Abstract
Mycobacterium tuberculosis (Mtb) signals through Toll-like receptor 2 (TLR2) to regulate antigen presenting cells (APCs). Mtb lipoproteins, including LpqH, LprA, LprG and PhoS1, are TLR2 agonists, but their co-receptor requirements are unknown. We studied Mtb lipoprotein-induced responses in TLR2−/−, TLR1−/−, TLR6−/−, CD14−/− and CD36−/− macrophages. Responses to LprA, LprG, LpqH and PhoS1 were completely dependent on TLR2. LprG, LpqH, and PhoS1 were dependent on TLR1, but LprA did not require TLR1. None of the lipoproteins required TLR6, although a redundant contribution by TLR6 cannot be excluded. CD14 contributed to detection of LprA, LprG and LpqH, whereas CD36 contributed only to detection of LprA. Studies of lung APC subsets revealed lower TLR2 expression by CD11bhigh/CD11clow lung macrophages than CD11blow/CD11chigh alveolar macrophages, which correlated with hyporesponsiveness of lung macrophages to LpqH. Thus, lung APC subsets differ in TLR expression, which may determine differences in responses to Mtb.
Keywords: Toll-like receptor 2, lipoprotein, Mycobacterium tuberculosis, CD14, CD36, antigen presenting cell
Introduction
Toll-like receptor 2 (TLR2) is important for control of mycobacterial infections, including infections by Mycobacterium tuberculosis (Mtb) [1; 2; 3; 4] and M. leprae [5]. Mycobacterial agonists of TLR2 include lipoproteins and glycolipids. The genome of Mtb is predicted to encode 48–100 lipoproteins of largely unknown function, nearly half of which share no conserved domains with lipoproteins of species outside the genus Mycobacteria [6]. LprA [7], LprG [8], LpqH (19-kDa lipoprotein) [9; 10; 11] and PhoS1 (38-kDa lipoprotein) [12] are Mtb lipoproteins with TLR2 agonist activity that modulate antigen presenting cell (APC) functions, which we define to include cytokine production and other innate immune responses of APCs as well as antigen presentation, recapitulating many effects of APC infection with Mtb. Despite the importance of understanding how TLR2 recognizes its ligands, many questions remain about the mechanisms whereby Mtb lipoproteins are recognized. In this study we used four well-characterized mycobacterial lipoproteins to dissect the contributions of TLR2, its co-receptors and accessory receptors to recognition of lipoproteins.
TLR2 cooperates with other surface receptors, including “co-receptors” with which it forms heterodimers and “accessory receptors” that may enhance ligand delivery or recognition. Co-receptors of TLR2 are genetically related to TLR2 and include TLR1 and TLR6. TLR1 contributes to the recognition of triacylated lipopeptides [13] and lipoarabinomannan [14], whereas TLR6 contributes to the detection of diacylated lipopeptides [15; 16]. Although there is now crystallographic data for the structure of a synthetic triacylated lipopetide (Pam3CSK4) bound to TLR2 and TLR1 [17], a number of questions remain about how TLR2 and its co-receptors recognize the diverse set of physiological ligands with TLR2 agonist activity. For example, the mechanisms by which non-acylated ligands, such as neisserial porins [18] and capsular carbohydrates [19], are recognized by TLR2 and its co-receptors remain unclear. For full-length, natural lipoproteins, it remains unknown how the peptide component [20] or glycan component [21] may influence signaling by TLR2, TLR1 and TLR6. Our studies used co-receptor knock-out cells to assess TLR2 co-receptor requirements for responses to four different Mtb lipoproteins, revealing variable dependence on TLR1 and a lack of dependence on TLR6.
In addition to contributions of the co-receptors that form heterodimers with TLR2, recognition of TLR2 agonists is influenced by several accessory receptors, including CD14 [22], CD36 [23], lipopolysaccharide binding protein [24], CD11b-CD18 integrin [25] and ganglioside GD1a [26]. All of these accessory receptors are hypothesized to deliver agonist to TLR2, and some are also thought to contribute to cytoplasmic signaling. CD14, a GPI-linked protein, directly binds Pam3CSK4 and delivers it to TLR2 [27], increasing the sensitivity of TLR2 responses by approximately 100-fold [28]. CD36 enhances the sensitivity of macrophages to lipoteichoic acid and MALP-2, but not Pam3CSK4 [23]. Although biochemical evidence of direct binding of CD36 to lipoteichoic acid or MALP-2 is lacking, there are reports that it signals with its C-terminal cytoplasmic tail and that CD36-dependent phagocytosis is important for its ability to augment TLR2 responses [29; 30]. One caveat for some of these studies is their reliance on small synthetic lipopeptide agonists, which may differ from full-length lipoproteins in solubility, mode of interaction with TLR2 and reliance on specific accessory receptors. While there is data that triacylated OspA of Borrelia burgdorferi is sensed in a TLR2 and TLR1 dependent manner [31], there has been little assessment and comparison of accessory receptor usage among different physiological TLR2 ligands, e.g. lipoproteins, so potential heterogeneity in physiological accessory receptor usage remains unexplored. Our studies assessed accessory receptor requirements for responses to four different Mtb lipoproteins, revealing both overlaps and differences in accessory receptors used to recognize these lipoproteins.
Mtb is an intracellular pathogen that infects APCs, and the ability of different APC types to respond to Mtb infection may be governed by their relative expression of TLR2 and its co-receptors and accessory receptors. The primary site of infection by Mtb is the lung. Lung APC populations include lung macrophages and lung dendritic cells (DCs) in the lung parenchyma, as well as alveolar macrophages [32]. Mtb is harbored within alveolar macrophages early in infection and within lung macrophages and lung DCs later in infection [33; 34; 35]. Potential differences between these APC subsets in responsiveness to Mtb or its lipoproteins are largely unexplored and could have significant implications for host responses to Mtb infection. For example, the relative expression of TLR2 has not been assessed on lung APC subsets. We demonstrate that purified APC subsets from murine lungs differ in expression of TLR2, and the subset with lowest TLR2 expression, lung macrophages, is hyporesponsive to Mtb lipoprotein.
Materials and Methods
Cloning and expression of Histidine-tagged LprG (Rv1411c) and LprA (Rv1270c)
LprA was cloned previously [7]. LprG was amplified from Mtb H37Rv genomic DNA by PCR using the following primers: 5′GCATATCCATATGCGGACCCCCAGACGCCACTG 3′GTACAAGCTTGCTCACCGGGGGCTTCG. The 5′ primers included an NdeI site and the 3′ primer included a HindIII site. The PCR product was digested with NdeI and HindIII (NEB, Ipswitch, MA) and ligated into the shuttle vector pVV16 (provided by J. Belisle, Colorado State University, Fort Collins, CO) behind the mycobacterial hsp60 promoter and in-frame with a C-terminal 6x His tag. All constructs were verified by sequencing and analyzed using Clone Manager (SciEd software, Cary, NC). Chemically competent E. coli (Invitrogen, Carlsbad, CA) was transformed according to the manufacturer’s protocol. M. smegmatis was transformed by electroporation with a Gene Pulser (Bio-Rad, Hercules, CA) set at 2.5 kV, 25 μF, and 800 Ohms. LprA and LprG were expressed in M. smegmatis MC2 1-2C (R. Wilkinson, Imperial College, London, U.K.) cultivated in Middlebrook 7H9 broth (Difco, Lawrence, KS) supplemented with 1% casamino acids (Fisher, Pittsburgh, PA, BP1424), 0.2% glycerol (Fisher G33-1), 0.2% glucose, and 0.05% Tween 80. Kanamycin was used at 30 μg/ml for selection of both E. coli and M. smegmatis. Bacteria were isolated by centrifugation at 6000 × g for 20 min at 4°C.
Mtb strain H37Ra (ATCC 25177) was cultured with shaking at 37°C to late log phase growth (2.5 weeks) in Mtb 7H9 broth (4.7 g/l 7H9 (Difco 271310), 5 ml/l glycerol, 0.5 ml/l Tween-80 (Sigma, St. Louis, MO, P4780) supplemented with 10% albumin/dextrose/catalase (BD, Franklin Lakes, NJ 212352). Bacilli were harvested by centrifugation at 5,000 × g for 20 min at 4°C.
Lysis and Purification of His6-tagged proteins
Purification of LprA and LprG was accomplished as reported previously [7]. Cells were resuspended in lysis buffer (10 ml/liter of bacterial culture) consisting of 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0, 2.5% protease inhibitor cocktail (Sigma P8849), 75 U/ml benzonase (Novagen, Madison, WI, 70664-3), and 2.5 mg lysozyme (Sigma, L-3790) and incubated for 15 min at 37°C. Bacteria were disrupted mechanically by 4 passages through a French press (2000 psi). Insoluble material was removed from the lysate by ultracentrifugation at 100,000 × g for 1 h at 4°C, and supernatant was incubated directly with Ni-NTA beads (Qiagen, Valencia, CA, 1018244) for 2–4 h at 4°C. Ni-NTA beads were transferred to polypropylene columns, washed with 75 volumes of wash buffer (50 mM NaH2PO4, 1 M NaCl, 20 mM imidazole, 10% glycerol, pH 8.0), and bound protein was dissociated with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 450 mM imidazole, pH 8.0). To prepare for anion exchange chromatography, sample was desalted into 20 mM Tris, pH 8.0 using PD-10 columns (GE Healthcare, Uppsala, Sweden 17-085-01). Samples were subjected to anion exchange chromatography using quaternary ammonium columns (GE Healthcare, 17-5053-01), and eluted with the addition of NaCl in the following steps: 50, 100, 200, 400, 1000 mM. Presence and purity of desired protein was verified by SDS-PAGE and visualized as single bands by both silver stain and anti-His Western blot; yields were estimated by BCA protein assay (Pierce, Rockford, IL, 23225). Material eluted by 100 mM NaCl was used for all experiments.
Lysis of Mtb H37Ra and purification of LpqH
Bacilli from 4 liters of Mtb H37Ra culture were collected by centrifugation as above, resuspended in 40 ml of water supplemented with protease inhibitor cocktail (Sigma P8849), 75 U/ml benzonase (Novagen 70664-3), and 2.5 mg of lysozyme, incubated for 15 min at 37°C, and mechanically disrupted via 4 passages through a French Press (2000 psi). To extract hydrophobic molecules, lysate was combined with 6% Triton X-114 (Sigma, X-114) in Tris buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.5) to achieve a concentration of 4% Triton X-114. After overnight incubation at 4°C, insoluble material was removed by centrifugation at 100,000 × g for 1 h at 4°C, and supernatant was stored at 4°C overnight. Insoluble material was resuspended in 4% Triton X-114 in TBS at 4°C and incubated overnight at 4°C. Insoluble material was again removed by centrifugation at 100,000 × g for 1 h at 4°C. Soluble material from the two sequential extractions was used for further purification. To remove hydrophilic contaminants, soluble material from Triton X-114 extractions was back-extracted three times by addition of a two-fold excess of ice-cold TBS, incubation on ice for 15 min, incubation at 37°C for 15 min to allow separation of detergent and aqueous phases, centrifugation (3,000 × g for 15 min at 37°C) and removal of the upper aqueous phase by aspiration. To remove TBS, the detergent phase was similarly back-extracted once with ice-cold water. To remove detergent, protein was precipitated by the addition of 5 volumes of acetone at −20°C and stored overnight at −20°C. The acetone precipitate was collected by centrifugation, washed 3 times with −20°C acetone, and air-dried to yield approximately 100 mg dry weight of a white powder per liter of culture. To exclude glycolipids, the acetone precipitate was solubilized in PBS-saturated phenol and subjected to six sequential extractions by the addition of PBS (1:1 v/v) at room temperature for 4–15 h (rotating in Teflon-coated tubes with sealing caps, Fisher scientific 05-562-16B, 05-563-1c), centrifugation at 15,000 × g for 30 min at room temperature, and removal of the upper aqueous phase. The final phenol phase was dialyzed against distilled water using a 3.5 kD molecular weight cut-off membrane (Corning, Lowell, MA, 132720). The precipitate was removed from dialysis tubing, washed extensively with distilled water at room temperature, and lyophilized. The yield from one liter of culture was approximately 25 mg of precipitate. Eight mg of lyophilized phenol precipitate was subjected to SDS-PAGE through a preparative 20 × 20 cm, 1.5 mm thick 13% polyacrylamide Tris-Cl gel using Protean IIxi electrophoresis apparatus (BioRad, Hercules, CA). The sample was eluted in fractions using a Whole-Gel Eluter (BioRad) into imidazole-HEPES buffer (43 mM imidazole, 35 mM HEPES, pH 7.4). To identify fractions containing LpqH, a portion of each fraction was analyzed by SDS-PAGE with silver staining or Western blotting using polyclonal anti-BCG Ab (DAKO, B0124, 1:30,000) or the LpqH-specific monoclonal antibody IT-19 (1:3000) [36]. Fractions containing LpqH and no other bands by silver gel were pooled and concentrated using 10,000 molecular weight cut-off tubes (Millipore, Billerica, MA, UFC801096). Yields were determined by BCA protein assay.
Purified PhoS1 was provided by John T. Belisle under NIAID contract HHSN266200400091c “TB Vaccine Testing and Research Materials”; it was purified from Mtb culture supernatant by ammonium sulfate precipitation, lectin chromatography, and hydrophobic interaction chromatography.
Mammalian cell culture
Unless otherwise specified, incubations with eukaryotic cells were performed at 37°C in 5% CO2 atmosphere. Standard medium was DMEM (Hyclone, Logan, UT, ASK30773) supplemented with 10% heat-inactivated FCS, 50 μM 2-ME, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, pH 7.4, and penicillin/streptomycin (Hyclone). Female C57BL/6J mice (8–16 weeks old) were obtained from the Jackson Laboratory, housed under specific pathogen-free conditions and used to produce macrophages. TLR1−/−, TLR2−/− and TLR6−/− mice were generously provided by Shizuo Akira (Research Institute for Microbial Disease, Osaka University, Osaka, Japan) and were back-crossed to C57BL/6J mice a minimum of eight times. CD14−/− mice (B6.129S-Cd14tm1Frm/J) were obtained from the Jackson Laboratory, maintained under specific pathogen-free conditions and used to produce macrophages. CD14−/− mice were compared to C57BL/6J mice and F2 hybrids of C57BL/6J and 129sv. CD36−/− mice were obtained and backcrossed onto C57BL/6 background a minimum of eight times as described [37]. Bone marrow cells were cultured for 7–12 d in standard medium supplemented with 25% LADMAC cell-conditioned medium [38]. HEK293 cells stably expressing TLR2-YFP (HEK293.TLR2) were produced previously [39; 40]. HEK293 cells (ATCC CRL-1573) were stably transfected with the empty vector to produce a control HEK293.pcDNA3 cell line. Transfected HEK293 cell lines were maintained in HEK medium (DMEM supplemented with 10% heat-inactivated FCS (HyClone), ciprofloxacin (10 μg/ml) and geneticin (500 μg/ml)).
Cytokine ELISAs
HEK293.TLR2 or HEK293.pcDNA3 cells were incubated in 96-well plates (20,000 cells/well) for 8 h in 90 μl of HEK medium and then for an additional 16 h with or without TLR2 agonist. Supernatant IL-8 concentration was quantified by ELISA (R&D, Minneapolis, MN, DY208). Bone marrow-derived macrophages were incubated overnight at 100,000 cells/well in standard medium and then with or without lipoprotein for 12 h. Supernatants were collected and stored at −80°C. TNF-alpha in the supernatant was quantified by ELISA (BD Biosciences #558874, R&D DY410). The following synthetic TLR agonists were also used: Ultrapure E. coli LPS (Invivogen, San Diego, CA, tlrl-pelps), FSL-1 (Invivogen, tlrl-fsl) and Pam3CSK4 (Invivogen, tlrl-pms).
Lung APC preparation and flow cytometry
Mice were anesthetized with 1.25% 2,2,2 tribromoethanol (Sigma, T4840-2) solution dissolved in 40% 2-methyl-2-butanol (tert-amyl alcohol; Sigma, A-1685). The pulmonary circulation was perfused with ~10 ml PBS with 2 mM EDTA. Lungs were removed, minced, and incubated in standard medium supplemented with 2.5 mg/ml collagenase D (Roche, Indianapolis, IN) and 75 U/ml benzonase for 45 min at 37°C. Tissue was then passed through a 70-μm filter and rinsed extensively with standard medium. Cells were resuspended in ACK lysis buffer for 5 min at room temperature. Dead cells and tissue debris were removed with Dead Cell Removal kit (Miltenyi Biotec, Auburn, CA, 130-090-101). Crude lung APCs were prepared using a 1:1 mixture of magnetic beads selective for CD11b (Miltenyi biotec, 130-049-601) and CD11c (Miltenyi Biotec, 130-052-001) (100 μl of each slurry per 107 viable cells). For flow cytometry, crude lung APCs were stained using the following antibodies: APC-AF750-conjugated anti-CD11b (eBioscience, San Diego, CA, M1/70, #27-0112, used at 0.4 μg/1×106 cells), PB-conjugated anti-CD11c (eBioscience, N418, #57-0114, used at 0.4 μg/1×106 cells) or APC-conjugated anti-CD11c (eBioscience, N418, #17-0114-82, used at 0.4 μg/1×106 cells), PECy7-conjugated anti-TLR2 (eBioscience, T2.5, #25-9024, used at 0.4 μg/1×106 cells), FITC-conjugated anti-CD14 (eBioscience, Sa 2-8, #11-141-82, used at 1.0 μg/1×106 cells), PE-conjugated anti-CD36 (eBioscience, No.72-1, #12-0261, used at 0.4 μg/1×106 cells) and biotin-conjugated anti-GR1 (eBioscience, RB6-8C5, #13-5931, used at 1.0 μg/1×106 cells). Flow analysis and sorting were performed on a BD FACSAria cytometer. Specific mean fluorescence intensity (MFI) was calculated as MFI with specific Ab minus MFI with isotype control Ab. Samples to be sorted were stained only with antibodies specific for CD11b, CD11c, and GR-1. Data was analyzed using Winlist 5.0 software (Verity, Topsham, ME). Events representing single cells were gated by forward and side scatter. Of these, only GR1-negative events were included in the analysis and sort. GR1-negative events were sorted by FACS to isolate lung APC subsets based on surface expression of CD11b and CD11c as previously described [32; 41]. Lung APC populations included alveolar macrophages (AM, CD11blow/CD11chigh), lung macrophages (LM, CD11bhigh/CD11clow), and lung DCs (CD11bhigh/CD11chigh). To assess relative responsiveness to a TLR2 agonist, FACS-purified lung macrophages and alveolar macrophages were incubated in 96-well plates (30,000 cells/well) in standard medium for 3 h and then in serum-free medium (Macrophage SFM, Invitrogen, 12065074) with or without LpqH for 16 h. Supernatants were collected, stored at −80°C and assessed for TNF-alpha by ELISA.
Statistical analysis
Two-tailed Student’s T-test was used for comparisons of responses of two cell types (e.g. HEK293.TLR2 vs. HEK293.pcDNA3, wild-type vs. CD14−/− macrophages, or lung macrophages vs. alveolar macrophages). Two-way analysis of variance (ANOVA) with Bonferroni post-test was used comparisons of multiple cell types with different TLR deficiencies.
Results
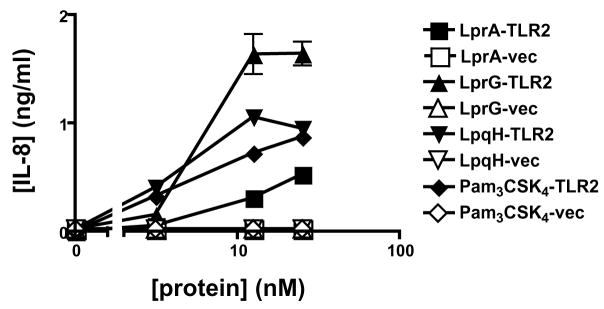
These studies were designed to enhance our understanding of TLR recognition of Mtb lipoproteins, including the potential contributions of co-receptors or accessory receptors. To assess potential variation in recognition mechanisms and determine general principles, we studied multiple Mtb lipoproteins, including LprA, LprG, and LpqH; a fourth Mtb lipoprotein, PhoS1, was included in some studies. To compare their TLR2 agonist function, Mtb lipoproteins and a TLR2 model agonist (Pam3CSK4) were incubated with HEK293.TLR2 cells (transfected with human TLR2) or HEK293.pcDNA3 cells (transfected with control vector), and induction of IL-8 was assessed by ELISA. Mtb lipoproteins induced expression of IL-8 by HEK293.TLR2 cells but not HEK293.pcDNA3 cells (Fig. 1). A concentration of approximately 3–10 nM was sufficient to induce responses by LprA, LprG, LpqH and Pam3CSK4 (Fig. 1). These data confirmed that LprA, LprG and LpqH are all agonists of human TLR2, consistent with other data from studies with murine TLR2−/− macrophages (below) that indicate that these lipoproteins, as well as PhoS1, are agonists of TLR2.
Figure 1. Mycobacterial lipoproteins are TLR2 agonists.
HEK293.TLR2 cells (filled symbols) or control HEK293.pcDNA3 cells (empty symbols) were exposed to mycobacterial lipoproteins for 15 h, and the concentration of IL-8 in culture supernatant was assayed by ELISA. Data represent mean +/− SD for triplicate samples, except where error is too small to be visualized. For concentrations above 10 nM, LprG, LpqH, LprA and Pam3CSK4 produced responses in HEK293.TLR2 cells that were significantly different from responses in HEK293.pcDNA3 cells (p < 0.01 for all such comparisons).
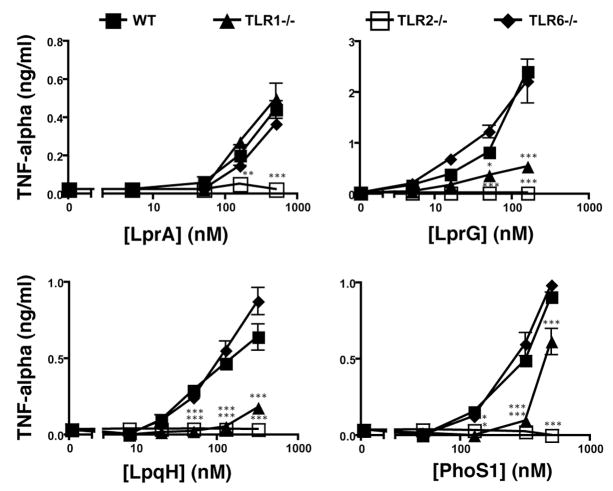
Since TLR2 is thought to signal as a heterodimer with either TLR1 or TLR6, we investigated contributions of these co-receptors to recognition of Mtb lipoproteins. TLR dependence of TNF-alpha induction by Mtb lipoproteins was studied with bone marrow-derived macrophages from wild-type, TLR2−/−, TLR1−/− or TLR6−/− mice (Fig. 2). TNF-alpha induction was completely dependent on TLR2 expression for LprA, LprG, LpqH, and PhoS1. With the exception of LprA, TNF-alpha induction by these lipoproteins was dependent on TLR1 expression (responses were significantly reduced with TLR1−/− macrophages, although to a lesser degree than with TLR2−/− macrophages). Lipoprotein-induced responses were not reduced in TLR6−/− cells, although a redundant contribution of TLR6 cannot be excluded. As predicted, TNF-alpha induction by the synthetic diacylated lipopeptide FSL-1 was dependent on expression of TLR2 and TLR6 but not TLR1, and the response to Pam3CSK4 lipopeptide was dependent on expression of TLR2 and TLR1 but not TLR6 (data not shown). Induction of TNF-alpha by LprA was not reduced by single knockout of either TLR1 or TLR6; possible explanations include redundant contributions of TLR1 and TLR6 for LprA. These data suggest that Mtb lipoproteins are generally more dependent on TLR1 than TLR6, although TLR2 co-receptor dependence varies to some degree with different lipoproteins.
Figure 2. TLR1 is more important than TLR6 for response to mycobacterial lipoproteins.
Bone marrow-derived macrophages from wild-type, TLR1−/−, TLR2−/−, or TLR6−/− mice were stimulated for 12 h with mycobacterial lipoproteins in standard medium. Culture supernatants were assayed for TNF-alpha by ELISA. Data are representative of three independent experiments (two for PhoS1) that used independent preparations of lipoproteins and different preparations of bone marrow derived macrophages. Data are shown as the mean +/− SD of triplicate assays (*, p < 0.05; **, p < 0.01; and ***, p < 0.001 for comparison of wild-type vs. TLR1−/− or TLR2−/− cells).
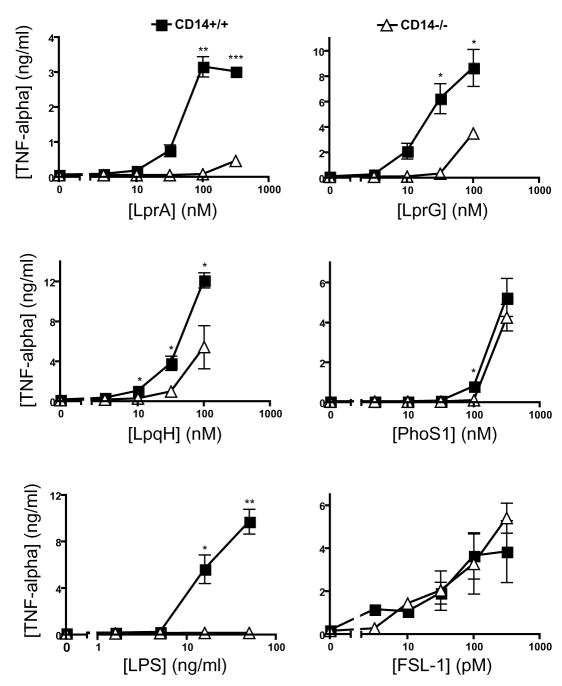
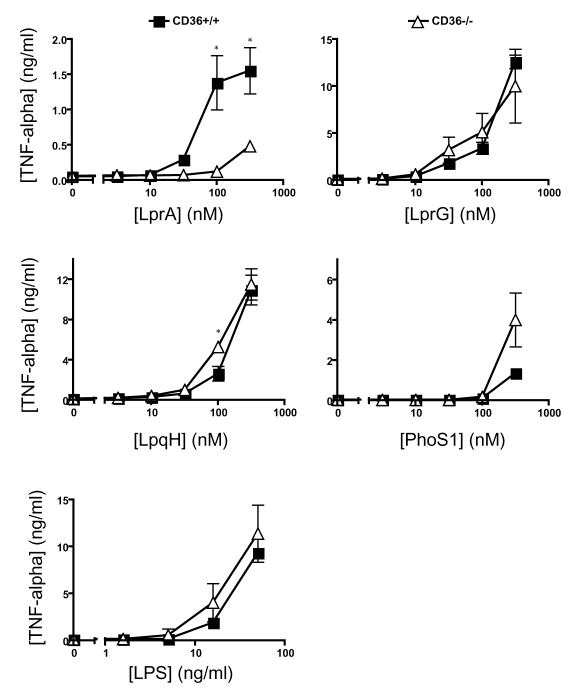
In addition to the co-receptors that form heterodimers with TLR2, lipoprotein recognition may be influenced by accessory receptors that assist in recognition or delivery of ligands. CD14 and CD36 have been reported to contribute to recognition of other ligands by TLR2 [22; 23]. To assess CD14 contribution to TLR2-mediated recognition of Mtb lipoproteins, we tested lipoprotein induction of TNF-alpha expression by bone marrow-derived macrophages from CD14−/− and wild-type mice (Fig. 3). CD14−/− macrophages exhibited impaired cytokine responses relative to wild-type macrophages, although the magnitude of the deficit in CD14−/− cells varied with different TLR2 agonists. Among the Mtb lipoproteins, responses to LprA showed the greatest dependence on CD14; responses to LprG and LpqH showed intermediate dependence on CD14, and the response to PhoS1 showed no discernable dependence on CD14. LPS, a TLR4 agonist, showed a predicted dependence on CD14, while FSL-1 did not. Similar CD14 dependence was seen when bone marrow-derived macrophages from CD14−/− mice were compared with B6 × 129sv F2 hybrid mice (results not shown). To investigate the importance of CD36 to recognition of Mtb lipoproteins by TLR2, we compared Mtb lipoprotein induction of TNF-alpha by wild-type and CD36−/− mice (Fig. 4). While LprA responses were dependent on CD36, responses to PhoS1, LprG and LpqH were independent of CD36 expression. In agreement with previous literature [23], response to LPS was not dependent on CD36 expression. These results indicate that responses to different Mtb lipoproteins vary in their dependence on accessory receptors, with dependence on CD14 more common than dependence on CD36 in the set of studied lipoproteins.
Figure 3. CD14 contributes to the detection of LpqH, LprA and LprG, but not PhoS1 or FSL-1.
Bone marrow-derived macrophages derived from wild-type or CD14−/− mice were stimulated with a range of concentrations of agonist in standard medium (LPS) or SFM (all other agonists). TNF-alpha in culture supernatant was measured by ELISA. Data are representative of three independent experiments (two for PhoS1). Data represent mean +/− SD for triplicate samples (*, p < 0.05; **, p < 0.01 for comparison of CD14−/− vs. wild-type cells).
Figure 4. CD36 contributes to the detection LprA, but not LpqH, LprG, or PhoS1.
Bone marrow-derived macrophages derived from wild-type or CD36 knock-out mice were stimulated with a range of concentrations of agonist for 12 h in standard medium (LPS) or SFM (all other ligands). Culture supernatant was assayed for TNF-alpha by ELISA. Data are representative of three independent experiments (two for PhoS1). Data represent mean +/− SD for triplicate samples (*, p < 0.05 for comparison of CD36−/− vs. wild-type cells).
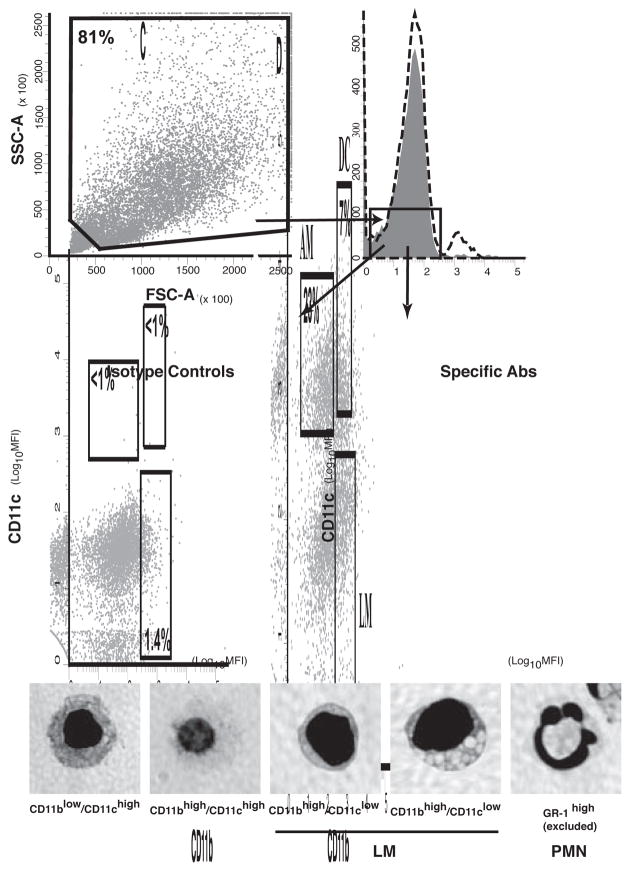
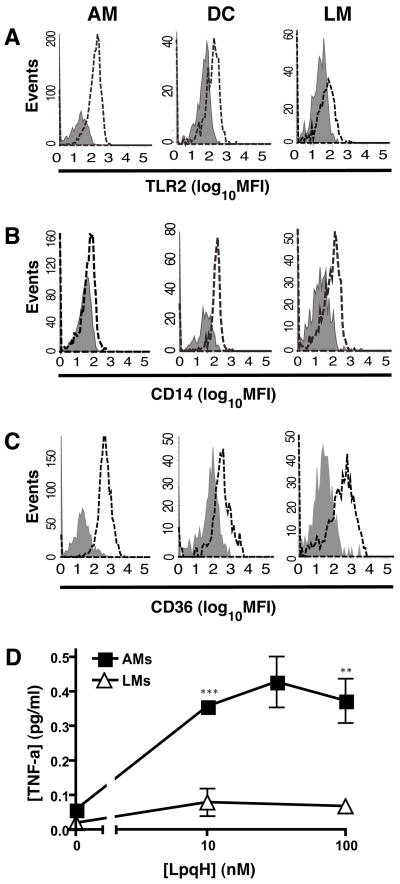
Since Mtb infection primarily occurs in the lung, differential expression of TLR2 or its co-receptors on lung APC subsets may influence the contributions of these cells to host defense in Mtb infection. Therefore, we assessed receptor expression in several APC populations prepared directly from lung tissue. Crude lung APCs were prepared from lung homogenates with a mixture of CD11b and CD11c affinity magnetic beads, and specific lung APC subpopulations were then purified by FACS to exclude Gr-1+ events (neutrophils) and prepare lung APC subsets, including CD11blow/CD11chigh alveolar macrophages, CD11bhigh/CD11clow lung macrophages and CD11bhigh/CD11chigh lung DCs. These APC subset definitions are supported by other published studies [32]; furthermore, bronchoalveolar lavage cells are predominantly CD11blow/CD11chigh, supporting the definition of alveolar macrophages by this marker phenotype, and CD11bhigh/CD11chigh lung DCs have high expression of MHC-II and C-type lectin receptors DC-SIGN and Dec-205 [42], consistent with this cell type assignment. By this approach and selective gating (Fig. 5), crude lung APCs were approximately 23% alveolar macrophages, 13% lung macrophages and 7% lung DCs, with other cells consisting of CD11blow/CD11clow cells or cells with intermediate phenotypes lying between the gates for the defined APC subsets. Interestingly, surface expression of TLR2, CD14 and CD36 varied among the APC subsets (Fig. 6). For TLR2, CD14 and CD36, respectively, alveolar macrophages had specific MFI of 422, 287 and 1175, lung DCs had specific MFI of 673, 917 and 744, and lung macrophages had specific MFI of 113, 247 and 1007. Thus, expression of TLR2, CD14 and CD36 varies on lung APC subsets, and lung macrophages have particularly low TLR2 surface expression.
Figure 5. Isolation and characterization of lung APCs by flow cytometry.
Crude lung APCs were gated by scatter properties (A) and GR1-negativity (shaded peak is isotype, dotted line is stain) (B). APC subsets were characterized by expression of CD11b and CD11c (C and D). Alveolar macrophages (AM) are defined as CD11blow/CD11chigh. Lung macrophages (LM) are defined as CD11bhigh/CD11clow and consisted of 2 morphologies. Lung DCs are defined as CD11bhigh/CD11chigh. GR1high events were excluded from analysis. PMN, morphology of GR1-positive events.
Figure 6. Differing levels of TLR2 on lung APC subsets.
Surface staining for TLR2 (A), CD14 (B) and CD36 (C) is shown with dotted lines, and isotype control staining is shown in gray. Specific MFI values (see text) were calculated as MFI with specific Ab minus MFI with isotype control Ab. In panel D, cells were stimulated with LpqH for 16 h, and TNF-alpha in culture supernatant was measured by ELISA. Results are representative of 3 independent experiments. Data points represent the mean +/− SD of results from at least triplicate cell cultures. Due to limited cell numbers, fewer LpqH concentrations were assessed with lung macrophages. At the LpqH concentration points that were common to both cell types (10 and 100 nM), the responses were significantly different (**, p < 0.01; ***, p < 0.001).
We investigated whether low TLR2 expression by lung macrophages correlated with a lack of responsiveness to the well-characterized mycobacterial TLR2 ligand, LpqH (the 19-kD lipoprotein). Lung APC populations were purified by FACS with populations defined by expression of CD11b and CD11c (see Methods). Since cell yields were too low to obtain adequate numbers of all three lung APC types by three-way sorting, our analysis was focused on comparison of alveolar macrophages and lung macrophages. Macrophages were incubated with LpqH for 16 h, and TNF-alpha production was then assessed by ELISA. While alveolar macrophages produced TNF-alpha in response to LpqH, lung macrophages were minimally responsive (Fig. 6D). Thus, the low level of TLR2 expression on lung macrophages was associated with a decreased ability of these cells to respond to LpqH by induction of TNF-alpha. These data suggest that differences in TLR2 expression have functional consequences for lung APCs.
Discussion
In this work, we assessed the contributions of co-receptors and accessory receptors to the recognition of several Mtb lipoproteins by TLR2. Mtb lipoproteins were generally more dependent on TLR1 than TLR6 as a TLR2 co-receptor, and more dependent on CD14 than CD36 as an accessory receptor. Furthermore, expression of TLR2 and its co-receptors differed between lung APC subsets, and the very low expression of TLR2 by lung macrophages was associated with low responsiveness of these cells to LpqH. These data suggest that variation in expression of TLR2 may influence the ability of different APC types to respond to infection with Mtb.
While TLR1 and TLR6 are both characterized as co-receptors of TLR2, the degree to which each contributes and the mechanisms by which each contributes remain unclear for physiological TLR2 ligands, e.g. Mtb lipoproteins. TLR1 and TLR6 are thought to participate directly in both ligand recognition and adaptor recruitment. Investigations with synthetic lipopeptides suggested that TLR2/TLR1 heterodimers sense triacylated lipopeptides [13] whereas TLR2/TLR6 heterodimers sense diacylated lipopeptides [15]. However, the ability of TLR2 to detect nonacylated ligands [18] and capsular carbohydrates [19] indicates that other structural determinants affect TLR2 co-receptor usage. We compared the responses of TLR2−/−, TLR1−/− or TLR6−/− macrophages to several lipoproteins (Fig. 2). These studies revealed three major characteristics of responses to Mtb lipoproteins. First, TLR2 was absolutely required for response to all of the Mtb lipoproteins, consistent with previous studies with LpqH [13; 43]. Second, lipoproteins differ significantly in their efficacy for cytokine induction in both the HEK293.TLR2 cell line and murine bone marrow-derived macrophages. Third, TLR6 is not important for response to these lipoproteins. Finally, TLR1 contributes significantly to recognition of most of the studied Mtb lipoproteins, although low responses to high concentrations of the lipoproteins may still be achieved in the absence of TLR1; similar results were observed for Pam3CSK4 lipopeptide in our studies and others [13; 44]. Interestingly, LprA showed markedly less dependence on TLR1 than the other lipoproteins. It is possible that some TLR2 agonists have cross-reactivity for TLR1 and another TLR2 co-receptor, e.g. TLR6, TLR10 (which is closely related to TLR1 and TLR6) or a non-TLR receptor. Since Mtb lipoproteins show variability in their co-receptor usage, they may be useful tools for further studies of the structural determinants of TLR2 co-receptor dependence.
The mechanisms by which CD14 contributes as an accessory receptor to promote TLR2 signaling are incompletely understood; prevalent hypotheses include a role in trafficking and/or increasing bioavailability of TLR ligands. Like the TLRs, CD14 is a leucine rich repeat protein that is GPI-linked to the cell surface and cannot signal directly. CD14 can also be released from the cell surface and is present in soluble forms in mammalian serum. The presence of either membrane-associated CD14 or recombinant soluble CD14 enhances Pam3CSK4 binding to TLR2 [27] and resultant signaling [28]. Our data show that not all lipoproteins require CD14 (Fig. 3). Among the Mtb lipoproteins, responses to LprA showed the greatest dependence on CD14; responses to LprG and LpqH showed intermediate dependence on CD14, and the response to PhoS1 showed no discernable dependence on CD14. PhoS1 is particularly striking in its ability to signal in CD14−/− macrophages. Further studies of these lipoproteins may yield insight into the basis of CD14 dependent recognition of bacterial lipoproteins by TLR2.
The role for CD36 in TLR2 signaling is less clear. As a prototypic class B scavenger receptor, CD36 performs homeostatic functions, including recognition and phagocytosis of apoptotic cells, senescent cells, cellular debris, and oxidized LDL [45]. It is involved in the detection and phagocytosis of pathogens, including Staphylococcus aureus and, to a lesser extent, E. coli [29]. More specifically, CD36 is thought to associate with the TLR2 receptor complex and contribute selectively to the detection of some diacylglycerol motifs by TLR2 [23; 46]. Of the Mtb lipoproteins examined in this work, only LprA induced a response that was dependent on CD36 expression. Therefore, LprA may be useful in future studies examining structural components responsible for CD36 accessory function.
It is interesting that LprA and LprG differ in their receptor usage despite being closely related (~34% identical amino acid sequence). LprA was partially dependent on both CD14 and CD36, whereas LprG showed partial dependence on CD14 but not CD36. LprA, unlike the other three Mtb lipoproteins in this study showed no dependence on TLR1. The structural characteristics of LprA that determine its unusual receptor usage are as yet undefined.
Lung APCs that are infected during the course of aerosol infection with mycobacteria include alveolar macrophages, lung macrophages and lung dendritic cells. We investigated the expression of TLR2 and two potential accessory receptors (CD14 and CD36) by these lung APC subsets. Lung macrophages expressed less TLR2 than the other lung APC subsets (Fig. 6A–C). Consistent with their low level of TLR2 expression, lung macrophages were hyporesponsive to LpqH. Since lung macrophages may be infected by mycobacteria, the low level of TLR2 expression on these cells may limit TLR2-dependent responses to mycobacteria that these cells harbor. Thus, low levels of receptor expression on certain APC types may become a limiting factor for responses to pathogens. Due to the likely contribution of multiple receptors in host cell interactions with a pathogen, reduced expression of a single receptor may not ablate host cell responses to the pathogen, but limitation in expression of innate immune receptors by subsets of APCs may alter the quality or magnitude of responses, potentially affecting pathogenesis of infection.
Acknowledgments
This work was supported by NIH grants AI069085, AI034343 and AI035726 to CVH, AI27243 and HL55967 to WHB, and NIH contract HHSN266200700022C/NO1-AI-70022 for Tuberculosis Research Unit (TBRU) to WHB. MGD was supported in part by NIH T32 AI007024 and T32 GM007250. The authors are grateful to Dr. Shizuo Akira (Osaka University, Osaka, Japan) for TLR2, TLR1 and TLR6 deficient mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Drennan MB, Nicolle D, Quesniaux VJ, Jacobs M, Allie N, Mpagi J, Fremond C, Wagner H, Kirschning C, Ryffel B. Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am J Pathol. 2004;164:49–57. doi: 10.1016/S0002-9440(10)63095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–24. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara I, Yamada H, Li C, Mizuno S, Takeuchi O, Akira S. Mycobacterial infection in TLR2 and TLR6 knockout mice. Microbiol Immunol. 2003;47:327–36. doi: 10.1111/j.1348-0421.2003.tb03404.x. [DOI] [PubMed] [Google Scholar]
- 4.Reiling N, Holscher C, Fehrenbach A, Kroger S, Kirschning CJ, Goyert S, Ehlers S. Cutting edge: toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to airborne infection with Mycobacterium tuberculosis. J Immunol. 2002;169:3480–4. doi: 10.4049/jimmunol.169.7.3480. [DOI] [PubMed] [Google Scholar]
- 5.Bochud PY, Hawn TR, Aderem A. Cutting edge: a toll-like receptor 2 polymorphism that is associated with lepromatous leprosy is unable to mediate mycobacterial signaling. J Immunol. 2003;170:3451–4. doi: 10.4049/jimmunol.170.7.3451. [DOI] [PubMed] [Google Scholar]
- 6.Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28:645–59. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Pecora ND, Gehring AJ, Canaday DH, Boom WH, Harding CV. Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol. 2006;177:422–429. doi: 10.4049/jimmunol.177.1.422. [DOI] [PubMed] [Google Scholar]
- 8.Gehring AJ, Dobos KM, Belisle JT, Harding CV, Boom WH. Mycobacterium tuberculosis LprG (Rv1411c): A novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J Immunol. 2004;173:2660–8. doi: 10.4049/jimmunol.173.4.2660. [DOI] [PubMed] [Google Scholar]
- 9.Noss EH, Pai RK, Sellati TJ, Radolf JD, Belisle J, Golenbock DT, Boom WH, Harding CV. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19 kD lipoprotein of Mycobacterium tuberculosis. J Immunol. 2001;167:910–918. doi: 10.4049/jimmunol.167.2.910. [DOI] [PubMed] [Google Scholar]
- 10.Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol. 2003;171:175–184. doi: 10.4049/jimmunol.171.1.175. [DOI] [PubMed] [Google Scholar]
- 11.Pai RK, Pennini ME, Tobian AA, Canaday DH, Boom WH, Harding CV. Prolonged toll-like receptor signaling by Mycobacterium tuberculosis and its 19-kilodalton lipoprotein inhibits gamma interferon-induced regulation of selected genes in macrophages. Infect Immun. 2004;72:6603–14. doi: 10.1128/IAI.72.11.6603-6614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung SB, Yang CS, Lee JS, Shin AR, Jung SS, Son JW, Harding CV, Kim HJ, Park JK, Paik TH, Song CH, Jo EK. The mycobacterial 38-kilodalton glycolipoprotein antigen activates the mitogen-activated protein kinase pathway and release of proinflammatory cytokines through Toll-like receptors 2 and 4 in human monocytes. Infect Immun. 2006;74:2686–96. doi: 10.1128/IAI.74.5.2686-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–4. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 14.Tapping RI, Tobias PS. Mycobacterial lipoarabinomannan mediates physical interactions between TLR1 and TLR2 to induce signaling. J Endotoxin Res. 2003;9:264–8. doi: 10.1179/096805103225001477. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–40. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 16.Takeda K, Takeuchi O, Akira S. Recognition of lipopeptides by Toll-like receptors. J Endotoxin Res. 2002;8:459–63. doi: 10.1179/096805102125001073. [DOI] [PubMed] [Google Scholar]
- 17.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Massari P, Visintin A, Gunawardana J, Halmen KA, King CA, Golenbock DT, Wetzler LM. Meningococcal Porin PorB Binds to TLR2 and Requires TLR1 for Signaling. J Immunol. 2006;176:2373–80. doi: 10.4049/jimmunol.176.4.2373. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, McLoughlin RM, Cobb BA, Charrel-Dennis M, Zaleski KJ, Golenbock D, Tzianabos AO, Kasper DL. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J Exp Med. 2006;203:2853–63. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita Y, Maeda Y, Takeshita F, Brennan PJ, Makino M. Role of the polypeptide region of a 33kDa mycobacterial lipoprotein for efficient IL-12 production. Cell Immunol. 2004;229:13–20. doi: 10.1016/j.cellimm.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.VanderVen BC, Harder JD, Crick DC, Belisle JT. Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science. 2005;309:941–3. doi: 10.1126/science.1114347. [DOI] [PubMed] [Google Scholar]
- 22.Henneke P, Takeuchi O, van Strijp JA, Guttormsen HK, Smith JA, Schromm AB, Espevik TA, Akira S, Nizet V, Kasper DL, Golenbock DT. Novel engagement of CD14 and multiple toll-like receptors by group B streptococci. J Immunol. 2001;167:7069–76. doi: 10.4049/jimmunol.167.12.7069. [DOI] [PubMed] [Google Scholar]
- 23.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–7. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 24.Schroder NW, Heine H, Alexander C, Manukyan M, Eckert J, Hamann L, Gobel UB, Schumann RR. Lipopolysaccharide binding protein binds to triacylated and diacylated lipopeptides and mediates innate immune responses. J Immunol. 2004;173:2683–91. doi: 10.4049/jimmunol.173.4.2683. [DOI] [PubMed] [Google Scholar]
- 25.Hajishengallis G, Ratti P, Harokopakis E. Peptide mapping of bacterial fimbrial epitopes interacting with pattern recognition receptors. J Biol Chem. 2005;280:38902–13. doi: 10.1074/jbc.M507326200. [DOI] [PubMed] [Google Scholar]
- 26.Liang S, Wang M, Tapping RI, Stepensky V, Nawar HF, Triantafilou M, Triantafilou K, Connell TD, Hajishengallis G. Ganglioside GD1a is an essential coreceptor for Toll-like receptor 2 signaling in response to the B subunit of type IIb enterotoxin. J Biol Chem. 2007;282:7532–42. doi: 10.1074/jbc.M611722200. [DOI] [PubMed] [Google Scholar]
- 27.Vasselon T, Detmers PA, Charron D, Haziot A. TLR2 recognizes a bacterial lipopeptide through direct binding. J Immunol. 2004;173:7401–5. doi: 10.4049/jimmunol.173.12.7401. [DOI] [PubMed] [Google Scholar]
- 28.Vasselon T, Hanlon WA, Wright SD, Detmers PA. Toll-like receptor 2 (TLR2) mediates activation of stress-activated MAP kinase p38. J Leukoc Biol. 2002;71:503–10. [PubMed] [Google Scholar]
- 29.Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170:477–85. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahaman SO, Lennon DJ, Febbraio M, Podrez EA, Hazen SL, Silverstein RL. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 2006;4:211–21. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexopoulou L, Thomas V, Schnare M, Lobet Y, Anguita J, Schoen RT, Medzhitov R, Fikrig E, Flavell RA. Hyporesponsiveness to vaccination with Borrelia burgdorferi OspA in humans and in TLR1- and TLR2-deficient mice. Nat Med. 2002;8:878–84. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Juarrero M, Shim TS, Kipnis A, Junqueira-Kipnis AP, Orme IM. Dynamics of macrophage cell populations during murine pulmonary tuberculosis. J Immunol. 2003;171:3128–35. doi: 10.4049/jimmunol.171.6.3128. [DOI] [PubMed] [Google Scholar]
- 33.Reljic R, Di Sano C, Crawford C, Dieli F, Challacombe S, Ivanyi J. Time course of mycobacterial infection of dendritic cells in the lungs of intranasally infected mice. Tuberculosis. 2005;85:81–8. doi: 10.1016/j.tube.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Humphreys IR, Stewart GR, Turner DJ, Patel J, Karamanou D, Snelgrove RJ, Young DB. A role for dendritic cells in the dissemination of mycobacterial infection. Microbes Infect. 2006;8:1339–46. doi: 10.1016/j.micinf.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 35.Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K, Ernst JD. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 36.Ashbridge KR, Prestidge RL, Booth RJ, Watson JD. The mapping of an antibody-binding region on the Mycobacterium tuberculosis 19 kilodalton antigen. J Immunol. 1990;144:3137–42. [PubMed] [Google Scholar]
- 37.Febbraio M, Podrez EA, Smith JD, Hajjar DP, Hazen SL, Hoff HF, Sharma K, Silverstein RL. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105:1049–56. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sklar MD, Tereba A, Chen BD, Walker WS. Transformation of mouse bone marrow cells by transfection with a human oncogene related to c-myc is associated with the endogenous production of macrophage colony stimulating factor 1. J Cell Physiol. 1985;125:403–12. doi: 10.1002/jcp.1041250307. [DOI] [PubMed] [Google Scholar]
- 39.Flo TH, Ryan L, Latz E, Takeuchi O, Monks BG, Lien E, Halaas O, Akira S, Skjak-Braek G, Golenbock DT, Espevik T. Involvement of toll-like receptor (TLR) 2 and TLR4 in cell activation by mannuronic acid polymers. J Biol Chem. 2002;277:35489–95. doi: 10.1074/jbc.M201366200. [DOI] [PubMed] [Google Scholar]
- 40.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277:47834–43. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 41.Pecora ND, Fulton SA, Reba SM, Drage MG, Simmons DP, Urankar-Nagy NJ, Boom WH, Harding CV. Mycobacterium bovis BCG decreases MHC-II expression in vivo on murine lung macrophages and dendritic cells during aerosol infection. Cell Immunol. 2008 doi: 10.1016/j.cellimm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ordway D, Henao-Tamayo M, Orme IM, Gonzalez-Juarrero M. Foamy macrophages within lung granulomas of mice infected with Mycobacterium tuberculosis express molecules characteristic of dendritic cells and antiapoptotic markers of the TNF receptor-associated factor family. J Immunol. 2005;175:3873–81. doi: 10.4049/jimmunol.175.6.3873. [DOI] [PubMed] [Google Scholar]
- 43.Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, Maitland M, Norgard MV, Plevy SE, Smale ST, Brennan PJ, Bloom BR, Godowski PJ, Modlin RL. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 44.Buwitt-Beckmann U, Heine H, Wiesmuller KH, Jung G, Brock R, Akira S, Ulmer AJ. Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur J Immunol. 2005;35:282–9. doi: 10.1002/eji.200424955. [DOI] [PubMed] [Google Scholar]
- 45.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108:785–91. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triantafilou M, Gamper FG, Haston RM, Mouratis MA, Morath S, Hartung T, Triantafilou K. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281:31002–11. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]