Abstract
Natural killer (NK) cells play critical roles in antiviral immunity. While the importance of effector mechanisms such as interferons have been demonstrated through knockout mice, specific mechanisms of how viruses are recognized and controlled by NK cells are less well-defined. Previous genetic studies have mapped the resistance genes for murine cytomegalovirus (MCMV), herpes simplex virus-1 (HSV-1) and ectromelia virus, to the NK gene complex (NKC) on murine chromosome 6, a region containing the polymorphic Ly49 and Nkrp1 families. Genetic resistance to MCMV in C57BL/6 has been attributed to Ly49H, an activation receptor, through susceptibility of the recombinant inbred strain BXD-8 that lacks Ly49h (also known as Klra8) but derived about half of its genome from its DBA/2 progenitor. However, it remained possible that epigenetic effects could account for the MCMV phenotype in BXD-8 mice. Herein we report the generation of a novel congenic murine strain, B6.BXD8-Klra8Cmv1-del/Wum, on the C57BL/6 genetic background to evaluate the effect of deletion of a single NK activation receptor, Ly49H. Deletion of Ly49H rendered mice much more susceptible to MCMV infection. This increase in susceptibility did not appear to be a result of a difference in NK cell expansion or interferon-γ (IFN-γ) production between the C57BL/6 and the B6.BXD8 strains. On the other hand, the deletion of Ly49h did not otherwise affect NK cell maturation or Ly49D expression, and had no effect on susceptibility to HSV-1 or ectromelia virus. In conclusion, Ly49h is necessary for genetic resistance to MCMV, but not HSV-1 or ectromelia virus.
Keywords: MCMV, Ly49H, cytomegalovirus, Cmv1, natural killer cells
INTRODUCTION
Natural killer (NK) cells play important roles in innate immunity, particularly to viruses (French and Yokoyama 2003; Gazit et al. 2006). This is best illustrated by the clinical course of patients with natural killer cell defects which are characteristically manifested as recurrent infections with herpesviruses, such as cytomegalovirus (CMV) and herpes simplex virus-1 (HSV-1) (Biron et al. 1989). Thus, greater understanding of how natural killer cells specifically recognize and control viral infections will have important clinical applications.
Given that CMV viruses are species-specific and that murine CMV (MCMV) share significant biologic and genomic similarity with human CMV, MCMV has been used extensively as an animal model for HCMV (Gonczol et al. 1985; Rawlinson et al. 1997). Several host factors, both MHC-dependent and MHC-independent, account for MCMV resistance (Adam et al. 2006; Dighe et al. 2005; Grundy et al. 1981; Rodriguez et al. 2004; Scalzo et al. 1990). The first identified MHC-independent resistance gene, Cmv1, was mapped to distal mouse chromosome 6, in the vicinity of the NK gene complex (NKC) which encodes receptors expressed on NK cells (Scalzo et al. 1995). Identification of the Ly49H activation receptor as the product of the Cmv1 locus was aided by the BXD-8 recombinant inbred (RI) strain (Brown et al. 2001b; Dokun et al. 2001; Lee et al. 2001).
Ly49H is a member of the Ly49 family of C-type lectin-like receptors, which are encoded in the NKC region on distal mouse chromosome 6. The Ly49 family is extremely polymorphic, displaying haplotypes with variations in gene number and allelic polymorphism for each gene (Proteau et al. 2004; Wilhelm et al. 2002). The significant difference in viral susceptibility between the resistant C57BL/6 and susceptible BALB/c has been attributed to the expression of Ly49H on NK cells in C57BL/6 whereas BALB/c mice lack the Ly49h gene (Brown et al. 2001a; Lee et al. 2001; Lee et al. 2003). Interestingly, loci for genetic resistance to two other viruses, HSV-1 and ectromelia virus, an orthopoxvirus, have also been mapped to the NKC (Brown et al. 2001a; Delano and Brownstein 1995). Although antibody-blocking experiments do not support a role for Ly49H in resistance to these viruses (data not shown), Ly49H has not been extensively studied in resistance by genetic approaches.
The BXD-8 RI strain arose from the consanguineous mating between C57BL/6 and DBA/2 progenitor strains for twenty generations. Consequently, half of the genome of this recombinant inbred strain is derived from the resistant C57BL/6 progenitor strain, while the other half from the susceptible DBA/2. BXD-8 mice carried C57BL/6 markers for distal chromosome 6 flanking the NKC region, but demonstrated increased susceptibility to MCMV compared to the C57BL/6 strain. This increased susceptibility has previously been shown to correlate with the absence of Ly49H expression by staining with the monoclonal antibody 3D10 on BXD-8 NK cells and a selective deletion in Ly49h by Southern analysis (Brown et al. 2001a). However, since approximately half of the BXD-8 genome is derived from the DBA/2 strain, it was not clear whether any DBA/2 alleles had an epistatic genetic effect on its increased susceptibility. Transgenic expression of a bacterial artificial chromosome containing the C57BL/6 allele of Ly49h into the susceptible BALB/c genetic background also conferred resistance to MCMV (Lee et al. 2003) but it remained possible that other background BALB/c alleles may modify the Ly49h effect.
To further analyze the role of Ly49h in NK cell function, particularly in infections, we backcrossed the BXD-8 RI strain onto C57BL/6 strain by marker assisted backcrossing (speed congenics). We termed this novel strain B6.BXD8-Klra8Cmv1-del/Wum, referred to by the shorthand of B6.BXD8 in this paper, and characterized the NK cell function of this strain.
MATERIALS AND METHODS
Animals
C57BL/6NCR mice were purchased from National Cancer Institute. The RI strain BXD-8 was purchased from The Jackson Laboratory (Bar Harbor, Maine) and was backcrossed to C57BL/6 for nine generations to generate the novel strain B6.BXD8-Klra8Cmv1-del/Wum. At each successive generation, heterozygous pups carrying the deleted Ly49h allele were identified by flow cytometry. Microsatellite genotyping was performed by the Speed Congenics Core of the Rheumatic Diseases Core Center at Washington University to screen for C57BL/6 markers for the initial seven generations. Subsequently, two more generations were backcrossed onto C57BL/6 while following the heterozygous expression of Ly49H phenotype by flow cytometry. All animals were maintained in a specific-pathogen free facility at Washington University School of Medicine and all experiments were approved by the Animal Studies Committee.
Southern Analysis
Genomic DNA for BXD-8 was purchased from Jackson Laboratory. Splenic genomic DNA for C57BL/6 and B6.BXD8 were extracted using Gene Pure following manufacturer’s protocol. Digested DNA was separated on a 1% agarose gel and transferred onto a nitrocellulose membrane. Membrane was subsequently hybridized with the BglII-Pst I fragment of Ly49H cDNA labeled with 32αP-dCTP using Rediprime II System (Amersham, United Kingdom) following manufacturer’s protocol, and image acquired using the Typhoon system (Amersham, United Kingdom).
Plaque assays
Viral titers were quantified using methods previously published (French et al. 2004). Briefly, whole spleens were harvested and frozen in 1 ml aliquots of D10 (DMEM supplemented with 10% newborn calf serum (Hyclone), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM glutamine.) Liver sections were weighed in 1 ml aliquots of D10 medium. Tissues were subsequently thawed, homogenized via dounce homogenizers and used to infect NIH-3T12 fibroblast monolayers in triplicates in 6-well plates. Titers were calculated as Log (PFU/spleen) or Log (PFU/100 mg of liver).
Flow cytometric analysis of natural killer cells
Single-cell suspensions were prepared from spleens as previously described (French et al. 2004). Splenocytes were stained with APC-PK136 (NK1.1), PerCP-Cy5.5-2C11 (CD3), and PerCP-Cy5.5-1D3 (CD19) in supernatant produced by 2.4G2 hybridoma to block FcRII/III. The following antibodies were used to identify respective cell surface markers: PE-eBio12AB (Ly49A/D), PE-5E6 (Ly49C/I), FITC-4E4 (Ly49D), FITC-3D10 (Ly49H), PE-16a11 (NKG2AB6), and PE-CX5 (NKG2D). Except for the Ly49H antibody, which was made in house, the other antibodies were purchased from BD Biosciences (San Jose, CA). For intracellular IFN-γ staining, cells were permeabilized with Cytofix/Cytoperm (PharMingen, La Jolla, CA) per manufacturer’s protocol and stained with PE-conjugated anti-IFN-γ (clone XMG1.2), purchased from BD Biosciences. To evaluate BrdU incorporation, each mouse was injected with 2 mg of BrdU intraperitoneally three hours prior to spleen harvest. Splenocytes were stained with surface markers and then permeabilized using Cytofix/Cytoperm following manufacturer’s protocol. Intracellular BrdU was determined by staining with FITC-anti-BrdU antibody (BD Biosciences) as previously described (Dokun et al. 2001).
Quantitative RT-PCR
Total RNA was extracted from spleens using Trizol (Invitrogen, Carlsbad, CA). cDNA was subsequently generated using oligo-dT primers while following the protocol for Superscript III First Strand Synthesis (Invitrogen). PCR primers for Ly49D were: Forward 5’-TTCAGGGTTGCAGAACGAGATGAG-3’; reverse 5’-AGGATCCCGAGAGCTATCACAATG-3’. The internal probe sequence was 5’-AAAGCTCGCCTCAGAGTTCCCTGGCA-3’. The primers for Ly49H were: Forward 5’-CACAAGTCTTCAGGGTTGAACAGC-3’; reverse 5’-CAACAATTACCAGCCGAAGGGAAC-3’; internal probe 5’-TGGCCTAAGAGTCCCTTGGCAGCTCATTGT-3’. mRNA expression was normalized against β-actin expression and then compared to levels expressed in C57BL/6 splenocytes.
RESULTS
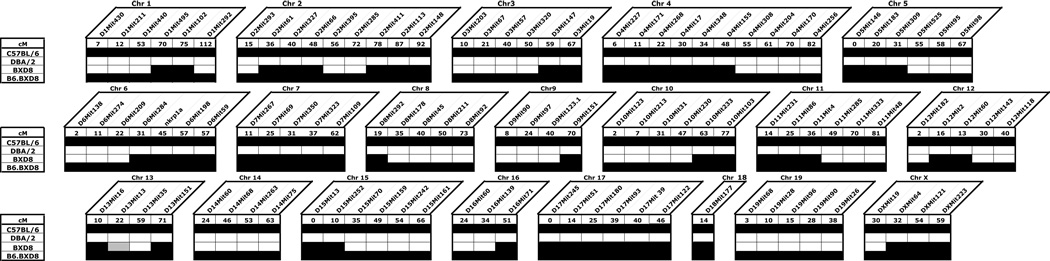
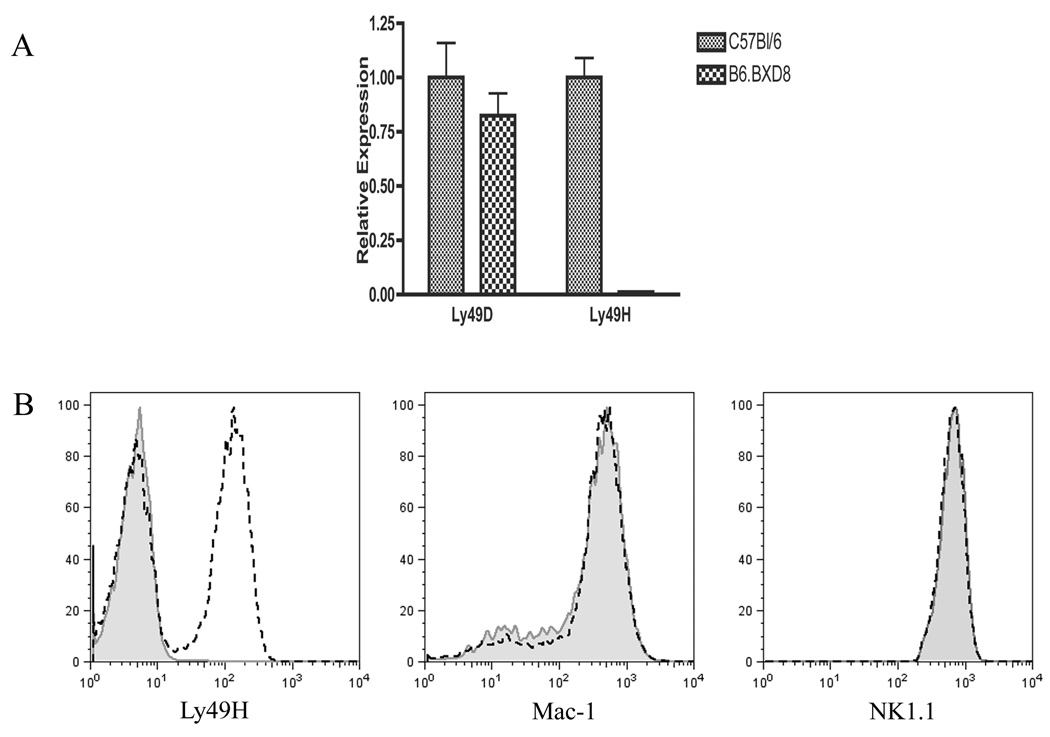
Analysis of 110 microsatellite markers polymorphic between C57BL/6 and DBA/2 revealed that whereas the original recombinant inbred strain BXD-8 carried 52% of C57BL/6-derived genome, B6.BXD8 demonstrated 100% of C57BL/6-derived genome except for Ly49h (Figure 1). Previous study from our laboratory demonstrated that BXD-8 carried a genomic deletion at the Ly49h locus in the midst of a chromosomal segment derived from C57BL/6 (Brown et al., 2001). Southern analysis of B6.BXD8 genomic DNA digested with BglII and EcoRI, when hybridized with the 177 base-pair BglII-PstI fragment of Ly49h cDNA, revealed the same patterns as those of BXD-8 genome (data not shown). The absence of a 3.5-kb band in B6.BXD8 in the BglII digest likely represented the genomic locus 82385903 to 82389340 in the C57BL/6 reference sequence (GeneBank accession number Mm6_39393_37). Furthermore, the absence of a 5-kb band in B6.BXD8 in the EcoRI digest likely represented the genomic locus 82385831 to 82390624 in C57BL/6. The exact junctions where this deletion occurred, however, was difficult to define given the high degree of sequence homology amongst Ly49 family members and the frequency of repeat sequences in this region. Repeated attempts to amplify genomic sequences in this region with PCR suggested that B6.BXD8 contained minimally a 20-kb genomic deletion spanning Ly49h, including the exon encoding the first start codon for Ly49h (data not shown). Consistent with this idea was the absence of detectable Ly49H mRNA by quantitative RT-PCR (Figure 2A). Moreover, mRNA expression level of Ly49d, the gene immediately proximal to Ly49h in the NKC was similar between C57BL/6 and B6.BXD8. Taken together, these data demonstrate that B6.BXD8 preserved the genomic Ly49h deletion from the original recombinant inbred strain BXD-8 while the rest of the genome is derived from C57BL/6.
Figure 1.
Microsatellite genotyping analysis of B6.BXD8 strain. Numbers below microsatellite markers denote the distance of markers in centiMorgans according to the WI-Genetic Map (Hudson et al. 2001). Black boxes denote C57BL/6 alleles whereas white boxes indicate DBA/2 alleles. One shaded box (Chr 13) denotes a mixed-marker type.
Figure 2.
Absence of Ly49h expression in B6.BXD8. A) mRNA expression of Ly49H in C57BL/6 and B6.BXD8. As determined by quantitative RT-PCR, the relative expression represents ratios of Ly49D or Ly49H mRNA expression in B6.BXD8 splenocytes, normalized by β-actin, compared to those in C57BL/6 splenocytes. These results are representative of two independent experiments. B) Flow cytometric analysis of C57BL/6 and B6.BXD8 NK cells. Fresh splenocytes were labeled with a combination of anti-CD3, CD19 and NK1.1 antibodies in the presence of 2.4G2 hybridoma supernatant to block non-specific FcγRII/III binding. NK cell gate was defined as CD3−/CD19−/NK1.1+. The dashed lines represent NK cells from C57BL/6 while the solid shaded curves represent B6.BXD8. These results are representative of a minimum of two independent experiments.
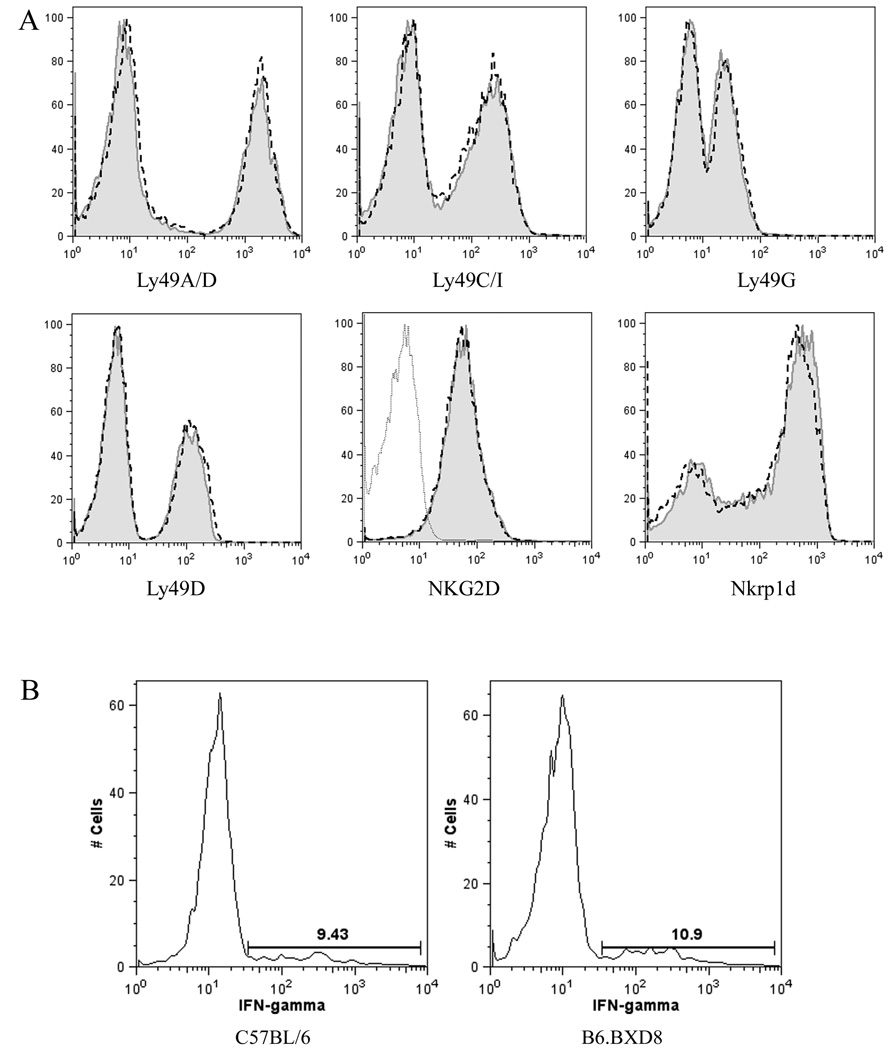
At the protein level, flow cytometric analysis of NK cells from B6.BXD8 demonstrated no detectable surface staining with the anti-Ly49H-specific 3D10 monoclonal antibody as compared to 40% of NK cells from C57BL/6 (Figure 2B, left panel). On the other hand, both C57BL/6 and B6.BXD8 NK cells expressed comparable levels of NK1.1, an NK cell-specific marker encoded by Nkrp1c in the neighboring NKC, adjacent to the Ly49 locus. Furthermore, other receptors encoded in the NKC, such as Nkrp1d and NKG2D, were also expressed at similar levels in C57BL/6 and B6.BXD8. Finally, both B6.BXD8 and C57BL/6 NK cells expressed comparable levels of other Ly49 family members, Ly49A/D, Ly49D, Ly49C/I and Ly49G (Figure 3A), further supporting that B6.BXD8 specifically lacks Ly49H expression, while demonstrating intact expression of known NKC surface receptors.
Figure 3.
Normal NKC receptor expression and function of NK cells in B6.BXD8. A) Cell surface expression of NKC ligands on C57BL/6 and B6.BXD8 NK cells. The dashed lines represent NK cells from C57BL/6 while the solid shaded curves represent B6.BXD8. For the NKG2D staining, the dotted, unshaded curve represents staining with the isotype control. Results were representative of two independent experiments. B) NK cell activation after NK1.1 activation in vitro. Freshly isolated splenocytes were plated in 6-well plates coated with PK136 (anti-NK1.1 monoclonal antibody) at 5 µg/well for 7 hours in the presence of GolgiPlug (BD). Cells were subsequently permeabilized and intracellular IFN-γ expression analyzed following manufacturer’s protocol (BD Biosciences). These results were representative of two independent experiments.
Further characterization of NK cells from B6.BXD8 mice demonstrated comparable levels of the maturation marker, Mac-1, as NK cells from C57BL/6 (Figure 2B, middle panel). Inasmuch as Mac-1 is expressed on functionally mature NK cells, these data suggest full functional maturation of B6.BXD8 NK cells. Indeed, both B6.BXD8 and C57BL/6 NK cells produced similar levels of IFN-γ after NK1.1 activation (Figure 3B). Thus, B6.BXD8 NK cells do not have any gross defect in IFN-γ production.
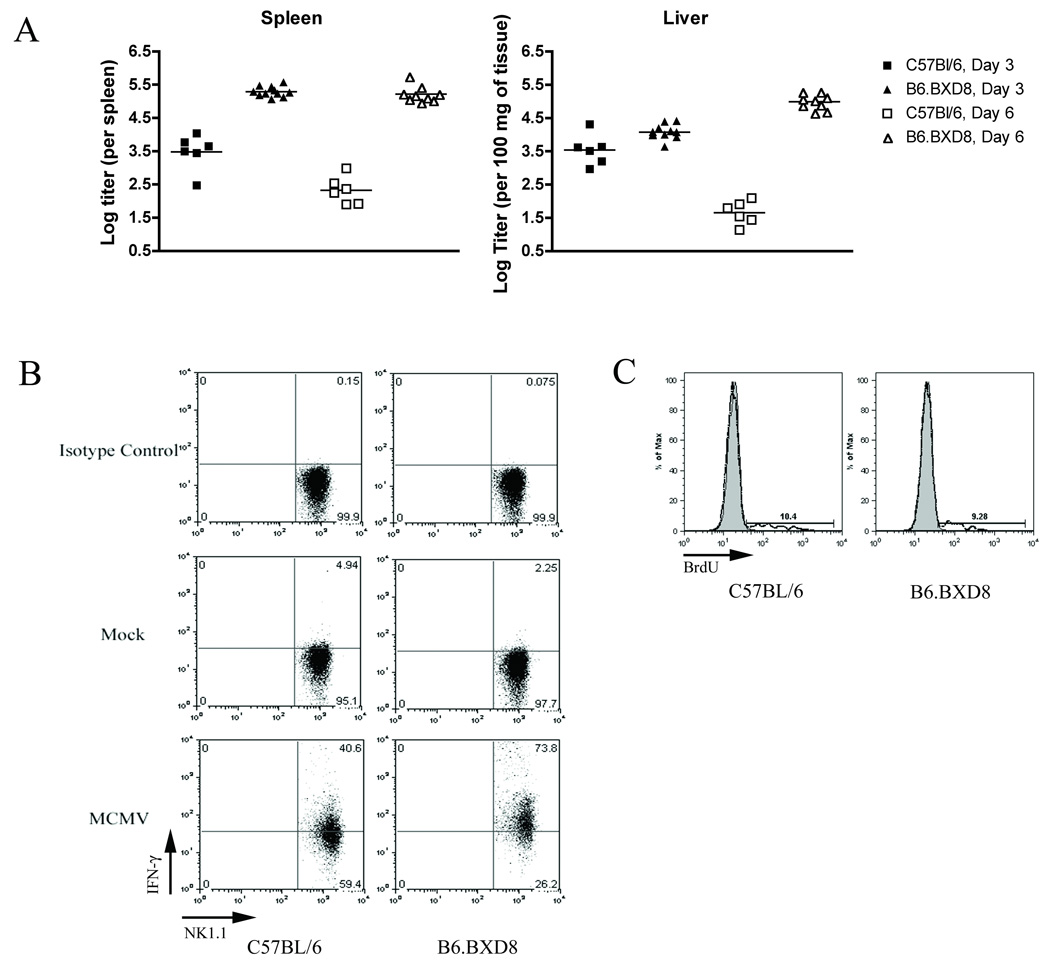
We next sought to further delineate the MHC-independent contribution of Ly49H in controlling MCMV infection. We measured splenic and liver viral titers after intraperitoneally infecting both C57BL/6 and B6.BXD8 mice with Smith strain MCMV (Figure 4A). Indeed, at day 3 post MCMV infection, the splenic titers from B6.BXD8 mice were nearly two logs higher than those of C57BL/6 (mean log titers for C57BL/6 and B6.BXD8 were 3.5 and 5.3 PFU/spleen respectively, p<0.0001 by unpaired t test). Moreover, consistent with previous findings (Brown et al. 2001a), a small but statistically significant difference was found between hepatic viral titers between C57BL/6 and B6.BXD8 on day 3 post infection (p=0.0046 by unpaired t test). After infection with Smith-strain MCMV, B6.BXD8 mice demonstrated a LD50 of 7 × 104 PFU, approximately three-fold less than that for C57BL/6 (data not shown). Thus, these data indicate that Ly49h plays a significant role in C57BL/6 in controlling MCMV infection, and rule out any epistatic effect by DBA/2 alleles in BXD-8.
Figure 4.
MCMV infection in B6.BXD8. A) Splenic and liver MCMV viral titers in C57BL/6 and B6.BXD8 mice. Each mouse was infected with 5x104 PFU of Smith strain MCMV intraperitoneally, and spleens and livers were harvested on day 3 post infection. MCMV viral titers were quantified using methods described previously. B) IFN-γ production in C57BL/6 and B6.BXD8 NK cells after MCMV infection. Mice were infected with 5×104 PFU of MCMV intraperitoneally, and spleens were harvested at 38 hours post infection. Intracellular IFN-γ staining was performed on freshly isolated splenocytes following manufacturer’s protocol. The results shown were gated on the total NK cell population (CD3−/CD19−/NK1.1+). The upper two panels show isotype controls for anti-IFN-γ antibody. The middle two panels are NK cells from mock-infected mice. The lowest two panels show NK cells after MCMV infection. This result is representative of two independent experiments. C) NK cell proliferation after MCMV infection. C57BL/6 and B6.BXD8 mice were infected with 5×104 PFU of MCMV, and spleens harvested on day 2 post infection. Three hours prior to splenocyte harvest, each mouse was intraperitoneally injected with 2 mg of BrdU, and subsequent intracellular BrdU staining was performed following manufacturer’s protocol. The histograms display total NK cell populations, defined as CD3−/CD19−/NK1.1+. The shaded curves denote unstained controls, while the dotted lines denote cells stained with anti-Brdu antibody. The percentages of cells positive for anti-BrdU are quantified in the gates.
During MCMV infection, we previously reported that NK cells are both non-specifically and specifically activated when considering Ly49H expression (Dokun et al. 2001). However, it was not clear if specific activation of NK cells through Ly49H contributed to non-specific activation. When we infected B6.BXD8 mice with MCMV, we found that B6.BXD8 NK cells express IFN-γ at similar levels as those of C57BL/6 NK cells at 38 hours post MCMV infection (Figure 4B). This finding indicated that the early, non-specific IFN-γ production occurs normally during MCMV infection in the absence of Ly49h. Another non-specific effect is the stimulation of NK cell proliferation early during MCMV infection. We observed no difference in the incorporation of BrdU in C57BL/6 versus B6.BXD8 splenocytes at 38 hours after MCMV infection (Figure 4C). Thus, the “non-specific” (with respect to Ly49H expression) activation of NK cells does not require triggering through Ly49H itself (Dokun et al. 2001).
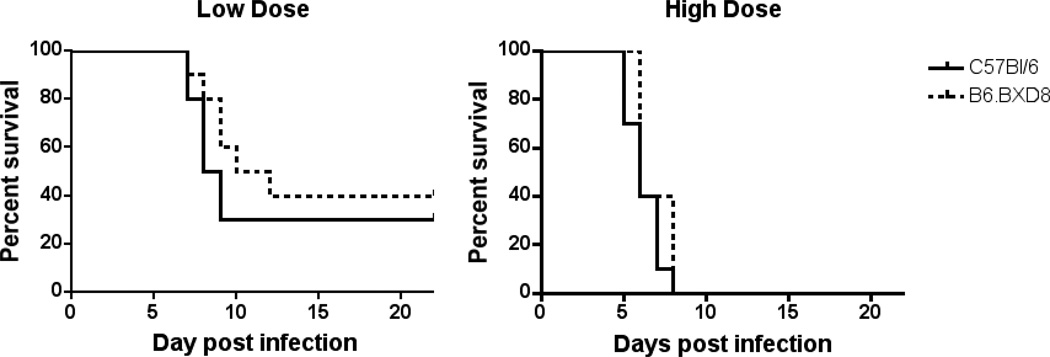
Previous genetic studies mapped the resistance to two other DNA viruses, HSV-1 and ectromelia virus, to NKC, but genes for host resistance have not been further elucidated (Brown et al. 2001a; Delano and Brownstein 1995). We thus sought to evaluate whether the deletion of Ly49h on the C57BL/6 background altered host susceptibility to these viral infections. At both high and low doses of intraperitoneal HSV-1 infection, B6.BXD8 mice demonstrated similar rates of survival as C57BL/6 (Figure 5). Similarly, infection with ectromelia virus resulted in comparable viral titers in C57BL/6 and B6.BXD8 mice on days 5, 6, and 7 post-infection as measured by quantitative PCR from two independent experiments (data not shown). Together, these data suggest that Ly49h does not account for NKC-linked genetic resistance to HSV or ectromelia virus infections.
Figure 5.
Survival curves of C57BL/6 and B6.BXD8 mice after HSV infection. Mice were intraperitoneally inoculated with either low dose (3×106 PFU) or high dose (3×107 PFU) of strain 17 HSV-1. Mice were observed for survival for 22 days.
DISCUSSION
Natural killer cells play important roles in controlling viral infections, and how NK cells are able to recognize and control diverse viral pathogens with a limited repertoire of innate receptors is still poorly defined. In the case of C57BL/6 mice and control of MCMV viremia, we show here that the deletion of a single NK cell receptor, namely Ly49h, in the C57BL/6 genetic background greatly increased host susceptibility. This current paper further extends previous findings that Ly49H, a single NK activation receptor, plays a critical role in the control of MCMV infection in C57BL/6 mice because the present data exclude any epistatic genetic effect of DBA/2 alleles in the phenotype of BXD-8 mice. Taken together with previous work (Brown et al. 2001b; Lee et al. 2001; Lee et al. 2003), these data provide strong genetic evidence that Ly49h plays a major role in the ability of C57BL/6 mice to resist MCMV infection.
Although we were not able to pinpoint the genetic lesion in B6.BXD8, our novel strain demonstrated a specific deficiency of Ly49H at the protein level but intact expression of other examined receptors in the NKC region. Ly49n and Ly49k, two loci juxtaposing Ly49h, were previously identified as pseudogenes (McQueen et al. 1999). Their presence or absence is thus unlikely to account for the increased susceptibility of B6.BXD8 to MCMV, as compared to C57BL/6. Furthermore, while it is theoretically plausible that DBA/2 genes could have recombined into the Ly49h region in B6.BXD8 during the recombinant inbreeding process, this is very unlikely because a double crossover event would be needed in a very tight region. Regardless, B6.BXD8 has a specific Ly49h deletion on the C57BL/6 genetic background.
Interestingly, we previously detected non-stochastic expression of Ly49H with Ly49D on NK cells in normal B6 mice (Smith et al. 2000). These activation receptors both signal through the DAP12 signaling chain. One possible explanation for their co-expression is their interdependent transcriptional regulation. However, our current studies indicate that the absence of Ly49h did not appear to affect Ly49D expression, indicating that Ly49D expression is independent of Ly49H expression, raising the possibility that their tendency to be expressed with each other in normal mice may be related to their common requirement for DAP12.
Our current study found no gross defect with NK cell maturation markers, expression of other NKC-encoded receptors, or NK cell function in B6.BXD8 mice, indicating that Ly49H is not necessary for NK cell development. Importantly, unlike other Ly49 members, which interact with host MHC class I molecules and modulate NK activation, Ly49H has no known endogenous ligand. Whether Ly49H plays roles other than antiviral protection against MCMV remains to be determined, and it is possible that Ly49H enables NK cells to recognize pathogens other than MCMV. However, in our current study, we found that B6.BXD8 and C57BL/6 mice are equally susceptible to HSV-1 and ectromelia virus, two pathogens whose resistance is encoded in the NKC region.
Our previous studies indicated that NK cells appear to undergo two phases of activation during MCMV infection, an early (day 1–2) “non-specific” (independent of Ly49H expression) and a later (days 3–6) “specific” phase whereby Ly49H+ NK cells undergo selective proliferation (Dokun et al. 2001). However, it was not clear if triggering through Ly49H itself contributed to the non-specific phase. Herein, we found that NK cells in B6.BXD8 mice have no defect in their non-specific responses indicating that Ly49h is dispensable for early non-specific NK cell stimulation during MCMV infections.
Given the importance of Ly49 family in modulating NK cell functions, the novel strain B6.BXD8 may serve as a useful tool for further delineation of NK activation receptors. Previously, functional analysis of Ly49 activation receptors were derived from examining mice deficient or defective in the adaptor molecule DAP12 since activation Ly49 receptors do not contain intrinsic signaling motifs and signal downstream by association with the DAP12 adaptor molecule (Bakker et al. 2000; Kaifu et al. 2003; Smith et al. 1998). However, DAP12 can also associate with other activation receptors and is expressed in several different cell types (Bouchon et al. 2001), indicating that DAP12 deficient mice have broader defects. Therefore, B6.BXD8 will allow the evaluation of the endogenous function of Ly49H and indirectly other activation receptors with more precision.
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01AI051345 to W.M.Y. Microsatellite typing was supported by the Rheumatic Diseases Core Center grant P30AR048335. W.M.Y. is an investigator of the Howard Hughes Medical Institute. T.P.C. is supported by the Abbott Research Scholars fellowship.
Contributor Information
Tammy P. Cheng, Division of Rheumatology, Washington University School of Medicine, St Louis MO 63110, USA
Anthony R. French, Division of Pediatric Rheumatology, Department of Pediatrics, Washington University School of Medicine, St Louis MO 63110, USA
Beatrice Plougastel, Division of Rheumatology, Washington University School of Medicine, St Louis MO 63110, USA.
Jeanette T. Pingel, Howard Hughes Medical Institute Washington University School of Medicine, St Louis MO 63110, USA
Michael M. Orihuela, Department of Medicine, Washington University School of Medicine, St Louis MO 63110, USA
Mark L. Buller, St. Louis University Health Sciences Center, Department of Molecular Microbiology and Immunology, 1402 S Grand Blvd, St. Louis, MO 63104, USA
Wayne M. Yokoyama, Division of Rheumatology, Washington University School of Medicine, St Louis MO 63110, USA Howard Hughes Medical Institute Washington University School of Medicine, St Louis MO 63110, USA.
REFERENCE
- Adam SG, Caraux A, Fodil-Cornu N, Loredo-Osti JC, Lesjean-Pottier S, Jaubert J, Bubic I, Jonjic S, Guenet JL, Vidal SM, Colucci F. Cmv4, a new locus linked to the NK cell gene complex, controls innate resistance to cytomegalovirus in wild-derived mice. J Immunol. 2006;176:5478–5485. doi: 10.4049/jimmunol.176.9.5478. [DOI] [PubMed] [Google Scholar]
- Bakker AB, Hoek RM, Cerwenka A, Blom B, Lucian L, McNeil T, Murray R, Phillips LH, Sedgwick JD, Lanier LL. DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity. 2000;13:345–353. doi: 10.1016/s1074-7613(00)00034-0. [DOI] [PubMed] [Google Scholar]
- Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001a;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- Brown MG, Scalzo AA, Stone LR, Clark PY, Du Y, Palanca B, Yokoyama WM. Natural killer gene complex (Nkc) allelic variability in inbred mice: evidence for Nkc haplotypes. Immunogenetics. 2001b;53:584–591. doi: 10.1007/s002510100365. [DOI] [PubMed] [Google Scholar]
- Delano ML, Brownstein DG. Innate resistance to lethal mousepox is genetically linked to the NK gene complex on chromosome 6 and correlates with early restriction of virus replication by cells with an NK phenotype. J Virol. 1995;69:5875–5877. doi: 10.1128/jvi.69.9.5875-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe A, Rodriguez M, Sabastian P, Xie X, McVoy M, Brown MG. Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice. J Immunol. 2005;175:6820–6828. doi: 10.4049/jimmunol.175.10.6820. [DOI] [PubMed] [Google Scholar]
- Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- French AR, Pingel JT, Wagner M, Bubic I, Yang L, Kim S, Koszinowski U, Jonjic S, Yokoyama WM. Escape of mutant double-stranded DNA virus from innate immune control. Immunity. 2004;20:747–756. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol. 2003;15:45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, Zakay-Rones Z, Porgador A, Mandelboim O. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- Gonczol E, Danczig E, Boldogh I, Toth T, Vaczi L. In vivo model for the acute, latent and reactivated phases of cytomegalovirus infection. Acta Microbiol Hung. 1985;32:39–47. [PubMed] [Google Scholar]
- Grundy JE, Mackenzie JS, Stanley NF. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect Immun. 1981;32:277–286. doi: 10.1128/iai.32.1.277-286.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson TJ, Church DM, Greenaway S, Nguyen H, Cook A, Steen RG, Van Etten WJ, Castle AB, Strivens MA, Trickett P, Heuston C, Davison C, Southwell A, Hardisty R, Varela-Carver A, Haynes AR, Rodriguez-Tome P, Doi H, Ko MS, Pontius J, Schriml L, Wagner L, Maglott D, Brown SD, Lander ES, Schuler G, Denny P. A radiation hybrid map of mouse genes. Nat Genet. 2001;29:201–205. doi: 10.1038/ng1001-201. [DOI] [PubMed] [Google Scholar]
- Kaifu T, Nakahara J, Inui M, Mishima K, Momiyama T, Kaji M, Sugahara A, Koito H, Ujike-Asai A, Nakamura A, Kanazawa K, Tan-Takeuchi K, Iwasaki K, Yokoyama WM, Kudo A, Fujiwara M, Asou H, Takai T. Osteopetrosis and thalamic hypomyelinosis with synaptic degeneration in DAP12-deficient mice. J Clin Invest. 2003;111:323–332. doi: 10.1172/JCI16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- Lee SH, Zafer A, de Repentigny Y, Kothary R, Tremblay ML, Gros P, Duplay P, Webb JR, Vidal SM. Transgenic expression of the activating natural killer receptor Ly49H confers resistance to cytomegalovirus in genetically susceptible mice. J Exp Med. 2003;197:515–526. doi: 10.1084/jem.20021713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen KL, Lohwasser S, Takei F, Mager DL. Expression analysis of new Ly49 genes: most transcripts of Ly49j lack the transmembrane domain. Immunogenetics. 1999;49:685–691. doi: 10.1007/s002510050665. [DOI] [PubMed] [Google Scholar]
- Proteau MF, Rousselle E, Makrigiannis AP. Mapping of the BALB/c Ly49 cluster defines a minimal natural killer cell receptor gene repertoire. Genomics. 2004;84:669–677. doi: 10.1016/j.ygeno.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD, Zeng F, Farrell HE, Cunningham AL, Scalzo AA, Booth TW, Scott GM. The murine cytomegalovirus (MCMV) homolog of the HCMV phosphotransferase (UL97(pk)) gene. Virology. 1997;233:358–363. doi: 10.1006/viro.1997.8593. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Sabastian P, Clark P, Brown MG. Cmv1-independent antiviral role of NK cells revealed in murine cytomegalovirus-infected New Zealand White mice. J Immunol. 2004;173:6312–6318. doi: 10.4049/jimmunol.173.10.6312. [DOI] [PubMed] [Google Scholar]
- Scalzo AA, Fitzgerald NA, Simmons A, La Vista AB, Shellam GR. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J Exp Med. 1990;171:1469–1483. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalzo AA, Lyons PA, Fitzgerald NA, Forbes CA, Yokoyama WM, Shellam GR. Genetic mapping of Cmv1 in the region of mouse chromosome 6 encoding the NK gene complex-associated loci Ly49 and musNKR-P1. Genomics. 1995;27:435–441. doi: 10.1006/geno.1995.1074. [DOI] [PubMed] [Google Scholar]
- Smith KM, Wu J, Bakker AB, Phillips JH, Lanier LL. Ly-49D and Ly-49H associate with mouse DAP12 and form activating receptors. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- Wilhelm BT, Gagnier L, Mager DL. Sequence analysis of the ly49 cluster in C57BL/6 mice: a rapidly evolving multigene family in the immune system. Genomics. 2002;80:646–661. doi: 10.1006/geno.2002.7004. [DOI] [PubMed] [Google Scholar]