Abstract
In few years our understanding of microRNA (miRNA) biogenesis, molecular mechanisms by which miRNAs regulate gene expression, and the functional roles of miRNAs has been expanded. Interestingly, numerous miRNAs are expressed in a spatially and temporally controlled manner in the nervous system, suggesting that their posttrascriptional regulation may be particularly relevant in neural development and function. MiRNA studies in neurobiology showed their involvement in synaptic plasticity and brain diseases. In this review ,correlations between miRNA-mediated gene silencing and Alzheimer's, Parkinson's, and other neurodegenerative diseases will be discussed. Molecular and cellular neurobiological studies of the miRNAs in neurodegeneration represent the exploration of a new Frontier of miRNAs biology and the potential development of new diagnostic tests and genetic therapies for neurodegenerative diseases.
1. Introduction
Neurodegenerative diseases represent a large group of neurological disorders with heterogeneous clinical and pathological expressions, affecting specific groups of neurons, in specialized functional anatomic systems. A mixture of environmental and genetic factors seems to engender neurodegenerative diseases, and aging has been found a common risk factor. Neurodegenerative diseases result from the gradual and progressive loss of neuronal cells, leading to nervous system dysfunction. They are characterized by the formation of distinct pathological changes in the brain, including extracellular protein deposits, cellular inclusions, and remodelling of cell morphology. However, while many different forms of neurodegenerative disease are recognized, the lines that separate one from another are often unclear. For instance, symptoms such as motor impairment and memory loss may occur in many different types of neurodegenerative disease. Alzheimer's disease, Parkinson's disease, prion diseases, and polyglutamine disorders, including Huntington's disease and various spinocerebellar ataxias, are well-known neurodegenerative disorders [1]. To date, with few exceptions, no diagnostic laboratory tools exist that can clearly indicate the presence, absence, or category of a neurodegenerative disease. Diagnoses are usually based on clinical evaluation of the symptoms.
The microRNA-(miRNA-) guided RNA silencing pathway is a recently discovered process found to regulate gene expression acting on messenger RNA (mRNA). MiRNA biogenesis is mediated by Dicer which catalyzes the processing of double-stranded RNAs (dsRNAs) into ≈22 nt-long small miRNAs. These small noncoding RNA molecules operate as guides for RISC (RNA Induced Silencing Complex) to cleave a target mRNA in case of a perfect complementarity (siRNA) or to block the target mRNA translation (miRNA) when there is an imperfect pairing between miRNAs and the targets. In mammalian cells the repression of translation by miRNA is mediated by an imperfect pairing with 3′UTR of the mRNA target [2]. MiRNAs are conserved throughout the evolution, and their expression may be constitutive or spatially and temporally regulated. Increasing efforts to identify the specific targets of miRNAs lead to speculate that miRNAs can regulate more than 90% of human genes. Specific miRNA subsets were expressed in specific brain area and in neuronal and glial cell subtypes [3]. The studies of microRNAs expression profiles in nervous system represent the first step in understanding how, where, and when miRNAs are involved in the regulation of neurodevelopment, differentiation, dendritic spine development, local protein synthesis, and synaptic plasticity [4]. Several works have shown spatially and/or temporally restricted distribution of miRNAs, suggesting that they might regulate neuronal gene expression. By comparative analysis of miRNA expression in the normal and pathologic brain, the microRNA signatures in several neurodegenerative diseases, including polyglutamine expansions, Parkinson's and Alzheimer's diseases are coming up. To date we have few pieces of information about the expression profiles, and the complex composition of the brain, containing several neuronal and glial istotypes, while representing its main biological characteristic, is also the principal obstacle for an accurate analysis. Furthermore in order to interpretate miRNA expression data from post-mortem human brains affected by neurodegenerative diseases, the source of neural tissues, together with the RNA isolation techniques used, need to be carefully considered. Overall, the identification of miRNA's physiological target genes should be a primary approach to reveal the specific contribution of microRNAs to neural function. Recent studies of microRNA in nonneuronal cellular systems could drive future research in primary neuronal cells. In fact, prediction of microRNA targets utilizing different algorithms based on the general rule of the seed region has been complemented by an elegant proteomic approach which uses a mass spectrometric method called stable-isotope labelling with aminoacid in cell culture (SILAC) to measure changes in protein levels in response to miRNA induction or knockdown. SILAC approach showed that individual microRNA can reduce the production of hundred proteins [5, 6]. Many targets are repressed at both mRNA and protein level and others are predominantly regulated at protein level. Although several miRNA-induced changes in the proteome correlate with the presence of seeds in the mRNA of the affected proteins, some changes remain to be explained. They might be due both to indirect effects and/or to miRNA direct targeting mediated by still unknown rules. Remarkably an increasing level of complexity of species-specific miRNA expression during evolution emerged. However, 447 new miRNA genes expressed in human fetal and chimpanzee adult brains were identified. Many of them are not conserved beyond primates, indicating a recent evolutionary origin. Since 8% of miRNA were found to be human-specific, they might play a role in the human brain evolution. However, expression levels of miRNAs common to human and chimpanzee were not determined, because different regions were analyzed at various ages. Several features in cognitive functions of humans and chimpanzees are probably elucidated by variations in cortical structures. Therefore, the diversity of miRNA repertoire in the brain likely contributes to the dissimilarities between human and chimpanzee, arguing for a role of miRNA in brain evolution and function [7].
In this review we discuss the recent studies on the involvement of miRNA-mediated gene silencing in neurodegenerative diseases.
2. MicroRNAs in Neurodegenerative Diseases
2.1. MicroRNAs in Alzheimer's Disease
Alzheimer's Disease (AD) is the best known degenerative disease affecting the central nervous system [8]. AD is a chronic progressive disease characterized by early memory impairments followed by these cognitive deficits: aphasia (language disturbances), agnosia (failure to recognize people or objects in presence of intact sensory function), apraxia (inability to perform motor acts in presence of intact motor system). Neuropathologically the areas of brain most affected are the hippocampus followed by association cortices and subcortical structures. The neurodegeneration is characterised by synapse and cellular loss, β-amyloid plaques, and neurofibrillary lesions. The major component of plaques is the Aβ peptide which derives from the proteolytic processing of its precursor protein (APP). The neurofibrillary lesions contain aggregates of hyperphosphorylated microtubule-associated protein tau. This histopathological hallmark is used in Braak's Alzheimer's system [9] to describe postmortem AD brain samples in six stages: in the transentorhinal stage (Stages I and II), the neurofibrillary pathology is essentially confined to the transentorhinal and entorhinal cortex and slightly to the CA1/CA2 sections of the hippocampus; the limbic stage (Stages III and IV) frames a severe involvement of the entorhinal areas and a moderate engagement of the hippocampus; the hallmark of the neocortical stage (Stages V and VI) was the dismantlement of the neocortex. AD is the most common cause of dementia in aged populations. About 1% early onset familial form of the disease (onset before 60 to 65 years of age) is due to mutations in three genes, APP, presenilin 1 (PSEN1), and presenilin 2 (PSEN2) all of which cause Aβ overproduction. Aβ production is initiated by the processing of APP by the β-amyloid cleavage enzyme 1 (BACE1) which generated a C-terminal fragment of APP, labelled as C99. This fragment is further cleaved by γ-secretase complex, which includes the presenilins, and generates the more abundant Aβ40 and the less abundant, but more pathogenic, Aβ42 [8]. Aβ load in AD brain was suggested to trigger neuronal dysfunctions. For the majority of AD cases, which shows less obvious familial aggregation (hence they are also called sporadic AD), the molecular bases of the disease are matter of intensive research.
A discrete number of studies has suggested that a dysregulated microRNA expression could be aging-associated and could contribute to AD. miRNA expression profiles are changed in pathological conditions in all studies published until now. Lukiw's laboratory evaluated the expression of 12 miRNAs in hippocampal region of fetal, adult, and AD brain [10]. They found that miR-9 was upregulated in both fetal and AD hippocampus, and miR-128 was increased specifically in AD hippocampus. However, the translational changes induced by these specific miRNAs in AD hippocampus remain to be investigated. More recently espression profiles of 328 microRNAs in anterior temporal cortex from five sporadic AD patients and five age-mathced controls showed 13 microRNAs significantly downregulated in cortex of sporadic AD [11]. Using various prediction algorithms to identify AD-related potential target genes, 7 of the 13 microRNAs had candidate binding sites in the 3′UTR of BACE (miR-15a, -29b-1, -9, and -19b) or APP (let-7, miR-101, miR-15a, and miR-106b,) or in the 3′UTR of PSEN1 (miR-9). On the other hand, 6 microRNAs do not seem to be related to obvious targets (miR-210, -181c, -22, -26b, -363, -93). In particular the relationship between BACE1 and miR-29 was deeply investigated (mentioned in what follows). The changes of miRNA profiles might be specific for sporadic AD and might cause or exacerbate the neuropathology. Furthermore, linking microRNA expression to their specific targets could suggest novel pathways of the disease.
A recent study evaluated the importance of deregulation of miRNA expression in brains and cerebrospinal fluid (CSF) of Alzheimer's patients [12]. Over 300 microRNAs were determined in the hippocampus, medial frontal gyrus, and cerebellum from early and late stage AD compared to age-matched control. Deregulated microRNAs have been associated to known and novel molecular pathways in AD pathogenesis such as neurogenesis, oxidative stress, insulin resistance, and innate immunity. For example, miR-9 and miR-132 downregulation was correlated to impaired neurogenesis and neuronal differentiation. The finding that miR-423 was upregulated in hippocampus while miR-98 was decreased in cerebellum was appealing. In fact both miRNAs modulate IDH2 (isocitrate dehydrogenase 2) expression and IDH2 reduction was described to be involved in oxidative stress in AD prefrontal cortex. These observations suggest a mechanism for the specific susceptibility of particular AD brain areas such as the hippocampus and the relative sparing of others such as the cerebellum.
Sixty miRNAs were differentially expressed in the CSF of patients between Braak stage 5 and stage 1. Among these miRNAs few were brain enriched while several were not correlated to the miRNAs changes observed in AD brain regions. Therefore, CSF microRNAs were suggested to derive from T lymphocytes present in the CSF. The altered expression of miRNA in the CSF of patients affected by Alzheimer's disease, opens a new scenario on the use of these expression profiles as putative AD biomarkers. To date, CSF analysis from AD patients produced some of the most reproducible biomarkers, such as decreased Aβ42, increased total tau (ttau), and increased phosphorylated tau (p-tau) [13]. Combinations of these CSF markers have been also proposed to diagnosticate AD. Future work might be to evaluate if the CSF miRNAs profile correlates with Aβ, total and phosphorylated tau protein presently carried out in the CSF of AD patients.
Specific molecular mechanisms involving microRNAs and expression of BACE1 and APP are emerging in the AD field. By microarray and in situ hybridization of superior-medial frontal cortex of AD, Nelson's laboratory showed that the expression of miR-107 decreased during progression of the disease in parallel to BACE1 mRNA increase. In addition cell culture experiments showed that the expression of a luciferase reporter gene fused to a 3′UTR containing BACE1 microRNA 107 binding site is modulated by miR-107 [14]. Another study, investigating changes in microRNA expression profiles of anterior temporal cortex from sporadic AD patients found that the expression of the cluster miR-29a/b-1 was significantly decreased in a subgroup of AD patients in which BACE 1 protein was abnormally upregulated while BACE 1 mRNA levels were unchanged [11]. Consistently, during mouse brain development from E17 to 1 year, BACE1 protein level decrease was correlated to miR-29a/b-1 upregulation while BACE1 mRNA level was stable. In cell culture experiments, BACE1 target validation was demonstrated by monitoring the effects of miR-29a/b-1 on the translation of a BACE1 3′UTR luciferase reporter carrying wilde-type or mutated miR29a/b-1 responsive site. Finally, upon either overexpression or downregulation of miR-29a/b-1 in human cell culture both BACE1 protein levels and APP cleavage product Aβ were, respectively, reduced and increased.
In both previous studies [11–14], the loss of microRNA in AD is not specific for a certain brain area more susceptible to the disease, that is, miR-107 is also downregulated in motor cortex of AD patients and miR-29a/b-1 expression also decreases in AD cerebellum. Therefore, altered BACE expression due to microRNA deregulation is not responsible for increased sensitivity of particular brain regions. However also in the AD familial cases, mutations of APP and PSEN are present in all cells of the brain and only specific regions are affected from AD.
Interestingly other two microRNAs, miR-298 and miR-328, regulate BACE 1 protein expression in cultured neuronal cells [15]. It is relevant that in APPswe/Psen1 transgenic mice, an AD mouse model which recapitulates some features of the disease, it was observed that BACE1 mRNA decreased and protein levels increased in the hippocampus at 19 months of age. In transgenic mice, the expression of miR-298 and miR-328 decreased in the granular neurons of the hippocampus during aging. However, while the miR-328 sequence is perfectly conserved between mouse and human, that of miR-298 is only 72% identical. Clearly, additional work will be needed to determine whether all of these microRNAs are really active in human brain and their relative contribution to BACE expression in physiological and pathological conditions and in different neuronal populations.
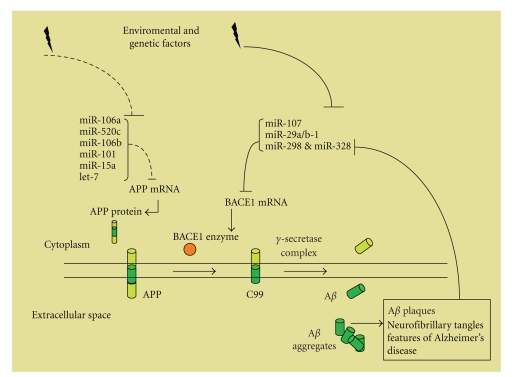
It has been shown that AD can be caused by increased expression of the APP gene due to either genomic duplication or regulatory sequence alterations. In C. elegans, APP orthologue APL-1 is regulated by developmentally timed microRNA [16]. In particular, apl-1 expression in seam cells is indirectly repressed by let-7 family microRNA, and apl-1 transcription is regulated by downstream targets of let-7 microRNA. This study opens new insights into the time-dependent progression of AD. The 3′UTR of APP mRNA is a potential target for several microRNAs. Recently, utilizing human HEK-293 cells, it has been demonstrated that miR106a and miR-520c negatively regulate expression of reporter genes containing their predicted target sequences present in the APP 3′UTR [17]. In addition, overexpression of miR-106a or miR-520c (which is not expressed in brain) reduces APP levels by 50%. It will be important to translate these results in a cellular context relevant for AD pathology. Interestingly, miR-106b is one of the four microRNA that have been predicted to target the 3′UTR of APP (let-7, miR-101, miR-15a, and miR-106b) and that were found to be downregulated in anterior temporal cortex from five sporadic AD patients [11]. All these investigations suggest that dysregulation of miRNAs, by modulation of APP and BACE1 expression, might be a cause or a consequence of AD (Figure 1).
Figure 1.
APP or BACE1 upregulation might lead to Aβ overproduction in Alzheimer's Disease. The picture shows molecular pathways modulating APP and BACE1 expression and amyloidogenic processing of APP by BACE1 and the γ-secretase complex leading to Aβ production. Reduction of miRNA/RISC posttranscriptional regulation of APP and/or BACE1 mRNA induces the increase of the relative proteins, which drive to Aβ accumulation. Changes of miRNA expression might trigger molecular events inducing AD pathology or generate a feed-forward mechanism during AD progression (as suggested for miR-298 and miR-328).
2.2. MicroRNAs in Parkinson's Disease
Parkinson's disease (PD) is associated with progressive neurodegeneration of dopaminergic neurons (DNs) in the substantia nigra and leads to tremor, rigidity, and bradykinesia. Futrhermore, widespread neuronal modifications lead to complex and variable non-motor symptoms. Lewy bodies are a neropathological feature of PD and are cellular inclusions comprising a dense core of filamentous material surrounded by a halo of fibrils, which mainly consists of a-synuclein. Mutations in genes coding for synuclein (SNCA), parkin, pink1, DJ-I, Lrrk2, can explain only a limited number of familial PD cases, while the molecular bases of vast numbers of non familial cases are not yet understood [18]. A recent study shed some light on the role of microRNA in DNs differentiation and raises the question whether microRNA are involved in etiology of PD. Deletion of Dicer impairs the ability of ES cells to differentiate into DNs. Since Dicer deletion was partially rescued by transfection of small RNA derived from embryonic mouse midbrain, it is likely that microRNAs are involved in DNs differentiation and survival [19]. In addition, specific deletion of Dicer in vivo, in mouse midbrain dopaminergic neurons, leads to cell death in the substantia nigra. Behavioural studies of the animals revealed reduced locomotion in an-open-field assay, reminiscent of the phenotype of human patients with PD. MiRNA expression profiles of normal adult midbrain compared with the profiles of midbrain depleted of DNs from PD patients revealed alterations of certain midbrain-enriched miRNA, in PD brain [19]. The role of miR-133b, enriched in midbrain and absent in the brains of PD patients, was further investigated. It was demonstrated that miR-133b constitutes a negative feedback loop with the transcription factor Ptx-3: Ptx-3 transcribes miR-133b which in turn represses Ptx-3 translation. In vitro experiments showed that depletion of miR-133b increases the expression of DN markers and depolarization-induced dopamine release while miR-133b overexpression suppresses the full differentiation of DN neurons and produces a significant decrease in dopamine release [19]. Thus, although miR-133b is involved in differentiation and function of DN neurons, additional microRNAs should be responsible for Dicer deletion phenotype in DN.
Polymorphisms affecting the interactions between microRNAs and their targets are emerging in various studies on neurodegenerative disease. Genetic analyses showed that relevant polymorphic variations in the fibroblast growth factor 20 gene (FGF20) are associated with the risk of developing PD. FGF20 is preferentially expressed in the substantia nigra and promotes survival of dopaminergic neurons. More recently, one SNP (rs 127202208), located within the FGF20 3′UTR, was strongly associated with PD. SNP rs 127202208 lies within a predicted binding site for microRNA 433 which is highly expressed in the brain [20]. Through several functional assays, it was demonstrated that the risk allele rs 127202208 damped a binding site for microRNA 433 and increased translation of FGF20. In cell culture experiments and in PD brains, the increased FGF20 translation was correlated with increased synuclein expression. Synuclein is included into the genes responsible for familial cases of PD and althought the function of this protein is not yet defined, it has been demonstrated that overexpression and point mutations can cause PD.
2.3. MicroRNAs in Polyglutamine Diseases
Polyglutamine (polyQ) disorders constitute a family of dominantly inherited neurodegenerative diseases caused by the expansion of CAG triplet repeats in a specific gene. A common signature is the accumulation of the mutant protein in large intranuclear inclusions. The clinical features include spasticity and cognitive impairments. To date, ten such neurodegenerative disorders known to be caused by expansion of the CAG repeat in the coding region of the respective genes have been identified [21]. These prototypical protein misfolding disorders include Huntington disease (HD), six distinct forms of spinocerebellar ataxia (SCA-1, 2, 3, 6, 7 and 17), dentatorubropallidoluysian atrophy (DRPLA), and spinobulbar muscular atrophy (SBMA).
2.3.1. Huntington Disease
Huntington's disease (HD) is the most common and well-studied polyglutamine neurodegenerative disorder [22]. It is an hereditary autosomal dominant disease characterized by motor, cognitive, and psychiatric symptoms. It affects about 3 in 100 000 individuals. The onset of symptoms typically occurs between the ages 35 and 50 years, though it may appear at any age. The molecular basis of the disease is the expansion of the trinucleotide CAG in the first exon of a gene on chromosome four (4p 16.3). This gene encodes the protein huntingtin (Htt) of 3136 amino acids. The mutation of huntingtin produces an expanded stretch of glutamine (Gln) residues. This CAG/polyGln expansion has 6–39 units in normal individuals and 36 to 180 units in HD patients. Huntingtin appears to be associated to protein trafficking, transcriptional regulation, synaptic signalling, vesicle transport, and apoptosis. HD patients show progressive loss of cortical and striatal neurons associated with choreic movement and dementia. The neuropathological hallmark is the gradual atrophy of the striatum (caudate nucleus and putamen), observed in 95% of the HD brains. Mechanisms of neurodegeneration implicated in HD pathology are excitotoxicity, dopamine toxicity, mithocondrial dysfunction, oxidative stress, apoptosis, and autophagy.
Several observations suggested microRNAs dysregulation in HD. Interaction between wild type Htt and Repressor Element1 Silencing Transcription (REST) factor was described. In pathological conditions Htt mutation inhibits its interaction with REST and provokes REST build-up in the nucleus of HD neurons, decreasing neuronal gene expression. REST is a transcriptional repressor of neural genes, including several microRNAs [23].
Recently, Johnson et al. [24] identified miRNA regulated by REST in neurons, and measured the expression of these miRNAs in the brains of HD mouse models and in postmortem tissue of HD patients. Several changes in microRNAs expression profile were allocated to species-specific differences, and others to the comparative analysis of a specific human cortex, area versus whole mouse cortex. Both in HD mouse model and in human HD cortex, miR-132 was downregulated, and its mRNA target p250 GAP, which modulates dendritic plasticity, was increased. Since REST is highly involved in HD, microRNAs are likely expected to play an important role in the disease pathogenesis. In addition, it was demonstrated that huntingtin protein co-purified with Argonaute proteins, fundamental components of RISC complex. Argonaute proteins have been shown to localize to cytoplasmic foci, named P bodies. Htt, colocalized with Argonaute2 in P bodies, and depletion of Htt showed compromised RNA-mediated gene silencing. Thereafter, in mouse striatal neurons expressing Htt mutation, P bodies formation and translation miRNA-mediated repression were impaired. These data suggest that Htt play a role in miRNA processes [25].
2.3.2. Spinocerebellar Ataxia Type 3
The polyglutamine (polyQ) protein Ataxin-3 is mutated in the human polyglutamine disease spinocerebellar ataxia type 3 (SCA3), resulting in a progressive dysfunction of the cerebellum. SCA3 is typically a late-onset fatal autsosomal dominant neurodegenerative disease that, like all ataxias, is characterized by loss of motor coordination and balance. In SCA-3 Drosophila model, the suppression of miRNA processing by dicer mutation increases ataxin-3 toxicity, inducing a neurodegenerative phenotype. Moreover, depletion of R3D1, a dsRNA-binding protein, that forms a stable complex with Dicer-1, causes accumulation of precursor miRNA, increasing ataxin-3-induced toxicity [26, 27]. In HeLa cell line, dicer reduction by RNAi enhances polyQ protein toxicity only in cells expressing pathogenic Ataxin-3, causing loss of 70% of the cultured cells. These findings suggest a neuroprotective role of miRNAs in these neurodegenerative diseases [27].
2.3.3. Spinocerebellar Ataxia Type 1
Spinocerebellar ataxia type 1 (SCA1) is a dominant inherited disease caused by expanded trinucleotide repeats resulting in an increased polyglutamine tract in the gene product ataxin-1 (ATXN-1). SCA1 patients loose motor coordination and develop slurred speech, spasticity, and cognitive impairments. A typical feature of SCA1 pathology is the atrophy and loss of Purkinje cells from the cerebellar cortex. Purkinje cells are the major integrative neurons of the cerebellar cortex, projecting their axons onto the deep cerebellar nuclei. A recent study showed that a conditional Purkinje (PK) cell-specific ablation of Dicer leads to PK cell death, cerebellar dysfunction, and ataxia indicating an involvement of miRNAs in cerebellar neurodegeneration [28].
Another line of evidence suggesting a role of miRNAs in SCA1 pathogenesis comes from the observation that miR-19, miR-101, and miR-130 cooperatively regulate ATXN1 levels [29]. When miR-19, miR-101 and miR-130 were inhibited by 2′-O-methyl oligonucleotides, an increase of ATXN1 protein level was observed. Moreover, it was demonstrated that miR-19, miR-101, and miR-130 were expressed in mouse cerebellum and Purkinje cells by Northern blot analysis and in situ hybridization. These miRNAs regulate the cell toxicity of the polyQ-expanded ATXN1, suggesting to investigate miRNAs-mediated regulation in SCA1 neurodegenerative disorder.
2.4. MicroRNAs in Frontotemporal Dementia
Frontotemporal dementia (FTD) is a neurodegenerative disease representing ~5% of all dementia patients, characterized by the progressive degeneration of the frontal and anterior temporal cortex. Considering the involvement of the frontal lobe, the clinical picture is cognitive and memory impairment, language dysfunction, and/or changes in personality or behavioural disorders. FTD can be divided into two main neuropathological subtypes: frontotemporal lobar degeneration (FTLD) with neuronal and glial tau inclusions (FTLD-tau), and FTLD with neuronal cytoplasmic inclusions (NCIs) that are positive for ubiquitin and TAR DNA-binding protein (TDP-43) (FTLD-U). However, 20%–30% of cases of FTD follow an autosomal dominant pattern of inheritance, and half of them are caused by defects in microtubule-associated protein tau (MAPT), multi-vesicular body protein 2B (CHMP2B), and valosin-containing protein (VCP) [30].
Mutations in the progranulin gene (GRN), encoding a secreted growth factor, on chromosome 17q21, have recently been identified as a major cause of familial FTLD-U. These cases have a characteristic pattern of neuropathology that is a distinct subtype of frontotemporal lobar degeneration with ubiquitinated inclusions (FTLD-U), with NCIs in layer II of the cortex and lentiform neuronal intranuclear inclusions (NIIs). To date, more than 60 different mutations in GRN were mapped, and a recent breakthrough was the identification of a genetic variant (rs5848), located in the 3′UTR of GRN mRNA, in a binding site for miR-659 [32]. This research showed that miR-659 targets GRN suppressing its translation, and demonstrated a decrease of GRN protein levels of ~30% in FTLD-U rs5848 homozygous TT carriers compared to CC carriers. Consistently, in FTLD-U patients heterozygous for rs5848, an intermediate dosage of GRN protein was determined. miR-659 seems to be a specie-specific human microRNA, which expressed in brain, including frontal and temporal neocortex. In addition “seed” sequence for miR-659 in the GRN 3′UTR is only present in humans, and is not found in other mammals. Although a small number of FTLD-U patients were examined, the enhanced binding of miR-659 to the 3′UTR of the GRN gene is an important risk for TDP-43-positive FTLD-U. Future studies on specific human cortical miRNAs might be relevant to decrypt human neurodegenerative disease.
2.5. MicroRNAs in Prion Disease
Prion diseases or transmissible spongiform encephalopathies (TSEs) are a family of rare progressive neurodegenerative disorders that affect both humans and animals. They are distinguished by long incubation periods, characteristic spongiform changes associated with neuronal loss, and a failure to induce inflammatory response. The causative agent of TSEs is believed to be a prion, a transmissible agent, which is able to induce abnormal folding of normal cellular prion proteins in the brain [33]. According to the protein-only hypothesis, the central event in the pathogenesis of prion diseases is the conversion of a normal cellular protein termed PrP(C) to PrP(Sc), a conformational isoform. Prion diseases impair brain function, causing memory impairment, personality changes, dementia, and movement disorders and the characteristic signs and symptoms of the disease. TSEs begin in adulthood are rapidly progressive and lead to death within a few months to several years. Familial prion diseases of humans include classic Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträussler-Scheinker syndrome (GSS), and fatal insomnia (FI). To explain TSE pathogenesis, it is important to identify disease-associated alterations in gene expression. Recently, by microarrays and RT-PCR, the analysis of miRNA expression was made [31]. Brain miRNAs expression of mice infected with mouse-adapted scrapie showed changes of 15 miRNAs. Among these, only two, miR-338-3p and miR337-3p, were downregulated, whereas the others were up-regulated. Several predictions of the theoretical mRNA targets of changed miRNAs during prion disease were performed, using web-based computational algorithms. From this in silico analysis, genes involved both in transcription, cell cycle, ubiquitin-proteasome pathway, and in normal functioning of synapses, neuronal activity, neurogenesis, and neurites growth were identified. Lastly, only one target, the transcriptional regulator EGR1, was experimentally validated by luciferase assay in vitro. In particular, the authors suggested that the prion disease upregulates miR-191 which represses the EGR1 mRNA translation. The transcriptional regulators EGR1 and CREB1 were already identified as downregulated prion-related genes with a central role in biologically relevant networks in prion infection [34]. As a consequence, miRNAs mediated regulation of these prion-related genes could contribute to neuronal death and neurodegeneration. Finally, the miRNA expression profile was proposed as potential biomarker of prion diseases.
3. Concluding Remarks
The studies on miRNA in neurodegenerative diseases (Table 1) are only now coming to light. Until now, both changes of several miRNA expression profiles and polymorphisms affecting the interactions between miRNAs and their targets are emerging in various studies on neurodegenerative disease. It is difficult to determine if the changes in miRNA expression detected in the brains or CSF of patients are primary or secondary events, or both. Nevertheless early or late in the evolution of the disease, they could contribute to the pathogenesis of the observed lesions and neuronal loss. Unique patterns of miRNA expression profile in the CSF of particular neurodegenerative disease could be useful as molecular biomarkers for disease diagnosis and eventually prediction of therapeutic responses. The identification of miRNA causing a specific pathology could open new therapeutic perspectives to block endogenous miRNAs or deliver exogenous miRNAs. Until now either antisense oligonucleotides chemically modified [35] or expressed sequences corresponding to multiple miRNA seed target (miRNA sponge) [36] have been used as microRNA inhibitors. Delivery of these molecules to the CNS, avoiding toxicities, could be the challenge of future research. Furthermore since in several neurodegenerative disorders specific nuclear or cytoplasmic protein accumulation is causative of the neuropathological picture, the identification of microRNAs regulating the translation of these targets could represent the first step aimed to therapeutic applications. The second step might be to evaluate the quantitative effects on the proteome of specific amounts of the “therapeutic” microRNAs.
Table 1.
MIND: MicroRNAs in neurodegenerative diseases.
| miRNA | Neurodegenerative disease | mRNA target | Reference |
|---|---|---|---|
| miR-298; miR328 ↓ | Alzheimer's Disease mouse model | BACE1 | [15] |
| miR-107 ↓ | Alzheimer's Disease | BACE1 | [14] |
| miR-29a/b-1 ↓ | Alzheimer's Disease | BACE1 | [11] |
| miR-133b ↓ | Parkinson's disease | Pitx3 | [19] |
| miR-433 | Parkinson's disease | FGF20 (SNP rs127202208) ↑ | [20] |
| miR-191 ↑ | Prion disease | EGR1 | [31] |
| miR-132 ↓ | Huntington disease | P250GAP | [24] |
| miR-659 | Frontotemporal dementia | GRN (SNP s5848) ↓ | [32] |
Acknowledgments
This work was supported by Italian Institute of Technology (IIT), REGIONE LAZIO grant—‘Studio delle basi molecolari della neurodegenerazione nella malattia di Alzheimer's—(to C. Cogoni), National Research Council (CNR) grant DG.RSTL.059.012 (to F. Ruberti), and by “Fondazione Alazio Award 2007” (to C. Barbato) (http://www.fondazionealazio.org/). The first and second author contributed equally to the preparation of this manuscript.
References
- 1.Skovronsky DM, Lee VM-Y, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annual Review of Pathology. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Kosik KS. The neuronal microRNA system. Nature Reviews Neuroscience. 2006;7(12):911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 4.Barbato C, Giorgi C, Catalanotto C, Cogoni C. Thinking about RNA? MicroRNAs in the brain. Mammalian Genome. 2008;19(7-8):541–551. doi: 10.1007/s00335-008-9129-6. [DOI] [PubMed] [Google Scholar]
- 5.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berezikov E, Thuemmler F, van Laake LW, et al. Diversity of microRNAs in human and chimpanzee brain. Nature Genetics. 2006;38(12):1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 8.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid β-peptide. Nature Reviews Molecular Cell Biology. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 9.Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathologica. 1991;(82):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 10.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. NeuroReport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 11.Hébert SS, Horré K, Nicolaï L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/β-secretase expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. Journal of Alzheimer's Disease. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 13.Formichi P, Battisti C, Radi E, Federico A. Cerebrospinal fluid tau, Aβ, and phosphorylated tau protein for the diagnosis of Alzheimer's disease. Journal of Cellular Physiology. 2006;208(1):39–46. doi: 10.1002/jcp.20602. [DOI] [PubMed] [Google Scholar]
- 14.Wang W-X, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. Journal of Neuroscience. 2008;28(5):1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse β-amyloid precursor protein-converting enzyme 1. Journal of Biological Chemistry. 2009;284(4):1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Developmental Biology. 2008;315(2):418–425. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel N, Hoang D, Miller N, et al. MicroRNAs can regulate human APP levels. Molecular Neurodegeneration. 2008;3(1):p. 10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandemakers W, Morais VA, De Strooper B. A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. Journal of Cell Science. 2007;120(10):1707–1716. doi: 10.1242/jcs.03443. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Inoue K, Ishii J, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang G, van der Walt JM, Mayhew G, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-Synuclein. American Journal of Human Genetics. 2008;82(2):283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siwach P, Ganesh S. Tandem repeats in human disorders: mechanisms and evolution. Frontiers in Bioscience. 2008;13(12):4467–4484. doi: 10.2741/3017. [DOI] [PubMed] [Google Scholar]
- 22.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington's disease. European Journal of Neuroscience. 2008;27(11):2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 23.Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiology of Disease. 2008;29(3):438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Savas JN, Makusky A, Ottosen S, et al. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(31):10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes and Development. 2005;19(14):1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Molecular Cell. 2006;24(1):157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer A, O'Carroll D, Tan CL, et al. Cerebellar neurodegeneration in the absence of microRNAs. Journal of Experimental Medicine. 2007;204(7):1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Samaco RC, Gatchel JR, Thaller C, Orr HT, Zoghbi HY. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nature Neuroscience. 2008;11(10):1137–1139. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns NJ, Bigio EH, Mackenzie IRA, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathologica. 2007;114(1):5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saba R, Goodman CD, Huzarewich RLCH, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS ONE. 2008;3(11):p. e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rademakers R, Eriksen JL, Baker M, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Human Molecular Genetics. 2008;17(23):3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguzzi A, Baumann F, Bremer J. The prion's elusive reason for being. Annual Review of Neuroscience. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 34.Sorensen G, Medina S, Parchaliuk D, Phillipson C, Robertson C, Booth SA. Comprehensive transcriptional profiling of prion infection in mouse models reveals networks of responsive genes. BMC Genomics. 2008;9, article 114 doi: 10.1186/1471-2164-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10(3):544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]