Abstract
Brassinosteroids bind to the extracellular domain of the receptor kinase BRI1 to activate a signal transduction cascade that regulates nuclear gene expression and plant development. Many components of the brassinosteroid signaling pathway have been identified and studied in detail. However, the substrate of BRI1 kinase that transduces the signal to downstream components remains unknown. Proteomic studies of plasma membrane proteins lead to the identification of three homologous BR-signaling kinases (BSK1, BSK2 and BSK3). The BSKs are phosphorylated by BRI1 in vitro and interact with BRI1 in vivo. Genetic and transgenic studies demonstrate that the BSKs represent a small family of kinases that activate BR signaling downstream of BRI1. These results demonstrate that BSKs are the substrates of BRI1 kinase that activate downstream BR signal transduction.
One-sentence summary: Brassinosteroid signaling kinases identified by proteomics
Cell-surface receptor kinases activate cellular signal transduction pathways upon perception of extracellular signals, thereby mediating cellular responses to the environment and to other cells. The Arabidopsis genome encodes over 400 receptor-like kinases (RLKs) (1). Some of these RLKs function in growth regulation and plant responses to hormonal and environmental signals. However, the molecular mechanism of RLK signaling to immediate downstream components remains poorly understood, as no RLK substrate that mediates signal transduction has been established in Arabidopsis (2). BRI1 is an RLK that functions as the major receptor for the steroid hormones brassinosteroids (2). Brassinosteroids bind the extracellular domain of BRI1 to activate its kinase activity, initiating a signal transduction cascade that regulates nuclear gene expression and a wide range of developmental and physiological processes (fig. S1) (3). Many components of the BR signaling pathway have been identified and much detail has been revealed about how BR activates BRI1(4–8) and how phosphorylation by downstream GSK3-like kinase BIN2 regulates the activity of the nuclear transcription factors that mediate BR-responsive gene expression (fig. S1) (3, 9–13). However, no direct interaction has been observed between BRI1 and BIN2, and it remains unclear how BRI1 kinase at the plasma membrane transduce the signal to cytoplasmic components of the BR pathway (14).
To identify additional components of the BR signaling pathway, we performed quantitative proteomic studies of BR-responsive proteins using two-dimensional difference gel electrophoresis (2-D DIGE). Seedlings of BR-deficient det2-1 mutant were treated with brassinolide (the most active form of brassinosteroids) or mock solution, and proteins were labeled with Cy3 or Cy5 dyes, mixed together, and separated in the same gel by two-dimensional gel electrophoresis (2-DE). Brassinolide-induced BAK1 phosphorylation and BZR1 dephosphorylation were detected in the plasma membrane and phosphoprotein fractions, respectively (15), but not in total proteins (16). Similar to BAK1, two additional rows of spots showed BR-induced increase of the acidic forms and decrease of the basic forms (Fig. 1A and B), which is consistent with BR-induced phosphorylation. Mass spectrometry analysis of these spots identified two kinases encoded by Arabidopsis genes At4g35230 and At5g46570, which we named BR-Signaling kinase 1 and 2 (BSK1 and BSK2) (Fig. 1B and fig. S2). BSK1 and BSK2 share 60% amino acid sequence identity (fig. S3), and are members of the receptor-like cytoplasmic kinase sub-family RLCK-XII (1). The RLCK-XII sub-family includes twelve Arabidopsis proteins that each contains a kinase domain at the N-terminal side and tetratricopeptide repeat (TPR) domains at the C-terminus (fig. S3) (1). TPR domains are known to mediate protein-protein interactions and are present in components of steroid receptor complexes in animals (17). BSK1 and BSK2 do not contain predicted transmembrane domains but have putative N-terminal myristylation sites (glycine 2) that could mediate their membrane localization (fig. S3).
Figure 1. Identification of BSK1 and BSK2 as early BR regulated plasma membrane proteins.
(A) 2-D DIGE image of plasma membrane proteins isolated from 7-day-old det2 seedlings treated for 2 hr with either 100 nM BL (labeled with Cy5, red) or mock solution (Cy3, green). (B) Zoom-in view of an area of panel A shows BR-induced (black arrows, red spots) and BR-repressed (white arrows, green spots) protein spots. The table summarizes the protein identity, number of unique peptides, and the percentage of protein sequence coverage of mass spectrometry data for the spots numbered in the upper panel. (C-E) 2-dimensional gel immunoblotting analysis of BR-regulation of post-translational modification of BSK1 in det2 (C) and bri1-5 (D) background. Transgenic det2 or bri1-5 mutant seedlings expressing BSK1-YFP fusion protein were treated for 15 min with mock solution (−BL) or 100 nM BL (+BL). The proteins were separated by 2-DE and immunoblotted using anti-YFP antibody. (E) Quantitation of relative spot intensity along the IEF dimension in panel C and D. (F) Confocal microcopy images show localization of BSK1-YFP in hypocotyl cells of 3-day-old dark-grown transgenic det2 seedlings before (−BL) and 2 hr after treatment with 100 nM brassinolide (+BL). Scale bar, 10 µm.
The BR-induced shift of BSK1 from basic to acidic side in 2-DE gels was confirmed by immunoblotting of transgenic plants expressing a BSK1-YFP (yellow fluorescence protein) fusion protein (Fig. 1C and 1E). The response was significantly weaker in the bri1-5 mutant background (Fig. 1D and 1E), suggesting that BR regulation of BSK1 is BRI1 dependent. Consistent with their identification in the plasma membrane fractions, BSK1-YFP fusion proteins showed localization on the cell surface and the localization is not affected by brassinolide treatment (Fig. 1F).
The plasma membrane localization and BR-induced modification of BSKs suggest that they might be substrates of BRI1 or BRI1’s co-receptor kinase BAK1 (18, 19). In vitro kinase assays demonstrated that BRI1, but not BAK1, phosphorylates BSK1 (Fig. 2A). Mass spectrometry analysis of BRI1-phosphorylated BSK1 identified serine 230 of BSK1 as a BRI1 phosphorylation site (fig. S4). This same residue is also phosphorylated in vivo (20). While deletion of the C-terminal TPR domain has no effect on BSK1 phosphorylation by BRI1, a S230A mutation reduced the phosphorylation by 82% (Fig. 2B), indicating that S230 is the major site for BRI1 phosphorylation.
Figure 2. BSK1 is a substrate of BRI1.
(A, B) BRI1 phosphorylates serine230 of BSK1 in vitro. (A) Autoradiography of in vitro kinase assays performed with wild type (BRI1 and BAK1) or kinase-dead mutant (mBRI1 and mBAK1) forms of the kinase domain of BRI1 and BAK1 as GST fusion proteins (B). In vitro kinase assays of BRI1 phosphorylation of full-length BSK1, BSK1 with deletion of TPR domain (ΔTPR), and S230A mutant BSK1. (C) BiFC assay shows BRI1 interaction with BSK1. YFP fluorescence images of N. benthamiana leaf epidermal cells co-transformed with the indicated constructs. (D) Co-immunoprecipitation of BRI1 with BSK1. Arabidopsis plants expressing BSK1-myc only (lanes 1 and 4) or co-expressing BSK1-myc and BRI1-GFP (lanes 2, 3, 5, 6) were treated with 100 nM brassinolide (BL+) or mock solution (BL-) for 30 min. Microsomal proteins (lanes 1 to 3) were immunprecipitated with anti-GFP antibodies (lanes 4 to 6), and the immunoblot was probed using anti-GFP antibodies or anti-myc antibodies.
In vivo interactions with BRI1 were demonstrated using bimolecular fluorescence complementation (BiFC) and co-immunoprecipitation assays. While cells co-expressing BSK1 fused to the C-terminal half of YFP (BSK1-cYFP) and non-fusion N-terminal half of YFP (nYFP) or BAK1-nYFP fusion showed no or weak fluorescence signals (Fig. 2C), cells co-expressing BRI1-nYFP and BSK1-cYFP showed strong BiFC fluorescence at the plasma membrane (Fig. 2C). Anti-BSK1 antibodies immunoprecipitated the BRI1-GFP protein expressed from the BRI1 promoter (fig. S5), and a BSK1-myc protein was immunoprecipitated by anti-GFP antibodies only in transgenic Arabidopsis plants expressing both BRI1-GFP and BSK1-myc (Fig. 2D). BR-treatment reduced the amount of the co-immunoprecipitated BSK1-myc to 46% of the untreated sample (Fig. 2D), suggesting that BSK1 might be released from BRI1 upon phosphorylation. These results indicate that BSK1 is a BRI1 kinase substrate that is phosphorylated upon BR activation of BRI1.
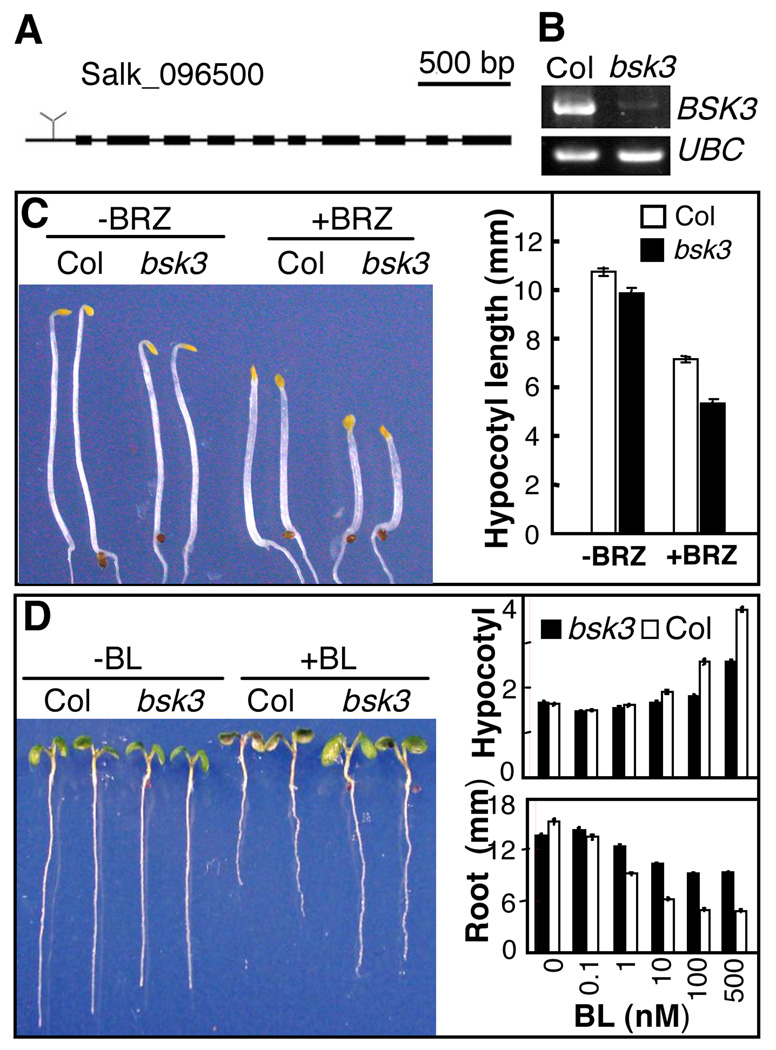
To determine the functions of BSK and their homologs in BR signaling, T-DNA insertion mutants were obtained for BSK2, BSK3, BSK4, BSK5, and BSK12 genes (21). Of these, only the bsk3-1 mutant showed an obvious phenotype (fig. S6). The bsk3-1 mutant contains a T-DNA insertion in the 5’ untranscribed region and expresses much reduced level of the BSK3 RNA (Fig. 3A and 3B). The bsk3-1 mutant seedlings grown in the dark on regular medium or medium containing the BR biosynthetic inhibitor brassinazole (BRZ) showed shorter hypocotyl length than wild type seedlings (Fig. 3C). Brassinolide treatment increases hypocotyl elongation and inhibits root growth in wild type plants grown in the light. Compared to wild type, the bsk3-1 mutant showed reduced responses to BL in hypocotyl elongation, root inhibition, and expression of BZR1-target gene DWF4 and BES1 target-gene SAUR-Ac (Fig. 3D and S7). These results demonstrate that loss-of-function mutation of bsk3 reduces BR sensitivity, indicating essential role of BSK3 in BR signaling. Similar to BSK1, the BSK3 protein is also regulated by brassinosteroid (fig. S8), phosphorylated by BRI1 kinase in vitro (fig. S9), and interacts with BRI1 in a BR-dependent manner in vivo (fig. S10). BSK1 and BSK3 are expressed in similar tissues as BRI1 (fig. S11). These results suggest that BSK3 and its homologs play redundant or overlapping roles in BR signaling, which could explain the weak BR-insensitive phenotypes of bsk3-1.
Figure 3. The bsk3-1 mutant has reduced BR sensitivity.
(A) T-DNA insertion site of bsk3-1 knockout mutant (T-DNA line SALK_096500). (B) RT-PCR analysis of BSK3 RNA expression in seedlings of wild type Columbia ecotype (Col) and the bsk3-1 mutant, with UBC RNA as control. (C) Wild type (Col) and bsk3-1 seedlings grown in the dark for 4 days on regular medium (−BRZ) or medium containing 1 µM brassinazole (+BRZ). The right panel shows average hypocotyl length of at least 25 seedlings. Error bars represent standard error. (D) The bsk3-1 mutant shows reduced sensitivity to brassinolide (BL). Left panel shows representative seedlings of Col or bsk3-1 grown in the absence (−BL) or presence (+BL) of 50 nM BL for 7 days under constant light. The right panel shows hypocotyl and root lengths (average of at least 60 seedlings) of wild type (Col) and bsk3-1 seedlings grown on various concentrations of BL under continuous light. Error bars represent standard error.
When overexpressed in the BR-insensitive bri1-5 mutant (Fig. 4A, 4B and fig. S12) or BR-deficient det2-1 (fig. S13) mutant backgrounds, BSK1, BSK3, and BSK5 obviously suppressed the dwarf phenotypes of the mutants. Consistent with reduced BR sensitivity of the bsk3-1 mutant, over expression of BSK3 is most effective in rescuing the bri1 phenotypes. The growth phenotypes correlated with altered expression of the BZR1-target gene DWF4 (Fig. 4C and S12), indicating that overexpression of the BSKs activates downstream BR signaling. Overexpression of BSK3 partly suppressed the dwarf phenotype of the null allele bri1-116 (Fig. 4D), but not that of the bin2-1 mutant (Fig. 4E), indicating that BSK3 functions downstream of BRI1 but upstream of BIN2, which is consistent with BSK3 being a substrate of the BRI1 kinase.
Figure 4. BSKs function downstream of BRI1 but upstream of BIN2 in the BR signaling pathway.
Phenotype of light grown three-week-old (A) or five-week-old (B) wild type, bri1-5, or transgenic bri1-5 overexpressing BSK3, BSK5, or BSK1 as YFP fusion proteins. (C) Quantitative RT-PCR analysis of DWF4 RNA expression in plants represented in panel A. Error bars represent standard deviation. (D) Overexpression of BSK3 partly suppresses the bri1-116 mutant. (E) Overexpression of BSK3 cannot suppress the bin2-1 mutant. (F) A model of BR signal transduction. Components in the inactive and active states are shown in blue and red colors, respectively. In the absence of BR (−BR), BRI1 associates with BSKs in an inactive state; BIN2 phosphorylates BZR1 and BZR2 to inhibit their DNA binding activity and promote their cytoplasmic retention by the 14-3-3 proteins. BR-binding (+BR) to BRI1 induces its dimerization with BAK1 and activation of BRI1 kinase, which phosphorylates BSKs. Phosphorylated BSKs dissociate from BRI1 and presumably inhibit BIN2 kinase and/or activate BSU1 phosphatase through yet unknown mechanisms, leading to dephosphorylation of BZR1 and BZR2, which regulate BR-responsive gene expression.
Using quantitative proteomics, we identified BSKs as new BR signal transduction components. This study demonstrates that sample pre-fractionation followed by 2-D DIGE is a powerful proteomic approach for dissecting signaling pathways. While only BSK1 and BSK2 were identified in the proteomic study, additional members (BSK3 and BSK5) of this family of RLCKs appear to play similar role in BR signaling. Our results support a model for the function of BSKs in BR signaling (Fig. 4F). In the absence of BR, BSKs are associated with BRI1. Upon BR activation of BRI1, BSKs are phosphorylated and then disassociate from the receptor complex to activate downstream signaling. Such ligand-induced disassociation from a pre-existing receptor complex potentially provides faster signaling than ligand-induced recruitment of a free component into the receptor complex.
Both BSKs and BAK1 are substrates of the BRI1 kinase, but several lines of evidence support that they play distinct roles in BR signaling. First, BR induces BRI1-BAK1 interaction (6) but reduces BRI1-BSK1 and BRI1-BSK3 interactions. Second, overexpression of BSK3 suppresses the bri1-116 null allele, whereas overexpression of BAK1 only suppresses weak alleles but not a strong allele of bri1 nor a double mutant containing the weak bri1-5 allele and the BR-biosynthetic mutation det2-1 (19), suggesting that BSK3 functions downstream of BRI1 whereas BAK1’s action on downstream BR response requires a functional BRI1. BAK1 and its homolog BKK1 are required in additional signaling pathways and BAK1 is also a co-receptor for the FLS2 receptor kinase, a receptor for flagelin, suggesting that BAK1 is not a specific component of the BR pathway (22–25). BAK1 most likely mediates activation of BRI1 kinase rather than signal transduction to specific downstream components in the BR signaling pathway. In contrast, the BSKs directly mediate signal transduction from BRI1 to downstream BR responses (Fig. 4F). Identification of the downstream direct targets of BSKs will be the key to fully understanding how BR signal is transduced from the cell surface to the nuclear transcription factors.
Supplementary Material
References and Notes
- 1.Shiu SH, et al. Plant Cell. 2004 May;16:1220. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson KL, Ingram GC. Curr Opin Plant Biol. 2005 Dec;8:648. doi: 10.1016/j.pbi.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Annu Rev Cell Dev Biol. 2005;21:177. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 4.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. Nature. 2001 Mar 15;410:380. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita T, et al. Nature. 2005 Jan 13;433:167. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, et al. Plant Cell. 2005 Jun;17:1685. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Chory J. Science. 2006 Aug 25;313:1118. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, et al. Dev Cell. 2005 Jun;8:855. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZY, et al. Dev Cell. 2002 Apr;2:505. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 10.He J-X, et al. Science. 2005;307:1634. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Y, et al. Cell. 2005 Jan 28;120:249. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 12.Vert G, Chory J. Nature. 2006 May 4;441:96. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 13.Gampala SS, et al. Dev Cell. 2007 Aug;13:177. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendron JM, Wang ZY. Curr Opin Plant Biol. 2007 Oct;10:436. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang W, et al. Mol Cell Proteomics. 2008;7:728. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Z, et al. Mol Cell Proteomics. 2007 Dec;6:2058. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DF. Cell Stress Chaperones. 2004 Summer;9:109. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam KH, Li J. Cell. 2002 Jul 26;110:203. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 19.Li J, et al. Cell. 2002 Jul 26;110:213. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 20.Niittyla T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. Mol Cell Proteomics. 2007 Oct;6:1711. doi: 10.1074/mcp.M700164-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Alonso JM, et al. Science. 2003 Aug 1;301:653. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 22.He K, et al. Curr Biol. 2007 Jul 3;17:1109. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Kemmerling B, et al. Curr Biol. 2007 Jul 3;17:1116. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 24.Chinchilla D, et al. Nature. 2007 Jul 26;448:497. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 25.Heese A, et al. Proc Natl Acad Sci U S A. 2007 July 17;104:12217. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.We thank Dr. Winslow Briggs for comment on the manuscript. Research was supported by grants from the National Science Foundation (NSF 0724688), U.S. Department of Energy (DE-FG02-04ER15525), and NIH (R01GM066258). S.Z and R.W are supported by the Chinese Scholarship Council. The UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) is supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR RR01614, RR012961 and RR019934.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.