SUMMARY
Mitochondrial pathologies underlie a number of life-shortening diseases in humans. In the nematode Caenorhabditis elegans, severely reduced expression of mitochondrial proteins involved in electron transport chain-mediated energy production also leads to pathological phenotypes, including arrested development and/or shorter life; in sharp contrast, mild suppression of these same proteins extends lifespan. Here we show that the C. elegans p53 ortholog cep-1 mediates these opposite effects. We find that cep-1 is required to extend longevity in response to mild suppression of several bioenergetically relevant mitochondrial proteins, including frataxin - the protein defective in patients with Friedreich’s Ataxia. Importantly we show that cep-1 also mediates both the developmental arrest and life shortening induced by severe mitochondrial stress. Our findings support an evolutionarily conserved function for p53 in modulating organismal responses to mitochondrial dysfunction and suggest that metabolic checkpoint responses may play a role in longevity control and in human mitochondrial-associated diseases.
Keywords: p53/cep-1, mitochondria, frataxin, C. elegans, Mit mutants, aging, metabolic checkpoint
INTRODUCTION
Mutations in genes that affect mitochondrial functionality lead to a variety of life-shortening, degenerative disorders in humans (Wallace 2005). At the root of most of these diseases are reduced energy production and increased free radical production, resulting from direct or indirect impairment of the mitochondrial electron transport chain (ETC). Tissues most affected in these diseases are those with high oxidative energy requirements such as neuronal, cardiac, endocrine and renal systems. Friedreich’s Ataxia (FRDA), the most frequently inherited ataxia, is one such disorder (Campuzano et al. 1996), and it is ascribed to severely defective expression of frataxin, a nuclear-encoded mitochondrial protein that plays a role in the biogenesis of Fe-S cluster containing proteins and thus in the functionality of mitochondrial proteins such as aconitase and complexes I, II and III of the ETC (Puccio et al. 2001).
In the nematode Caenorhabditis elegans, a collection of mutants defined by RNAi or genetic disruption of proteins involved in Mitochondrial ETC functionality (hereafter referred to as Mit mutants) are unexpectedly long-lived; they also have slower rates of development and decreased fertility (Rea 2005; Ventura et al. 2006). In our attempts to generate a nematode model of FRDA, we surprisingly found that decreasing the expression of the frataxin C. elegans ortholog, frh-1, also prolonged animal longevity (Ventura et al. 2005). Reduced expression of the NDUSF3 subunit of complex I of the ETC also leads to lifespan extension in C. elegans. In humans, mutation in complex I subunits including NDUSF3, cause Leigh Syndrome, another degenerative disorder. How can loss of genes critical for both cellular energy production and proper mitochondrial function, and which when mutated in humans lead to life-shortening debilitative diseases, result in life extension in C. elegans? To reconcile this human-nematode paradox we recently suggested that down to a certain threshold, cells have the ability to counteract a reduction in mitochondrial ETC function by invoking compensatory mechanisms, but beyond this threshold cell viability is severely compromised (Ventura et al. 2006). Consistent with this model, we previously showed that C. elegans Mit mutants exhibit lifespan extension only in a defined window of mitochondrial protein suppression; indeed when mitochondrial dysfunction becomes too severe and passes a critical threshold, these mutants exhibit pathological phenotypes including arrested development and lifespan reduction, perhaps similar to mitochondrial disease states in humans (Rea et al. 2007; Ventura & Rea 2007). The molecular mechanisms underlying this transition in phenotype in response to different levels of mitochondrial stress are still unknown.
Here we show that, cep-1, the C. elegans p53 ortholog, modulates Mit mutant phenotypes. Our findings are in agreement with a growing body of evidence that reveal a role for p53 as a metabolic checkpoint sensor (Bensaad & Vousden 2007), and support an evolutionarily conserved function for the p53 family in specifying organismal response to mitochondrial stress. Importantly they suggest that p53 in humans may also play a role in controlling the presentation of FRDA, Leigh Syndrome and possibly other Human Mitochondrial Associated Diseases (HMADs).
RESULTS
Frataxin modulation of stress response genes and longevity
Previous findings suggest that the C. elegans Mit mutants experience endogenous stress (Rea et al. 2007; Ventura & Rea 2007). We tested if hormetic-like responses are responsible for life extension in these worms. Feeding frh-1 RNAi to wild type animals for three consecutive generations results in a 75% decrease in frataxin mRNA level and a maximal increase in mean lifespan (Ventura et al. 2005). We found that this treatment also robustly induced the expression of multiple stress-responsive reporter genes (including gst-4::GFP, hsp-16.2::GFP (Link et al. 1999) and hsp-6::GFP (Yoneda et al. 2004)) when compared with animals fed empty vector (Fig. 1A). We quantified this induction by western blotting and found that, relative to control-treated animals, frh-1 RNAi increased the expression of GST-4 and of HSP-6, 30 and 25 fold, respectively. HSP-16.2 is only expressed after heat shock. In control animals, HSP-16.2 was induced 30 fold by heat shock but almost 50 fold in frh-1 RNAi-treated animals (Fig. 1B and Fig. S1A). frh-1 RNAi progressively increased the expression of the GST-4 and the HSP-6 throughout the three generations of feeding (data not shown). frh-1 RNAi also weakly induced the expression of the antioxidant gene sod-3 over control (Fig. S1B). These data clearly indicate that mild reduction of frataxin expression induces a robust stress response.
Figure 1. Frataxin suppression induces stress response genes.
(A) Nomarski (DIC, top panels) and fluorescence (GFP, bottom panels) images of gst-4, hsp-6 and hsp-16.2 GFP reporter strains fed for three consecutive generations (F3) on vector control (con) or frataxin RNAi (frh-1). GFP expression reveals the induction of GST-4 and HSP-6 in the parental generation (P0) compare to the third generation (F3). For HSP16.2, GFP expression has been analyzed in animals cultured for three generations on frh-1 RNAi (untreated F3) or after 12 hours recovery from a 1.5h heatshock (35°C) (F3+HS). (B) GFP induction of animals treated as in A was quantified by western blotting and normalized against actin. Shown are results from one experiment out of three performed with similar results.
We next sought to determine if known genetic modulators of reaction to stress and lifespan in worms, also controlled frh-1 RNAi-mediated longevity. SKN-1 is the C. elegans functional ortholog of the mammalian redox transcription factor Nrf2. SKN-1 is activated following oxidative stress and, like Nrf2 in other species, it controls the induction of antioxidant responsive element (ARE)-containing genes, such as the Phase II enzymes gcs-1 (An & Blackwell 2003), and gst-4 (Kahn et al. 2008). Mild oxidative stress increases C. elegans lifespan ((Cypser et al. 2006) and Fig. S2A). We tested whether frh-1 RNAi-induced oxidative stress might extend lifespan through skn-1. While a strain carrying a skn-1(zu67) mutant allele is short-lived compared to wild-type (An & Blackwell 2003), we found that frh-1 suppression by RNAi still increased its lifespan (Fig. 2A and Table I). Moreover, while the weak increase in lifespan induced by mild oxidative stress is completely blocked in skn-1 mutants, these animals were still long-lived after treatment with frh-1 RNAi, even when exposed to oxidative stress (Fig. S2B and Table I). The skn-1 gene encodes three different SKN-1 isoforms, SKN-1A, SKN-1B and SKN-1C, which are expressed in different tissues. The skn-1(zu67) mutant allele does not knock out isoform B (Fig. S2C), which is primarily expressed in the ASI neurons and which was recently shown to specifically mediate lifespan extension by caloric restriction, via effects on mitochondrial respiration (Bishop & Guarente 2007). Nonetheless, even in a skn-1(zu135) mutant background, which disrupts the expression of all three SKN-1 isoforms, frh-1 RNAi was still able to evoke a lifespan increase (Table I).
Figure 2. Loss of cep-1, but not skn-1, daf-16 or sir-2.1 KO, significantly shortened the increase in lifespan induced by frh-1 RNAi.
Survival analysis of different knock-out strains, skn-1(zu67), daf-16(mgDf50), sir2.1(ok434) and cep-1(gk138) carried out, along with their wild-type (WT) control strains, after feeding for three consecutive generations with vector alone (con) or RNAi targeting frataxin (frh-1). frh-1 RNAi increased lifespan on wild-type strains (closed vs open circles) as well as on skn-1(zu67) (A), daf-16(mgDf50) (B) and sir2.1(ok434) (C) (closed vs open triangles). (D) cep-1(gk138) significantly shortened the increase in lifespan induced by frh-1 RNAi (closed vs open triangles). Compare red arrows. Shown are cumulative data from at least 2 experiments.
Table I.
Lifespan Analyses – Summary Statistics
| Genotype | RNAi* | Mean LS | SEM | p vs con ^ | p vs N2 ^ | Sample size (censored)^^ |
Replicates (N) |
|---|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 15.9 | 0.2 | 393 (34) | 5 | ||
| frh-1 | 20.4 | 0.3 | 4.4E-25 | 406 (44) | 5 | ||
| skn-1(zu67) | con | 12.3 | 0.3 | 8.4E-16 | 140 (20) | 2 | |
| frh-1 | 20.2 | 0.5 | 2.5E-26 | 0.644 | 136 (18) | 2 | |
| gst-4::gfp | con | 18.1 | 0.4 | 165 (18) | 2 | ||
| con+ O2 | 20.7 | 0.4 | 0.0034 | 181 (8) | 2 | ||
| frh-1 | 23.3 | 0.5 | 3.5E-1 1 | 125 (13) | 2 | ||
| skn-1(zu67); | |||||||
| gst-4::gfp | con | 12.3 | 0.3 | 120 (20) | 2 | ||
| con+O2 | 12.4 | 0.3 | 0.8 | 146 (18) | 2 | ||
| frh-1 | 20.1 | 0.5 | 9.3E-27 | 136 (18) | 2 | ||
| frh-1+O2# | 19.5 | 0.8 | 3.9E-14 | 76 (2) | 1 | ||
| skn-1(zu135) | con | 11.3 | 0.2 | 3.39E-29 | 121 (2) | 2 | |
| frh-1 | 16.8 | 0.8 | 4.9E-11 | 0.0002 | 65 (3) | 1 | |
| age-1(hx546) | con | 22.2 | 1.1 | 5.5E-09 | 97 (10) | 1 | |
| frh-1 | 31.3 | 0.9 | 1.0E-06 | 2.8E-26 | 96 (11) | 1 | |
| daf-16(mgDf50) | con | 11.3 | 0.2 | 4.4E-21 | 121 (13) | 2 | |
| frh-1 | 17.8 | 0.5 | 5.8E-19 | 0.003 | 124 (24) | 2 | |
| sir2.1(ok434) | con | 14.2 | 0.2 | 9.5E-13 | 178 (37) | 2 | |
| frh-1 | 17.1 | 0.4 | 2.7E-07 | 0.005 | 191 (27) | 2 | |
| sir2.3(ok444) | con | 12.3 | 0.3 | 9.5E-09 | 81 (21) | 1 | |
| frh-1 | 16.7 | 0.4 | 2.4E-11 | 0.25 | 80 (28) | 1 | |
| cep-1(gk138)BX10 | con | 17.6 | 0.3 | 0.001 | 343 (130) | 5 | |
| frh-1 | 19.1 | 0.3 | 2.2E-07 | 0.002 | 414 (45) | 5 | |
| cep-1(lg12501) | con | 17.8 | 0.4 | 5.5E-05 | 184 (27) | 3 |
Lifespan analyses were performed on 3rd generation worms cultured continuously on frh-1 RNAi. Unless specified, all other lifespan analyses were derived using worms cultured for a single generation on undiluted isp-1, atp-3, cco-1 and nuo-2 RNAi; O2 is treatment with Hyperbaric Oxygen
p-values were calculated using the log-rank test
Pooled worms from N replicate experiments (censored individuals were included in the log rank analysis)
p-values against frh-1 RNAi on the same strain is 0.9
indicates out-crossed ten times and refers to the cep-1(gk138) allele used in this work
Daf-16, a C. elegans homolog of the human forkhead redox transcription factor FOXO family, is part of the insulin/IGF-1-like pathway and is required for increased stress resistance and longevity in many C. elegans Age mutants (Henderson et al. 2005). Daf-16 modulates lifespan in parallel with skn-1 (Tullet et al. 2008), and along with other Daf genes is involved in the hormetic response to heat shock (Cypser et al. 2006). Compared to control-fed animals, frh-1 RNAi increased lifespan of the daf-16(mgDf50) short-lived mutant (Fig. 2B and Table I). This result is in agreement with previous work showing other mitochondrial mutants increase longevity independently of daf-16 (Dillin et al. 2002). Moreover, consistent with frh-1 RNAi specifying longevity independently of the IGF/insulin like pathway, we also found that RNAi against frh-1 further increased the lifespan of the long-lived PI3K mutant age-1(hx546) (Table I).
SIRT1 and FOXO interact to modulate longevity and gene expression in response to oxidative stress and caloric restriction both in mammalian cells and in C. elegans (Wang & Tissenbaum 2006; Wang et al. 2007). We found that frh-1 RNAi still increased longevity in two different sirtuin knock-out mutants, sir-2.1(ok434) and sir-2.3(ok444) (Fig. 2C and Table I). Our findings indicate that known lifespan-modulating pathways, which are often activated in response to stress, do not mediate the lifespan extension induced by reduced expression of frataxin.
Mit mutant longevity is dependent upon p53/cep-1
The tumor suppressor p53 is activated in response to a broad range of stressors, including genotoxic, metabolic and oxidative challenges (Vousden & Lane 2007). In C. elegans, there is one p53 homolog, cep-1, already known to be required for germ-line apoptosis in response to DNA damage and for normal resistance in the soma to environmental stressors such as hypoxia and starvation (Derry et al. 2001). Consistent with the role of p53 in modulating cell senescence and aging in different species (Bauer & Helfand 2006), it was recently shown that loss of cep-1 in C. elegans increases lifespan (Arum & Johnson 2007). We have now confirmed that a different cep-1 allele, cep-1(lg12501) (Schumacher et al. 2001) is also long lived (Table I).
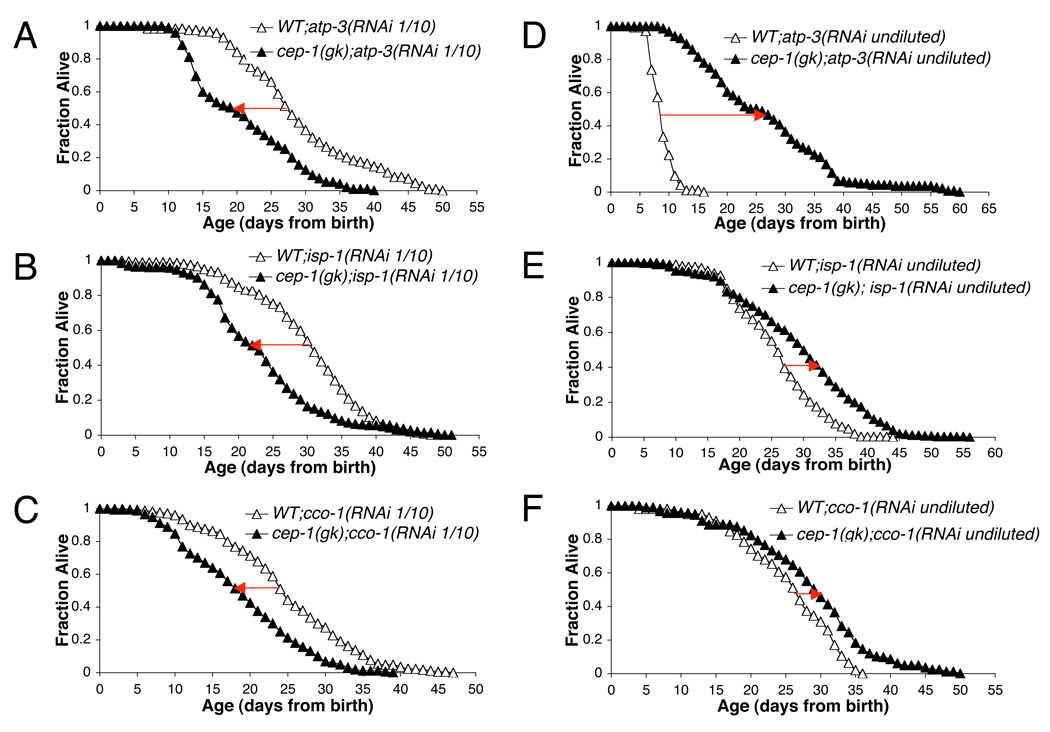
We tested if p53/cep-1-modulated C. elegans longevity following reduced frh-1 expression. We fed the C. elegans p53 knock-out strain carrying the cep-1(gk138) allele, previously out-crossed ten times (Arum & Johnson 2007), with control or frataxin RNAi for three consecutive generations, and found that the absence of cep-1 significantly suppressed the increase in lifespan induced by frh-1 RNAi (Fig. 2D and Table I). We next investigated whether our observation with p53/cep-1 could be translated to other Mit mutants. Many of these mutants display life extension only in a discrete range of mitochondrial protein suppression. When mitochondrial protein expression is reduced too severely, lifespan begins to shorten and other pathological phenotypes appear. We previously demonstrated this phenomenon using an RNAi feeding dilution strategy ((Rea et al. 2007) and Fig.S3). We therefore followed the lifespan of wild-type and cep-1(gk138) strains cultured on RNAi to one of three Mit targets, at concentrations found to maximally increase lifespan: we decreased the expression of the following mitochondrial components - ATP-3, the ATP5O/OSCP subunit of the F1FoATPase; ISP-1, the Rieske iron-sulfur protein of ubiquinol-cytochrome c oxidoreductase; CCO-1, the COX5B subunit of cytochrome c oxidoreductase. Wild-type animals fed with a 1/10 dilution of atp-3, isp-1 and cco-1 RNAi are all long lived compared to control RNAi-treated animals (Rea et al. 2007). Interestingly, the lifespan extension induced by the same Mit RNAi concentration was significantly suppressed in the cep-1(gk138) strain (Fig. 3A–C and Table II). Nevertheless, these Mit mutants remained significantly longer-lived than vector-treated animals. These results indicate a role for cep-1 in sensing mitochondrial alterations and consequently modulating C. elegans longevity.
Figure 3. cep-1 modulates lifespan in opposite directions depending on the level of mitochondrial protein suppression.
Survival curves of wild-type (WT) and cep-1(gk138) animals fed bacteria containing RNAi against atp-3 (A), isp-1 (B) or cco-1 (C) diluted 1/10 with empty vector-containing bacteria (RNAi 1/10); or RNAi against atp-3 (D), isp-1 (E) or cco-1 (F) left undiluted (RNAi undiluted). Shown are cumulative data from at least 2 experiments.
Table II.
Lifespan Analyses – Summary Statistics
| Genotype | RNAi | Mean LS | SEM | p vs con ^ | p vs N2 ^ | Sample Size (censored)^^ |
Replicates (N) |
|---|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 15.9 | 0.2 | 393 (34) | 5 | ||
| atp-3 1/10 | 28.5 | 0.8 | 2.8E-33 | 124 (14) | 2 | ||
| atp-3 1/2 | 14.2 | 0.3 | 8.6E-05 | 157 (34) | 2 | ||
| atp-3 | 8.4 | 0.2 | 4E-254 | 160 (55) | 2 | ||
| isp-1 1/10 | 29.7 | 0.4 | 4.2E-91 | 375 (51) | 5 | ||
| isp-1 | 25.3 | 0.4 | 2E-53 | 280 (29) | 4 | ||
| cco-1 1/10 | 24.4 | 0.6 | 2.9E-30 | 181 (14) | 3 | ||
| cco-1 | 24.9 | 0.7 | 2.5E-24 | 118 (23) | 2 | ||
| cep-1(gk138)BX10 | con | 17.6 | 0.3 | 0.0004 | 343 (130) | 5 | |
| atp-3 1/10 | 24.7 | 0.8 | 5.7E-13 | 0.008 | 118 (19) | 2 | |
| atp-3 1/2 | 20.3 | 0.8 | 6.8E-05 | 1.3E-09 | 160 (65) | 2 | |
| atp-3 | 25.7 | 0.9 | 8.6E-16 | 8E-256 | 160 (21) | 2 | |
| isp-1 1/10 | 22.7 | 0.5 | 6.4E-17 | 2.0E-14 | 339 (58) | 5 | |
| isp-1 | 29 | 0.6 | 1.1E-41 | 3.2E-10 | 282 (40) | 4 | |
| cco-1 1/10 | 18.3 | 0.6 | 0.005 | 2.3E-09 | 185 (23) | 3 | |
| cco-1 | 27 | 1 | 2.5E-18 | 0.009 | 119 (36) | 2 |
p-values were calculated using the log-rank test
Pooled worms from N replicate experiments (censored individuals were included in the log rank analysis)
indicates out-crossed ten times and refer to the cep-1(gk138) allele used in this work
Janus-faced role of p53 is evolutionary conserved
p53 responds to different levels of stress by inducing opposite cellular outcomes. This effect is mediated, in part, by the action of differential gene activation and p53 modification (Vousden & Lane 2007). We wondered whether cep-1 would have opposite effects on the specification of Mit mutant longevity, depending on the severity of mitochondrial bioenergetic stress. We thus fed wild-type and cep-1(gk138) strains with more severe preparations (undiluted) of RNAi against atp-3, isp-1 or cco-1 and found that cep-1 mutants displayed longer lifespan than wild-type animals (Fig. 3 D–F and Table II). Specifically, undiluted atp-3 RNAi decreased the lifespan of the wild-type strain below that of control RNAi-treated animals ((Rea et al. 2007) and Fig. S3A) while in the cep-1 mutant strain lifespan was increased with the same RNAi treatment (Fig. 3D and Fig. S4A). We also tested a 1:2 atp-3 to empty-vector RNAi ratio and consistently found the same results (Fig. S4A). Undiluted isp-1 RNAi shortened lifespan of the wild-type strain when compared to the 1/10 dilution, though to control-treated animals this treatment still resulted in some life extension ((Rea et al. 2007) and Fig. S3B). Undiluted cco-1, more severely affected animals’ phenotype, which appeared more fragile and almost sterile, but it did not shorten animal lifespan compared to the 1/10 dilution. Nonetheless, consistent to what with saw with more severe mitochondrial damage induced by undiluted atp-3 RNAi, treatment of cep-1 mutant strain with undiluted isp-1 and cco-1 RNAi increased lifespan relative to wild-type animals fed with the same RNAi concentration (Fig. 3 E, F and Fig. S4B).
One trivial explanation for the differential effects of cep-1(gk138) on Mit mutant longevity is that the efficacy of RNAi in this line is reduced relative to wild type (N2) animals. RT-PCR was used to compare atp-3 mRNA levels in cep-1 and N2 animals following RNAi treatment (Fig 4A). No difference in RNAi knockdown efficacy was observed. Importantly, we obtained similar lifespan outcomes upon feeding different atp-3 RNAi dilutions to a different cep-1 allele, cep-1(lg12501) (Fig. 4B, Fig. S4C and Table III).
Figure 4. cep-1(gk138) effects on Mit mutant longevity are not due RNAi efficacy reduction, and phenocopy in cep-1(lg12501).
(A) Quantitation of atp-3 mRNA levels in wild-type (WT) and cep-1 (gk138) animals after growth on 1/10 diluted atp-3 RNAi (atp-3 1/10) or undiluted atp-3 RNAi (atp-3 undiluted) relative to vector only treated animals (con). Samples have been normalized to a housekeeper gene (F23B12.3). Shown are the mean and SEM of three independent experiments. (B) Survival curves of wild-type (WT) and cep-1 (lg12501) animals fed atp-3 RNAi diluted 1/10 with vector –only bacteria, or left undiluted, and compared to vector-only fed animals (con). Shown are cumulative data from at least 2 experiments.
Table III.
Lifespan Analyses – Summary Statistics
| Genotype | RNAi | Mean LS | SEM | p vs con ^ | p vs N2 ^ | Sample Size (censored)^^ |
Replicates (N) |
|---|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 17 | 0.4 | 120 (10) | 2 | ||
| atp-3 1/10 | 26.5 | 1 | 4.6E-14 | 121 (9) | 2 | ||
| atp-3 | 7 | 0.4 | 4.2E-24 | 119 (9) | 2 | ||
| cep-1(lg12501) | con | 19.1 | 0.5 | 0.006 | 124 (24) | 2 | |
| atp-3 1/10 | 22.9 | 0.9 | 0.02 | 0.02 | 124 (11) | 2 | |
| atp-3 | 12.3 | 0.7 | 4.0E-06 | 1.5E-08 | 117 (19) | 2 |
p-values were calculated using the log-rank test
Pooled worms from N replicate experiments (censored individuals were included in the log rank analysis)
In summary, our results are consistent with a Janus-faced role of p53 in response to different level of stress (Vousden & Lane 2007) and suggest that cep-1 may sense mitochondria alterations and subsequently induce opposite effects on animal longevity depending on the level of mitochondrial stress.
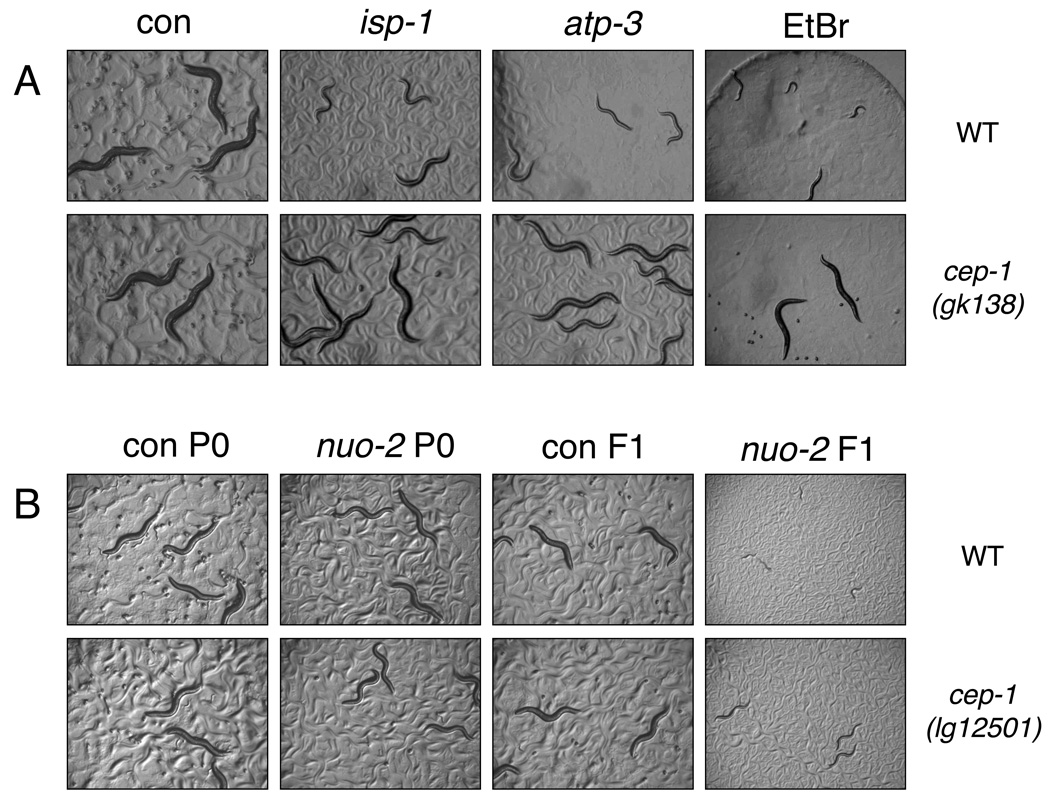
cep-1 modulates Mit mutant development
Besides changes in lifespan, Mit mutants display other phenotypic alterations that correlate with degree of mitochondrial disruption (Rea et al. 2007). Severe reduction of either atp-3 or isp-1 by feeding RNAi is sufficient to arrest development in wild-type animals (Rea et al. 2007). Developmental arrest is also observed in animals fed for two consecutive generations with RNAi against nuo-2 (which encodes the NDUFS3 subunit of NADH-ubiquinone oxidoreductase) (Ventura & Rea 2007). cep-1(gk138) mutant strain had no effect on the developmental rate of animals fed control vector but it clearly prevented the developmental arrest induced by either undiluted atp-3 or isp-1 RNAi (Fig. 5A), although this effect was not fully penetrant. Similar results were obtained using the cep-1(lg12501) allele fed undiluted atp-3 (Fig. S6), and when undiluted nuo-2 RNAi was fed for two consecutive generations (Fig. 5B). These findings suggest cep-1 can modulate development under conditions of bioenergetic stress.
Figure 5. cep-1 regulates animal development in response to severe mitochondrial disruption.
(A) Digital images of four day old wild-type (WT) and cep-1(gk138) animals fed from the time of hatching with either empty vector (con), undiluted isp-1 or atp-3 RNAi, or EtBr (75µg/ml). (B) Digital images of five day old animals fed from the time of hatching with empty vector (con) or undiluted nuo-2 RNAi (‘P0’ indicates starting generation animals, ‘F1’ specifies progeny animals derived from P0 hermaphrodites and maintained under the same feeding RNAi conditions).
The DNA intercalating agent ethidium bromide (EtBr) inhibits mitochondrial (mt) DNA replication and transcription at low concentrations without causing detectable effects on nuclear DNA (Nass 1972). For this reason it is widely used in mammalian cells to mimic human mitochondrial diseases (Biswas et al. 1999). Previous studies have also shown that EtBr induces both developmental arrest and increase lifespan in C. elegans (Tsang & Lemire 2002). We found that the developmental arrest induced by 75 µg/ml of EtBr is partially reverted by loss of cep-1 (Fig. 5A), lending further support to a role for CEP-1 in sensing mitochondrial bioenergetic alterations and modulating the coordinate expression of multiple Mit mutant phenotypes. This finding again underscores the notion that the effect of cep-1 in our above experiments is not due simply to subtle differences in RNAi efficacy.
Life shortening and developmental arrest induced by severe electron transport chain disruption involves egl-1
Mitochondrial ETC disruption can lead to DNA damage, which is a major p53-activating stressor in human cells and, depending on its severity, can in turn lead either to transient cell-cycle arrest or apoptosis. In worms, atm-1 (the C. elegans ortholog of the human Ataxia Teleangectasia gene), and egl-1 (the C. elegans Bcl-2 homology domain-3 (BH3)-only protein), act upstream and downstream, respectively, of cep-1 to induce germline apoptosis in response to DNA damage (Hofmann et al. 2002; Garcia-Muse & Boulton 2005). EGL-1 is also required to induce apoptosis and mitochondrial fragmentation of the 131 out of 1090 somatic cells that normally die during C. elegans development (Conradt & Horvitz 1998; Jagasia et al. 2005), and it is required in the embryo to induce autophagy in response to starvation (Maiuri et al. 2007). We tested if either atm-1 or egl-1 were essential to the life-lengthening or the life-shortening effects of mitochondrial ETC disruption.
We found that atm-1(gk186) is long lived while egl-1(n487) is short lived compared to wild-type animals (Table IV). We observed that both atm-1 and egl-1 play no apparent role in the life extension that occurs following mild frh-1 RNAi treatment (Fig. S5 A, B and Table IV), or following feeding with 1/10 diluted atp-3 RNAi (Fig. S5C, Fig. 6A and Table IV). Furthermore, atm-1 plays no obvious role in the life shortening that occurs following treatment with undiluted atp-3 RNAi (Fig. S5D and Table IV). Loss of egl-1, on the other hand, shortens the lifespan of control fed animal but partially reverted the lifespan shortening effect caused by growth on undiluted atp-3 RNAi (Fig. 6B and Table IV). This effect was never complete, illustrating that other factors await identification or that egl-1(n487) is not fully penetrant. Consistently, we observed that egl-1 knock-out animals suppressed the developmental arrest induced by undiluted atp-3 RNAi, but this phenotype was also not fully penetrant (data not shown). We conclude that mild mitochondrial disruption extends lifespan in a cep-1 dependent manner but independently of the DNA-damage apoptotic pathway, while severe reduction of mitochondrial function requires, in part, the cep-1/egl-1 pathway to decrease lifespan and arrest development in the Mit mutants.
Table IV.
Lifespan Analyses – Summary Statistics
| Genotype | RNAi | Mean LS | SEM | p vs con ^ | p vs N2 ^ | Sample Size (censored)^^ |
Replicates (N) |
|---|---|---|---|---|---|---|---|
| Wild-type (N2) | con | 16.3 | 0.3 | 241 (29) | 4 | ||
| frh-1 | 21.3 | 0.5 | 4.2E-15 | 234 (66) | 4 | ||
| atp-3 1/10 | 20.2 | 0.6 | 4.3E-09 | 119 (9) | 2 | ||
| atp-3 | 5.3 | 0.3 | 1.2E-69 | 123 (12) | 2 | ||
| atm-1(gk186) | con | 19.5 | 0.4 | 9.8E-12 | 237 (55) | 4 | |
| frh-1 | 26.2 | 0.7 | 5.7E-13 | 2.6E-06 | 121 (25) | 2 | |
| atp-3 1/10 | 25.5 | 0.9 | 2.5E-09 | 1.2E-06 | 121 (45) | 2 | |
| atp-3 | 5.1 | 0.3 | 4.4E-51 | 0.6 | 120 (28) | 2 | |
| egl-1(n487) | con | 14.6* | 0.4 | 0.12 | 241 (74) | 4 | |
| frh-1 | 25 | 0.9 | 1.1E-15 | 0.0007 | 120 (56) | 2 | |
| atp-3 1/10 | 18.8 | 0.8 | 4.2E-06 | 0.9 | 115 (19) | 2 | |
| atp-3 | 7.7 | 0.5 | 1.1E-16 | 0.0002 | 115 (21) | 2 |
p-values were calculated using the log-rank test
Pooled worms from N replicate experiments (censored individuals were included in the log rank analysis)
Mean LS is significantly shorter compared to N2, t-test = 0.002
Figure 6. egl-1 is a downstream target of cep-1 that contributes to lifespan modulation after severe atp-3 disruption.
(A, B) Survival analysis of wild-type (WT) and egl-1(n487) animals fed atp-3 RNAi diluted 1/10 with vector –only bacteria, or left undiluted, and compared to vector-only fed animals (con). Shown are cumulative data from 2 experiments. (C) Quantitative measurement of egl-1 mRNA in wild-type (WT) and cep-1 (gk138) animals following growth on 1/10 diluted atp-3 RNAi (atp-3 1/10) or atp-3 RNAi left undiluted (atp-3 undiluted). Data is shown relative to vector-control fed animals (con) and is normalized to a housekeeping gene (F23B12.3). Shown are mean and SEM of three different experiments.
In agreement with our above observations showing a functional requirement for egl-1 only at strong atp-3 RNAi concentrations, we observed that growth of wild type animals on treatments that increased lifespan, namely mild frh-1 RNAi or a 1/10 dilution of atp-3 RNAi, only marginally induced egl-1 mRNA (Fig. 6C). Neither treatment affected the expression of an egl-1::gfp translational reporter strain (data not shown). On the contrary, growth of wild type animals on a treatment that shortens lifespan, namely undiluted atp-3 RNAi, strongly induced egl-1 mRNA (Fig. 6C), although not at the classical levels shown in response to DNA-damage inducing agents (Hofmann et al. 2002; Garcia-Muse & Boulton 2005). Induction of egl-1 by atp-3 RNAi at 1/10 and undiluted potency are significantly suppressed by loss of cep-1. These findings reveal a new role for the cep-1/egl-1 pathway acting during C. elegans development to regulate animal lifespan.
DISCUSSION
Friedreich’s Ataxia (FRDA), as well as many other disorders associated with mitochondrial dysfunction, is a chronic, degenerative disease that often becomes clinically apparent later in life. It is probable that protective pathways are activated in the pre-symptomatic phase of FRDA to adapt and cope with dysfunctional mitochondria, thus delaying its clinical appearance. Similar pathways may be induced in C. elegans by a mild decrease of the frataxin protein (Ventura et al. 2006). We have now shown that protective stress-response genes, such as heat shock proteins and antioxidants, are strongly induced in worms in response to mild frataxin silencing. In addition we found that the tumor suppressor protein p53 has a key role in modulating the phenotypic effects resulting from mitochondrial disruption. Our findings hence invoke new players in the etiology of human mitochondrial associated diseases such as FRDA and Leigh Syndrome.
Protective Responses to Mitochondrial Dysfunction
Hormetic responses to mild heat shock or oxidative stress have been associated with prolonged longevity in C. elegans. When we looked at the contribution of daf-16 and skn-1 (which encode transcription factors that modulate heat shock and oxidative stress responses, respectively), to the longevity increase induced by frh-1 RNAi, we found that loss of neither transcription factor prevented life extension. Our results indicate that oxidative stress response, at least that controlled by these two transcription factors, is not the limiting factor regulating Mit mutant lifespan. Importantly if, as we have previously hypothesized (Ventura et al. 2006), similar pathways counteract mitochondrial dysfunction in both long-lived animals and in the pre-symptomatic phase of FRDA, our result imply that oxidative stress responses may also play a role in the human disease, but that it might not be the only player in eliciting protective pathways to help delay the established pathology.
Caloric Restriction (CR) extends C. elegans longevity and increases respiratory rate in a SIR2.1 and SKN-1 dependent manner (Wang & Tissenbaum 2006; Bishop & Guarente 2007). We found that frh-1 RNAi increased C. elegans lifespan independently of sir2.1 and, as mentioned, skn-1. Our findings do not, however, discount a role for increased mitochondrial respiration resulting from a compensatory induction of mitochondrial biogenesis in response to frh-1 RNAi. Indeed, induction of mitochondrial biogenesis in response to mild mitochondrial dysfunction in the pre-symptomatic phase of FRDA could help sustain cellular viability and thus act to clinically delay disease onset. In humans, p53 is known to regulate both mitochondrial biogenesis and mitochondrial respiration (Donahue et al. 2001; Matoba et al. 2006) and we have now found that p53/cep-1 is required for life extension in the Mit mutants. Derry and colleagues have reported that loss of cep-1, per se, causes significant transcriptional alterations (Derry et al. 2007), although these findings were not confirmed in a subsequent study (Greiss et al. 2008). Nevertheless, among the purportedly altered genes of the former analysis, there were many encoding for mitochondrial proteins (listed in Table S1). We have previously shown that mitochondrial ETC protein expression is pliant and responds uniquely in different Mit mutants (Ventura & Rea 2007). Additional work is warranted in order to understand the role played by p53 in the regulation of mitochondrial biogenesis, mitochondrial proteins composition and its relationship with longevity specification and disease prevention in response to mitochondrial dysfunction.
The Janus-faced role of p53
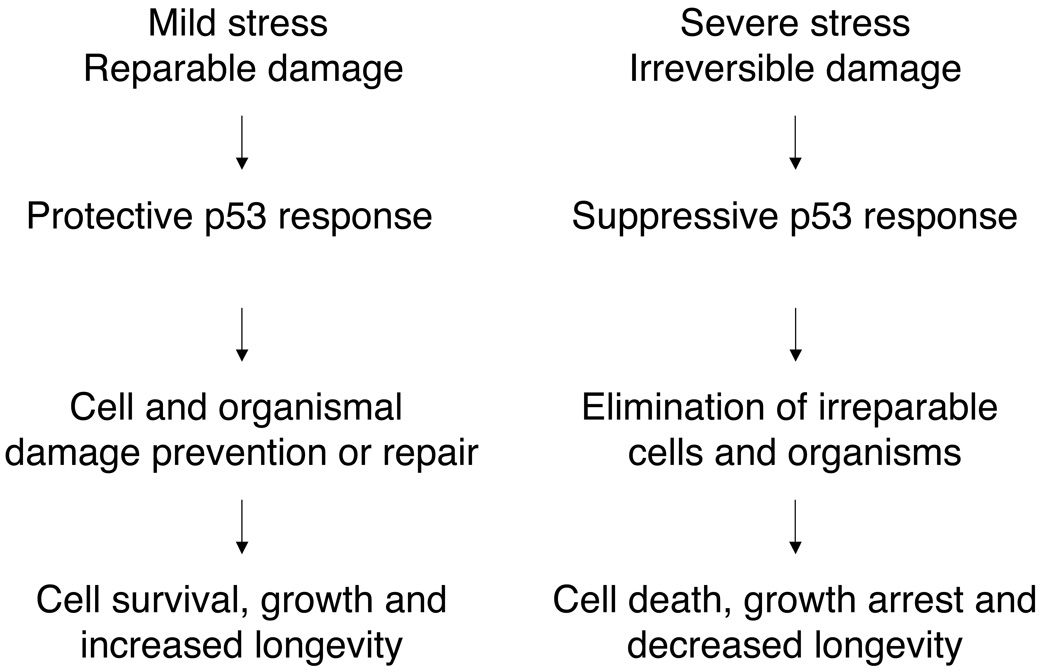
Considerable evidence supports a dual role for p53 in eliciting different responses depending on level of cellular stress (Vousden & Lane 2007). Mild or transient cellular damage, such as mild increase in free radical production, minor DNA damage, and transitory glucose or ATP depletion, induce protective p53 pathways that improve antioxidant defenses and DNA repair mechanisms, and fulfill energy requirements. In these instances p53 temporarily arrests the cell-cycle until the stress has been resolved. More severe oxidative stress, irreparable DNA damage, or complete ATP deprivation leads to p53-dependent cell death or irreversible cell-cycle arrest (replicative cell senescence). Both p53-dependent responses account for its tumor suppressor activity in humans. Consistent with the Janus-faced activity of p53 (Vousden & Lane 2007), and with p53’s role as a sensor and mediator of mitochondrial metabolism (Jones et al. 2005; Mandal et al. 2005), we have shown that cep-1, the C. elegans p53 ortholog, modulates Mit mutant lifespan in an opposite manner, depending on the level of mitochondrial stress experienced. cep-1 is required for increased longevity under mild mitochondrial disruption, and for mediating the detrimental effect on lifespan when mitochondrial damage is more severe.
Recently, two Mit mutations that increase lifespan, isp-1(qm150) and clk-1(qm30), were shown to protect against tumor growth in the C. elegans gld-1 tumor-like mutant, in a cep-1-dependent fashion (Pinkston et al. 2006). In conjunction with our present findings, these results imply that modulating mitochondrial function, by way of p53, can both regulate longevity and concurrently serve as an anti-tumor strategy. A similar situation seems to exist in mice where increased, but otherwise normally-regulated, levels of p53 lead to both tumor suppression and anti-aging effects (Matheu et al. 2007).
p53-activating stressors can lead to transient cell-cycle arrest or apoptosis. While we can not rule out the possibility that apoptosis via CEP-1 independent pathways play a role in the appearance of Mit mutant phenotypes, our results with egl-1 and atm-1 knock out strains, suggest that apoptosis is not the main mechanism through which CEP-1 is extending longevity in the Mit mutants. On the other hand, deregulated induction of a CEP-1 dependent apoptotic pathway may explain the short lifespan induced by severe mitochondrial stress (this study and (Senoo-Matsuda et al. 2003). Independent of its pro-apoptotic activity, EGL-1 is known to also induce mitochondrial fragmentation (Delivani et al. 2006), a process that may contributes to its lifespan shortening effect (Chan & Mattson 1999).
In worms the pro-apoptotic and anti-proliferative effects of cep-1 can follow different pathways (Derry et al. 2007). Moreover, in the gld-1 mutant, the two long-lived Mit mutants, isp-1 (qm150) and clk-1(qm30), reduce germ cell hyperproliferation without inducing germ cell apoptosis (Pinkston et al. 2006). Furthermore, aak-2 is an AMP kinase recently shown to be required for isp-1 and clk-1 Mit mutant longevity (Curtis et al. 2006). This same kinase is also necessary for the longevity increase induced by mild treatment of atp-3 RNAi (our unpublished observation). Two papers have shown that, in response to mitochondrial dysfunction and energy deprivation, p53 induces cell-cycle arrest via AMP kinase-dependent phosphorylation (Jones et al. 2005; Mandal et al. 2005). These observation suggest that an AMPK/CEP-1 metabolic checkpoint may play an integral role in specifying Mit mutant longevity.
Another role of p53, which is Janus-faced and therefore potentially relevant to mitochondrial stress, is the control of autophagy (Maiuri et al. 2007; Levine & Abrams 2008). Autophagy is a form of organelle and cellular digestion that can be either necessary or detrimental for tissue homeostasis and organismal survival. It has been associated both with prevention and causation of diseases in human, including neurodegenerative disorders (Vellai et al. 2007; Levine & Kroemer 2008). Autophagic genes are required during normal development in C. elegans and also for the increased lifespan observed in several mutant backgrounds, including a p53/cep-1 knock out strain (Melendez et al. 2003; Tavernarakis et al. 2008; Toth et al. 2008). Moreover, gain-of function mutation of the sole C. elegans BH3-only protein EGL-1, induces autophagy, while deletion of EGL-1 compromises starvation-induced autophagy (Maiuri et al. 2007). Based on our results it will be interesting to determine whether autophagy is associated and/or necessary to specify longevity resulting from different degrees of mitochondrial dysfunction.
A developmental checkpoint role for cep-1: the whole p53 family in one gene?
In our studies we found that the absence of cep-1 partially rescued the developmental arrest induced by severe mitochondrial damage. Importantly, we showed that cep-1 mutants also prevented the developmental arrest induced by treatment with EtBr, ruling out the possibility that lack of cep-1 is simply modulating RNAi potency rather then the response to mitochondrial stress. A developmental role of cep-1 in response to stress is consistent with a cep-1-dependent metabolic checkpoint acting during C. elegans development (Rea et al. 2007) and/or by cep-1 functions reminiscent of other p53 family members, such as those of p63 and p73 in differentiation and development (D'Erchia et al. 2006; Ou et al. 2007). Under normal, well-fed conditions, cep-1 appears to be dispensable for C. elegans development. Only under conditions of increased energy and nucleotide demand, when its function may be required for mitochondrial biogenesis, we observed a requirement for cep-1. These conditions may exist when cells are rapidly proliferating, such as during embryonic C. elegans development or at the transition between L3–L4 stages, the point of gonad formation. The regulation of animal lifespan through a mitochondrial checkpoint in proliferating cells during animal development is an interesting possibility.
Concluding Remarks
Our data suggest a model in which mild disruption of ETC function in C. elegans induces transient cellular damage (perhaps genotoxic stress, proliferation errors and/ or organelle dysfunction), which is readily repaired or eliminated by sub-lethal activation of cep-1. A temporary halt of the cell-cycle would allow for metabolic adaptation to mild mitochondrial stress and to elicit proper defenses against oxidative and other stressors; cep-1 activation also mediates increased animal lifespan. When mitochondrial dysfunction surpasses a critical threshold, cep-1 functionality is altered in an attempt to eliminate irreparably damaged cell leading to the detrimental phenotypes observed in the Mit mutants (Fig. 5). Our current findings suggest that p53 has a previously unrecognized role in modulating animal lifespan and development in response to mitochondrial stress. p53 activity is now a prime target for investigations into the pathogenesis and treatment of human mitochondrial disorders.
EXPERIMENTAL PROCEDURES
Nematode Strains and Maintenance
Standard nematode culturing techniques were employed (Wood 1988). The following strains were utilized: N2 (wild-type), SJ4100 [hsp-6::gfp(zcIs13)], TJ375 [hsp-16.2::gfp(gpIs1)], CL2166 [dvIs19[pAF15(gst-4::gfp::NLS)] III], CL691 [dvIs19[pAF15(gst-4::gfp::NLS) III]; skn-1(zu67) IV/nT1[unc-?(n754);let-?](IV;V)], EU1 [skn-1(zu67)IV/nT1[unc-?(n754) let-?](IV;V)], EU31[skn-1(zu135)IV/nT1[unc-?(n754)let-?](IV;V)], GR1307 [daf-16(mgDf50)], TJ1052 [age-1(hx546)], VC199 [sir-2.1(ok434)], RB654 [sir-2.3(ok444)], VC172 [cep-1(gk138)], TJ1 [cep-1(gk138)], XY1054 [cep-1(lg12501)], VC381 [atm-1(gk186)], MT1082 [egl-1(n487)], MD166 [egl-1::gfp(bcIs1)].
Feeding RNAi
RNAi feeding techniques were previously described (Ventura et al. 2005; Rea et al. 2007). The following RNAi feeding constructs were derived from the Ahringer C. elegans RNAi library (Kamath & Ahringer 2003): atp-3 (F27C1.7), cco-1 (F26E4.9), nuo-2 (T10E9.7) (Kamath & Ahringer 2003) while feeding RNAi constructs against frh-1 and isp-1 have been previously described (Ventura et al. 2005; Rea et al. 2007).
Lifespan and Statistical Analysis
Survival analyses began from hatching and were recorded at 20°C using synchronous populations of 60–100 animals per strain. We calculated mean and standard deviation of the mean from pooled population of animals and used the log-rank test to assess differences in survival curves between pooled populations as previously described (Rea et al. 2007).
Developmental Analysis
Development was followed at 20°C on populations of synchronous animals from the time of hatching after a 4 hour synchronized egg lay, which was carried out directly on RNAi plates or EtBr plates. Images were collected 4 and 5 days after the time of egg lay for the parental and first generation animals, respectively. Live worm images, to record development, were captured with a digital camera connected to a dissecting microscope.
Microscopy
For fluorescence images, individual worms were randomly picked using a regular dissecting scope from a population of 200 worms for each condition. Remaining worms were then collected for western blotting or followed for lifespan analysis. Worms were arranged for image analysis on 2% agarose pads and Nomarski and fluorescence images were captured via a Ziess Axioskop running SlideMaker 4.0 software retrofitted with deconvolution capabilities and a digital fluorescent camera containing a Sedat (Quad) filter set. GFP fluorescence was collected using a FITC 490/528nm absorption/emission filter pair (peak transmission with a 20/38 nm bandpass). All images were collected using identical exposure settings.
Quantitative RT-PCR
Quantitation of egl-1 and atp-3 mRNA in frh-1 and atp-3 RNAi-treated animals relative to vector-only treated animals was undertaken using primers specific for egl-1 (Schumacher et al. 2005) and atp-3 (Rea et al. 2007). Data was normalized to a non-variable control gene (F23B2.13). mRNA was extracted from approximately 5000 synchronized worms per sample, using a Total RNA and Protein Isolation Kit (Macherey-Nagel). Oligo(dT) primed cDNA was synthesized using a SuperScript cDNA Synthesis Kit (Invitrogen). Quantitative PCR (qPCR) was undertaken on an ABI Prism® 7000 instrument with SYBR Green PCR Master Mix kit (Applied Biosystems).
Western Blot Analysis
Western Blot analysis of whole-worm protein extracts were prepared from ~200 adult worms. Animals were boiled in 5% SDS, 0,02% β-mercaptoethanol and 1mM protease inhibitor cocktail (SIGMA P2714). Following protein quantitation (BCA, Pierce), protein levels were measured by western analysis, quantified using densitometry, and normalized against actin. Antibodies employed were anti-GFP (MSA02, MitoSciences, Oregon, 1:1000 dilution) and anti-Actin (AC-15, Sigma, 1:5000).
Supplementary Material
Figure 7. Evolutionary conserved functionality of Janus-faced p53/cep-1 in C. elegans.
Mild or transient cellular and organismal damage induces p53 protective pathways leading to damage repair, growth, survival and increased longevity. More severe or irreversible damage induces p53 suppressive responses that eliminate cells and irreversibly compromise the organsim. Such drastic actions lead to growth arrest and/or decreased survival probability.
ACKNOWLEDGEMENTS
We thank A. Kell for technical assistance and Dr. D. Wu for help with statistical analysis. Most of the strains used in this work were provided by the C. elegans Gene Knockout Consortium, which is funded by the National Institute of Health and National Center for Research Resource. Dr. C. Link constructed strain CL2166 and CL691. Strain XY1054 was a kind gift of Dr. A. Gartner, SJ4100 of Dr. D. Ron, and MD166 of Dr. B. Conradt. This work was supported by NIA grant RO1-AG16219 to T.E.J.; NIA grant R21-AG025207-01A1 to S.L.R.; Telethon grant GGP060059, ASI-MoMa, FARA and Ataxia UK to R.T.; NAF and FIRC post doctoral fellowship to N.V.
ABBREVIATIONS
- CEP
C. elegans p53
- ETC
Electron Transport Chain
- FRDA
Friedreich’s Ataxia
- Mit mutants
Mitochondrial mutants
- NDUFS3
NADH Dehydrogenase [Ubiquinone] iron (Fe)-Sulfur protein 3
- HMAD
Human Mitochondrial-Associated Disease
- FRH
frataxin
- gst
Gluthathione-S-Transferase
- hsp
heat shock protein
- SKN
SkiNhead
- Nrf
Nuclear factor erythroid-2 Related Factor
- ARE
Antioxidant Responsive Element
- Daf
Abnormal Dauer Formation
- SIRT
sirtuins
- FOXO
FOrkhead boX
- ATM
Ataxia Telangectasia Mutated
- EGL
EGg Laying defective
- ATP
adenosine triphosphate
- CCO
Cytochrome C Oxidase
- ISP
Iron-Sulfur Protein
- EtBr
ethidium bromide
- GLD
defective in Germ Line Development
- AMPK
adenosine triphosphate (AMP)-activated protein kinase.
Footnotes
COMPETING INTEREST
The authors have declared that no competing interests exist.
REFERENCES
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arum O, Johnson TE. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J Gerontol A Biol Sci Med Sci. 2007;62:951–959. doi: 10.1093/gerona/62.9.951. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Helfand SL. New tricks of an old molecule: lifespan regulation by p53. Aging Cell. 2006;5:437–440. doi: 10.1111/j.1474-9726.2006.00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. Embo J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Chan SL, Mattson MP. Caspase and calpain substrates: roles in synaptic plasticity and cell death. J Neurosci Res. 1999;58:167–190. [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Curtis R, O'Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Cypser JR, Tedesco P, Johnson TE. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Erchia AM, Tullo A, Lefkimmiatis K, Saccone C, Sbisa E. The fatty acid synthase gene is a conserved p53 family target from worm to human. Cell Cycle. 2006;5:750–758. doi: 10.4161/cc.5.7.2622. [DOI] [PubMed] [Google Scholar]
- Delivani P, Adrain C, Taylor RC, Duriez PJ, Martin SJ. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. Mol Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Derry WB, Bierings R, van Iersel M, Satkunendran T, Reinke V, Rothman JH. Regulation of developmental rate and germ cell proliferation in Caenorhabditis elegans by the p53 gene network. Cell Death Differ. 2007;14:662–670. doi: 10.1038/sj.cdd.4402075. [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Donahue RJ, Razmara M, Hoek JB, Knudsen TB. Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. Faseb J. 2001;15:635–644. doi: 10.1096/fj.00-0262com. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. Embo J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics. 2008;9:334. doi: 10.1186/1471-2164-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Rea SL, Johnson TE. Dissecting the Processes of Aging Using the Nematode Caenorhabditis elegans. New York: Academic Press; 2005. [Google Scholar]
- Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M, Hengartner MO. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. Embo J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Nass MM. Differential effects of ethidium bromide on mitochondrial and nuclear DNA synthesis in vivo in cultured mammalian cells. Exp Cell Res. 1972;72:211–222. doi: 10.1016/0014-4827(72)90583-6. [DOI] [PubMed] [Google Scholar]
- Ou HD, Lohr F, Vogel V, Mantele W, Dotsch V. Structural evolution of C-terminal domains in the p53 family. Embo J. 2007;26:3463–3473. doi: 10.1038/sj.emboj.7601764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- Rea SL. Metabolism in the Caenorhabditis elegans Mit mutants. Exp Gerontol. 2005;40:841–849. doi: 10.1016/j.exger.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, Conradt B, Shaham S. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 2005;12:153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Hartman PS, Akatsuka A, Yoshimura S, Ishii N. A complex II defect affects mitochondrial structure, leading to ced-3- and ced-4-dependent apoptosis and aging. J Biol Chem. 2003;278:22031–22036. doi: 10.1074/jbc.M211377200. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4 doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochem Biophys Res Commun. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Toth ML, Kovacs AL. Janus-faced autophagy: a dual role of cellular self-eating in neurodegeneration? Autophagy. 2007;3:461–463. doi: 10.4161/auto.4282. [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea S, Henderson ST, Condo I, Johnson TE, Testi R. Reduced expression of frataxin extends the lifespan of Caenorhabditis elegans. Aging Cell. 2005;4:109–112. doi: 10.1111/j.1474-9726.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea SL. Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnol J. 2007;2:584–595. doi: 10.1002/biot.200600248. [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Testi R. Long-lived C. elegans mitochondrial mutants as a model for human mitochondrial-associated diseases. Exp Gerontol. 2006;41:974–991. doi: 10.1016/j.exger.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wood WB. The Nematode Caenorhabditis elegansed^. New York: Cold Spring Harbor Laboratory; 1988. p. 667. [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.