CDC28 and PHO85 Function in Parallel Pathways to Regulate Whi5 Function
To determine if Pcl-Pho85 and Cln3-Cdc28 might function in parallel to regulate Start, we first tried to test whether pcl9Δ cln3Δ or pcl1Δ pcl9Δ cln3Δ strains showed any synthetic growth defects. As expected, no growth defects were observed, probably because of the redundant effects of other Pcls [27]. Unlike the Cdc28 cyclins, which shows distinct cell cycle expression patterns, most Pcls are expressed at all cell cycle stages [25]. We then examined the phenotype of a pho85Δ cln3Δ double mutant. Cells lacking cln3Δ are larger than wt cells but do not display overt defects in growth rate while pho85Δ mutants are slow growing (Figure 5A). However, pho85Δcln3Δ double mutants exhibited a more pronounced growth defect compared to single mutants and analysis of DNA content revealed that the pho85Δ cln3Δ double mutant cells accumulated in G1 phase with predominantly unreplicated DNA (Figure 5A). Importantly, deleting WHI5 overcame both the cell cycle progression and growth defects observed in the absence of both CLN3 and PHO85. Notably, a pho85Δ cln3Δ whi5Δ triple mutant exhibited a growth rate similar to a cln3Δ single mutant indicating that Pcl-Pho85 and Cln3-Cdc28 function in separate yet converging pathways to regulate Whi5 function and, by extension, G1 cell cycle progression (Figure 5A). These observations also hold true under liquid growth conditions as shown. WHI5-dependent suppression appears to be specific to the pho85Δ cln3Δ phenotype because WHI5 deletion was unable to rescue 53 additional synthetic lethal interactions involving PHO85 (Table S1; D.Q. Huang and B.J. Andrews, unpublished data).
Figure 5. PHO85 regulates G1 transcription via WHI5.
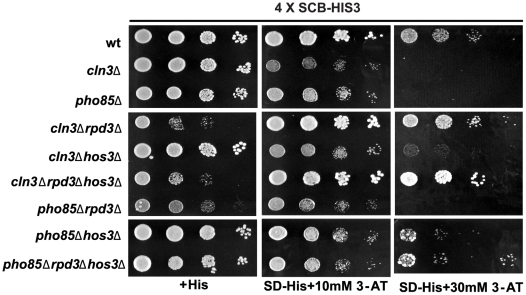
(A) The G1 delay phenotype associated with a cln3Δ pho85Δ strain is dependent on WHI5. Wt (BY263), cln3Δ (BY653), pho85Δ (BY391), cln3Δ pho85Δ (BY4291), and cln3Δ pho85Δ whi5Δ (BY4292) strains were spotted in serial 10-fold dilutions on rich media (YPED) and incubated for 24 h at 30°C. DNA content of exponentially growing cultures was determined by FACS analysis. Liquid growth assays were also performed for these strains and growth rate is reported relative to wt as shown in the bar graph. Graphical representations of growth rates are shown above the bar graph as line plots, where the upper red line represents the growth of WT and the black line shows the growth of each mutant. (B) A cln3Δ pho85Δ strain exhibits defects in SCB-driven gene expression. Wt (BY4302), cln3Δ (BY4303), pho85Δ (BY4304), cln3Δ pho85Δ (BY4305), cln3Δ pho85Δ whi5Δ (BY4306), pho85Δwhi5Δ (BY4308), and cln3Δwhi5Δ (BY4307) strains harboring an integrated SCB-HIS3 reporter were spotted in serial 10-fold dilutions on histidine-containing medium or media lacking histidine and supplemented with 10 or 30 mM 3-AT. Plates were incubated at 30°C for 48 h. We note that the synthetic growth defect of a cln3 pho85 mutant is most pronounced on rich medium (A), and is not as evident when strains are grown on minimal medium.
Given that Whi5 represses SBF- and MBF-specific transcription, we asked whether PHO85 affects SBF-driven reporter gene expression. A reporter gene consisting of tandem SCB consensus element repeats fused upstream of the HIS3 coding region was constructed and integrated into wt, cln3Δ, and pho85Δ strains. Previous work has shown that this reporter provides a highly specific read-out for SBF-dependent transcription [13],[52]. Growth on medium lacking histidine supplemented with 3-aminotriazole (3-AT) was used to assess SBF transcriptional activity (Figure 5B). Even though cells lacking PHO85 were moderately sensitive to higher concentration (5 mM) of 3-AT (unpublished data), both cln3Δ and pho85Δ mutants showed no growth in media containing 30 mM 3-AT indicating that SBF transcription is impaired in these mutants, whereas growth of wt cells was unaffected [13]. Furthermore, defects in SCB-driven gene expression were more pronounced in the pho85Δ cln3Δ double mutant (at 10 mM 3-AT, Figure 5B). Consistent with the genetic interactions described above (Figure 5A), SBF-dependent reporter activity was restored in pho85Δ cln3Δ mutants when WHI5 was deleted (Figure 5B). However, WHI5 deletion only partially rescued the growth defect in pho85Δ cells at 30 mM of 3-AT (Figure 5B). The Whi5-independent 3-AT sensitivity of pho85Δ cells may be due to unregulated Gcn4 in the absence of PHO85, since GCN4 is induced by 3-AT and Pho85 has been shown to regulate Gcn4 stability [53],[54]. Nonetheless, these data suggest that, like Cln3-Cdc28, Pcl-Pho85 modulates SBF activity through Whi5.
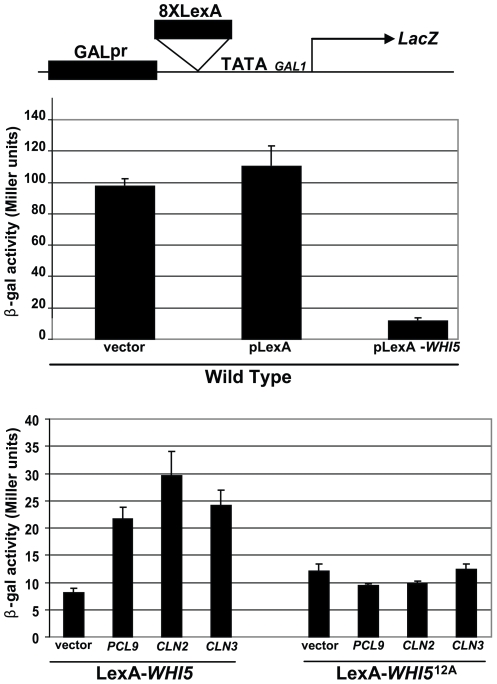
We next interrogated the effects of CDK activity on Whi5-mediated transcriptional repression (Figure 6). A construct expressing a LexA DNA binding domain fused to WHI5 was introduced into a strain harboring a LacZ reporter gene containing LexA binding sites in its promoter (Figure 6). Consistent with its role as a negative regulator of G1-specific transcription, a ∼10-fold reduction in β-galactosidase activity was observed in cells expressing the LexA-Whi5 fusion protein compared to a vector control (Figure 6). Overexpression of PCL9, CLN3, or CLN2 restored LacZ expression to intermediate levels indicating that activation of either CDC28 or PHO85 was capable of antagonizing Whi5 function in this assay (Figure 6). Consistent with suppression of WHI5-mediated growth defects (Figure 4), inhibition of Whi5 activity was dependent on phosphorylation since LacZ expression could not be restored in cells harboring an unphosphorylatable LexA-Whi512A fusion protein (Figure 6).
Figure 6. Whi5-mediated transcriptional repression is antagonized by PHO85 and CDC28.
A reporter gene consisting of eight LexA binding sites flanked by the GAL1 promoter and the LacZ coding sequence was constructed (pBA1976). β-galactosidase activity (upper histogram) was measured in a wt strain (BY263) bearing the LacZ reporter along with one of the following: a vector control (pBA230v); a LexA expressing plasmid (pLexA; pBA1977); or a construct expressing a LexA-Whi5 fusion protein (pLexA-WHI5; pBA1978). β-galactosidase activities were also assayed (lower histogram) in a wt strain harboring the LacZ reporter construct alone (vector control; pBA1976) or overexpressing the G1 cyclins, PCL9 (pBA1974), CLN2 (pBA2247), or CLN3 (pBA2248) in the presence of LexA-Whi5 (pBA1978) or LexA-Whi512A (pBA1979) fusion proteins.
Pho85 Does Not Regulate Whi5 Localization or Its Interactions with G1-Specific Transcription Complexes
Cln2-Cdc28 activity was previously shown to disrupt recombinant Whi5-SBF complexes in vitro [13], but Cln3-Cdc28 and Pho85 kinases had not been assessed for this activity. A preassembled recombinant Whi5-Swi4FLAG-Swi6 complex bound to anti-FLAG resin was incubated with purified kinases in the presence of radiolabeled ATP and separated into soluble (Figure 7B, labeled “S”) and bound fractions (Figure 7B, labeled “B”). Equivalent amounts of kinase were approximated on the basis of in vitro kinase activity (Figure 7A, and Materials and Methods). As expected, Cln2-Cdc28 phosphorylation caused most of the SBF-bound Whi5 to be released into the soluble fraction (Figure 7B, lanes 3 and 4). In contrast, we failed to observe dissociation of Whi5 from SBF in the presence of Cln3- or Pcl9-CDK complexes (Figure 7B, lanes 5–10). In addition to negatively regulating the interaction of Whi5 with SBF, Cdc28 also controls its localization [13]. Unlike Cln-Cdc28 phosphorylation, which promotes Whi5 export from the nucleus, deletion of PHO85 did not dramatically affect the subcellular localization of Whi5 (Figure 7C). Together, these results suggest that Pho85 must regulate Whi5 function through alternate mechanisms.
Figure 7. Pho85 does not affect known Whi5 regulatory mechanisms.
(A) Determination of relative Cdc28 and Pho85 kinase activity. In vitro kinase assays using varying amounts of recombinant Cln2-Cdc28, Cln3-Cdc28, and Pcl9-Pho85 in the absence (lane 1–3) or presence of purified Whi5 (lanes 5–8) were conducted and the degree of Whi5 phosphorylation was determined by SDS-PAGE and autoradiography. Purified Whi5 and γ-32P-ATP were incubated in the absence of kinase in lane 8, and lane 4 is empty. A 3 µM final concentration of Cln3-Cdc28 and Pcl9-Pho85 and a 60 nM final concentration of Cln2-Cdc28 give similar amounts of 32P-incorporation in Whi5, although phosphorylation by Cln2-Cdc28 caused Whi5 to migrate more slowly than Whi5 phosphorylated by Cln3-Cdc28 or Pcl9-Pho85. The concentration of kinase used in (B) was based on these experiments. (B) Cln3-Cdc28 and Pcl9-Pho85 do not influence Whi5-SBF complex stability. A preassembled recombinant Whi5-Swi4FLAG-Swi6 complex bound to anti-FLAG resin was incubated with Cln2-Cdc28, Cln3-Cdc28, Pcl9-Pho85, or both Cln3-Cdc28 and Pcl9-Pho85 in the presence of radiolabeled ATP. After washing, proteins in the bound and supernatant fractions were identified by autoradiography. (C) Subcellular localization of Whi5 in cdk mutant strains. Wt (BY263), pho85Δ (BY391), and cdc28-4 strains (BY465) expressing WHI5GFP from a methionine-repressible promoter (pBA1981) were examined for Whi5GFP fluorescence. Representative fields are shown.
Mechanism for Whi5-Mediated Transcriptional Repression by Pho85
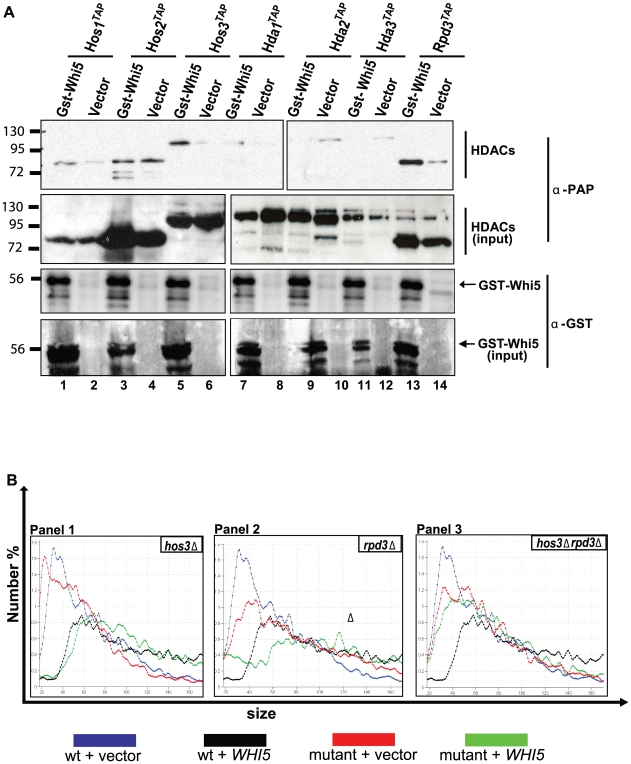
We next explored what additional mechanism might explain Pcl- and Cln3-mediated regulation of Whi5 activity. Functional conservation clearly extends to Whi5 and its metazoan analogue Rb [14]. Since Rb represses transcription, in part, through recruitment of HDACs, we used a batch affinity chromatography assay to test for physical interactions between a Whi5GST ligand and tandem affinity tagged HDACs (Figure 8A). Specific interactions between Whi5 and Hos3, Rpd3, and, to a lesser extent, Hos1 were identified (Figure 8A, lanes 1, 5, 13) suggesting that, like Rb, Whi5-dependent transcriptional repression involves recruitment of HDACs. This observation is consistent with previous work that detected Rpd3 at the PCL1 promoter using a ChIP assay [55]. Furthermore, HOS3 and RPD3 were required for WHI5 dose-dependent effects on cell size. Like wt cells, strains lacking either HOS3 (Figure 8B, panel 1) or RPD3 (Figure 8B, panel 2) also exhibited a dose-dependent increase in cell size in response to WHI5 overexpression. However, additional cell size effects were not observed in strains lacking both HDACs, suggesting that Hos3 and Rpd3 regulate Whi5 function synergistically (Figure 8B, panel 3).
Figure 8. Whi5 function is dependent on HDAC activity.
(A) Whi5 associates with Hos3 and Rpd3. Lysates prepared from the indicated epitope-tagged HDAC strains (BY4309–4315) harboring a vector control (pEG-H) or construct expressing WHI5GST (pBA1973) were incubated with glutathione sepharose beads. Whi5GST -HDAC interactions were detected by immunoblot using α-GST and α-PAP antibodies. (B) Hos3 and Rpd3 modulate Whi5 cell size effects. A plasmid expressing WHI5 (pBA1980) or vector control (pBA230v) were introduced into wt (BY263), hos3Δ (BY4293), rpd3Δ (BY4294), and hos3Δ rpd3Δ (BY4295) strains, and cell size distributions were measured. Each panel corresponds to a specific mutant and wt distributions are superimposed in each panel. The median cell volume based on three replicates was: 42.06 fl±1.09 (wt+vector control, blue); 73.12 fl±1.16 (wt+WHI5, black); 30.57 fl±1.23 (hos3Δ+vector control, panel 1, red); 71.35 fl±1.59 (hos3Δ+WHI5, panel 1, green); 51.20 fl±1.73 (rpd3Δ+vector control, panel 2, red); 69.75 fl±2.79 (rpd3Δ+WHI5, panel 2, green); 45.62 fl±1.22 (hos3Δ rpd3Δ+vector control; panel 3, red); 50.26 fl±1.14 (hos3Δ rpd3Δ+WHI5, panel 3, green).
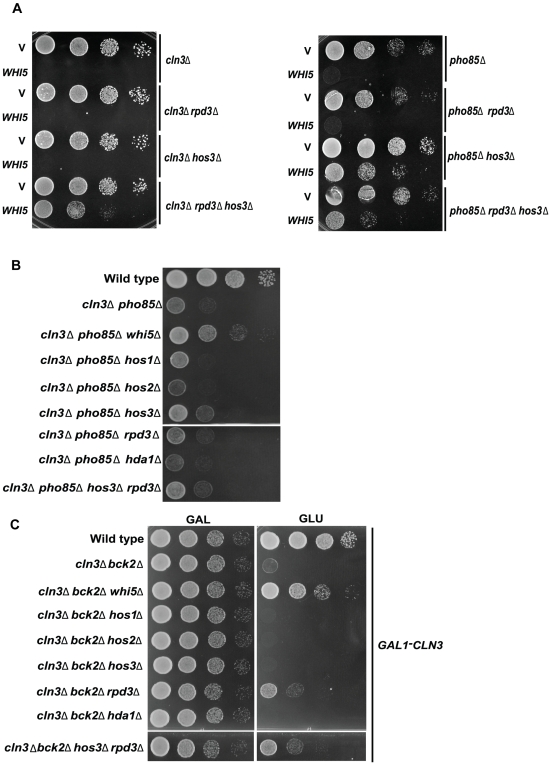
If HDACs are required for Whi5 function, then strains lacking HDAC function should be resistant to toxic effects associated with WHI5 overexpression. Consistent with this prediction, the growth defect caused by WHI5 overproduction in a cln3Δ was alleviated by the deletion of HOS3 and RPD3 (Figure 9A). Deletion of HOS3 alone rescued WHI5 toxicity in a pho85Δ strain while a cln3Δ mutant required deletion of both HOS3 and RPD3 in order to tolerate increased dosage of WHI5 (Figure 9A).
Figure 9. WHI5 toxicity is dependent on HOS3 and RPD3.
(A) cln3Δ (BY4290), cln3Δ rpd3Δ (BY4297), cln3Δ hos3Δ (BY4296), and cln3Δ rpd3Δ hos3Δ (BY4298) strains harboring a methionine-repressible WHI5 construct (pBA1975) or vector control (pBA228v) were spotted in serial 10-fold dilutions on medium lacking methionine. In a similar experiment, pho85Δ (BY391), pho85Δ rpd3Δ (BY4300), pho85Δ hos3Δ (BY4299), and pho85Δ rpd3Δ hos3Δ (BY4301) strains bearing a galactose-inducible WHI5 plasmid (pBA1973) or appropriate vector control (pEG-H) were spotted in serial 10-fold dilutions on galactose-containing medium. Plates were incubated at 30°C for 48 h. (B) Deletion of HOS3 partially restores growth of a cln3Δ pho85Δ strain. The indicated strains (BY263; BY4291, BY4292, BY4455–4461) were spotted in serial 10-fold dilutions on rich medium (YPED) and incubated at 30°C for 48 h. (C) Deletion of RPD3 and HOS3 partially restore viability of a cln3Δ bck2Δ strain. The indicated strains (BY4741; BY2948, BY4462–4468) were spotted in serial 10-fold dilutions on glucose-containing medium (YPED) to repress CLN3 expression. Strains were also spotted on medium containing galactose as a control. Plates were incubated at 30°C for 72 h.
Given that Whi5 appears to be acting through HDACs, we predicted that deletion of HOS3 and RPD3 should phenocopy those genetic interactions seen in whi5Δ mutants. We first tested various HDAC deletion strains for suppression of the slow growth phenotype of a pho85Δcln3Δ mutant. As for WHI5, deletion of HOS3 and RPD3 partially suppressed the growth defect seen in the pho85Δcln3Δ double mutant strain (Figure 9B). Suppression was specific to HOS3 and RPD3 because deletion of other HDACs showed no suppression, and the growth rate of the pho85Δcln3Δhos3Δ strain was not improved by subsequent deletion of RPD3 and vice versa (Figure 9B).
We next asked if deletion of HDACs might overcome the Start arrest seen in cells lacking both CLN3 and BCK2, another regulator of G1 transcription that functions in parallel with CLN3 [56]. A cln3Δbck2Δwhi5Δ triple mutant grows as vigorously as wt, placing WHI5 downstream of both upstream activators of G1 transcription [13]. Interestingly, deletion of RPD3 partially restored growth in the cln3Δbck2Δ strain providing further evidence for an HDAC requirement in Whi5-mediated transcriptional repression (Figure 9C). Neither subsequent deletion of HOS3 nor deletion of other HDACs affected growth appreciably (Figure 9C). We also employed the SCB-HIS3 assays used above to explore SBF-driven reporter gene expression in the HDAC mutants (Figure 10). As expected, deletion of RPD3 rescued the growth defects of cln3Δ SCB-HIS3 cells in the presence of both 10 mM and 30 mM of 3-AT, whereas HOS3 gene knockout had a marginal but additive effect. In contrast, the growth of pho85Δ cells was slightly rescued by deletion of HOS3 but not RPD3 providing further evidence for Pho85 acting specifically through Hos3. Because of difficulties in detecting HDACs at promoters, we were unable to confirm these observations in vivo.
Figure 10. Repression of gene expression by Whi5 is dependent on HOS3 and RPD3.
The growth defects of cln3Δ and pho85Δ strains can be rescued by removing RPD3 and HOS3 in SCB-driven gene expression. Wt (BY4302), cln3Δ (BY4303), pho85Δ (BY4304), cln3Δ rpd3Δ (BY4297), cln3Δ hos3Δ (BY4296), cln3Δrpd3Δhos3Δ (BY4298), pho85Δrpd3Δ (BY4300), pho85Δhos3Δ (BY4299), and pho85Δrpd3Δhos3Δ (BY4301) strains harboring an integrated SCB-HIS3 reporter were spotted in serial 10-fold dilutions on histidine-containing medium or media lacking histidine and supplemented with 10 or 30 mM 3-AT. Plates were incubated at 30°C for 48 h.
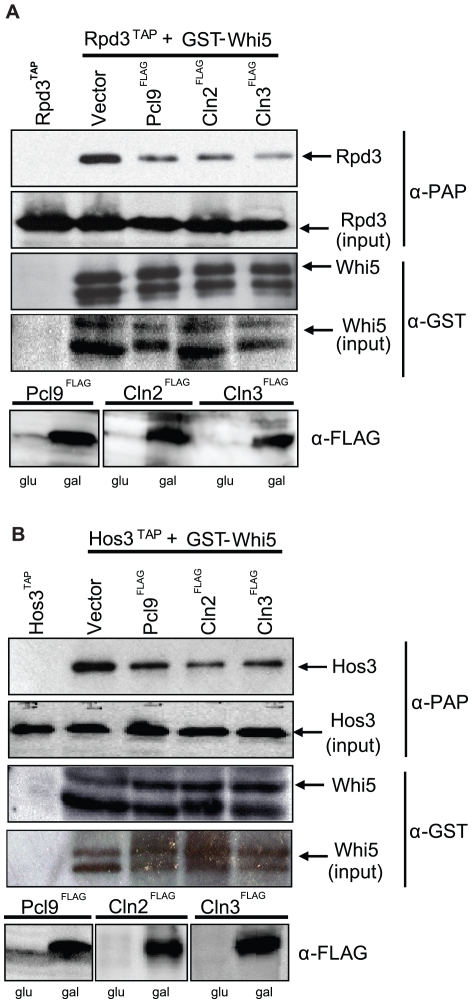
We also performed co-immunoprecipitation assays using affinity tagged RPD3 and HOS3 strains and observed an obvious decrease in Rpd3 and Hos3 in Whi5 precipitates from strains harboring increased levels of Pcl9, Cln2, or Cln3 cyclins (Figure 11A and 11B). Together, our genetic and biochemical results suggest that Pho85 may preferentially influence Whi5-Hos3 activity, whereas Cln3-Cdc28 is required for inhibition of both Rpd3 and Hos3.
Figure 11. CDK activity antagonizes Whi5-HDAC interactions.
(A) Pho85 and Cdc28 activity inhibits interaction between Whi5 and Rpd3. PCL9FLAG (pBA1974), CLN2FLAG (pBA2247), CLN3FLAG (pBA2248), or a vector control (pMT3164) were introduced into a strain harboring RPD3TAP at the chromosomal locus (BY4315) and a WHI5GST plasmid (pBA1973). Cyclin expression was confirmed by immunoblot using anti-FLAG antibodies. Lysates were incubated with glutathione sepharose beads. Whi5GST-Rpd3TAP interactions were detected by immunoblot using α-GST and α-PAP antibodies, (B) Pho85 and Cdc28 activity inhibits interaction of Whi5 and Hos3. Experiments were conducted as described in (A) but using a strain bearing HOS3TAP at the chromosomal locus.
Discussion
Whi5 is a critical cell cycle regulator that links CDK activity in G1 phase to the broad transcriptional program that accompanies commitment to cell division. We provide substantial evidence that the multifunctional Pho85 CDK is an important regulator of Whi5 activity and G1 phase-specific transcription including: (1) Whi5 is phosphorylated and antagonized by Pho85 and is the first reported substrate for the G1-specific CDK complex, Pcl9-Pho85; (2) the activity of an SBF-dependent promoter is influenced by PHO85; (3) the Pcl9 cyclin binds to SBF-regulated promoters; (4) the repressor function of Whi5 is mediated through the HDACs Hos3 and Rpd3; and (5) HDAC-Whi5 association is regulated by G1-specific forms of both the Pho85 and Cdc28 CDKs. We therefore conclude that timely and efficient release from Whi5 inhibition and subsequent G1/S cell cycle progression requires the concerted activity of both Cdc28 and Pho85.
Several lines of evidence point to common roles for Pho85 and Cdc28. For example, a burst of both G1-specific Cdc28 and Pho85 activity is essential for cellular morphogenesis. A strain lacking the G1-specific cyclins, CLN1, CLN2, PCL1, and PCL2, undergoes a catastrophic morphogenic change and fails to establish polarized cell growth and cytokinesis [30]. Consistent with these observations, a chemical genomic analysis demonstrated that expression of genes involved in polarized cell growth was sensitive to simultaneous inhibition of both kinases, but not either single kinase [31]. A functional connection between Pho85 and Cdc28 is further supported by independent genetic and biochemical analyses that identify common targets phosphorylated by both kinases [45],[46],[48],[57]–[59].
Despite the clear functional overlap for G1-specific forms of Cdc28 and Pho85 in controlling morphogenesis, up to now, a direct role for Pho85 in cell cycle commitment and G1 phase-specific transcription has remained unclear. We discovered that, like Cdc28, Pho85 activates G1 transcription through inhibition of the Whi5 repressor. While the two kinases collaborate to control certain facets of Whi5 regulation, they are also specialized to modulate Whi5 function by distinct mechanisms. We have defined a novel HDAC-dependent mechanism that impinges on Whi5 function and implicates both Pho85 and Cdc28 as regulators of this process.
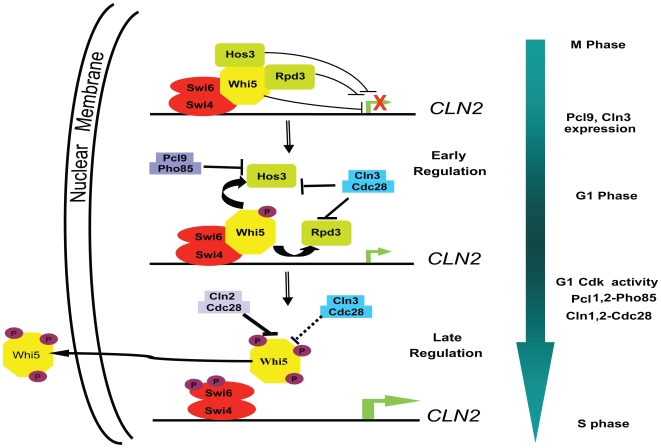
On the basis of these and other observations, we propose that Whi5 functional regulation involves perturbation of specific HDAC-Whi5 interactions and requires the concerted activity of both Cdc28 and Pho85 (summarized in Figure 12). Interestingly, our genetic observations support a model whereby Pcl-Pho85 preferentially targets the Hos3-Whi5 interaction illustrating a functional distinction between the two CDKs. While Pho85 associates with several cyclin subunits, only Pcl9 exhibits temporal expression and localization patterns compatible with such a function. PCL9 is expressed at the M/G1 phase transition and encodes a short-lived protein localized exclusively to the nucleus in early G1 phase [27],[60],[61]. Cln3 is also present in early G1 cells, but shows a complex localization pattern, with significant retention to the ER in early G1 cells, followed by chaperone-mediated release into the nucleus in late G1 phase [62]. How the specific features of Pcl9 and Cln3 localization might influence the timing of HDAC inhibition remains to be explored.
Figure 12. Model for CDK-dependent regulation of Whi5 activity and G1/S-specific transcription.
Shown is a schematic of the disruption of interactions between Whi5 and the HDACs, Hos3 and Rpd3, by Cln3-Cdc28 and Pcl9-Pho85-dependent phosphorylation, leading to transcription of G1 genes, including the CLN1 and CLN2 cyclins. Whi5 is then further phosphorylated by Cln1- and Cln2-Cdc28 complexes leading to complete disassembly of the Whi5-SBF complex, Whi5 nuclear export and a burst in gene expression necessary for the G1/S phase transition.
The second component of Whi5 regulation is predicated on previous studies indicating that G1/S gene expression is preceded by Whi5-SBF complex dissociation and subsequent nuclear export of Whi5 (Figure 12) [13]. Unlike early regulatory events, Cdc28 activity is both necessary and sufficient to drive these events since neither SBF binding to Whi5 nor nuclear localization of Whi5 was adversely affected in a pho85Δ mutant (Figure 7). Also, we are able to detect binding of SBF in vivo to CLN2 promoters when PHO85 is deleted (Figure 3C). However, both purified Cln3-Cdc28 and Pcl9-Pho85 failed to affect Whi5-SBF stability in vitro, while complex disruption was effectively achieved in the presence of Cln2-Cdc28 kinases (Figure 7). Cln3-Cdc28 and Pcl9-Pho85 may have a more pronounced effect on the Whi5-SBF complex in vivo. Alternatively, Cln3- and Pcl9-CDKs may act primarily as agonists of HDAC interactions while physical interactions with SBF and nuclear export are optimally mediated by the late G1 CDKs, Cln1- and Cln2-Cdc28. Indeed, recent work reveals activation of CLN2 expression while Whi5 remains bound to the promoter (H. Wang, L.B. Carey, Y. Cai, H. Wijnen, and B. Futcher, personal communication). Such a mechanism may serve to sharpen the onset, as opposed to the timing, of G1/S gene expression thus ensuring a sustained transcriptional burst and irreversible commitment to cell division [13]. Consistent with this idea, recent analysis of cyclin gene expression using a single cell assay affirms that positive feedback involving the Cln1 and Cln2 cyclins induces the G1/S regulon, and that this regulatory feedback is important for maintaining coherence of gene expression at Start [63].
SBF promoter recruitment depends on a series of well-organized chromatin remodeling events [7],[36]. SBF, in turn, regulates the recruitment of the general transcription machinery via a two-step process beginning with the mediator complex followed by CDK-dependent recruitment of RNA PolII, TFIIB, and TFIIH [9]. Previous studies suggested that this CDK requirement stems from Whi5, which in its unphosphorylated state, remains bound to SBF and occludes the basal transcription machinery from binding specific promoters [13]. We have extended this model to include a role for HDAC activity. We predict that Hos3 and Rpd3 contribute to Whi5 repression by preventing holoenzyme access to chromatin. During states of high CDK activity, Cdc28 and Pho85 abrogate Whi5-HDAC and Whi5-SBF interactions and initiate transcription. Consistent with our model, Pcl9 and Cln3 cyclins localize to G1 promoters and Whi5 remains associated with G1-specific promoters in the absence of HDAC-promoter interactions (Figure 3; H. Wang, L.B. Carey, Y. Cai, H. Wijnen and B. Futcher, personal communication). However, Whi5 may also repress transcription by additional mechanisms since its activity is partially retained in hos3Δ rpd3Δ mutants (Figure 9).
Rpd3 is a well-characterized HDAC that accomplishes most of its functions as part of a large protein complex [37]. The Rpd3-Sin3 deacetylase complex has long been implicated as a cell cycle regulator required for silencing HO gene expression to prevent mating type switching in newly budded cells [64],[65]. Our observations that Whi5 associates with Rpd3 and our genetic data linking G1 Cdks, Whi5, and Rpd3 reveal a more general role for Rpd3 in G1/S-phase specific transcription. These data are consistent with observations from Futcher and colleagues that the Rpd3 protein can be detected at the CLN2 promoter and that the amount of Rpd3 at the promoter is decreased when CLN3 is induced (H. Wang, L.B. Carey, Y. Cai, H. Wijnen and B. Futcher, personal communication). The Rpd3-Sin3 HDAC has also been connected to G1 transcription factors through the interaction of Sin3 with Stb1, a Swi6-binding protein [66]–[68]. Both Stb1 and Sin3 are required for repression of G1 transcription early in G1 phase [68]. Unlike Rpd3, Hos3 is largely uncharacterized, although a recent study suggests a role for Hos3 in yeast apoptosis upon exposure to oxidative radicals [69]. We have uncovered an additional role for Hos3 in Whi5-mediated transcriptional repression.
A question that arises from our observations is what advantage does combinatorial kinase regulation impart on specific biological processes such as G1/S cell cycle progression? Contributions from multiple CDKs may provide the precision and accuracy necessary for rapid definitive decisions that irreversibly affect cellular fate. Indeed, distributive multisite phosphorylation mechanisms exhibit ultrasensitivity with respect to kinase concentration, thereby creating a “switch-like” behavior in biological circuits [70]. Since cell cycle transitions typically display switch-like attributes, multisite phosphorylation by various kinase combinations may prove to be a rule rather than the exception amongst CDK targets, including key cell cycle regulators such as Whi5. In fact, a recent computational analysis showed enrichment of multiple closely spaced consensus sites for Cdc28 substrates in yeast, a pattern that proved predictive of likely CDK targets [71].
Although kinase combinations are likely necessary for cell cycle regulation, the contribution of each individual kinase may vary depending on specific signals and environmental stimuli. In certain environments, Pcl-Pho85 may have more dramatic, condition-specific effects on Whi5 function than Cdc28 analogous, perhaps, to the regulation of Rb that is required for quiescence and prevention of apoptosis [72],[73]. Previous studies indicate that Whi5 localizes to nuclei in stationary phase cells suggesting that Whi5 may also play a role in G0 [13]. Interestingly, Pho85 is required for survival in starvation conditions and plays an important role during stationary phase [74]–[76]. Furthermore, CDK5, the mammalian Pho85 homolog, induces apoptosis in neuronal cells via Rb phosphorylation [77]. Whether Whi5 activity is more prominently affected by Pcl-Pho85 in response to stationary or stress conditions requires additional investigation.
Similarities between metazoan and yeast cell cycle regulation are increasingly evident as we continue to characterize Whi5 function. For example, similar to proposed Pcl9/Cln3 “early” phase regulation (Figure 12), cyclinD-CDK4/6 phosphorylates Rb to promote HDAC dissociation and E2F transcriptional activation. E2F activation then leads to cyclin E expression, which, similar to Cln1/2 “late” phase regulation (Figure 12), may establish a positive feedback loop whereby cyclinE-CDK2 activity disrupts Rb-promoter interactions and stimulates G1-transcription further [15]. Despite these similarities, the importance of multiple regulatory components in both yeast and mammalian systems remains poorly understood and may be most fruitfully dissected using the yeast model.
Materials and Methods
Yeast Strains, Growth Conditions, and Plasmids
The S. cerevisiae strains used are listed in Table 1. All gene disruptions and integrations were achieved by homologous recombination at their chromosomal loci by standard PCR-based methods and confirmed by PCR with flanking primers [78]. Standard methods and media were used for yeast growth and transformation. Two percent of galactose in the media was used to induce the expression of genes under the GAL1 promoter. Synthetic minimal medium with appropriate amino acid supplements was used for cells containing plasmids. Appropriate amounts of 3-AT were added to SD-HIS plates to assess the expression of HIS3 reporter genes. 10-fold serial dilutions (5–10 µl) of yeast cells were spotted onto plates with appropriate nutrition conditions to assess growth. Plasmids used in this study are listed in Table 2. In most cases, a DNA insert was amplified by PCR and inserted into a linearized vector by homologous recombination in yeast. Details of construction will be provided upon request.
Table 1. Yeast strains.
| Strain | Genotype | Source or Reference |
| BY186 | BY263 MATa swi4ΔHIS3 | [82] |
| BY263 | MATa trp1 leu2 his3 ura3 lys2 ade2 | [29] |
| BY391 | BY263 MATa pho85ΔLEU2 | [29] |
| BY451 | BY263 MATa pcl2ΔLYS2 | [25] |
| BY462 | MATa leu2 his3 ura3 cdc28-13 | M. Tyers |
| BY465 | MATa leu2 his3 ura3 cdc28-4 | M. Tyers |
| BY490 | BY263 MATa pho80ΔHIS3 | [25] |
| BY628 | BY263 MATa pcl1ΔLEU2 | [25] |
| BY653 | BY263 MATa cln3ΔURA3 | This study |
| BY694 | BY263 MATa pcl9ΔHIS3 | [25] |
| BY760 | BY263 MATa pcl1ΔLEU2 pcl9ΔHIS3 | [25] |
| BY764 | BY263 MATa pcl1ΔLEU2 pcl2ΔLYS2 pcl9ΔHIS3 | [25] |
| BY867 | BY263 MATa pho85ΔTRP1 | [25] |
| BY1446 | BY263 MATα cln3ΔURA3 pho85ΔLEU2 whi5ΔKANR | This study |
| BY1502 | Y2454 MATα pho85ΔLEU2 | [74] |
| BY2507 | BY4741 MATa WHI5myc::KANR | M. Tyers |
| BY2948 | BY4741 MATa cln3ΔHPHR bck2ΔNATR pGAL-CLN3 URA3 | This study |
| BY4148 | BY4741 MATa GALpr-HA-CDC20::KANR pho85ΔNATR PCL9myc | This study |
| BY4151 | BY4741 MATa GALpr-HA-CDC20::KANR | This study |
| BY4152 | BY4741 MATa WHI5myc::KANR pho85ΔNATR | This study |
| BY4153 | BY4741 MATa WHI5myc::KANR cdc28-4 | This study |
| BY4154 | BY4741 MATa WHI5myc::KANR cdc28-4 pho85ΔNATR | This study |
| BY4242 | BY4741 MATa GALpr-HA-CDC20::KANRcln1ΔNATR cln2ΔHPHR | This study |
| BY4269 | BY4741 MATα GALpr-HA-CDC20::KANRcln3ΔURA3 pho85ΔLEU2R | This study |
| BY4270 | BY4741 MATα GALpr-HA-CDC20::KANRcln3ΔURA3 pho85ΔLEU2 whi5ΔKANR | This study |
| BY4273 | BY4741 MATa GALpr-HA-CDC20::KANRcln3ΔURA3 | This study |
| BY4274 | BY4741 MATa GALpr-HA- CDC20::KANR pho85ΔLEU2 | This study |
| BY4288 | BY4741 MATa WHI5myc::KANR cln3ΔLEU2 | This study |
| BY4289 | BY4741 MATa WHI5myc::KANR cln1ΔNATR cln2ΔHPHR | This study |
| BY4290 | BY263 MATa cln3ΔTRP1 | This study |
| BY4292 | BY263 MATα cln3ΔURA3 pho85ΔLEU2 whi5ΔKANR | This study |
| BY4293 | BY263 MATa hos3ΔKANR | This study |
| BY4294 | BY263 MATa rpd3ΔNATR | This study |
| BY4295 | BY263 MATa hos3ΔKANR rpd3ΔNATR | This study |
| BY4296 | BY263 MATa cln3ΔTRP1 hos3ΔKANR | This study |
| BY4297 | BY263 MATa cln3ΔTRP1 rpd3ΔNATR | This study |
| BY4298 | BY263 MATa cln3ΔTRP1 hos3ΔKANR rpd3ΔNATR | This study |
| BY4299 | BY263 MATa pho85ΔLEU2 hos3ΔKANR | This study |
| BY4300 | BY263 MATa pho85ΔLEU2 rpd3ΔNATR | This study |
| BY4301 | BY263 MATa pho85ΔLEU2 hos3ΔKANR rpd3ΔNATR | This study |
| BY4302 | BY4741 MATa hoΔ::SCB:HIS3::URA3 | This study |
| BY4303 | BY4741 MATa hoΔ::SCB:HIS3::URA3 cln3ΔNATR | This study |
| BY4304 | BY4741 MATa hoΔ::SCB:HIS3::URA3 pho85ΔLEU2 | This study |
| BY4305 | BY4741 MATa hoΔ::SCB:HIS3::URA3 cln3ΔNATR pho85ΔLEU2 | This study |
| BY4306 | BY4741 MATa hoΔ::SCB:HIS3::URA3 cln3ΔNATR pho85ΔLEU2 whi5ΔKANR | This study |
| BY4307 | BY4741 MATa hoΔ::SCB:HIS3::URA3 cln3ΔNATR whi5ΔKANR | This study |
| BY4308 | BY4741 MATa hoΔ::SCB:HIS3::URA3 pho85ΔLEU2 whi5ΔKANR | This study |
| BY4309 | BY4741 MATa HOS1TAP::HIS3 | This study |
| BY4310 | BY4741 MATa HOS2TAP::HIS3 | This study |
| BY4311 | BY4741 MATa HOS3TAP::HIS3 | This study |
| BY4312 | BY4741 MATa HDA1TAP::HIS3 | This study |
| BY4313 | BY4741 MATa HDA2TAP::HIS3 | This study |
| BY4314 | BY4741 MATa HDA3TAP::HIS3 | This study |
| BY4315 | BY4741 MATa RPD3TAP::HIS3 | This study |
| BY4454 | BY263 MATa whi5ΔKANR | This study |
| BY4455 | BY263 MATa cln3ΔURA3 pho85ΔLEU2 hos1ΔHIS5 | This study |
| BY4456 | BY263 MATa cln3ΔURA3 pho85ΔLEU2 hos2ΔHIS5 | This study |
| BY4457 | BY263 MATα cln3ΔURA3 pho85ΔLEU2 hos3ΔNATR | This study |
| BY4458 | BY263 MATα cln3ΔURA3 pho85ΔLEU2 rpd3ΔNATR | This study |
| BY4459 | BY263 MATa cln3ΔURA3 pho85ΔLEU2 hda1ΔHIS5 | This study |
| BY4461 | BY263 MATa cln3ΔURA3 pho85ΔLEU2 hos3ΔKANR rpd3ΔNATR | This study |
| BY4462 | BY2948 whi5ΔKANR | This study |
| BY4463 | BY2948 hos1ΔKANR | This study |
| BY4464 | BY2948 hos2ΔKANR | This study |
| BY4465 | BY2948 hos3ΔKANR | This study |
| BY4466 | BY2948 rpd3ΔHIS5 | This study |
| BY4467 | BY2948 hda1ΔHIS5 | This study |
| BY4468 | BY2948 hos3ΔKANR rpd3ΔHIS5 | This study |
| BY4541 | BY263 pho85ΔLEU2 whi5ΔKANR | This study |
| BY4542 | BY263 cln3ΔTRP1 whi5ΔKANR | This study |
| BY4741 | MATa leu2Δ0 his3Δ1 ura3Δ0 met15Δ0 | — |
| Y2454 | MATα mfa1Δ MFApr-HIS3 can1Δ his3Δ1 leu2Δ0 lys2Δ0 | [83] |
Of the wt strains used in this study, both BY263 and BY4741 are derived from S288C background. All the other strains are derived from these two strains. BY263 is an ssd1-d strain; BY4741 is an SSD1-V strain and is the parent strain for the yeast deletion consortium. Y2454 is congenic to BY4741 and is the parent for query strains used in synthetic genetic array (SGA) experiments.
Table 2. Plasmids used in this study.
| Name | Relevant Genotype | Source |
| pEG-H | pGAL1-GST URA3 2 µm | M. Snyder |
| pMT3164 | pGAL-c-FLAG LEU2 CEN | Y. Ho |
| pMT3446 | GST-WHI5 in pGEX4T1 (Escherichia coli expression vector) | M. Tyers |
| pMT3586 | pGAL-WHI5-FLAG LEU2 CEN | Y. Ho |
| pBA230v | pGPD TRP1 2 µm | M. Funk |
| pBA330v | pGPD LEU2 2 µm | M. Funk |
| pBA1820 | pGPD-HA-PCL1 LEU2 2 µm | This study |
| pBA1821 | pGPD-HA-PCL2 LEU2 2 µm | This study |
| pBA1822 | pGPD-HA-PCL9 LEU2 2 µm | This study |
| pBA1823 | pGPD-HA-PHO80 LEU2 2 µm | This study |
| pBA1973 | GST-WHI5 in pEG-H | M. Snyder |
| pBA1974 | pGAL-PCL9-FLAG LEU2 CEN | Y. Ho |
| pBA1975 | pMET-GST-WHI5 HIS3 CEN | This study |
| pBA1976 | pGAL-8XLexAop-LacZ URA3 2 µm | This study |
| pBA1977 | pGPD-LexA TRP1 2 µm | This study |
| pBA1978 | pGPD-LexA-WHI5 TRP1 2 µm | This study |
| pBA1979 | pGPD-LexA-WHI512A TRP1 2 µm | This study |
| pBA1980 | pGPD-WHI5 TRP1 2 µm | This study |
| pBA1981 | pMET-WHI5-GFP HIS3 CEN | This study |
| pBA2112 | pGAL-HA-PCL9 URA3 2 µm | J. Moffat |
| pBA2239 | GST-PCL1 in pGEX4T1 (E.coli expression vector) | This study |
| pBA2240 | GST-PCL2 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2241 | GST-PCL9 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2242 | GST-PHO80 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2243 | GST-PHO85 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2244 | GST-CLN2 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2245 | GST-CLN3 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2246 | GST-CDC28 in pAcGHLT (baculovirus transfer vector) | This study |
| pBA2247 | pGAL-CLN2-FLAG LEU2 CEN | Y. Ho |
| pBA2248 | pGAL-CLN3-FLAG LEU2 CEN | Y. Ho |
| pBA2249 | pMET-GST-WHI512A HIS3 CEN | This study |
Kinase Assays
The in vitro protein kinase assays monitored the incorporation of [32P] transferred from γ-32P-ATP to purified recombinant GST-Whi5. The reaction mixture for assays shown in Figure 2A contained 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 10 mM MgCl2, and 1 µM ATP (including 20 µCi γ-32P-ATP) and 0.2 µg GST-Whi5 in 20 µl of total volume. 2 µl of a purified recombinant kinase (0.4 µg–0.8 µg) was added to the mixture and incubated at 30°C for 30 min. Purification of Cln and Pcl CDKs from insect cell expression systems have been previously described [13],[46]. Whi5 was then analyzed by SDS-PAGE and autoradiography. Kinase assays on immunoprecipitated proteins from yeast cell extracts were performed as described [13]. Kinase assays preceding the Whi5-SBF dissociation assay (Figure 7) were performed as described above except that 200 µM γ-32P-ATP was used instead of 1 µM. The final concentration of Cln3 and Pcl9 was 3 µM, and the final concentration of Cln2 was 60 nM (50-fold less).
Quantitative β-Galactosidase Assays
Liquid β-galactosidase assays were performed as described [29]. Strains carrying appropriate plasmids were grown in synthetic minimal medium to mid-log phase, transferred to synthetic galactose medium, and incubated for 4 h. Cells were harvested and broken in lysis buffer (100 mM Tris-HCl [pH 8.0], 1 mM DTT, and 20% glycerol with protease inhibitors) with glass beads. The β-galactosidase activity was determined by adding 100 µl of total cell extract to 0.9 ml of Z buffer (100 mM Na2PO4, 40 mM NaHPO4, 10 mM KCl, 1 mM MgSO4, and 0.027% β-mercaptoethanol) and 200 µl ONPG (4 mg/ml) (Sigma). Units of β-galactosidase activity were determined as described [29].
Whi5 Dissociation with SBF Complex In Vitro
The protein binding assay essentially followed the procedures described previously [13]. Briefly, 1 µl of insect cell lysate expressing SBF (Swi6-Swi4FLAG) was mixed with 1 µl of purified GST-Whi5 (∼0.1 µg) and 7 µl of M2 anti-FLAG resin (Sigma) in 8 µl of kinase buffer (50 mM Tris-HCl [pH 7.5], 1 mM DTT, and 10 mM MgCl2). The mixture was incubated at 4°C for 1 h with mixing. The beads bound to the SBF-Whi5 complex were then washed three times with kinase buffer, and mixed with various cyclin dependent kinases in kinase buffer with 0.2 mM ATP in a 20 µl volume. The kinase reaction was incubated at 30°C for 1 h. The soluble portion was taken out and mixed with 20 µl of 2×SDS-PAGE loading buffer. The beads in the tube were washed three times with kinase buffer before mixing with 15 µl of 2×SDS-PAGE loading buffer.
Liquid Growth Assays
Strains containing galactose-inducible plasmids were grown to saturation in 2% raffinose media for 48 h. Expression of plasmids were induced by transferring into 2% raffinose 2% galactose media and liquid growth assays were performed as previously described over 36 h using a Tecan GENios microplate reader (Tecan) [79]. Average doubling (AveG) for each culture was calculated as previously described [79]. Growth rate for each mutant was calculated relative to the AvgG of the wt strain.
Whi5-GFP Localization
The localization of Whi5-GFP was monitored in wt, cdc28-4, and pho85Δ strains. Cells expressing pMET-GFP-WHI5 were grown to log phase in synthetic glucose medium without methionine. Cells were observed at a magnification of 1,000× using Nomarski optics and fluorescence microscopy and photographed by a Cascade 512B high-speed digital camera (Roeper Scientific) mounted on a Leica DM-LB microscope. Images were captured and analyzed by MetaMorph software (Universal Imaging Media).
ChIP
The pho85Δ PCL9MYC GALpr-CDC20 and pho85Δwhi5ΔPCL9MYC GALpr-CDC20 cells were grown in YP-Galactose (YPG) medium to an optical density (OD600) of 0.4, blocked at M phase by growing in YPED medium for 3 h, and released into YPG medium. Samples were taken every 15 min after release and cross-linked with a final concentration of 1% formaldehyde. Wt and swi4Δ strains (for controls) were grown to OD600 of 0.6 in YPD. Formaldehyde cross-linking and preparation of whole-cell extracts were performed as previously described [80]. Immunoprecipitation were performed using 1∶200 dilution of α-myc monoclonal antibody (9E10), α-Swi6 or α-Swi4 polyclonal antibodies. The precipitates were washed twice with lysis buffer, once with LiCl detergent and once with Tris-buffered saline and processed for DNA purification. Enrichment at the CLN2 promoter sequence was quantified with real-time PCR, using a dual fluorogenic reporter TaqMan assay in an ABI PRISM 7500HT Sequence Detection System as previously described [13].
Other Materials and Methods
Recombinant GST-Pcl1 and GST-Whi5 were produced in a BL21 bacterial expression strain; other recombinant proteins were produced in insect cells infected with Baculovirus expression vectors [11],[46],[73]. Proteins were detected with 9E10 anti-Myc, 12C5 anti-HA, and M2 anti-FLAG monoclonal antibodies. FACS analysis of DNA content and cell size measurements were described previously [81].
Supporting Information
Expression levels of epitope-tagged Whi5 and Pcls. Whi5 and Pcl abundance in the indicated strains was determined by immunoblotting. Cyclin proteins were detected using 12CA5 anti-HA antibodies whereas Whi5 protein was detected using anti-GST antibodies.
(0.93 MB EPS)
List of 53 synthetic lethal interactions involving PHO85 that are not rescued by deletion of WHI5 .
(0.07 MB DOC)
Acknowledgments
We thank Corey Nislow, Mike Tyers, and Charles Boone for comments on the manuscript and useful discussion. We are grateful to Kim Nasmyth for providing strains.
Abbreviations
- 3-AT
3-aminotriazole
- ATP
adenosine triphosphate
- CDK
cyclin-dependent kinase
- ChIP
chromatin immunoprecipitation
- HDAC
histone deacetylase
- Rb
retinoblastoma
- Pcl
Pho85 cyclin
- SDL
synthetic dosage lethality
- wt
wild type
Footnotes
The authors have declared that no competing interests exist.
DvD is supported by a Postdoctoral Fellowship from the Best Foundation and RS was supported by a Terry Fox Foundation Research Studentship award from the National Cancer Institute of Canada. SK is partially supported by an Open Fellowship from the University of Toronto. This work was supported by a grant from the Canadian Institutes of Health Research to BJA. The functional genomics work in the Andrews laboratory is supported with funds from Genome Canada through the Ontario Genomics Institute (to BJA and Charles Boone). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bloom J, Cross F. R. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 2.Pardee A. B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 3.Cross F. R. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 4.Bahler J. Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet. 2005;39:69–94. doi: 10.1146/annurev.genet.39.110304.095808. [DOI] [PubMed] [Google Scholar]
- 5.Wittenberg C, Reed S. I. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 6.Cosma M. P. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 7.Cosma M. P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 8.Bhoite L. T, Yu Y, Stillman D. J. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosma M. P, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7:1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 10.Cross F. Transcriptional regulation by a cyclin-cdk. Trends Genet. 1995;11:209–211. doi: 10.1016/s0168-9525(00)89047-2. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 12.de Bruin R. A, McDonald W. H, Kalashnikova T. I, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 13.Costanzo M, Nishikawa J. L, Tang X, Millman J. S, Schub O, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Schaefer J. B, Breeden L. L. RB from a bud's eye view. Cell. 2004;117:849–850. doi: 10.1016/j.cell.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Hatakeyama M, Brill J. A, Fink G. R, Weinberg R. A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 16.Brehm A, Miska E. A, McCance D. J, Reid J. L, Bannister A. J, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 17.Luo R. X, Postigo A. A, Dean D. C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 18.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 19.Flemington E. K, Speck S. H, Kaelin W. G., Jr E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci U S A. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross J. F, Liu X, Dynlacht B. D. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- 21.Carroll A. S, O'Shea E. K. Pho85 and signaling environmental conditions. Trends Biochem Sci. 2002;27:87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- 22.Moffat J, Huang D, Andrews B. Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog Cell Cycle Res. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- 23.Sopko R, Huang D, Preston N, Chua G, Papp B, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Huang D, Moffat J, Wilson W. A, Moore L, Cheng C, et al. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, et al. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson W. A, Mahrenholz A. M, Roach P. J. Substrate targeting of the yeast cyclin-dependent kinase Pho85p by the cyclin Pcl10p. Mol Cell Biol. 1999;19:7020–7030. doi: 10.1128/mcb.19.10.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tennyson C. N, Lee J, Andrews B. J. A role for the Pcl9-Pho85 cyclin-cdk complex at the M/G1 boundary in Saccharomyces cerevisiae. Mol Microbiol. 1998;28:69–79. doi: 10.1046/j.1365-2958.1998.00773.x. [DOI] [PubMed] [Google Scholar]
- 28.Espinoza F. H, Ogas J, Herskowitz I, Morgan D. O. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- 29.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 30.Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- 31.Kung C, Kenski D. M, Dickerson S. H, Howson R. W, Kuyper L. F, et al. Chemical genomic profiling to identify intracellular targets of a multiplex kinase inhibitor. Proc Natl Acad Sci U S A. 2005;102:3587–3592. doi: 10.1073/pnas.0407170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feinberg A. P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 33.Goldberg A. D, Allis C. D, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Huang L. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. J Cell Physiol. 2006;209:611–616. doi: 10.1002/jcp.20781. [DOI] [PubMed] [Google Scholar]
- 35.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krebs J. E, Kuo M. H, Allis C. D, Peterson C. L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005;21:608–615. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 39.Rundlett S. E, Carmen A. A, Suka N, Turner B. M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 40.Suka N, Suka Y, Carmen A. A, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 41.Deckert J, Struhl K. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol Cell Biol. 2002;22:6458–6470. doi: 10.1128/MCB.22.18.6458-6470.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Suka N, Carlson M, Grunstein M. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 43.Kroll E. S, Hyland K. M, Hieter P, Li J. J. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Measday V, Hieter P. Synthetic dosage lethality. Methods Enzymol. 2002;350:316–326. doi: 10.1016/s0076-6879(02)50971-x. [DOI] [PubMed] [Google Scholar]
- 45.Sopko R, Huang D, Smith J. C, Figeys D, Andrews B. J. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. Embo J. 2007;26:4487–4500. doi: 10.1038/sj.emboj.7601847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 47.Wagner M. V, Smolka M. B, de Bruin R. A, Zhou H, Wittenberg C, et al. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS ONE. 2009;4:e4300. doi: 10.1371/journal.pone.0004300. doi: 10.1371/journal.pone.0004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson L. P, Reed S. I, Haase S. B. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol. 2006;26:2456–2466. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. Embo J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 51.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. Embo J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costanzo M, Schub O, Andrews B. G1 transcription factors are differentially regulated in Saccharomyces cerevisiae by the Swi6-binding protein Stb1. Mol Cell Biol. 2003;23:5064–5077. doi: 10.1128/MCB.23.14.5064-5077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meimoun A, Holtzman T, Weissman Z, McBride H. J, Stillman D. J, et al. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shemer R, Meimoun A, Holtzman T, Kornitzer D. Regulation of the transcription factor Gcn4 by Pho85 cyclin PCL5. Mol Cell Biol. 2002;22:5395–5404. doi: 10.1128/MCB.22.15.5395-5404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robert F, Pokholok D. K, Hannett N. M, Rinaldi N. J, Chandy M, et al. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wijnen H, Futcher B. Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics. 1999;153:1131–1143. doi: 10.1093/genetics/153.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishizawa M, Kawasumi M, Fujino M, Toh-e A. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol Biol Cell. 1998;9:2393–2405. doi: 10.1091/mbc.9.9.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBride H. J, Sil A, Measday V, Yu Y, Moffat J, et al. The protein kinase Pho85 is required for asymmetric accumulation of the Ash1 protein in Saccharomyces cerevisiae. Mol Microbiol. 2001;42:345–353. doi: 10.1046/j.1365-2958.2001.02601.x. [DOI] [PubMed] [Google Scholar]
- 59.Measday V, McBride H, Moffat J, Stillman D, Andrews B. Interactions between Pho85 cyclin-dependent kinase complexes and the Swi5 transcription factor in budding yeast. Mol Microbiol. 2000;35:825–834. doi: 10.1046/j.1365-2958.2000.01754.x. [DOI] [PubMed] [Google Scholar]
- 60.McInerny C. J, Partridge J. F, Mikesell G. E, Creemer D. P, Breeden L. L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 61.Miller M. E, Cross F. R. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verges E, Colomina N, Gari E, Gallego C, Aldea M. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol Cell. 2007;26:649–662. doi: 10.1016/j.molcel.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 63.Skotheim J. M, Di Talia S, Siggia E. D, Cross F. R. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sternberg P. W, Stern M. J, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 65.Stillman D. J, Bankier A. T, Seddon A, Groenhout E. G, Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. Embo J. 1988;7:485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kasten M. M, Stillman D. J. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol Gen Genet. 1997;256:376–386. doi: 10.1007/s004380050581. [DOI] [PubMed] [Google Scholar]
- 67.Ho Y, Costanzo M, Moore L, Kobayashi R, Andrews B. J. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol Cell Biol. 1999;19:5267–5278. doi: 10.1128/mcb.19.8.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Bruin R. A, Kalashnikova T. I, Wittenberg C. Stb1 collaborates with other regulators to modulate the G1-specific transcriptional circuit. Mol Cell Biol. 2008;28:6919–6928. doi: 10.1128/MCB.00211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahn S. H, Diaz R. L, Grunstein M, Allis C. D. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol Cell. 2006;24:211–220. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 70.Ubersax J. A, Ferrell J. E., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 71.Moses A. M, Heriche J. K, Durbin R. Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 2007;8:R23. doi: 10.1186/gb-2007-8-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jori F. P, Napolitano M. A, Melone M. A, Cipollaro M, Cascino A, et al. Role of RB and RB2/P130 genes in marrow stromal stem cells plasticity. J Cell Physiol. 2004;200:201–212. doi: 10.1002/jcp.20026. [DOI] [PubMed] [Google Scholar]
- 73.Dasgupta P, Padmanabhan J, Chellappan S. Rb function in the apoptosis and senescence of non-neuronal and neuronal cells: role in oncogenesis. Curr Mol Med. 2006;6:719–729. doi: 10.2174/1566524010606070719. [DOI] [PubMed] [Google Scholar]
- 74.Huang D, Moffat J, Andrews B. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol Cell Biol. 2002;22:5076–5088. doi: 10.1128/MCB.22.14.5076-5088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J, Colwill K, Aneliunas V, Tennyson C, Moore L, et al. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr Biol. 1998;8:1310–1321. doi: 10.1016/s0960-9822(07)00561-1. [DOI] [PubMed] [Google Scholar]
- 76.Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. Embo J. 2005;24:4271–4278. doi: 10.1038/sj.emboj.7600889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamdane M, Bretteville A, Sambo A. V, Schindowski K, Begard S, et al. p25/Cdk5-mediated retinoblastoma phosphorylation is an early event in neuronal cell death. J Cell Sci. 2005;118:1291–1298. doi: 10.1242/jcs.01724. [DOI] [PubMed] [Google Scholar]
- 78.Longtine M. S, McKenzie A, 3rd, Demarini D. J, Shah N. G, Wach A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 79.Lee W, St Onge R. P, Proctor M, Flaherty P, Jordan M. I, et al. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baetz K, Moffat J, Haynes J, Chang M, Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol Cell Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jorgensen P, Nishikawa J. L, Breitkreutz B. J, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 82.Baetz K, Andrews B. Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol Cell Biol. 1999;19:6729–6741. doi: 10.1128/mcb.19.10.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tong A. H, Evangelista M, Parsons A. B, Xu H, Bader G. D, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of epitope-tagged Whi5 and Pcls. Whi5 and Pcl abundance in the indicated strains was determined by immunoblotting. Cyclin proteins were detected using 12CA5 anti-HA antibodies whereas Whi5 protein was detected using anti-GST antibodies.
(0.93 MB EPS)
List of 53 synthetic lethal interactions involving PHO85 that are not rescued by deletion of WHI5 .
(0.07 MB DOC)