Abstract
S-Adenosylmethionine decarboxylase (AdoMetDC) is a key enzyme in the polyamine biosynthetic pathway. Inhibition of this pathway and subsequent depletion of polyamine levels is a viable strategy for cancer chemotherapy and for the treatment of parasitic diseases. Substrate analogue inhibitors display an absolute requirement for a positive charge at the position equivalent to the sulfonium of S-adenosylmethionine. We investigated the ligand specificity of AdoMetDC through crystallography, quantum chemical calculations and stopped-flow experiments. We determined crystal structures of the enzyme cocrystallized with 5′-deoxy-5′-dimethylthioadenosine and 5′-deoxy-5′(N-dimethyl)amino-8-methyl adenosine. The crystal structures revealed a favorable cation-π interaction between the ligand and the aromatic side chains of Phe7 and Phe223. The estimated stabilization from this interaction is 4.5 kcal/mol as determined by quantum chemical calculations. Stopped-flow kinetic experiments showed that the rate of the substrate binding to the enzyme greatly depends on Phe7 and Phe223, thus supporting the importance of the cation-π interaction.
The polyamines putrescine, spermidine and spermine are aliphatic polycations that are critical for maintaining cell differentiation and proliferation (1–3). Elevated levels of polyamines are found in cancerous and tumor cell lines (4, 5). Thus, depleting polyamine levels by inhibition of the polyamine biosynthetic pathway is a promising approach for the treatment and prevention of cancer and also for the treatment of various parasitic diseases. S-adenosylmethionine decarboxylase (AdoMetDC) is a key enzyme in the polyamine biosynthetic pathway and depends on a pyruvoyl cofactor for the decarboxylation reaction (6–9). AdoMetDC catalyzes the decarboxylation of S-adenosylmethionine (AdoMet) to S-adenosyl-5′-(3-methylthiopropylamine) (dcAdoMet). The aminopropyl group from dcAdoMet is transferred to putrescine or spermidine to form spermidine or spermine, respectively. AdoMetDC catalyzes an early step in the pathway and dcAdoMet is completely committed to polyamine biosynthesis; thus, AdoMetDC is an attractive target for drug design.
The early inhibitors of AdoMetDC included the potent competitive inhibitor methylglyoxal bisguanylhydrazone (MGBG) (10). Clinical studies on this compound were hampered by an unexpected mitochondrial toxicity unrelated to its inhibition of AdoMetDC. An MGBG analogue, 4-amidinoindan-l-one-2′-amidinohydrazone (CGP48664A) showed promise in multiple phase I and phase II clinical trials (11–18). Irreversible (or slowly reversible) substrate analogue inhibitors, which form a Schiff base with the active site pyruvoyl group, have also been synthesized but are limited by their nonspecific activity towards cellular aldehydes and ketones (19). In vitro assays showed that a positive charge at the position of the sulfonium ion is essential for ligand binding and inhibition (20). Thioether and sulfoxide substrate analogues, which lack the positive charge, showed no activity. On the other hand, replacement of a sulfur atom with a nitrogen atom, which is protonated at physiological pH and retains the positive charge, resulted in AdoMetDC inhibition. This is consistent with the observation that S-adenosylhomocysteine (SAH) is not a substrate for AdoMetDC (21).
Previously, the structure of AdoMetDC and its complexes with inhibitors 5′-deoxy-5′-[N-methyl-N-(3-hydrazinopropyl)amino]adenosine (MHZPA), 5′-deoxy-5′-[N-methyl-N-[(2-aminooxy)ethyl]amino]adenosine (MAOEA), the methyl ester of AdoMet (MeAdoMet), MGBG and CGP48664A were determined (22). MGBG and CGP48664A act as competitive inhibitors of the enzyme and stack between Phe7 and Phe223, and form hydrogen bonds with Glu247, Ser229 and the backbone amide of Leu65. The substrate analogues MHZPA, MAOEA and MeAdoMet have positive charges at the sulfonium ion position and covalently bind to the enzyme acting as slowly reversible inhibitors. The adenine base of these inhibitors stacks between Phe7 and Phe223, the glycosidic bond adopts an unusual syn conformation, and both ribose hydroxyl groups hydrogen bond to Glu247. The requirement for a positive charge in substrate analogues remained puzzling because no negatively charged amino acid side chain was located nearby.
We chose to investigate the basis of the ligand specificity of AdoMetDC using crystallography, quantum chemical calculations and stopped-flow kinetic experiments. We determined crystal structures of the enzyme co-crystallized with 5′-deoxy-5′-(dimethylsulfonio)adenosine (MMTA) and 5′-deoxy-5′-(N-dimethyl)amino-8-methyladenosine (DMAMA). The energy difference between the syn and anti conformation of the ligands in solution and in the active site of the enzyme was obtained using quantum chemical calculations. Stopped-flow kinetic experiments were carried out to investigate the importance of Phe7 and Phe223, two residues located near the positive charge. Our results show that ligand specificity in human AdoMetDC is mainly due to cation-π interactions and to electrostatic interactions between N3 of adenine and the sulfonium ion, which stabilizes the syn conformation.
MATERIALS AND METHODS
Materials
The syntheses of MMTA and DMAMA were previously reported (23) and the compounds were the gift of Dr. Jack Secrist at Southern Research Institute.
Protein Expression and Purification
The gene encoding AdoMetDC was cloned into a pQE30 vector and transformed into JM109 E. coli cells. Cells were grown in LB media supplemented with 100 mg/mL ampicillin at 37 °C until the absorbance reached an O.D600 of 0.6, at which point the cells were induced with 100 mg/L isopropyl-l-β-D galactopyranoside and the temperature was reduced to 15 °C. The cells were allowed to grow overnight before being harvested by centrifugation, washed using a lysis buffer containing 20 mM Na2HP04, pH 7.0, 500 mM NaCl, 2.5 mM putrescine, 0.02% polyoxyethyleneglycol dodecyl ether (Brij-35) and 10 mM imidazole, and stored at −80°C. The frozen cell pellet was resuspended in the lysis buffer and lysed using a French press at 1500 psi. The cellular debris was separated from the lysate by centrifugation at 12000 g. The cell lysate was incubated with Talon metal affinity resins for 1.5 h. The protein-resin complex was packed into a column and washed with 15–20 column volumes of wash buffer, which is the lysis buffer supplemented with 25 mM imidazole. The protein was eluted with the wash buffer containing 200 mM imidazole. The elute was further purified through a Superdex 75 column using a buffer containing 10 mM N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.5, 2.5 mM putrescine, 5 mM DTT, 0.1 mM ethylene diamine tetraacetic acid (EDTA), 0.02 % Brij-35 and 300 mM NaCl. The fractions containing the protein were pooled, concentrated to 10 mg/mL and stored at −80 °C. The construction, expression, and purification of the mutants were described previously (22, 23).
Crystallization
The protein was buffer exchanged into 10 mM HEPES, pH 7.5, 200 mM NaCl and 1 mM DTT using Bio-Rad buffer exchange chromatography columns. The protein was incubated separately with a 4–6 molar excess of MMTA and DMAMA for 24 h prior to crystallization. The crystals were grown using the hanging drop method at 22 °C in 13–16% polyethylene glycol 8000, 100 mM tris(hydroxymethyl)aminomethane, pH 8.0–9.0 and 10 mM DTT. Crystals appeared overnight and were stable for 1–2 weeks but deteriorated after that.
Data Collection and Processing
The crystals were sequentially transferred to solutions containing the well solution with 2%, 5%, 8%, 15% and 18% glycerol with 1–2 min equilibration between each step. The crystals were flash frozen under liquid nitrogen before being placed in the liquid nitrogen stream. The data for the complex of AdoMetDC with MMTA were collected at NE-CAT beamline 8-BM at the Advanced Photon Source using a ADSC Quantum 315 detector (Area Detector Systems Corporation). Data were collected over a rotation range of 200° with an oscillation range of 1° and 60 s exposure per frame with a detector to crystal distance of 320 mm. The data for the complex of DMAMA were collected at the NE-CAT beamline 24-ID-C. Data were collected over a rotation range of 200° with an oscillation range of 1° and 1 s exposure per frame with a detector to crystal distance of 250 mm. The data for the complexes were indexed, integrated and scaled using the HKL2000 program suite (24). The data collection statistics for both complexes are summarized in Table 1.
Table 1.
Data Collection Statistics for AdoMetDC Complexes
| AdoMetDC + MMTA | AdoMetDC + DMAMA | |
|---|---|---|
| Wavelength (Å) | 0.9795 | 0.9795 |
| Space Group (Å) | C2 | C2 |
| a(Å) | 93.85 | 99.45 |
| b(Å) | 49.54 | 50.02 |
| c(Å) | 70.00 | 68.69 |
| β | 105.03 | 105.32 |
| Resolution (Å) | 2.24 | 1.81 |
| Total/Unique reflectionsa | 56742/15123 | 107489/29093 |
| Redundancy | 3.9(4.0) | 3.7(2.5) |
| % complete | 95.1(99.4) | 95.8(73.5) |
| I/σ | 8.9(4.26) | 17.7(3.4) |
| Rsymb | 10.5(31.7) | 7.9(21.5) |
| Matthews no | 2.05 | 2.19 |
| Solvent content | 38.9 | 42.9 |
Values for the highest resolution shell are given in parentheses.
Rsym = ΣΣi | Ii − <I> |/Σ<I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h, k, l.
Structure Determination and Refinement
The structures of the complexes were determined by molecular replacement with CNS (25) using the structure of the AdoMetDC/MeAdoMet complex (PDB code 1I7B) as the search model. Model building for the complex of MMTA was performed using the program O (26). The model building for the complex of DMAMA was performed using the program Coot (27). The initial model obtained from molecular replacement was adjusted using composite omit maps and refined using successive rounds of simulated annealing, minimization, B-factor refinement, generation of new composite omit maps, difference Fourier maps and model building. After a few rounds of refinement, the positions and the conformations of the ligand molecules were identified using the improved difference Fourier maps and composite omit maps. The ligands were included in the models and water molecules were added based on the peaks in the difference Fourier maps. The parameter and the topology files for the ligands were generated using the HIC-Up server (28). The difference maps also showed density for a molecule of putrescine bound in each of the structures. The final refinement statistics for both complexes are given in Table 2.
Table 2.
Refinement Statistics for AdoMetDC Complexes
| AdoMetDC+MMTA | AdoMetDC+DMAMA | |
|---|---|---|
| Resolution (Å) | 2.24 | 1.81 |
| R factora | 0.226 | 0.193 |
| Rfreeb | 0.274 | 0.218 |
| No of non-H atoms | ||
| Protein | 2405 | 2439 |
| Ligand | 21 | 22 |
| Water | 76 | 234 |
| B-factors | ||
| Protein (Å2) | 32.6 | 29.3 |
| Ligand (Å2) | 43.1 | 22.8 |
| Putrescine (Å2) | 35.0 | 47.7 |
| rms deviations | ||
| bonds (Å) | 0.006 | 0.008 |
| angles (°) | 1.3 | 1.3 |
| dihedrals (°) | 25.1 | 25.3 |
| Ramachandran plot | ||
| Most favored region (%) | 90.7 | 92.0 |
| Additional favored region (%) | 8.5 | 6.9 |
| Generously allowed region (%) | 0.8 | 0.8 |
| Disallowed region (%) | 0.0 | 0.4 |
R factor = Σhkl||Fobs|−k|Fcal|/Σhkl|Fobs|, where Fobs and Fcal are observed and calculated structure factors respectively.
For Rfree, the sum is extended over a subset of reflections (5%) excluded from all stages of refinement.
Quantum Mechanical Calculations
The quantum chemical calculations on the cation-π interactions were performed using Jaguar version 6.0 or 6.5 (Schrödinger). The X-ray structure of MMTA bound to AdoMetDC was employed for these calculations and single point energy calculations were performed using the LMP2/6-31G** method on the following: (CH3)3S+ (from MMTA) plus two benzene rings (from Phe7 and Phe223); (CH3)3S+ (from MMTA) plus one benzene ring (from Phe223); (CH3)3S+ (from MMTA) plus one benzene ring (from Phe7); (CH3)3S+ alone; benzene alone; (CH3)2S (from MMTA) plus two benzene rings (from Phe223 and Phe7); (CH3)2S alone. The level of theory for the calculations is sufficient to account for the polarization effect and hence cation-π interactions (29).
The energies and conformations of MMTA and 5′-deoxy-5′-methyl thioadenosine (MTA) in solution were determined starting with the NMR structure of AdoMet in solution (30) and truncating the molecule to MMTA or MTA. The syn conformation was obtained by adjusting the O-C1′-N9-C4 torsion angle. The structures were subjected to geometry optimization using the B3LYP/6-31G* density functional method and the SCRF implicit aqueous solvation model available in Jaguar. The single point energy of the geometry optimized structures was then obtained using the Local Moller-Plesset second-order perturbation (LMP2)/cc-PVTZ(-f) method with aqueous self-consistent reaction field (SCRF) solvation.
The energies of the syn and anti conformations of MMTA in complex with the enzyme were determined using the MMTA complex as the basis. The truncated model for the syn conformation consisted of MMTA and residues Phe223 and Phe7. Backbone atoms of the adjacent residues were also included in the calculations. In addition, an acetate moiety derived from Glu247 was included. Hydrogen atoms were added to the model. Constrained geometry optimization was performed at the Hartree- Fock (HF)/6–31G** level and the single point energy was then calculated at the LMP2//6–31G** level. All atoms were constrained during the geometry optimization except the adenine ring, the ribose C1′ atom and the ribose hydroxyl groups. The anti conformation was generated from this truncated model by adjusting the O-C1′-N9-C4 torsion angle to −140° followed by geometry optimization as described above. The partial charges after geometry optimization were obtained for each of atom.
Stopped-Flow Experiments
The stopped-flow experiments were performed using a KinTek Stopped-Flow apparatus (Model SF-2004, KinTek Corp., Austin, TX). Experiments were carried out at 25 °C in 10 mM HEPES, pH 7.5, 200 mM NaCl, and 1 mM DTT. The time dependence of binding was measured by monitoring changes in the intrinsic protein fluorescence. An excitation wavelength of 298 nm (rather than the excitation maximum of ~280 nm) was used to avoid problems associated with inner filtering caused by increasing the ligand concentration. Emission was observed using a band-pass filter centered at 340 nm (±10 nm). All reported concentrations refer to the final value after rapidly mixing ligand with enzyme at a ratio of 1:1 (v/v). The final enzyme concentration was 25 μM for the WT protein, F7A and E247A mutants and 100 μM for the F223A mutant. AdoMet was mixed with enzyme in at least a 4-fold excess in order to maintain pseudo-first order conditions. Linear and nonlinear regression of the data were performed using GraFit 5 (Erithacus Software, Horley, Surrey, UK).
Figure Preparation
Structural figures were generated using Pymol (31).
RESULTS
Cocrystallization of AdoMetDC with Ligands
The enzyme was cocrystallized in the presence of SAH, MMTA, DMAMA and MTA. The crystallization experiments show that only MMTA and DMAMA bound to the enzyme in the crystal while MTA and SAH did not. Cocrystallization experiments for MTA and SAH were performed at increased concentrations and incubation times to increase the possibility of binding at the active site; however, no active site density was observed.
Crystal Structure of AdoMetDC with MMTA
The overall fold of AdoMetDC is a four-layer αββα sandwich as previously described (32). AdoMetDC autoprocessing results in an α subunit with an N-terminal pyruvoyl group and a smaller β subunit (32, 33). One molecule of putrescine is bound between the β sheets for each monomer and is located about 15–20 Å from the active site.
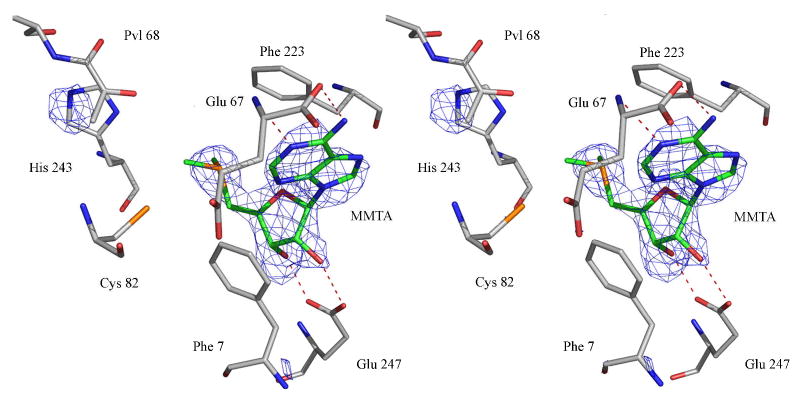
In the crystal structure of AdoMetDC with MMTA, loops containing residues 1–4, 22–26, 165–172, 288–299, 328–334 were disordered and were excluded from the model. MMTA ribose forms hydrogen bonds to Glu247; the O2′-Oε2 distance is 3.2 Å and the O3′-Oε1 distance is 2.7 Å. The sulfonium sulfur atom is 4.4 Å from the center of Phe233 ring and the closest contact of sulfur atom with Phe223 is 3.9 Å (Cε1). The methylene carbon atom adjacent to the sulfonium ion of MMTA is 4.4 Å from the center of Phe7 and 3.6 Å from the Cε2 carbon of Phe7. The glycosidic bond of MMTA adopts a syn conformation and the adenine ring stacks between Phe223 and Phe7. The stereoview of the electron density for MMTA in the active site is shown in Figure 1.
Figure 1.
Stereoview of the complex of MMTA with AdoMetDC. The difference Fo-Fc Fourier density is contoured at 2.5σ. The carbon atoms of the ligand are colored green. The hydrogen bonds are shown as red dashed lines.
Crystal Structure of AdoMetDC with DMAMA
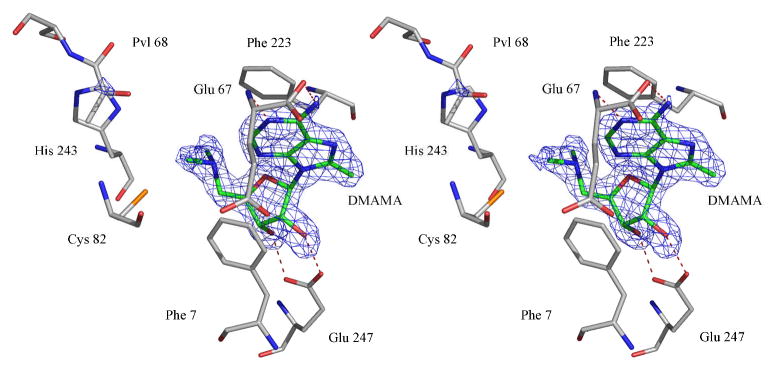
In the structure of AdoMetDC with DMAMA, the loops containing the residues 1–3, 24–26, 165–173, 288–299, 328–334 were missing in the crystal structure. The ribose makes two hydrogen bonds to Glu247 with the O2′-Oε2 distance being 2.9 Å and the O3′-Oε1 distance being 2.6 Å. The nitrogen is 4.7 Å from the center of Phe223 and 4.2 Å from the Cε1 atom of Phe223. The methylene carbon atom adjacent to the nitrogen atom is at a distance of 4.3 Å from the center of Phe7 and 3.7 Å from the Cε2 carbon of Phe7. The adenosine moiety of DMAMA is in a syn conformation with the adenine base stacking between Phe7 and Phe223. A stereoview of the electron density for DMAMA in the active site is shown in Figure 2.
Figure 2.
Stereoview of the complex of DMAMA with AdoMetDC. The difference Fo-Fc Fourier density is contoured at 4σ. The carbon atoms of the ligand are colored green. The hydrogen bonds are shown as red dashed lines.
Quantum Mechanical Calculations
The LMP2/6-31G** energies from the quantum mechanical calculations are shown in Table 3. The calculated gas phase binding energy of (CH3)3S+ between two benzene rings in the same geometrical orientations as Phe7 and Phe223 is −5.09 kcal/mol. On the other hand, the binding energy of (CH3)2S between two benzene rings in the same geometrical orientations as Phe7 and Phe223 is only −0.60 kcal/mol. These calculations suggest that the cation-π interaction provides an addition stabilization of approximately −4.5 kcal/mol. Similar calculations suggest that the binding energy of (CH3)3S+ to Phe223 is −3.15 kcal/mol and the binding energy to Phe7 is −3.41 kcal/mol.
Table 3.
Quantum Chemical Energies of Residues/Ligands
| Energy in hartrees | |
|---|---|
| Benzene | −231.47989407141 |
| (CH3)2S | −477.13908367149 |
| (CH3)2S + Phe223 + Phe 7 | −940.10191861550 |
| (CH3)3S+ + Phe223 | −748.08525375049 |
| (CH3)3S+ + Phe7 | −748.08567703979 |
| (CH3)3S+ | −516.60034059742 |
| (CH3)3S+ + Phe223 + Phe 7 | −979.57032658617 |
The ab inito energy of the syn conformation of MMTA in the complex with Phe223, Phe7 and Glu247 is −3039.28899965619 hartrees and the energy of the anti conformation is −3039.26998403002 hartrees. Thus, the energy difference between the syn and anti conformations of MMTA in the active site is −11.93 kcal/mol favoring the syn conformation. This would represent the stabilization of the syn conformation caused primarily by Phe223 and Phe7.
We also investigated the conformational energetics of MMTA and MTA in aqueous solution using quantum chemical calculations. The results are in agreement with the experimental results described by Markham et al. (30) These compounds prefer an anti conformation in aqueous solution. The LMP2/cc-PVTZ(-f)//B3LYP/6-31G* calculated energy difference between the anti and syn conformations was 1.06 kcal/mol for MMTA and 0.88 kcal/mol for MTA favoring the anti conformation in each case.
Stopped-Flow Experiments
An intrinsic fluorescence change was observed when AdoMetDC was rapidly mixed with AdoMet. However, when SAH was mixed with AdoMetDC, there was no significant change other than that determined to be due to photobleaching (compared to a control minus SAH). This indicates that SAH binding is much weaker and/or produces no discernable conformational change. The signal when MMTA was mixed with AdoMetDC was also too weak to measure the kinetics of the binding reaction.
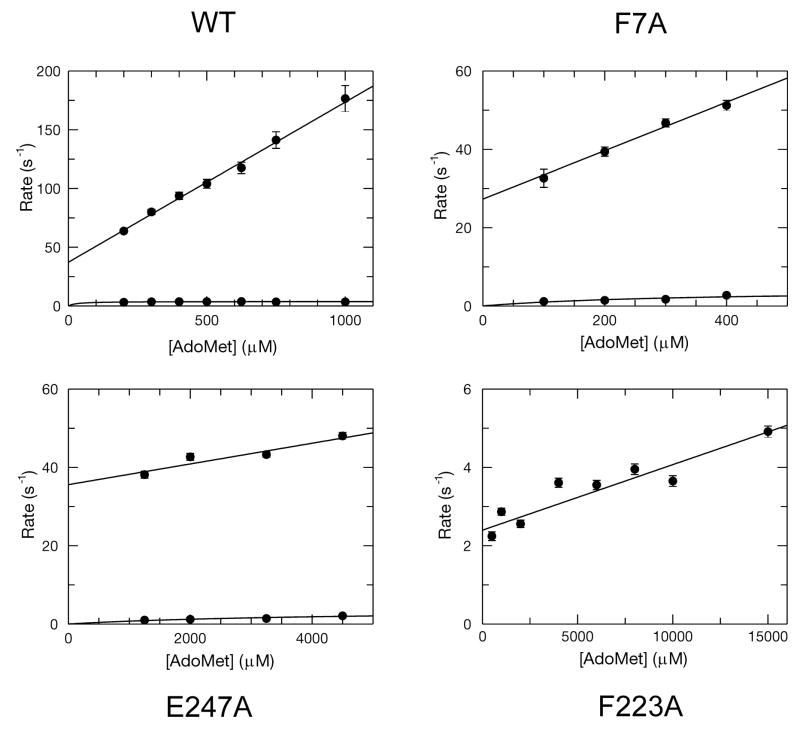
Kinetic data of AdoMet binding to WT AdoMetDC along with the F223A, F7A and E247A mutants were collected. For WT, F7A and E247A the data were best fit to a double exponential equation
to yield the observed rates (λ1 and λ2) and amplitudes (A1 and A2) of the two phases at each concentration of AdoMet. The concentration dependence of the fast and slow phases are plotted in Figure 3. The fast phase exhibited a linear concentration dependence, and did not saturate. The slow phase was approximately hyperbolic and saturated at relatively low concentrations. The data are best described by a minimal model of two sequential steps:
Figure 3.
Plots of observed rate vs. [AdoMet] for the WT, F223A, F7A and E247A mutants. The linear concentration dependence of the observed rates was fit using a line which allowed for the definition of k1 and k−1. A hyperbolic fit of the slow phase defined the sum of k2 and k−2, except in the case of F223A, where the kinetics were monophasic and indicative of a more simple one step binding model. In this case the slope of the best fit line defines the k1 and the y-intercept defines the k−1 directly.
According to this model, the fast phase of the reaction occurs at an observed rate approximately equal to the sum of all four intrinsic rate constants (34):
A linear fit of the fast phase (AdoMet binding to WT enzyme) results in a k1 = 0.136 ± 0.005 μM−1s−1, k−1 = 33 ± 2 s−1 and k2 + k−2 = 3.7 ± 0.2 s−1. Therefore, the dissociation constant (Kd) for AdoMet binding to AdoMetDC can be estimated to be 242 ± 17 μM from the ratio of k−1/k1.
For the F7A and E247A mutants, the data were analyzed in a similar fashion except that a lower overall signal change caused photobleaching to become a significant component of the data. Therefore, the decrease due to photobleaching was corrected by using data from a control reaction where substrate was omitted (resulting in a linear decrease in fluorescence). After correction, the kinetic parameters were obtained as for the WT enzyme. The values of k−1 and k2 + k−2 were similar for both the mutants but the value of the second order rate constant for substrate binding (k1) changed significantly. Based upon the values of k1 and k−1, the Kd for the F7A mutant was 370 ± 60 μM and the Kd for the E247A mutant was 12.2 ± 3.6 mM.
The fluorescence change seen upon mixing AdoMet with the F223A mutant was best fit using a single exponential equation. The concentration dependence of the observed rate varied linearly with a slope (k1) of 1.7 ± 0.3 × 10−4 μM−1s−1 and a y-intercept (k−1) of 2.4 ± 0.2 s−1 yielding a Kd of 14 ± 3 mM. Generally, the values measured here follow the same trend as the steady state kinetic parameters (kcat and Km) obtained previously (22). A comparison of Kd values for AdoMet binding to WT, F223A, F7A, and E247A mutants reveals that the importance of residues for substrate binding follows the order F223A ~ E247A ≫ F7A > WT.
DISCUSSION
Cation-π Interactions and Ligand Specificity of AdoMetDC
Cation-π interactions are ubiquitous in nature and aid in protein stability, ligand recognition, catalysis and ion channel function (35, 36). In the gas phase, the binding energy of cations to aromatic groups ranges from 10–40 kcal/mol which places the cation-π interaction among the strongest noncovalent forces (35, 37). The magnitude of the cation-π interaction depends on the geometry, distance and the nature of the cation and the aromatic group. In biological systems, the bulk of these interactions are seen with the amino acid side chains of proteins. Aromatic amino acids such as tryptophan, tyrosine and phenylalanine interact with positive charged amino acids such as lysine and arginine. The energetics of these interactions have been studied both experimentally and theoretically (38, 39). In a few cases, ligand recognition by an enzyme is attributed completely to the cation-π interaction (40).
It has been shown that AdoMetDC binds substrate analogues only if they have a positive charge at the sulfonium position (20). MHZPA and MAOEA, which do not contain a sulfonium center, have an ammonium group that is protonated at physiological pH. The inability to form MTA and SAH complexes in cocrystallization experiments is consistent with this observation.
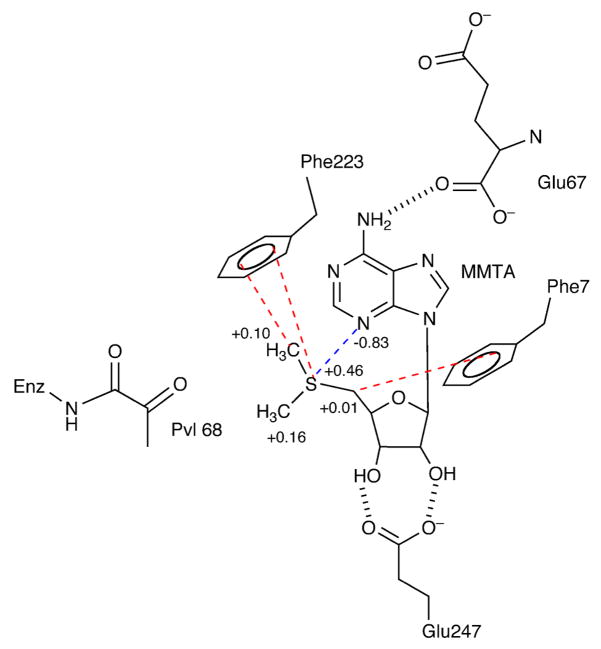
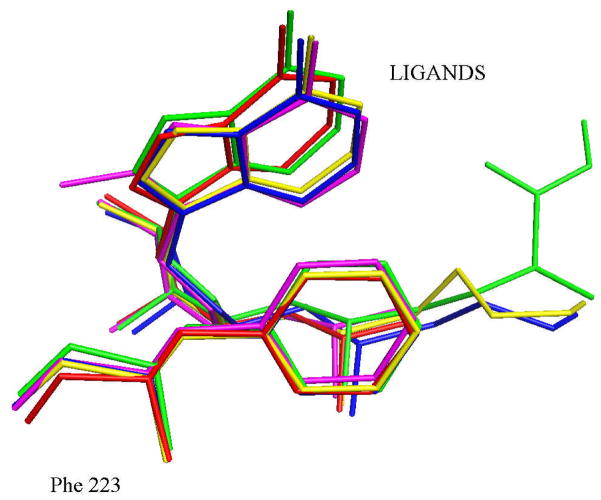
The results reported here suggest that cation-π interactions play a primary role in determining substrate specificity. The crystal structure of AdoMetDC with MMTA shows that the sulfonium center is at a favorable distance and geometry for a cation-π interaction with Phe223. The positive charge of the sulfonium ion is distributed to the adjacent methyl and the methylene groups (Figure 4). The methylene group of MMTA is at a favorable distance and geometry for a cation-π interaction with Phe7. The quantum chemical calculations suggest the stabilization of a (CH3)3S+ group in the active site is primarily due to its interaction with Phe223 and Phe7. Other ligands with a positive charge such as MHZPA, MAOEA, MeAdoMet and DMAMA show similar interactions with Phe223 and Phe7 (Figure 5). The magnitude of stabilization obtained due to the cation-π interaction is approximately −4.5 kcal/mol. The interaction may be weakened from its maximum value by distribution of positive charge to adjacent carbon and hydrogen atoms and because of non-ideal geometry.
Figure 4.
Schematic diagram of the key interactions MMTA makes in the active site of AdoMetDC. The adenine base stacks in the syn conformation aided by aromatic stacking interactions to Phe7 and Phe223. The N6 of the adenine base hydrogen bonds to the backbone carbonyl of Glu67. The sulfonium ion and the terminal methyl group have a cation-π interaction with Phe223 (shown in red) and the partially positively charged methylene group adjacent to sulfonium interacts with Phe7. The N3 atom is partially negative charged and interacts with the sulfonium. The partial charges on the relevant atoms are indicated. The ribose makes two hydrogen bonds to Glu247.
Figure 5.
The superposition of the complexes of AdoMetDC showing the geometry of the cation-π interaction between the positive sulfonium/nitrogen with Phe223. The color coding for the complexes is as follows; MMTA, red; DMAMA, magenta; MeAdoMet, green; MAOEA, blue; MHZPA, yellow.
The cation-π interaction as a theme for sulfonium recognition was debated by Markham et al. based on a survey of the crystal structures of AdoMet with various enzymes (30). The survey which spanned 20 crystal structures showed that the sulfonium of AdoMet interacts with negatively charged atoms and aromatic amino acids in very few cases. In glycine-N-methyltransferase (PDB code 1xva) and HhaI DNA methylase (PDB code 1hmy), the sulfonium interacts with negatively charged carboxylate atoms (41, 42). There are only two cases in the survey where the sulfonium ion has a close contact with an aromatic group. The sulfonium ion of AdoMet is 3.2 Å from the methylene carbon atom of Trp41 in HhaI DNA methylase (PDB code 1hmy) and in the structure of AdoMetDC with MeAdoMet bound (PDB code 1i7b) the sulfonium ion is located 4.1 Å from the center of Phe223. The survey concluded that in a broader picture, cation-π interactions are not a recurring theme for sulfonium recognition. However, the uniqueness of the AdoMet decarboxylation reaction in this study may explain the importance of cation-π interaction for AdoMetDC, which further is supported by theoretical calculations and stopped-flow kinetic experiments.
Stabilization of the syn Conformation by AdoMetDC
The conformational preference of AdoMet in solution and gas phase was studied by Markham et al. (30). According to the NMR studies, AdoMet prefers an anti conformation about the glycosidic bond in solution and a syn conformation in vacuum. The energy difference between the anti and syn conformations in solution was calculated to be −34 kcal/mol based on the modeling studies using NMR constraints. This energy difference was calculated based on molecular mechanics without polarization effects and it is likely that the actual difference is less negative (23). The crystal structure of AdoMetDC with MeAdoMet, MHZPA and MAOEA reported previously showed that the enzyme binds the ligands in the energetically unfavorable syn conformation (22).
The crystal structures of AdoMetDC with MMTA and DMAMA also show that the adenosine moiety binds in a syn conformation. The quantum chemical calculations for MMTA in a truncated model of the enzyme with only Phe7, Phe223 and Glu247 included suggest that the syn conformation of the ligand is stabilized by −12 kcal/mol compared to hypothetical anti conformation, which has never been observed in an AdoMetDC complex. The actual energy difference is likely to be greater than the above value since the anti conformation places the six-membered ring of the adenine in close proximity to Asn224 and Pro225. Moreover, the amino group on the adenine ring is in close proximity and makes hydrogen bonds to Glu67 in the syn conformation but forms no appreciable interactions with the enzyme in the anti conformation.
The syn conformation of the adenine base is primarily stabilized by stacking interactions with Phe7 and Phe223 and hydrogen bonds of the ribose to Glu247. The N3 atom of the adenine base carries a partial negative charge and interacts favorably with the sulfonium ion. This electrostatic effect further contributes in stabilizing the syn conformation of the ligand. The preference of AdoMetDC to bind ligands in the syn conformation was recently exploited in the design of substrate analogues with enhanced affinity. Substitution at the C8 position on the adenine base favored the syn conformation resulting a 5–18 fold increase in potency compared to the unsubstituted compounds (23).
Insights into Inhibitor Design
The design of substrate analogue inhibitors for AdoMetDC benefit from the presence of a positive charge required for the cation-π interaction. Unlike MAOEA and MHZPA, MMTA shows competitive inhibition because it lacks an amino terminus required to form a Schiff base with the active site pyruvoyl group. Studies on MMTA also show that the ribose, adenine base and the positive charge are sufficient for inhibition of AdoMetDC. The schematic diagram showing all of the stabilizing interactions of MMTA in the active site is shown in Figure 4.
The MMTA analog, DMAMA shows that the replacement of the sulfur by nitrogen also yields a competitive inhibitor because the nitrogen is protonated at physiological pH. DMAMA has a methyl substitution at the 8-position which should favor the syn conformation in solution and improve the binding to AdoMetDC (23). Because MMTA and DMAMA lack hydrazino or oxyamino groups, which react nonspecifically with cellular aldehydes and ketones, and because further evolution of their structures are possible, MMTA and DMAMA, with IC50 values of 15 μM and 600 nM (23), respectively, are promising lead compounds for inhibitor design.
Acknowledgments
We thank Ms. Leslie Kinsland for assistance in the preparation of this manuscript. We thank the Protein Production Facility, a Life Sciences Core Laboratory Center at Cornell University, for use of the stopped flow apparatus. This work is based upon research conducted at beamlines 24-ID-C and 8-BM of the Northeastern Collaborative Access Team of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
ABBREVIATIONS
- AdoMetDC
S-adenosylmethionine decarboxylase
- AdoMet
S-adenosylmethionine
- MeAdoMet
S-adenosylmethionine methyl ester
- dcAdoMet
S-adenosyl-5′-(3-methylthiopropylamine)
- SAH
S-adenosylhomocysteine
- MMTA
5′-deoxy-5′-(dimethylsulfonio)adenosine
- MTA
5′-deoxy-5′-methyl thioadenosine
- DMAMA
5′-deoxy-5′-(N-dimethyl)amino-8-methyl adenosine
- MGBG
methylglyoxal bis(guanylhydrazone)
- CGP48664A
4-amidinoindan-1 -one-2′-amidinohydrazone
- MAOEA
5′-deoxy-5′-[N-methyl-N-[(2-aminooxy)ethyl]amino]adenosine
- MHZPA
5′-deoxy-5′-[N-methyl-N-(3-hydrazinopropyl)amino]adenosine
- WT
wild type
- HEPES
N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid
- DTT
dithiothreitol
- Brij-35
polyoxyethyleneglycol dodecyl ether
- HF
Hartree-Fock
- LMP2
Local Moller-Plesset second-order perturbation
- SCRF
self-consistent reaction field
Footnotes
This work was supported by a National Cancer Institute Grant (CA-094000 to S.E.E.) from the National Institutes of Health. S.E.E. is also indebted to the W. M. Keck Foundation and the Lucille P. Markey Charitable Trust.
The Protein Data Bank codes for the complexes are under the following accession numbers: AdoMetDC + MMTA (3H0V) AdoMetDC + DMAMA (3H0W).
References
- 1.van Poelje PD, Snell EE. Pyruvoyl-dependent enzymes. Ann Rev Biochem. 1990;59:29–59. doi: 10.1146/annurev.bi.59.070190.000333. [DOI] [PubMed] [Google Scholar]
- 2.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casero RA, Jr, Celano P, Ervin SJ, Applegren NB, Wiest L, Pegg AE. Isolation and characterization of a cDNA clone that codes for human spermidine/spermine N1-acetyltransferase. J Biol Chem. 1991;266:810–814. [PubMed] [Google Scholar]
- 4.Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 5.Pegg AE, Feith DJ. Polyamines and neoplastic growth. Biochem Soc Trans. 2007;35:295–299. doi: 10.1042/BST0350295. [DOI] [PubMed] [Google Scholar]
- 6.Hackert ML, Pegg AE. Pyruvoyl-dependent enzymes. In: Sinnott ML, editor. Comprehensive Biological Catalysis. Academic Press; London: 1997. pp. 201–216. [Google Scholar]
- 7.Pegg AE, Xiong H, Feith D, Shantz LM. S-adenosylmethionine decarboxylase: structure, function and regulation by polyamines. Biochem Soc Trans. 1998;26:580–586. 526. doi: 10.1042/bst0260580. [DOI] [PubMed] [Google Scholar]
- 8.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 9.Tabor CW, Tabor H. Methionine adenosyltransferase (S-adenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Advan Enzymol Related Areas Mol Biol. 1984;56:251–282. doi: 10.1002/9780470123027.ch4. [DOI] [PubMed] [Google Scholar]
- 10.Williams-Ashman HG, Schenone A. Methylglyoxal bis(guanylhydrazone) as a potent inhibitor of mammalian and yeast S-adenosylmethionine decarboxylases. Biochem Biophys Res Commun. 1972;46:288–295. doi: 10.1016/0006-291x(72)90661-4. [DOI] [PubMed] [Google Scholar]
- 11.Regenass U, Mett H, Stanek J, Mueller M, Kramer D, Porter CW. CGP 48664, a new S-adenosylmethionine decarboxylase inhibitor with broad spectrum antiproliferative and antitumor activity. Cancer Res. 1994;54:3210–3217. [PubMed] [Google Scholar]
- 12.Eskens FA, Greim GA, van Zuylen C, Wolff I, Denis LJ, Planting AS, Muskiet FA, Wanders J, Barbet NC, Choi L, Capdeville R, Verweij J, Hanauske AR, Bruntsch U. Phase I and pharmacological study of the weekly administration of the polyamine synthesis inhibitor SAM 486A (CGP 48 664) in patients with solid tumors. European Organization for Research and Treatment of Cancer Early Clinical Studies Group. Clin Cancer Res. 2000;6:1736–1743. [PubMed] [Google Scholar]
- 13.Zhou H, Choi L, Lau H, Bruntsch U, Vries EE, Eckhardt G, Oosterom AT, Verweij J, Schran H, Barbet N, Linnartz R, Capdeville R. Population pharmacokinetics/toxicodynamics (PK/TD) relationships of SAM486A in phase I studies in patients with advanced cancers. J Clin Pharmacol. 2000;40:275–283. doi: 10.1177/00912700022008946. [DOI] [PubMed] [Google Scholar]
- 14.Paridaens R, Uges DRA, Barbet N, Choi L, Seeghers M, van der Graaf WTA, Groen JJM. A phase I study of a new polyamine biosynthesis inhibitor, SAM486A, in cancer patients with solid tumours. Br J Cancer. 2000;83:594–601. doi: 10.1054/bjoc.2000.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siu LL, Rowinsky EK, Hammond LA, Weiss GR, Hidalgo M, Clark GM, Moczygemba J, Choi L, Linnartz R, Barbet NC, Sklenar IT, Capdeville R, Gan G, Porter CW, Von Hoff DD, Eckhardt SG. A phase I and pharmacokinetic study of SAM486A, a novel polyamine biosynthesis inhibitor, administered on a daily-times-five every-three-week schedule in patients with Advanced solid malignancies. Clin Cancer Res. 2002;8:2157–2166. [PubMed] [Google Scholar]
- 16.van Zuylen L, Eskens F, Bridgewater J, Sparreboom A, Sklenar I, Planting A, Choi L, Mueller C, Capdeville R, Ledermann J, Verweij J. The Polyamine Synthesis Inhibitor SAM486A in Combination, with 5-FU/LV in Metastatic Colorectal Cancer (MCC): Results of a Phase I and Pharmacokinetic Study. Proc Am Soc Clin Oncol. 2000;36:751. [Google Scholar]
- 17.Pless M, Belhadj K, Kern W, Dumontet C, Chemnitz J, Menssen HD, Herrmann R, Barbet NC, Capdeville R. Clinical Efficacy of SAM486A, a Novel Polyamine Biosynthesis Inhibitor, in Patients with Refractory or Relapsed Non-Hodgkin’s Lymphoma. Proc Am Soc Clin Oncol. 2000;36:62. [Google Scholar]
- 18.Millward MJ, Joshua A, Kefford R, Aamdal S, Thomson D, Hersey P, Toner G, Lynch K. Multi-centre Phase II trial of the polyamine synthesis inhibitor SAM486A ( CGP48664) in patients with metastatic melanoma. Invest New Drugs. 2005;23:253–256. doi: 10.1007/s10637-005-6734-z. [DOI] [PubMed] [Google Scholar]
- 19.Shantz LM, Stanley BA, Secrist JA, Pegg AE. Purification of human S-adenosylmethionine decarboxylase expressed in Escherichia coli and use of this protein to investigate the mechanism of inhibition by the irreversible inhibitors, 5′-deoxy-5′-[(3-hydrazinopropyl)methylamino]adenosine and 5′{[(Z)-4-amino-2-butenyl]methylamino-5′-deoxyadenosine. Biochemistry. 1992;31:6848–6855. doi: 10.1021/bi00144a027. [DOI] [PubMed] [Google Scholar]
- 20.Pankaskie M, Abdel-Monem MM. Inhibitors of polyamine biosynthesis 8: Irreversible inhibition of mammalian S-adenosyl-L-methionine decarboxylase by substrate analogs. J Med Chem. 1980;23:121–127. doi: 10.1021/jm00176a004. [DOI] [PubMed] [Google Scholar]
- 21.Pegg AE, Jacobs G. Comparison of inhibitors of S-adenosylmethionine decarboxylase from different species. Biochem J. 1983;213:495–502. doi: 10.1042/bj2130495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolbert DW, Ekstrom JL, Mathews II, Secrist JAI, Kapoor P, Pegg AE, Ealick SE. The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase. Biochemistry. 2001;40:9484–9494. doi: 10.1021/bi010735w. [DOI] [PubMed] [Google Scholar]
- 23.McCloskey DE, Bale S, Secrist JA, Tiwari A, Moss TH, Valiyaveettil J, Brooks WH, Guida WC, Pegg AE, Ealick SE. New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine . J Med Chem. 2009;52:1388–1407. doi: 10.1021/jm801126a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt GJ, Jones TA. Databases in protein crystallography. Acta Crystallogr D. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 29.Mecozzi S, West AP, Dougherty DA. Cation-π interactions in simple aromatics: Electrostatics provide a predictive tool. J Am Chem Soc. 1996;118:2307–2308. [Google Scholar]
- 30.Markham GD, Norrby PO, Bock CW. S-adenosylmethionine conformations in solution and in protein complexes: conformational influences of the sulfonium group. Biochemistry. 2002;41:7636–7646. doi: 10.1021/bi025682c. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 32.Ekstrom JE, Matthews II, Stanley BA, Pegg AE, Ealick SE. The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 Å resolution reveals a novel fold. Structure. 1999;7:583–595. doi: 10.1016/s0969-2126(99)80074-4. [DOI] [PubMed] [Google Scholar]
- 33.Ekstrom JL, Tolbert WD, Xiong H, Pegg AE, Ealick SE. Structure of a human S-adenosylmethionine decarboxylase self-processing ester intermediate and mechanism of putrescine stimulation of processing as revealed by the H243A mutant. Biochemistry. 2001;40:9495–9504. doi: 10.1021/bi010736o. [DOI] [PubMed] [Google Scholar]
- 34.Johnson KA. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- 35.Ma JC, Dougherty DA. The Cation-π Interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 36.Gallivan JP, Dougherty DA. Cation-π interactions in structural biology. Proc Natl Acad Sci U S A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan C, Rodgers MT. Cation-π interactions: structures and energetics of complexation of Na+ and K+ with the aromatic amino acids, phenylalanine, tyrosine, and tryptophan. J Am Chem Soc. 2004;126:14600–14610. doi: 10.1021/ja048297e. [DOI] [PubMed] [Google Scholar]
- 38.Biot C, Buisine E, Kwasigroch JM, Wintjens R, Rooman M. Probing the energetic and structural role of amino acid/nucleobase cation-π interactions in protein-ligand complexes. J Biol Chem. 2002;277:40816–40822. doi: 10.1074/jbc.M205719200. [DOI] [PubMed] [Google Scholar]
- 39.Biot C, Buisine E, Rooman M. Free-energy calculations of protein-ligand cation-π and amino-π interactions: from vacuum to proteinlike environments. J Am Chem Soc. 2003;125:13988–13994. doi: 10.1021/ja035223e. [DOI] [PubMed] [Google Scholar]
- 40.Zacharias N, Dougherty DA. Cation-π interactions in ligand recognition and catalysis. Trends Pharmacol Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 41.Cheng X, Kumar S, Posfai J, Pflugrath JW, Roberts RJ. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 42.Fu Z, Hu Y, Konishi K, Takata Y, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry. 1996;35:11985–11993. doi: 10.1021/bi961068n. [DOI] [PubMed] [Google Scholar]