Abstract
The RB tumor suppressor gene is mutated in a broad range of human cancers, including pediatric retinoblastoma. Strikingly, however, Rb mutant mice develop tumors of the pituitary and thyroid glands, but not retinoblastoma. Mouse genetics experiments have demonstrated that p107, a protein related to pRB, is capable of preventing retinoblastoma, but not pituitary tumors, in Rb-deficient mice. Evidence suggests that the basis for this compensatory function of p107 is increased transcription of the p107 gene in response to Rb inactivation. To begin to address the context-dependency of this compensatory role of p107 and to follow p107 expression in vivo, we have generated transgenic mice carrying an enhanced GFP (eGFP) reporter inserted into a bacterial artificial chromosome (BAC) containing the mouse p107 gene. Expression of the eGFP transgene parallels that of p107 in these transgenic mice and identifies cells with a broad range of expression level for p107, even within particular organs or tissues. We also show that loss of Rb results in the upregulation of p107 transcription in specific cell populations in vivo, including subpopulations of hematopoietic cells. Thus, p107 BAC-eGFP transgenic mice serve as a useful tool to identify distinct cell types in which p107 is expressed and may have key functions in vivo, and to characterize changes in cellular networks accompanying Rb deficiency.
Keywords: p107, retinoblastoma, transgenic mice, GFP, reporter, hematopoiesis, BAC
Introduction
The retinoblastoma tumor suppressor protein, pRB, is the downstream mediator of a cellular pathway that controls numerous cellular functions, including cell cycle progression, cellular differentiation and cell death. In mammalian cells, the p107 and p130 genes code for two proteins that are structurally and functionally related to pRB. The analysis of mice and cells derived from these mice carrying single and combined mutations in Rb family genes has shown that the three family members have both distinct and overlapping functions (reviewed in refs. 1–6).
While mouse knockout experiments have suggested unique functions for p107 in pre-adipocytes and neural precursors,7–9 comprehensive studies of p107 function in vivo remain limited and the cellular functions are p107 are still unclear, probably due to the rapid compensation for loss of p107 function by pRB and p130 in mammalian cells. Strikingly, p107 is better known for its ability to compensate for loss of pRB than for its own cellular functions. In particular, single and double knock-out experiments have revealed that p107 functionally overlaps with Rb in order to prevent retinoblastoma formation (reviewed in ref. 10) and skin cancer in mice,11 but not pituitary or thyroid gland tumors. p107 also limits adipose and neuronal differentiation defects in Rb mutant mice.7,12,13 Overall, these observations in mutant mice have uncovered cell-type specific contexts for p107 compensation for loss of Rb (reviewed in refs. 6 and 14), although the mechanisms underlying this compensatory role of p107 are still only partly understood.
p107 is thought to be transcriptionally regulated, with the protein levels strongly correlating with the mRNA expression levels.15 Furthermore, p107 also demonstrates a cell cycle dependent expression pattern, with the highest levels of p107 being expressed during S-phase.15 This cell cycle dependency of p107 may contribute to its expression in highly cycling organs, such as the spleen and the thymus.16 Furthermore, evidence in fibroblasts and cell lines in culture suggests that loss of pRB directly results in increased levels of p107 expression presumably via an E2F-dependent transcriptional process.15,17 p107 levels also increase in pRB-mutant muscle cells18 and mouse retinal cells19 but not in mouse embryos20 or human retinal progenitors.19 This upregulation of p107 in cells lacking pRB function is thought to create a feedback loop, which, when active, may prevent cancer initiation in specific cell types,19,21 especially in cells expressing oncogenic forms of Ras.22 A better understanding of the molecular mechanisms underlying the functional redundancy within the Rb gene family in specific cellular contexts may uncover novel ways to compensate for loss of pRB in human tumors. Thus far, however, it has been difficult to understand p107 compensation within individual subpopulations in vivo as opposed to a whole organs, tissues or embryos.
In order to further determine the cellular roles of p107 in embryonic and adult cells, in a wild-type context and in the absence of pRB function, we sought to develop a novel tool to follow p107 expression in mice. Because p107 expression is strongly controlled at the transcriptional level, a reporter gene that is transcriptionally controlled by the p107 regulatory sequences should reflect p107 expression patterns. However, a p107 knock-in reporter would decrease the overall levels of p107, thereby potentially affecting the levels of other pocket proteins as well as cell cycle progression.23–26 Therefore, we chose instead to generate a reporter transgenic mouse strain expressing the eGFP reporter from a bacterial artificial chromosome (BAC) containing the entire p107 mouse gene and more than 50 kb of upstream and downstream regulatory elements. Here we describe a novel BAC-transgenic mouse line in which relative eGFP expression accurately reflects relative p107 expression, enabling the identification of single cells within an organ that express p107, in the presence and in the absence of functional pRB.
Results
Generation of p107 BAC-eGFP transgenic mice
Recombineering in bacteria was used to insert the cDNA coding for the eGFP reporter protein exactly at the first ATG codon within the first exon of mouse p107 in a bacterial artificial chromosome (BAC), enabling the expression of eGFP under the normal transcriptional control of the p107 gene (Fig. 1A). We also added a polyadenylation signal at the end of the eGFP cDNA to prevent the expression of potential fusion transcripts between eGFP and p107 from the BAC (Fig. 1A). This approach also prevents the expression of the shorter p68 isoform of p107 which is expressed from the same promoter.27
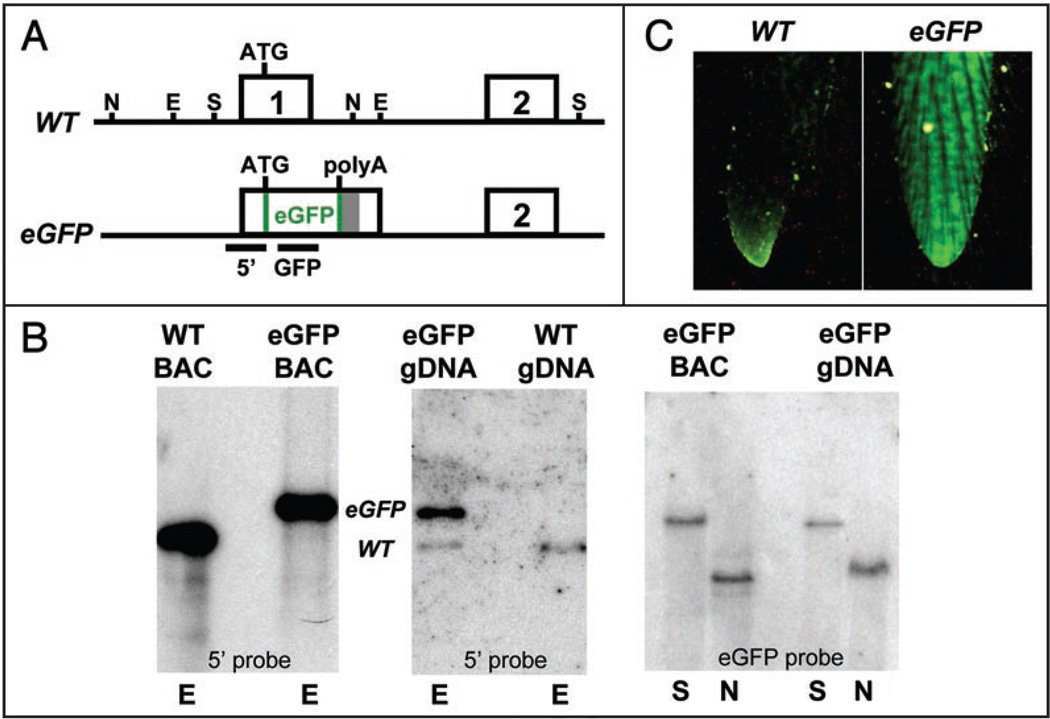
Figure 1.
Generation of p107 BAC-eGFP transgenic mice. (A) Schematic representation of the p107 BAC knock-in reporter. Exons are indicated by numbered boxes. A polyadenylation site (grey bar) is placed directly downstream of the eGFP cDNA. The 5' and GFP bars indicate DNA probes for Southern blots—E, EcoR1; N, Nde1; S, Sac1. (B) Southern blot analysis of the wild-type and the eGFP-transgenic allele of p107 demonstrates that multiple copies of a large fragment of the p107-eGFP BAC have integrated into the mouse genome. The left and middle panels are EcoR1 digests. WT represents the 4.6 kb wild-type band while eGFP represents the 5.6 kb transgene band obtained with a 5' probe—left panel, BAC DNA; middle panel, genomic DNA (gDNA). In the right panel, with an eGFP probe, a Sac1 digest of BAC and gDNA indicates the presence of a 15.5 kb band (top band) that corresponds to the transgene. The Nde1 digest of the eGFP-transgenic mouse gDNA shows the expected 7.5 kb band. (C) p107 BAC-eGFP transgenic mice are genotyped through direct visualization of eGFP expression in the tail under a UV light.
p107-eGFP knock-in BAC DNA was injected into fertilized oocytes to produce transgenic mice. Several founder mice tested positive for the transgene by direct visualization of eGFP in tail biopsies (Fig. 1C) and two independent lines, were generated from these founders. None of these p107 BAC-eGFP transgenic mice exhibit any gross phenotype at the heterozygous or homozygous state (data not shown). Southern blot analysis confirmed that the p107 region around the eGFP reporter gene was intact and that one transgenic line had ∼2 copies of the transgene while the other had ∼10 copies (Fig. 1B and data not shown). In a series of preliminary experiments, both p107 BAC-eGFP lines of transgenic mice produced similar expression patterns for eGFP. The line with more copies of the p107-eGFP BAC produced higher levels of the eGFP protein and was used in all subsequent experiments.
eGFP expression in p107 BAC-eGFP mice mimics endogenous p107 expression in whole organs and tissues
We first sought to determine if expression of the eGFP reporter accurately followed that of the mouse p107 gene. To this end, we performed real-time quantitative RT-PCR experiments (QPCR) on RNA isolated from various adult mouse tissues from p107 BAC-eGFP transgenic mice, between 10 and 12 weeks of age. These experiments showed that both the eGFP and p107 transcripts were low in slowly cycling tissues such as the heart, the kidney and the liver. In contrast, eGFP and p107 transcripts were more abundant in tissues with a higher proliferative index such as the thymus, spleen and intestines. Thus, with the exception of the testes where eGFP levels were much higher than p107 levels (data not shown), we consistently found that eGFP and p107 RNA molecules followed a similar pattern of expression in multiple mouse tissues and organs (Fig. 2A).
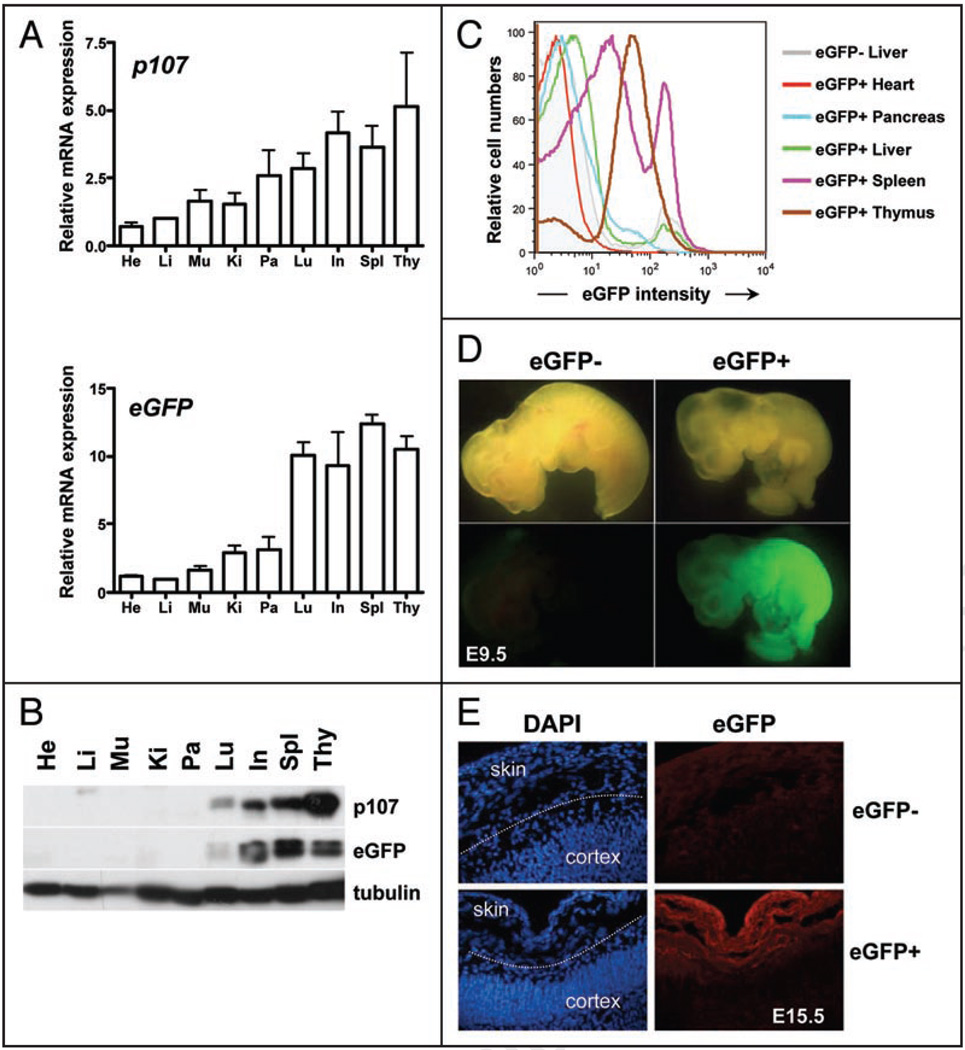
Figure 2.
Expression patterns of the eGFP transgene in adult mice and in embryos. (A) mRNA expression of p107 and eGFP relative to TBP (TATA Binding Protein) in whole organs from adult mice at 10–12 weeks of age (two mice—mean and standard error are graphed). (B) Representative immunoblot analysis of the p107 and eGFP proteins in extracts from whole adult mouse organs. He, heart; Li, liver; Mu, muscle; Ki, kidney; Pa, pancreas; Lu, lung; In, intestine; Spl, spleen; Thy, thymus. (C) Representative flow cytometric analysis of eGFP expression in single cell suspensions from whole organs obtained from adult p107 BAC-eGFP transgenic mice. The shaded grey area represents the eGFP signal for the liver of a non-transgenic mouse (eGFP−). Other organs are represented by lines of different colors, as indicated. (D) p107 BAC-eGFP is expressed throughout mid-gestation embryos (9.5 days of development, E9.5) by direct fluorescence. (E) Expression of the eGFP transgene in p107 BAC-eGFP mice becomes confined to proliferative regions such as the skin and not the maturing cortex in E15.5 embryos as demonstrated by indirect immunofluorescence using an anti-eGFP antibody (red)—DNA is visualized by DAPI staining (blue).
Immunoblot analysis further revealed that eGFP protein levels reflected the expression of the eGFP RNA molecules; tissues with high levels of p107 and eGFP RNA also contained the highest amount of eGFP protein (Fig. 2B). In addition, p107 protein levels followed the same trend as p107 transcripts in vivo. Both p107 and GFP protein levels are higher in extracts from the spleen, thymus, intestines and lungs of adult mice (Fig. 2B, upper) than in non-cycling organs. Thus, not only does eGFP expression in p107 BAC-eGFP mice reflect p107 transcription, but also it is largely representative of p107 expression at the protein level in vivo.
While the RNA and protein analyses described above characterize expression levels within whole organs, the use of eGFP as a reporter enables the additional use of flow cytometry, direct visualization of fluorescence in live cells, and immunostaining to identify eGFP/p107 expression patterns in individual cells within a whole organ. Overall, flow cytometric analysis of whole organs demonstrated similar patterns of eGFP expression to that observed at the protein level (Fig. 2C and data not shown—see below). These data confirm that the eGFP transgene accurately follows the expression of the endogenous p107 gene in adult tissues.27–30
In p107 BAC-eGFP embryos, the eGFP pattern of expression was also similar to what has been previously described by in situ hybridization for endogenous p107 transcripts.20 At 9.5 days of gestation (E9.5), when many embryonic cells are cycling, eGFP levels were high throughout the embryo (Fig. 2D). Gross eGFP expression in whole embryos decreases throughout development, with higher levels observed at E9.5 and gradually lower levels observed at E15.5 and E18.5 (data not shown). At E15.5, when some tissues in these transgenic embryos start to mature and increased number of cells exit the cell cycle, we observed through immunostaining of cryosections that eGFP remained high in some cells, such as the developing skin and decreased in others, such as the differentiated cortex (Fig. 2E).
Together, these experiments indicate that p107 BAC-eGFP mice represent a novel reporter mouse strain to faithfully monitor the expression pattern of mouse p107 in vivo.
Use of p107 BAC-eGFP transgenic mice to identify specific subpopulations of cells in mice
Flow cytometric analysis of splenocytes in adult p107 BAC-eGFP mice revealed two distinct populations of eGFP-expressing cells (Fig. 2C and Fig. 3A, left); these subpopulations were obviously not visible in immunoblot analysis of whole organs. Immunoblot analysis of splenocytes sorted based on their expression levels of eGFP showed that eGFPLow cells expressed lower levels of p107 compared to eGFPHigh populations (Fig. 3A, right), further confirming the validity of the BAC transgene.
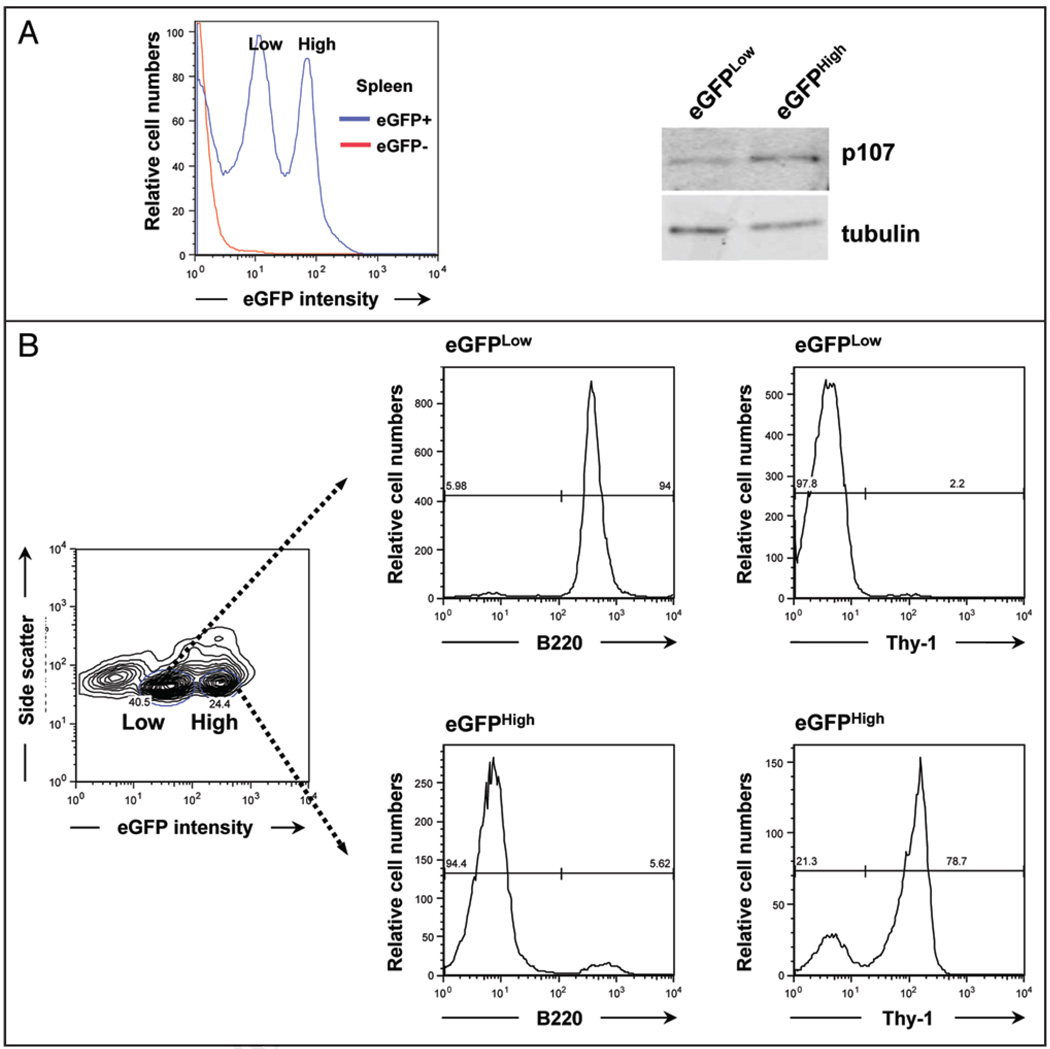
Figure 3.
eGFP expression identifies enriched populations of B cells and T cells in the spleen of adult transgenic mice. (A) The adult mouse spleen of p107 BAC-eGFP mice contains two distinct eGFP-expressing subpopulations as observed by flow cytometry (Low and High). Immunoblot analysis of cells sorted from these two populations reveals that the eGFP levels reflect different amounts of p107. Tubulin serves as a loading control. (B) The eGFPLow population is enriched for B cells as determined by B220 staining and is negative for T-cells as determined by Thy-1 staining. The eGFPHigh population is enriched for T-cells. A representative experiment is shown (n = 3 for B220, n = 2 for Thy-1).
We then asked if these populations that can be distinguished by different levels of eGFP/p107, represented distinct cell types among the multiple types present in the spleen of adult p107 BAC-eGFP mice. To this end, we incubated splenocytes with antibodies for B220, a marker of B lymphocytes, and Thy-1, a mouse pan-T cell marker, and analyzed these cells by FACS. Strikingly, we found that the eGFPLow splenic population was virtually completely comprised of B220+ cells (Fig. 3B). The eGFPHigh population is a mixed population, although the vast majority of these cells are Thy-1+ cells (Fig. 3B). Thus, eGFP expression in adult p107 BAC-eGFP mice can be used as a surrogate to enrich for tissue-specific subpopulations of cells.
Expression of the p107 BAC-eGFP transgene in response to acute loss of Rb function
p107 expression in quiescent mouse embryonic fibroblasts (MEFs) is upregulated in response to Rb deletion.17 To determine if eGFP expression in p107 BAC-eGFP mice also depended on pRB activity in MEFs, we generated p107 BAC-eGFP MEFs, and then we knocked-down Rb expression in these cells using a lentivirus expressing shRNA molecules against mouse Rb transcripts. We found that lower levels of Rb in these quiescent MEFs induced higher levels of eGFP mRNA and protein levels along with higher levels of endogenous p107 transcripts and protein, confirming that the p107 BAC-eGFP transgene is controlled by pRB activity similarly to the endogenous p107 gene (Fig. 4A and B).
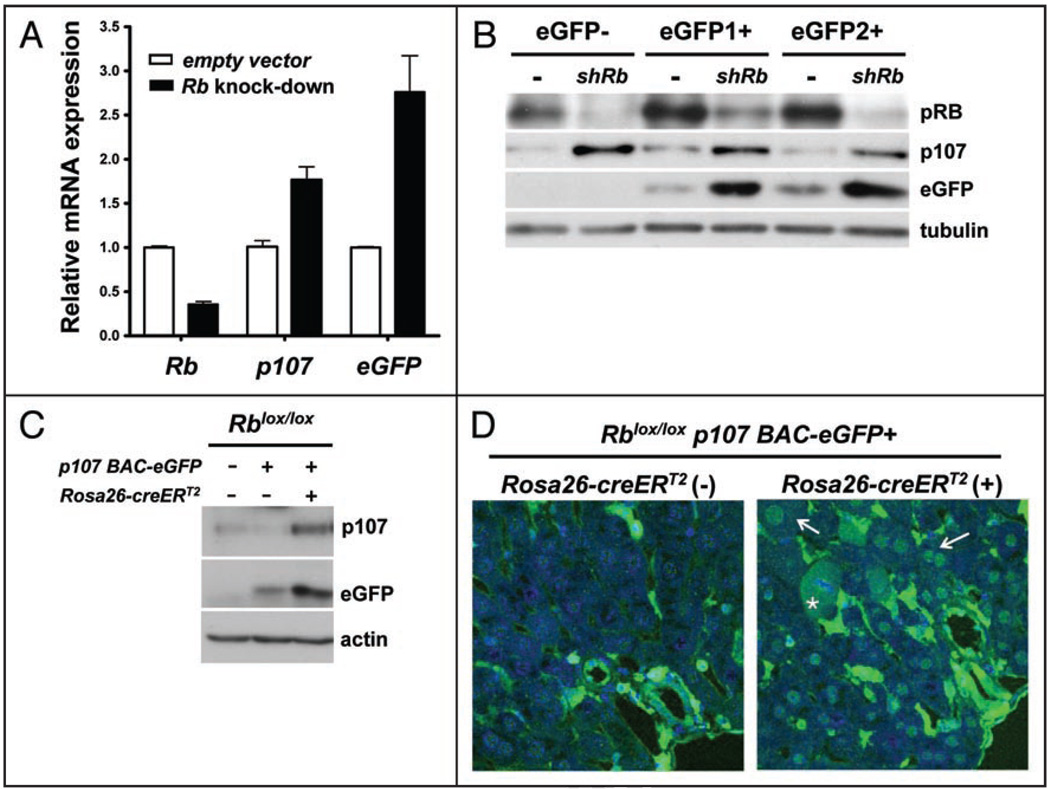
Figure 4.
p107-eGFP expression is upregulated after loss of pRB in MEFs and adult hepatocytes in vivo. (A and B) MEFs were infected with a lentivirus expressing short RNA molecules (shRNA) directed against the mouse Rb mRNA and then rendered quiescent through serum starvation. (A) mRNA expression analysis of Rb, p107 and eGFP, relative to TBP (n = 4), in MEFs infected with an empty vector or a vector to knock-down Rb. (B) Immunoblot analysis of pRB, p107 and eGFP in control and knock-down (shRb) MEFs—eGFP1+ and eGFP2+ are two MEF cultures from two independent transgenic embryos. Tubulin serves as a loading control. (C and D) Adult livers isolated from p107 BAC-eGFP;Rblox/lox;Rosa26CreERT2 and control mice. All samples were collected from mice three days following five days of treatment with tamoxifen. (C) Representative immunoblot analysis of p107 and eGFP proteins in liver extracts. Actin serves as a loading control. (D) eGFP expression only becomes detectable in the nucleus of adult mouse hepatocytes after Cre-mediated deletion of Rb, as demonstrated through confocal immunohistochemical detection of eGFP (green) in Rosa26CreERT2 negative and positive littermates injected with tamoxifen. The arrows point to the nuclei of two positive cells, the asterisk points to a positive cell undergoing mitosis. The blue signal is DAPI staining of DNA.
Next, we sought to investigate the consequences of deleting Rb function on p107 expression in vivo. To this end, we bred mice carrying Rb conditional mutant alleles17 to Rosa26CreERT2 knock-in mice,31 in which injection of tamoxifen conditionally induces Cre-mediated recombination of loxP sites. PCR and Southern blot analysis on genomic DNA isolated from Rosa26CreERT2;Rbloxlox mice five days after Cre induction showed deletion of the Rb conditional allele in all the organs tested with an estimated efficiency of 30–90% depending on the organs (data not shown), similar to what has been reported previously.32,33 We then generated p107 BAC-eGFP;Rblox/lox and p107 BAC-eGFP;Rosa26CreERT2;Rblox/lox mice. Acute deletion of Rb has been shown to increase p107 levels in the liver of adult mice.26 Accordingly, we detected increased expression of eGFP and p107 mRNA at three days following a five day injection schedule that induced efficient Rb deletion in the liver (data not shown and ref. 34). These changes were accompanied by increased levels of the eGFP and p107 proteins as analyzed by western blot on liver protein extracts (Fig. 4C). Despite testing different antibodies against p107 and different immunostaining methods, we failed to consistently detect p107 in liver sections. In contrast, immunostaining for eGFP on cryosections of transgenic livers readily confirmed that control hepatocytes do not express eGFP while Rb-deficient hepatocytes do express detectable levels of eGFP (Fig. 4D).
Thus, the expression of eGFP and p107 in p107 BAC-eGFP mice are similarly responsive to loss of Rb function, further validating these reporter mice, and suggesting that these transgenic mice may enable the detection of individual cell-types which upregulate p107 in response to Rb loss in vivo.
Variable expression of the p107 BAC-eGFP transgene in hematopoietic cells of wild-type and Rb deficient mice
The extent to which upregulation of p107 occurs in response to loss of pRB in individual cell types in multiple embryonic and adult organs is still largely unknown. We sought to begin this analysis focusing on the hematopoietic system because blood cells are constantly undergoing differentiation that is dependent upon carefully controlled periods of proliferation and quiescence. Furthermore, loss of pRB results in myeloid and erythroid developmental defects35–41 and p107 itself may play a role during hematopoiesis, as p107 deficiency results in a mild myeloid hyperplasia in a Balb/c genetic background.42
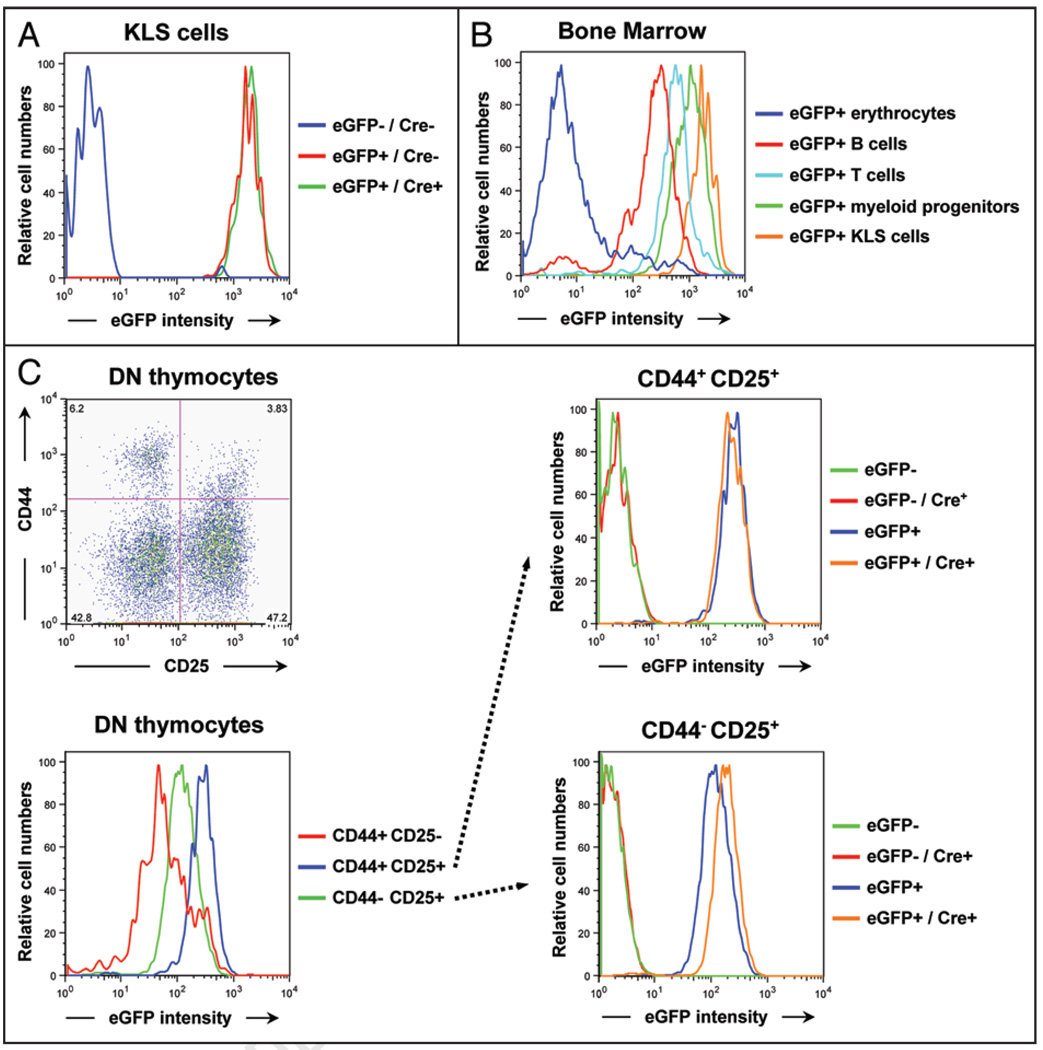
Recently, loss of Rb function in early hematopoietic progenitors in the bone marrow (“KLS” cells—negative for lineage markers, c-Kit+, Sca-1+) was shown to have no effect on p107 transcription.39 We isolated KLS progenitor cells from p107 BAC-eGFP;Rblox/lox and p107 BAC-eGFP;Rblox/lox;Rosa26CreERT2 mice by FACS and similarly found no increase in eGFP expression following induction with tamoxifen (Fig. 5A), despite efficient deletion of the Rb gene in this system (reviewed in ref. 43). Notably, compared to other hematopoietic cell types, KLS cells express relatively high amounts of eGFP (Fig. 5B). Thus, it is possible that p107 transcription levels are already too high in wild-type KLS cells to be further increased by the depletion of pRB.
Figure 5.
Changes in eGFP expression in specific hematopoietic subpopulations upon Rb deletion in p107 BAC-eGFP mice. (A) FACS analysis of eGFP expression in KLS early progenitors cells derived from Rosa26CreERT2 positive and negative p107 BAC-eGFP;Rblox/lox transgenic mice. Note that eGFP expression does not change in Rb-deficient KLS cells compared to wild-type cells. (B) Hematopoietic subpopulations that are present in the bone marrow of p107 BAC-eGFP mice express various levels of eGFP, with the erythrocytes expressing low eGFP levels and the KLS cells expressing the highest levels as observed by flow cytometry. (C) CD4− CD8− double negative (DN) thymocytes isolated from adult mice were stained with antibodies against the cell surface molecules CD25 and CD44 to mark subpopulations of progenitors and analyzed by FACS. The top left panel shows the four different subpopulations. CD44+ CD25+ progenitors from p107 BAC-eGFP mice express the highest amounts of eGFP (bottom left), and this level does not change when Rb is deleted (top right). CD44− CD25+ progenitors initially express a slightly lower amount of eGFP (bottom left), which increases reproducibly in Rb-deficient mice (bottom right). Data shown are representative of three independent experiments.
Within the thymus, careful control of cell proliferation during T-cell development is critical to enable expansion of the appropriate precursor cells to maximize the cells surviving the selection process. For example, early CD4− CD8− double negative (DN) thymocytes are initially CD44+ CD25− and largely quiescent. As these cells begin to differentiate, they express CD25 (CD44+ CD25+ cells) and proliferate extensively, until they downregulate CD44 expression; CD44− CD25+ cells are mostly quiescent.44 We found that proliferative CD44+ CD25+ DN progenitors from p107 BAC-eGFP mice express higher levels of eGFP than any other DN thymocytes, in correlation with the cell cycle status of these cells (Fig. 5C, left). The halt in cell proliferation that occurs between the CD44+ CD25+ and CD44− CD25+ subpopulations could potentially be mediated by pRB. This idea is supported by the observation that pRB is hypophosphorylated (active) in the CD44− CD25+ subpopulation.45 We deleted Rb in T cell populations using p107 BAC-eGFP;Rblox/lox;Rosa26CreERT2 mice and examined eGFP expression in control and mutant T-cell progenitor populations. In the CD44+ CD25+ cycling subpopulation, which already exhibits high amounts of eGFP, loss of pRB produced no change in eGFP expression (Fig. 5C, upper right). In contrast, in the non-cycling CD44− CD25+ subpopulation of T-cell progenitors, eGFP expression increased in response to loss of pRB (Fig. 5C, lower right), suggesting a previously unknown role for pRB in repressing p107 and potentially regulating the cell cycle of CD44− CD25+ T cell progenitors.
Discussion
The p107 cell cycle regulator belongs to the “pocket” family of proteins along with pRB and p130. Within this family, pRB is a potent tumor suppressor whose function is inactivated in a broad range of human tumors and has been extensively studied. In contrast, p107 and p130 are rarely found mutated in human tumors and their biological functions are still largely unknown, in part because of the ability of one family member to compensate for another. Here, we describe a novel tool to investigate p107 cellular functions in mice, without affecting the careful balance between pocket proteins. We first demonstrate that in p107 BAC-eGFP transgenic mice eGFP expression parallels that of the endogenous p107 gene, in wild-type cells and in response to loss of Rb function. We then use these transgenic mice to begin to characterize p107 expression in specific cell populations in vivo. Our data indicate that p107 transcription varies significantly between and within different organs and tissues, both in the presence and in the absence of a functional pRB protein. Our observations also suggest that p107 BAC-eGFP transgenic mice may be used to identify specific cell populations based on the levels of expression of eGFP.
Cell cycle progression and p107 expression
In cell lines, p107 expression is cell cycle dependent and thought to be largely controlled at the transcriptional level,15 following a model in which E2F repressor complexes prevent p107 transcription in quiescent cells and E2F activator complexes activate p107 transcription in the late G1 and S phases of the cell cycle.15,46 Our data analyzing the RNA and protein levels of p107 in whole mouse organs support this model, as we observed higher p107 levels in organs and tissues containing cycling cells. However, this approach may mask differences between different cell types or subpopulations of the same cell type within each organ and tissue, which has been challenging to test by immunostaining for p107. The use of transgenic mice expressing eGFP in lieu of p107 bypasses this technical difficulty and has allowed us to reveal striking differences within certain organs. In the spleen, for instance, FACS analysis shows that two subpopulations of cells express different levels of eGFP in p107 BAC-eGFP mice. These experiments also suggest that there is no strict correlation between the cell cycle of cells and the levels of expression of p107. For example, T cells and B cells in the spleen are normally largely non-cycling, but splenic T cells express higher levels of eGFP/p107 than B cells. Similarly, KLS cells in the bone marrow express the highest levels of eGFP of all of the cell types examined, although they are more quiescent than myeloid progenitors.47 Experiments to characterize p107 levels in additional specific populations of cells in different adult and embryonic organs and tissues will provide novel insights into the relationship between cell cycle progression and p107 expression in vivo.
Control of p107 expression
The BAC sequences that surround the eGFP transgene in p107 BAC-eGFP mice seem to contain most, if not all the regulatory elements required to transcribe eGFP similarly to p107. One notable exception is the testis where we found very high levels of eGFP transcripts and a pattern of expression of the eGFP protein that do not match with previous observations (reviewed in ref. 48). This suggests that eGFP transcripts and/or the eGFP protein are regulated differently in germ cells or that additional sequences may be required in these cells to repress p107 transcription. Nevertheless, the generation of novel mouse lines carrying a similar p107 BAC-eGFP but with specific mutations in the binding sites for transcription factors will allow in the future a better understanding of p107 transcription in vivo. Such a BAC transgenic approach would allow introducing mutations in the p107 promoter without affecting p107 levels, thereby ensuring that these studies are performed under normal cell cycle conditions. A similar approach with a plasmid containing proximal regulatory sequences has been successfully used to analyze the regulatory regions of Rb using lacZ reporter mice and has identified a key repressor role for an E2F binding site in the Rb promoter.49 It is important to note, however, that a similar experiment will be complicated for p107 due to the presence of two E2F binding sites.15,46 In addition, the identity of other transcription factors regulating p107 expression is still unknown.
p107 cellular functions
Although p107 has been demonstrated to be able to compensate for loss of pRB in preventing tumor formation in some specific mouse tissues, little is known about the normal cellular functions of p107 in cells with functional pRB. Given the capacity of p107 to inhibit cell cycle progression when ectopically expressed in mammalian cells,50,51 p107 may act as a tumor suppressor. However, there is little evidence that p107 is a potent tumor suppressor in humans52,53 and the initial characterization of p107-deficient mice indicated that these mutant mice were very similar to wild-type mice and did not develop tumors.10 Since then, however, p107 has been shown to have a more prominent role in restricting body growth, cell cycle progression in MEFs, and myeloid expansion in a Balb/c genetic background.42 Furthermore, p107 regulates the proliferation and the fate of precursors in the brain8,9 and the adipose tissue7 of mice. Some evidence in culture also suggests that p107 could play a role in response to DNA damage during S phase, when it is most highly expressed.54 As ongoing experiments test the roles of p107 in development and tumorigenesis in mice, p107 BAC-eGFP mice may help to identify individual populations of cells in which the biological function of p107 should be further characterized. For example, our results indicate that p107 levels of expression are very dynamic in blood cells, which suggests that specific types of blood cells may rely on p107 to control their cell cycle, making it possible that specific subsets of human leukemias or lymphomas would inactivate p107 as tumorigenesis progresses.
Compensatory functions of p107
Thus far, the only tumor suppressor role for p107 that has been clearly demonstrated is in cells that are Rb deficient (reviewed in refs. 6, 10, 11 and 14), suggesting that loss of p107 is not sufficient to initiate cancer but that p107 suppresses tumor development in Rb mutant cells. It is not yet clear if normal p107 levels are sufficient for this compensatory function or if, as suggested by experiments in culture,17,19 the upregulation of p107 induced by loss of pRB is required. However, p107 is clearly not capable of preventing tumorigenesis in the pituitary and thyroid glands of Rb mutant mice. Due to the small size of these glands in mice, we have not been able to characterize if p107 (or eGFP) levels increase in cells of these organs upon loss of Rb, but we have begun to analyze how the p107 promoter responds to loss of Rb in various cell types in p107 BAC-eGFP mice. While many organs showed no response at the level of the whole organ, such as the kidney and the pancreas, further experiments may uncover small populations of cells within these organs that do upregulate p107/eGFP upon loss of pRB. Even within organs that demonstrate a whole organ response, such as the thymus, our analysis of p107 BAC-eGFP mice has uncovered subpopulations in which eGFP expression does not change after Rb deletion. These data indicate that our understanding of p107 regulation and compensatory functions is still preliminary.
Understanding where and how p107 is upregulated in response to loss of Rb, as well as why it is not in other organs, and how this regulation contributes to its ability to function as a tumor suppressor in Rb-deficient may have eventual therapeutic potential. Because p107 is virtually never found mutated within human cancers, in the future it may be possible to take advantage of the transcriptional networks controlling p107 expression in order to serve as a way to halt the progress of Rb-deficient tumors.
Material and Methods
Generation of an eGFP knock-in p107 BAC
Recombineering was used to generate the p107-eGFP BAC, as described previously.55 Briefly, BAC clone RP23-163J20 ordered from BACPAC (http://bacpac.chori.org/) was transformed into the recombineering strain EL250 and the heat-inducible recombinase present in this strain enabled the insertion of an eGFP cDNA including a polyadenylation signal at the translation start site of p107 by homologous recombination.
Generation of p107 BAC-eGFP transgenic mice and other mouse strains
The p107 BAC-eGFP was purified using the Marligen Maxiprep Kit. BAC DNA was injected into F1 hybrid B6D2F1/J (The Jackson Laboratory, 100006) fertilized oocytes at the Stanford Transgenic Research Facility. This injection resulted in two germ line founders. Mice are maintained according to practices prescribed by the NIH, and housed in Stanford’s Research Animal Facility accredited by the AAALAC.
The Rb conditional knock-out allele and the Rosa26CreERT2 knock-in strain were described previously17,32 and are maintained in a mixed 129Sv/J:C57BL/6J background. Rosa26CreERT2; Rblox/lox; p107 BAC-eGFP mice are maintained in a mixed 129Sv/J:C57BL/6/J:DBA/2J background enriched for 129Sv/J and C57BL/6J.
Tamoxifen injections
Mice were injected with 1.5 mg of tamoxifen per injection, given by 5 consecutive intraperitoneal injections. Tamoxifen was resuspended in 100% ethanol at a concentration of 100 mg/ml. Once dissolved, corn oil was added to bring the final concentration to 10 mg/ml. The solution was sonicated for one minute before injection.
Cell culture
Mouse embryonic fibroblasts (MEFs) were derived from embryos 13.5 days after fertilization and cultured in DMEM supplemented with 10% bovine growth serum (BGS, Fisher Scientific). MEFs used in knockdown experiments were used between passage 3 and 5. For serum starvation experiments, MEFs were cultured in DMEM supplemented with 0.1% BGS for 3 days prior to collection.
Rb knock-down was achieved using the pSICOR lentivirus,56 as described.17 Briefly, exponentially growing MEFs were infected twice with viral supernatant from transfected 293T cells, and then selected with puromycin for three days (2 µg/ml, Sigma). The sequences in the Rb cDNA that is targeted by the shRNA molecules is 5'-TGAGAGCAAGGATGTCTCA-3'.
Histology
For paraffin sections, organs were fixed in 4% parafor-maldehyde for 4–8 hours at room temperature, and then transferred to 70% ethanol prior to processing and embedding. For cryosections, organs were fixed in 4% paraformaldehyde for 4–8 hours at room temperature and then transferred to 30% sucrose overnight. Organs were then embedded in OCT and frozen in OCT using liquid nitrogen-cooled isopentane.
Immunostaining and immunoblot
Deparaffinization and antigen retrieval of paraffin sections was performed using the Trilogy solution (Cell Marque) for 15 minutes in a pressure cooker. Cryosections were thawed at room temperature for at least 20 minutes. All sections were then briefly washed in PBS + 0.1% Tween (PBST), and then blocked for 1 hour at room temperature in PBST with 5% normal horse serum and 1% bovine serum albumin. Sections were incubated with primary antibodies diluted 1:200 in block solution overnight at 4°C, washed in PBST, and then incubated with secondary antibodies diluted 1:500 in block solution for 1 hour at room temperature. Primary antibodies used were rabbit anti-GFP (Molecular Probes, A11122), and mouse anti-GFP (Santa Cruz Biotechnology, sc-73556). Immunoblots of GFP and p107 were detected with sc-73556 and sc-318, respectively (Santa Cruz Biotechnology), as described before.57 Loading was verified with antibodies against mouse anti-alpha-tubulin (Sigma T9026) and mouse anti-beta-actin (Sigma A5441).
Microscopy
Tail biopsies and whole embryos were visualized using a Leica MZFLIII dissecting scope. Immunohistochemical analysis was performed using either a Leica DMLB or using confocal microscopy performed with a Leica SP2 DMIRE2 inverted confocal microscope.
FACS
For initial FACS analysis, whole organs were mechanically ground and filtered through a 100 µm cell strainer. Whole organ FACS analysis was performed using a BD FACScaliber. Bone marrow cells, splenocytes and thymocytes were filtered and red blood cells were lyzed with the ACK buffer (NH4Cl/KHCO3). Mature white blood cells were subsequently stained with fluorochrome conjugated antibodies against B220, Thy-1 or Ter119 for 30 minutes in the dark. Progenitors cells were stained with a cocktail of lineage-cy5PE antibodies and subsequently stained with antibodies against c-Kit and Sca-1 (Myeloid Progenitors and KLS cells) or CD44 and CD25 (T cell progenitors CD4/CD8 double negative populations). All the antibodies were purchased from eBioscience. Analysis was performed on an LSR at the Stanford Shared FACS Facility, using the CellQuest software for data acquisition (Becton Dickinson). Data were analyzed using the FlowJo software (Tree Star).
Quantitative RT-PCR
Organs were snap-frozen in liquid nitrogen and then ground with a disposable pestle. RNA was extracted with Trizol and cleaned using the Qiagen RNeasy kit. 800 ng–2 µg of RNA were reverse transcribed using the Qiagen Omniscript RT kit. Taqman quantitative PCR was performed using the Eurogentec Mastermix. p107: forward: 5'-CCGAAGCCCTGGATGACTT-3', reverse: 5'-GCATGCCAGCCAGTGTATAACTT-3', probe: 5'-FAM-CGCGGCAACTACAGCCTAGAGGGA-BHQ-3'; GFP: forward: 5'-ACTACAACAGCCACAACGTCTATATCA-3', reverse: 5'-GGCGGATCTTGAAGTTCACC-3', probe: 5'-FAM-CCGACAAGCAGAAGAACGGCATCA-BHQ-3'.
Acknowledgements
The authors thank Pauline Chu for her help with histology and Janelle Olson for discussions on T cell biology. This work was supported by the California Breast Cancer Research Program (D.B.), the Human Frontier Science Program Organization (P.V.), and the Damon Runyon Cancer Research Foundation and NIH-NCI RO1 CA114102 (J.S.).
Abbreviations
- eGFP
enhanced green fluorescent protein
- Rb
mouse retinoblastoma gene
- pRB
retinoblastoma protein
- BAC
bacterial artificial chromosome
- MEFs
mouse embryonic fibroblasts
- FACS
fluorescence-activated cell sorter
- KLS
c-Kit+, lineagenegative, Sca-1+ hematopoietic progenitors
References
- 1.Classon M, Dyson N. p107 and p130: versatile proteins with interesting pockets. Exp Cell Res. 2001;264:135–147. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- 2.Claudio PP, Tonini T, Giordano A. The retinoblastoma family: twins or distant cousins? Genome Biol. 2002:3. doi: 10.1186/gb-2002-3-9-reviews3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen DX, McCance DJ. Role of the retinoblastoma tumor suppressor protein in cellular differentiation. J Cell Biochem. 2005;94:870–879. doi: 10.1002/jcb.20375. [DOI] [PubMed] [Google Scholar]
- 5.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 6.Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci. 2006;63:767–780. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scime A, Grenier G, Huh MS, Gillespie MA, Bevilacqua L, Harper ME, Rudnicki MA. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2005;2:283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Vanderluit JL, Ferguson KL, Nikoletopoulou V, Parker M, Ruzhynsky V, Alexson T, McNamara SM, Park DS, Rudnicki M, Slack RS. p107 regulates neural precursor cells in the mammalian brain. J Cell Biol. 2004;166:853–863. doi: 10.1083/jcb.200403156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderluit JL, Wylie CA, McClellan KA, Ghanem N, Fortin A, Callaghan S, Maclaurin JG, Park DS, Slack RS. The Retinoblastoma family member p107 regulates the rate of progenitor commitment to a neuronal fate. J Cell Biol. 2007;178:129–139. doi: 10.1083/jcb.200703176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macpherson D. Insights from mouse models into human retinoblastoma. Cell Div. 2008;3:9. doi: 10.1186/1747-1028-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lara MF, Santos M, Ruiz S, Segrelles C, Moral M, Martinez-Cruz AB, Hernandez P, Martinez-Palacio J, Lorz C, Garcia-Escudero R, Paramio JM. p107 acts as a tumor suppressor in pRb-deficient epidermis. Mol Carcinog. 2008;47:105–113. doi: 10.1002/mc.20367. [DOI] [PubMed] [Google Scholar]
- 12.Batsche E, Moschopoulos P, Desroches J, Bilodeau S, Drouin J. Retinoblastoma and the related pocket protein p107 act as coactivators of NeuroD1 to enhance gene transcription. J Biol Chem. 2005;280:16088–16095. doi: 10.1074/jbc.M413427200. [DOI] [PubMed] [Google Scholar]
- 13.Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- 14.Dannenberg JH, te Riele HP. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl Cell Differ. 2006;42:183–225. doi: 10.1007/400_002. [DOI] [PubMed] [Google Scholar]
- 15.Smith EJ, Leone G, Nevins JR. Distinct mechanisms control the accumulation of the Rb-related p107 and p130 proteins during cell growth. Cell Growth Differ. 1998;9:297–303. [PubMed] [Google Scholar]
- 16.Garriga J, Limon A, Mayol X, Rane SG, Albrecht JH, Reddy EP, Andres V, Grana X. Differential regulation of the retinoblastoma family of proteins during cell proliferation and differentiation. Biochem J. 1998;333:645–654. doi: 10.1042/bj3330645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 18.Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 19.Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Z, Zacksenhaus E, Gallie BL, Phillips RA. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene. 1997;14:1789–1797. doi: 10.1038/sj.onc.1201014. [DOI] [PubMed] [Google Scholar]
- 21.Lee EY, Cam H, Ziebold U, Rayman JB, Lees JA, Dynlacht BD. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell. 2002;2:463–472. doi: 10.1016/s1535-6108(02)00207-6. [DOI] [PubMed] [Google Scholar]
- 22.Williams JP, Stewart T, Li B, Mulloy R, Dimova D, Classon M. The retinoblastoma protein is required for Ras-induced oncogenic transformation. Mol Cell Biol. 2006;26:1170–1182. doi: 10.1128/MCB.26.4.1170-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Classon M, Salama S, Gorka C, Mulloy R, Braun P, Harlow E. Combinatorial roles for pRB, p107 and p130 in E2F-mediated cell cycle control. Proc Natl Acad Sci USA. 2000;97:10820–10825. doi: 10.1073/pnas.190343497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulligan GJ, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and pRB. Mol Cell Biol. 1998;18:206–220. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novitch BG, Mulligan GJ, Jacks T, Lassar AB. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 27.Kim KK, Soonpaa MH, Wang H, Field LJ. Developmental expression of p107 mRNA and evidence for alternative splicing of the p107 (RBL1) gene product. Genomics. 1995;28:520–529. doi: 10.1006/geno.1995.1184. [DOI] [PubMed] [Google Scholar]
- 28.Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
- 29.Baldi A, Esposito V, De Luca A, Fu Y, Meoli I, Giordano GG, Caputi M, Baldi F, Giordano A. Differential expression of Rb2/p130 and p107 in normal human tissues and in primary lung cancer. Clin Cancer Res. 1997;3:1691–1697. [PubMed] [Google Scholar]
- 30.Jiang Z, Zacksenhaus E. Coordinated expression of Rb gene family in the mammary gland. Mech Dev. 2002;119:39–42. doi: 10.1016/s0925-4773(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin ME, Kruger GM, Slocum KL, Crowley D, Michaud NA, Huang J, Magendantz M, Jacks T. The Nf2 tumor suppressor regulates cell-cell adhesion during tissue fusion. Proc Natl Acad Sci USA. 2007;104:3261–3266. doi: 10.1073/pnas.0700044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007 doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 33.Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, Rijswijk L, Bex A, Berns A, Bockamp E. Toxicity of ligand-dependant Cre-recombinases and generation of a conditional Cre-deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 34.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Gorman JR, Stewart V, Williams B, Jacks T, Alt FW. Generation of normal lymphocyte populations by Rb-deficient embryonic stem cells. Curr Biol. 1993;3:405–413. doi: 10.1016/0960-9822(93)90347-q. [DOI] [PubMed] [Google Scholar]
- 36.Iavarone A, King ER, Dai XM, Leone G, Stanley ER, Lasorella A. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature. 2004;432:1040–1045. doi: 10.1038/nature03068. [DOI] [PubMed] [Google Scholar]
- 37.Spike BT, Dirlam A, Dibling BC, Marvin J, Williams BO, Jacks T, Macleod KF. The Rb tumor suppressor is required for stress erythropoiesis. Embo J. 2004;23:4319–4329. doi: 10.1038/sj.emboj.7600432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spike BT, Macleod KF. The Rb tumor suppressor in stress responses and hematopoietic homeostasis. Cell Cycle. 2005;4:42–45. doi: 10.4161/cc.4.1.1337. [DOI] [PubMed] [Google Scholar]
- 39.Walkley CR, Orkin SH. Rb is dispensable for self-renewal and multilineage differentiation of adult hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:9057–9062. doi: 10.1073/pnas.0603389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb Regulates Interactions between Hematopoietic Stem Cells and Their Bone Marrow Microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–475. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeCouter JE, Kablar B, Hardy WR, Ying C, Megeney LA, May LL, Rudnicki MA. Strain-dependent myeloid hyperplasia, growth deficiency and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol Cell Biol. 1998;18:7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, Rijswijk L, Bex A, Berns A, Bockamp E. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007;31:32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 44.Penit C, Lucas B, Vasseur F. Cell expansion and growth arrest phases during the transition from precursor (CD4−8−) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J Immunol. 1995;154:5103–5113. [PubMed] [Google Scholar]
- 45.Hoffman ES, Passoni L, Crompton T, Leu TM, Schatz DG, Koff A, Owen MJ, Hayday AC. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 46.Zhu L, Xie E, Chang LS. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol Cell Biol. 1995;15:3552–3562. doi: 10.1128/mcb.15.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan W, Kero J, Suominen J, Toppari J. Differential expression and regulation of the retinoblastoma family of proteins during testicular development and spermatogenesis: roles in the control of germ cell proliferation, differentiation and apoptosis. Oncogene. 2001;20:1343–1356. doi: 10.1038/sj.onc.1204254. [DOI] [PubMed] [Google Scholar]
- 49.Agromayor M, Wloga E, Naglieri B, Abrashkin J, Verma K, Yamasaki L. Visualizing dynamic E2F-mediated repression in vivo. Mol Cell Biol. 2006;26:4448–4461. doi: 10.1128/MCB.02101-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 51.Jiang H, Karnezis AN, Tao M, Guida PM, Zhu L. pRB and p107 have distinct effects when expressed in pRB-deficient tumor cells at physiologically relevant levels. Oncogene. 2000;19:3878–3887. doi: 10.1038/sj.onc.1203722. [DOI] [PubMed] [Google Scholar]
- 52.Takimoto H, Tsukuda K, Ichimura K, Hanafusa H, Nakamura A, Oda M, Harada M, Shimizu K. Genetic alterations in the retinoblastoma protein-related p107 gene in human hematologic malignancies. Biochem Biophys Res Commun. 1998;251:264–268. doi: 10.1006/bbrc.1998.9459. [DOI] [PubMed] [Google Scholar]
- 53.Ito Y, Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Kuma K, Miyauchi A. Decreased expression of p107 is correlated with anaplastic transformation in papillary carcinoma of the thyroid. Anticancer Res. 2003;23:3819–3824. [PubMed] [Google Scholar]
- 54.Kondo T, Higashi H, Nishizawa H, Ishikawa S, Ashizawa S, Yamada M, Makita Z, Koike T, Hatakeyama M. Involvement of pRB-related p107 protein in the inhibition of S phase progression in response to genotoxic stress. J Biol Chem. 2001;276:17559–17567. doi: 10.1074/jbc.M009911200. [DOI] [PubMed] [Google Scholar]
- 55.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 56.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]