Abstract
Intracellular degradation by autophagy plays a role in the maintenance of cellular homeostasis under normal conditions and during periods of cellular stress. Autophagy has also been implicated in several other cellular processes including immune recognition and responsiveness. More specifically, autophagy has been identified as a route by which cytoplasmic and nuclear Ag are delivered to MHC class II molecules for presentation to CD4+ T cells. Autophagy has also recently been implicated in MHC class I cross-presentation of tumor Ag and the activation of CD8+ T cells. This review discusses the role of autophagy in modulating MHC class I and class II Ag presentation as well as its implication in regulating autoimmunity and tolerance, tumor immunity, and host defense against intracellular pathogens.
Pathways of autophagy
Autophagy is one mechanism that cells utilize to degrade cytoplasmic proteins and organelles during the maintenance of cellular homeostasis (1). There are multiple pathways of autophagy. In microautophagy, small portions of the cytosol are internalized via lysosomal membrane invaginations, and proteins are continuously degraded in the lumen of this organelle even under resting conditions (2). Microautophagy is upregulated under conditions of cellular stress such as nutrient deprivation, but studies of this pathway have been limited to yeast and cell-free systems (3, 4). In mammalian cells, constitutively active autophagy pathways can also be upregulated as a mechanism to salvage amino acids during periods of cellular stress. Several types of selective autophagy have been identified in mammalian cells including macroautophagy and chaperone-mediated autophagy (CMA) (Fig. 1) as well as pexophagy and mitophagy (5). A related constitutive biosynthetic pathway, cytoplasm-to-vacuole targeting (Cvt), is involved in hydrolase sorting in yeast (6). Another pathway, xenophagy, involves the utilization of the autophagic machinery for the degradation of intracellular pathogens (7). Macroautophagy and CMA have been implicated in the presentation of Ag by MHC molecules, and thus, are the major focus of this review.
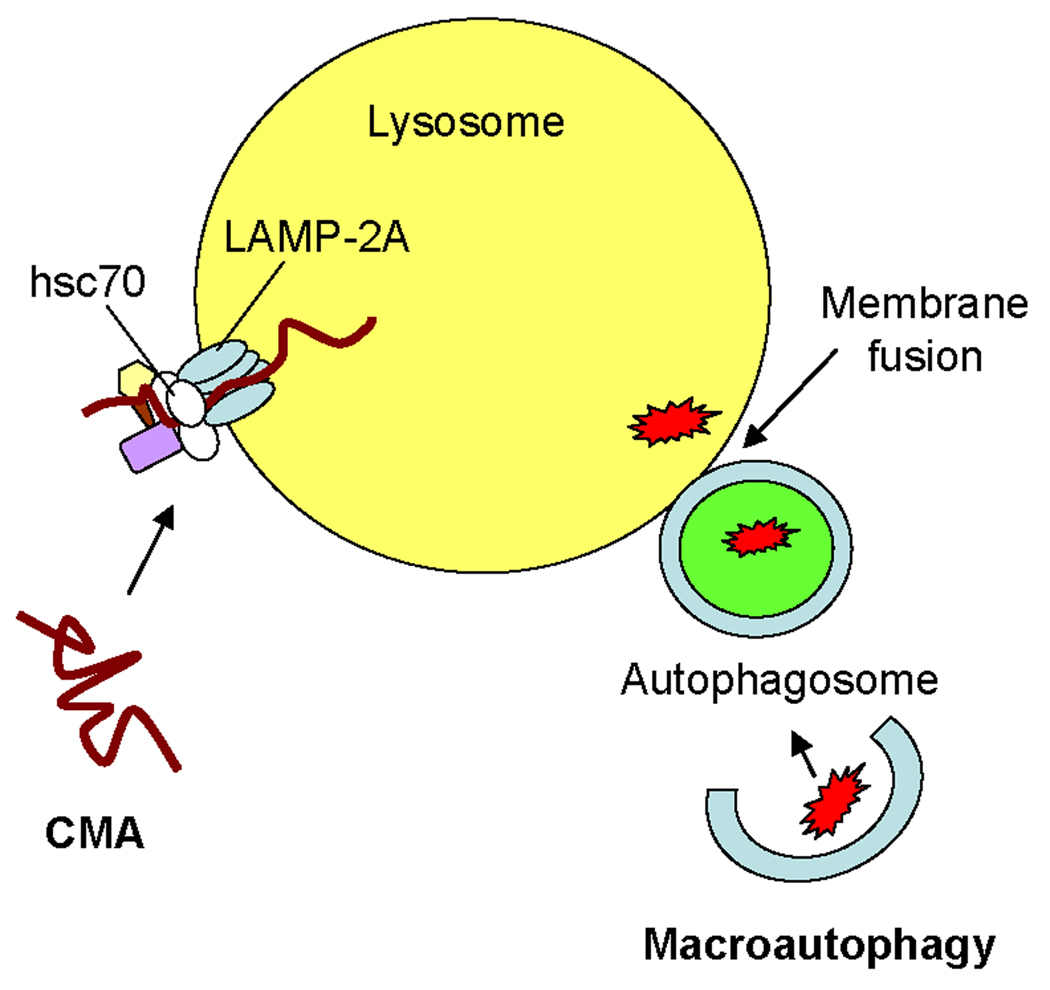
FIGURE 1. Pathways of autophagy in MHC class II-mediated Ag presentation.
In macroautophagy, the cytoplasm is sequestered into double-membraned structures known as autophagosomes which fuse with lysosomes. In chaperone-mediated autophagy (CMA), specific cytosolic proteins are transported into lysosomes via a molecular chaperone/receptor complex composed of hsc70 and LAMP-2A.
Macroautophagy is also a constitutively active cellular process modulating the degradation of long-lived proteins and organelles (1). During short periods of nutrient deprivation, this pathway is rapidly induced as a mechanism to salvage amino acids. The absence of growth factors or amino acids prevents signaling through phosphatidylinositol-3-kinase (PI3K) from activating the autophagy regulatory molecule, target of rapamycin (mTor); and thus, macroautophagy is induced (8). In macroautophagy, portions of the cytoplasm are sequestered into double-membrane structures known as autophagosomes. The formation of the autophagosome requires a number of autophagy-related gene (Atg) products which have been well-characterized in yeast (6) and are conserved in mammals; the specific details of autophagosome formation have been reviewed extensively elsewhere (2, 8). Briefly, the Atg12-Atg5-Atg16 complex in conjunction with Atg9 mediates the induction of autophagosome formation. During this process, LC3 (Atg8) is conjugated to phosphatidylethanolamine (PE) with the assistance of the Atg4, Atg7, and Atg3 molecules and incorporated into the autophagosomal membrane. This membrane next expands and engulfs portions of the cytosol, including entire organelles. Upon formation of the autophagosome, the Atg12-Atg5-Atg16 complex dissociates from this structure while LC3 (Atg8)-PE remains in the autophagic lumen as an autophagosomal marker. The outer membrane of the autophagosome fuses with the lysosomal membrane forming an autolysosome within which the single membrane structure, or autophagic body, is degraded by lysosomal esterases, lipases, and proteases.
During periods of prolonged cellular stress, bulk autophagy declines and the process of CMA is upregulated. In CMA, specific cytosolic proteins displaying homologous pentapeptide motifs bind to a molecular chaperone complex composed of multiple heat shock proteins including the heat shock 70 kDa protein (hsc70) (reviewed in (9)). This molecular chaperone complex transports the substrate protein to the lysosomal membrane where it associates with an isoform of the lysosome-associated membrane protein-2, LAMP-2A, which functions as part of the lysosomal receptor for CMA. The cytoplasmic domain of LAMP-2A serves as a potential docking site for the molecular chaperone-substrate complex, and chaperone-mediated substrate unfolding likely enables proteins to bind and translocate across the lysosomal membrane. Lysosomal hsc70 (ly-hsc70) assists with the transport of substrate proteins into the organelle lumen where these molecules are degraded by mature acidic proteases.
Autophagy and MHC class I Ag presentation
In the conventional paradigm, MHC class I molecules are restricted to surveying the cytosol for endogenous Ag from viruses, tumors, or self proteins for presentation to CD8+ T cells (reviewed in (10)). These endogenous Ag are degraded into peptide fragments by cytosolic proteases such as the proteasome and transported via a heterodimeric complex composed of Transporter associated with Antigen Processing 1 (TAP1) and TAP2 into the endoplasmic reticulum (ER). Once in the ER, peptides of the appropriate length (8 to 10 amino acids) bind nascent MHC class I heavy chains and β2-microglobulin and then transit through the secretory pathway to the cell surface for recognition by CD8+ T cells. A direct role for autophagy in the conventional pathway for MHC class I presentation has not been firmly established, yet recent studies discussed below demonstrate potential roles for autophagy in regulating class I-mediated cross-presentation of exogenous Ag.
During the process of cross-presentation, bone marrow-derived antigen presenting cells (APC) such as dendritic cells (DC) internalize and degrade Ag from the extracellular environment and display the resulting peptides in association with MHC class I molecules on their cell surface (11). The stimulation of naïve CD8+ T cells by these peptide-MHC class I complexes is known as cross-priming. The class I-restricted cross-presentation pathway plays an important role in the immune surveillance of infected cells as well as tumors and the subsequent development of an appropriate cytotoxic T cell response to these pathogens or malignancies. Precisely how peptides from exogenous Ag access the MHC class I presentation pathway is still under investigation although several routes have recently been proposed. These pathways have been described extensively elsewhere (11–13); thus, only a brief summary of the current models of class I-mediated cross-presentation is offered here.
Studies have suggested that MHC class I molecules may acquire peptides derived from exogenous Ag internalized into phagosomes in a TAP-dependent or TAP-independent manner. In the TAP-dependent phagosome-to-cytosol-to-phagosome pathway, Ag is transported to the cytosol by a yet unknown mechanism, possibly Sec61, and degraded by the proteasome. Peptides are then re-imported into the phagosome which has acquired TAP as well as other ER proteins such as MHC class I. The phagosome-to-cytosol pathway calls for peptides generated in the cytosol by the proteasome to be directly transported to MHC class I molecules in the ER via TAP. Another possible mechanism of class I-restricted cross-presentation may involve the ER associated degradation pathway (ERAD). Here, exogenous Ag internalized in endosomes is transported into the ER, translocated for degradation in the cytosol by the proteasome, and finally transported back into ER by TAP. It has also been reported that DC may acquire exogenous peptides from other cells via GAP-junctions in order to cross-prime naïve CD8+ T cells. Lastly, in the vacuolar pathway, MHC class I molecules may acquire peptides that are generated in phagosomes by cysteine proteases such as cathepsin S in a TAP-independent manner. The mechanisms which potentiate MHC class I peptide loading in phagosomes via this latter pathway have not yet been determined.
Autophagy has recently been proposed as another possible mechanism for the MHC class I-mediated cross-presentation of exogenous Ag with results pertinent to tumor Ag (14). In this study, the authors induced macroautophagy in HEK 293T cells expressing a model Ag ovalbumin (OVA) by starvation or treatment with the compound rapamycin, which inhibits the mTor molecule (15). Following these treatments in vitro, enhanced cross-presentation of OVA to epitope-specific CD8+ T cells was observed. Conversely, the treatment of the OVA-expressing HEK 293T cells with 3-methyladenine (3-MA), an inhibitor of PI3K and macroautophagy (16), significantly reduced OVA cross-presentation in vitro. These authors also used rapamycin, 3-MA, and another PI3K inhibitor wortmannin (17) to modulate macroautophagy in melanoma cells and then measured the MHC class I-restricted cross-presentation of the tumor Ag gp100 in vivo. Similar to the results with the OVA-expressing HEK 293T cells, induction of macroautophagy enhanced the cross-presentation of gp100 while inhibition of macroautophagy reduced the cross-presentation of this Ag to epitope-specific CD8+ T cells. Macroautophagy was also specifically inhibited through the use of siRNA against the autophagy gene Atg6/beclin-1 in melanoma cells, and a significant reduction in the in vivo MHC class I-mediated cross-presentation of gp100 was again observed.
These results support a potential role for macroautophagy in the MHC class I-restricted cross-presentation of tumor Ag, yet further mechanistic studies to define the role of autophagy pathways in class I-mediated Ag presentation are needed. The induction of macroautophagy by rapamycin in the OVA-expressing HEK 293T cells as well as the melanoma cells leads to the conversion of LC3-I to its LC3-II form, suggesting that the MHC class I-restricted cross-presentation of OVA and gp100 may require the initiation and formation of autophagosomes. Yet, treatment of cells with NH4Cl, which blocks the acidification of lysosomes and prevents their fusion with autophagosomes, enhances class I-mediated cross-presentation. These results suggest that the degradation of the contents of autolysosomes during the late phases of autophagy hinders cross-presentation perhaps by destroying the Ag that serve as the sources of peptide epitopes for MHC class I molecules. In support of this, Li et al. show that purified autophagosomes from OVA-expressing HEK 293T cells or melanoma cells serve as a source of Ag for DC to promote cross-presentation of OVA or gp100 to the appropriate epitope-specific CD8+ T cells. While the authors demonstrate that autophagosomes may function as efficient carriers of Ag from donor cells, how and where these structures and their antigenic contents intersect with MHC class I molecules has yet to be determined (Fig. 2). Further insights into the specific mechanisms by which autophagy plays a role in the presentation of tumor Ag by MHC class I molecules may lead to the development of new therapeutic strategies for the treatment of cancer.
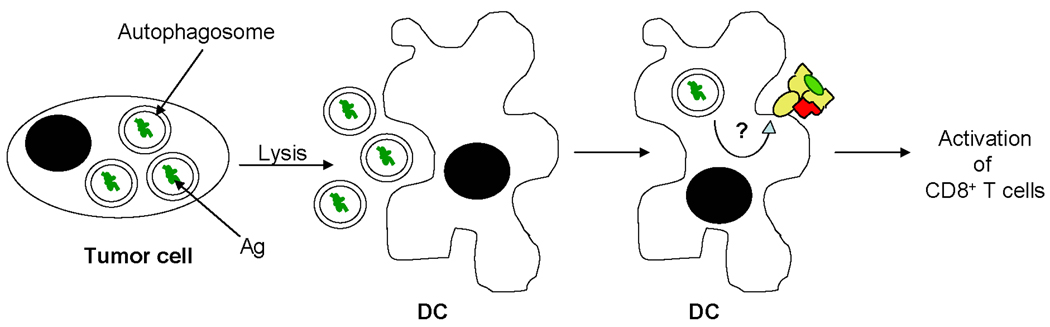
FIGURE 2. Autophagy as a potential mechanism for MHC class I-mediated presentation of tumor Ag.
Macroautophagy can be induced in tumor cells undergoing cellular stress as a result of nutrient depletion or chemotherapy. Autophagosomes containing tumor Ag may be released into the extracellular environment where they are internalized by DC. How and where tumor Ag in autophagosomes intersect with MHC class I molecules in DC is unknown. DC present tumor peptides in the context of class I for the activation of CD8+ T cells.
In addition to its role in tumor immunity, autophagy may also contribute to the CD8+ T cell response to intracellular pathogens such as bacteria, parasites, and viruses. For example, autophagy is required for the clearance of several pathogens (reviewed in (18–20), including the parasite Toxoplasma gondii which is localized in autophagosomes in infected macrophages (21, 22). Since CD8+ T cells are essential for protective immunity from T. gondii (23), it is possible that peptides derived from T. gondii access MHC class I molecules through autophagy. Several pathogens have been localized in autophagosomes that contain ER proteins (24), and perhaps components of the MHC class I presentation pathway such as class I molecules or TAP may also be found in autophagosomes. Furthermore, during periods of cellular stress, aggresome-like structures (ALIS) containing poly-ubiquitylated proteins are formed and serve as substrates for autophagy (25). ALIS can form in DC (DALIS) experiencing cellular stress, perhaps as a result of an infection by a pathogen, and the clearance of these DALIS by autophagy has been proposed as a source of peptides for MHC class I molecules (26).
Autophagy and MHC class II Ag presentation
Traditionally, MHC class II molecules present antigenic peptides derived from exogenous proteins to CD4+ T cells (27). MHC class II proteins are constitutively expressed on the surface of a number of professional APC such as DC, B cells, and macrophages as well as both cortical and medullary thymic epithelial cells (cTEC and mTEC, respectively). Treatment with an inflammatory signal such as interferon-gamma (IFN-γ) can induce MHC class II expression on the surface of non-professional APC such as endothelial cells, fibroblasts, epithelial cells, and many tumors. MHC class II complexes consist of α and β subunits which are first assembled in the ER with the chaperone molecule invariant chain (Ii) (28, 29). The cytoplasmic tail of Ii contains a motif that targets the Ii-MHC class II complexes to endosomal/lysosomal compartments where acidic proteases degrade Ii to a small fragment known as the class II-associated invariant chain peptide (CLIP) which remains associated with the MHC class II peptide binding groove (30, 31). Ag delivered into the endosomal/lysosomal network via receptor-mediated or fluid phase endocytosis are also exposed to proteases and denaturing reactions, yielding peptide ligands for class II molecules (32). CLIP removal and the capture of antigenic peptides by MHC class II proteins is catalyzed by the MHC-encoded molecule HLA-DM (33–35) and occurs in a highly specialized organelle resembling a late endosome/lysosome known as the MIIC (36). Resultant peptide-MHC class II complexes are ultimately trafficked to the cell surface for immune surveillance by CD4+ T cells.
Similar to the MHC class I-mediated cross-presentation of exogenous Ag to CD8+ T cells, recent studies have begun to reveal how cytoplasmic and nuclear Ag gain access to MHC class II molecules in lysosomal compartments for the presentation of peptides to CD4+ T cells (37, 38). Multiple pathways have been shown to contribute to the MHC class II-mediated presentation of cytoplasmic and nuclear viral, tumor, and self Ag in both professional and non-professional APC. These pathways include CMA, macroautophagy, a TAP-dependent pathway, and intercellular Ag transfer. The findings of these studies have been reviewed extensively elsewhere (39–42). Thus, here only a brief summary of these pathways for MHC class II-mediated endogenous Ag presentation is offered (Fig. 3).
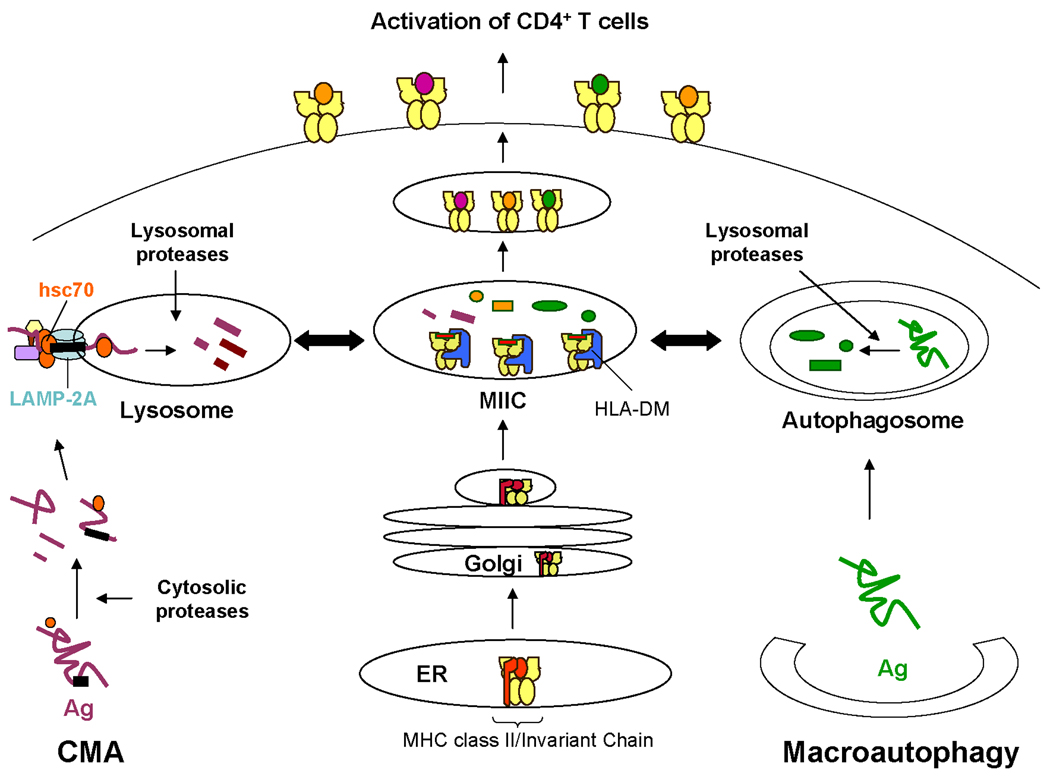
FIGURE 3. Autophagy pathways in the MHC class-mediated presentation of cytoplasmic and nuclear Ag.
Approximately 10–25% of MHC class II molecules display peptides from nuclear and cytoplasmic Ag likely accessing autophagy to reach class II. Left, Cytoplasmic Ag are first processed by cytosolic proteases and then utilize chaperone-mediated autophagy (CMA) to access lysosomes. These antigenic fragments intersect with MHC class II molecules in a mature endosomal/lysosomal compartment known as the MIIC prior to presentation to CD4+ T cells. Right, Cytoplasmic or nuclear Ag are sequestered into double-membrane structures known as autophagosomes which continuously fuse with mature endsosomes such as the MIIC and lysosomes, thus allowing Ag to associate with MHC class II molecules. Further processing of the cytoplasmic or nuclear Ag by lysosomal proteases may occur prior to binding to class II and presentation to CD4+ T cells.
Recently, our laboratory demonstrated a role for CMA in regulating cytoplasmic Ag presentation by MHC class II molecules by modulating the cellular levels of two components of the CMA pathway, LAMP-2A and hsc70, in human B cells expressing cytoplasmic Ag glutamic acid decarboxylase (GAD) or a mutant form of human Ig κ light chain, designated SMA (37). Expression of anti-sense cDNA for LAMP-2 reduced the MHC class II-restricted presentation of GAD while over-expression of the LAMP-2A isoform resulted in an increased presentation of epitopes from both GAD and SMA. In B cells, SMA, one of the causative agents of light chain amyloidosis, fails to fold properly after synthesis and is immediately translocated out of the ER to the cytosol (43), suggesting that CMA may contribute to the immune recognition of mis-folded ER proteins. Stress induced autophagy pathways also appear to play a role in the presentation of multiple cytoplasmic and nuclear Ag. Human B cells in which autophagy is induced by serum starvation display an increase in those MHC class II-associated peptides derived from intracellular proteins as measured by mass spectrometry (44). Studies using PI3K inhibitors 3-MA and wortmannin or siRNA targeting the Atg12 gene to block macroautophagy demonstrated that multiple epitopes from cytoplasmic and nuclear proteins are dependent on this pathway for their presentation by MHC class II molecules. These epitopes include the cytosolic and nuclear versions of the bacterial protein neomycin (45, 46), mucin-1 (47), and the EBV Nuclear Ag (EBNA) 1 (48). A recent report by Munz and colleagues demonstrates that in B cells and DCs, autophagosomes are constitutively formed and continuously fuse with MIIC (38). Furthermore, when the influenza matrix protein 1 (MP1) was targeted to autophagosomes by fusion with LC3 (Atg8), an increase in the MHC class II-restricted presentation of this epitope to CD4+ T cells was observed.
Two additional pathways have been shown to contribute to the MHC class II-mediated presentation of endogenous viral Ag in B cells and DC including a TAP-dependent pathway and intercellular Ag transfer. Both the proteasome and TAP were shown to be required for the generation of two MHC class II-restricted epitopes encoded within two distinct transmembrane glycoproteins from influenza virus using murine B cells and DC (49). These results differ from several reports suggesting that TAP plays no role in the presentation of cytoplasmic Ag to MHC class II molecules (37, 46, 50–53), thus further studies regarding this transport pathway in specialized immune cells such as DC are clearly needed. In the second pathway, two Epstein Barr virus (EBV) nuclear Ag, EBNA 2 and EBNA 3C, were shown to gain access to the MHC class II pathway via intercellular Ag transfer (54). Treatment of EBV-transformed human B cells with 3-MA to block macroautophagy failed to inhibit the MHC class II-restricted presentation of these endogenous viral Ag. Whether intact viral Ag or antigenic fragments complexed with other proteins or within exosomes are transferred to recipient APC for MHC class II-mediated presentation is unknown. However, Ag processing by lysosomal proteases within the recipient cell is required for the class II-restricted presentation of EBNA 2 and EBNA 3C viral Ag, strongly supporting intercellular or cross-presentation.
Autophagy and the regulation of immune responses
Autophagy pathways may be induced or altered during infection and immunity as cellular stress responses are initiated. In vitro studies have demonstrated recognition of viral, self, and tumor Ag dependent upon autophagy in the context of MHC class II Ag presentation. A role for autophagy in the cross-presentation of tumor Ag via MHC class I molecules has also been reported both in vitro and in vivo (14). Less is known about whether autophagy influences either CD4+ or CD8+ T cell responses to bacterial pathogens. A recent report on autophagy in the thymus provides a clue as to the importance of this process in the presentation of self Ag and its impact on the induction and maintenance of CD4+ T cell tolerance (55). The potential role of autophagy in regulating immune responses to pathogens, tumors, and self Ag has been reviewed extensively elsewhere (18, 41, 56, 57) with highlights of recent studies discussed here.
Intracellular pathogens
Studies have begun to examine the role of autophagy in a host cell’s defense against intracellular pathogens such as viruses and bacteria although the precise molecular steps involving autophagy or autophagy-linked gene products are still poorly understood (reviewed in (41, 58)). Autophagy limits the virus-induced encephalitis observed during Sindbis virus infection through the interaction of the Atg6 gene Beclin 1 with Bcl-2 (59). Additionally, during the process of xenophagy, autophagosomes engulf and destroy viruses (7). For example, the herpes simplex virus-1 (HSV-1) is degraded within autophagosomes in a dsRNA-activated protein kinase (PKR) dependent manner (60). Autophagy has recently been shown to play a role in the innate immune response to vesicular stomatitis virus (VSV) (61). In plasmacytoid DC (pDC), autophagy facilitates the transport of VSV replication intermediates from the cytosol to lysosomes potentially to promote Toll-like receptor 7 (TLR7) engagement. Activation of TLR7 in DC results in the induction of important mediators of anti-viral immunity such as IFN-α and IL-12. Plasmacytoid DC also recognize viruses through TLR9, localized in endosomes, and signaling through TLR9 recruits and activates the transcription factor interferon response factor 7 (IRF7) which is essential for the production of type I interferons (62). Using rapamycin and siRNA against mTOR, Cao et al recently demonstrated that virus-induced TLR9 dependent signaling through the mTOR pathways is also critical for the production of type I interferons in pDC as well as bone-marrow derived DC and macrophages (63). Whether signaling through mTOR following virus-induced activation of TLR9 affects the autophagy pathway during an innate anti-viral immune response has yet to be investigated. Lastly, as discussed above, the MHC class II-mediated presentation of the EBV Ag EBNA1 by autophagy suggests that this pathway may also play a role in the adaptive immune response to viruses (48). The demonstration that targeting influenza MP1 to autophagosomes enhanced MHC class II presentation of the MP1 epitope (38) may suggest that in virally infected cells, fusion of autophagosomes containing viral particles with lysosomes could promote viral Ag association with class II for presentation to CD4+ T cells.
Host cells also utilize autophagy pathways in their defense against bacteria and parasites. Autophagy limits bacterial replication by enveloping free bacteria such as group A Streptococcus in autophagosomes for delivery to lysosomes (64) or by targeting phagosomes containing bacteria such as Mycobacterium tuberculosis for fusion with lysosomes (65, 66). The innate immune response initiated as a result of pathogenic infections also influences autophagy pathways. For example, signaling through TLR7 during infection of macrophages with M. bovis bacilli Calmette-Guerin (BCG) induces macroautophagy resulting in the elimination of the intracellular bacteria (67). Furthermore, cytokine expression may modulate macroautophagy in order to limit pathogen replication. Type II IFN enhances the degradation of M. tuberculosis and Ricksettia conorii in infected cells (65, 66), and the TNF family member CD40L induces macroautophagy to facilitate the fusion of phagosomes containing T. gondii with lysosomes during parasite infection (21). Finally, although the role of autophagy in the adaptive immune response to bacteria and parasites has not yet been carefully examined, it is possible that in infected cells, fusion of autophagosomes containing the intracellular pathogens with lysosomes could be a mechanism by which bacterial or parasitic Ag associate with MHC class II molecules for presentation to CD4+ T cells.
Cancer
Similar to its role in infectious disease, autophagy is believed to contribute to both cancer development and tumor suppression (57). Autophagy may be induced during the later stages of oncogenesis as the rapid proliferation of the tumor depletes critical nutrients, thus suppressing further tumor growth. Additionally, following chemotherapy, tumor cells may initiate autophagy as a protective measure to recycle nutrients or remove damaged cellular components. For example, the induction of macroautophagy increases the survival of breast cancer cells undergoing treatment with 4-hydroxytamoxifen (68). Yet, how modulation of the autophagy pathways in tumor cells influences immune recognition and clearance has not yet been thoroughly investigated. As discussed above, one recent report suggests autophagy as a potential mechanism for the cross-presentation of tumor Ag onto MHC class I molecules. It is also possible that during the induction of autophagy within a tumor cell, the formation of autophagosomes might provide a source of tumor Ag which could intersect with MHC class II molecules upon fusion of the autophagosomes with lysosomes.
Autoimmunity and tolerance
Autophagy appears to also play a role in the breakdown of tolerance and the development of autoimmunity (reviewed in (41, 69)). During autophagy, contents of the cytosol and nucleus, including potential self Ag, are degraded in lysosomal compartments where MHC class II molecules reside. Thus, alterations in autophagy pathways may lead to a breakdown in tolerance or to insufficient tolerance induction and the development of autoimmunity. A recent report by Nedjic et al demonstrated that autophagy plays a key role in the generation and maintenance of tolerance in the thymus (55). In this study, autophagosomes were not readily detected in murine peripheral APC, yet cortical thymic epithelial cells (cTEC), which express MHC class II molecules, were found to display high levels of constitutive macroautophagy. The authors postulated that macroautophagy within cTEC may favor the presentation of a broader spectrum of self Ag via class II on these cells. Using thymi from Atg5−/− mice, the authors evaluated the positive and negative selection of several MHC class II-restricted transgenic TCRs and found that the loss of Atg5 altered the selection of some, but not all, TCRs. These results suggested that in the absence of autophagy, the composition of peptide-MHC class II complexes on the surface of cTECs was altered, thus skewing the T cell repertoire that was positively selected. Additionally, the lack of autophagy may also prevent MHC class II presentation of certain tissue specific Ag in medullary thymic epithelial cells (mTEC), thus impairing negative selection and allowing autoreactive T cells to exit into the periphery. In support of this, the authors observed severe colitis and inflammation of multiple organs in athymic mice which received thymi from Atg5−/− mice. Studies of T cells deficient in Atg5 revealed defects in cellular proliferation and survival (70), suggesting further analysis of the role of this gene product in regulating T cell responses as well as the thymic microenvironment are needed.
In humans, recent studies have identified ATG16L1 (autophagy-related 16-like 1), which is involved in autophagosome formation, and IRGM GTPase, which stimulates autophagy, as susceptibility loci for Crohn’s disease (71–74). It has been proposed that mutations in these genes may lead to defective macroautophagy, thus impairing the innate immune response to bacterial infection and increasing the bacterial load in the gut, resulting in enhanced mucosal inflammation and the development of disease. A recent study using Atg16L1-deficient mice demonstrated an increase in the production of the inflammatory cytokines IL-1β and IL-18 and in the susceptibility to colitis (75). Alternatively, as the data from the Atg5−/− mice would suggest, inhibition of autophagy pathways might also lead to the failure to induce tolerance to mucosal self Ag. Lastly, the tissue damage associated with certain autoimmune diseases such systemic lupus erthematosus (SLE) has been attributed to the defective clearance of apoptotic cells. Macroautophagy plays a role in the removal of these apoptotic cell corpses, and the impairment of this process may result in the build-up of necrotic material in tissues, and thus, inflammation.
Conclusions
Multiple mechanisms, including CMA and macroautophagy, have been implicated in the delivery of several endogenous Ag to lysosomes for their eventual presentation by MHC class II molecules. While substantial research has been conducted into the role of autophagy in the MHC class II presentation of endogenous Ag, far less is known about autophagy in MHC class I presentation and cross-presentation pathways. Further studies are also needed to more clearly define the relationship between the various autophagy pathways and MHC class I and class II Ag presentation during the immune response to intracellular pathogens, tumors, and self Ag. Inhibition or enhancement of autophagy may alter the MHC class I and class II presentation of Ag to CD8+ or CD4+ T cells, respectively, thus providing novel therapeutic strategies for the treatment of infectious diseases, cancer, and autoimmunity.
Abbreviations used in this paper
- CMA
chaperone-mediated autophagy
- Cvt
cytoplasm-to-vacuole targeting
- mTor
target of rapamycin
- Atg
autophagy-related gene
- PE
phosphatidylethanolamine
- hsc70
heat shock 70 kDa protein
- LAMP
lysosome-associated membrane protein
- ly-hsc70
lysosomal hsc70
- ER
endoplasmic reticulum
- DC
dendritic cells
- ERAD
ER associated degradation pathway
- 3-MA
3-methyladenine
- ALIS
aggresome-like structures
- DALIS
DC aggresome-like structures
- cTEC
cortical thymic epithelial cells
- mTEC
medullary thymic epithelial cells
- Ii
invariant chain
- GAD
glutamic acid decarboxylase
- EBNA
EBV Nuclear Ag
- MP1
influenza matrix protein 1
- PKR
dsRNA-activated protein kinase
- VSV
vesicular stomatitis virus
- pDC
plasmacytoid DC
- BCG
bacilli Calmette-Guerin
- ATG16L1
autophagy-related 16-like 1
- SLE
systemic lupus erthematosus.
Footnotes
Publisher's Disclaimer: The following disclaimer is a requirement of The Journal of Immunology: “This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the United States National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.”
V. L. Crotzer is supported by a Career Development Award from the Melanoma Research Foundation. NIH AI49589 supported the effort of J. S. Blum.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 Suppl 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattler T, Mayer A. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J Cell Biol. 2000;151:529–538. doi: 10.1083/jcb.151.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L, Strandberg L, Lenardo MJ. The selectivity of autophagy and its role in cell death and survival. Autophagy. 2008;4:567–573. doi: 10.4161/auto.5902. [DOI] [PubMed] [Google Scholar]
- 6.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 11.Shen L, Rock KL. Priming of T cells by exogenous antigen cross-presented on MHC class I molecules. Curr Opin Immunol. 2006;18:85–91. doi: 10.1016/j.coi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Raghavan M, Del Cid N, Rizvi SM, Peters LR. MHC class I assembly: out and about. Trends Immunol. 2008;29:436–443. doi: 10.1016/j.it.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–6895. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 16.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 19.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luder CG, Seeber F. Toxoplasma gondii and MHC-restricted antigen presentation: on degradation, transport and modulation. Int J Parasitol. 2001;31:1355–1369. doi: 10.1016/s0020-7519(01)00260-0. [DOI] [PubMed] [Google Scholar]
- 24.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Bacterial interactions with the autophagic pathway. Cell Microbiol. 2002;4:1–10. doi: 10.1046/j.1462-5822.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- 25.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, Bazett-Jones DP, Brumell JH. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 26.Pierre P. Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol Rev. 2005;207:184–190. doi: 10.1111/j.0105-2896.2005.00300.x. [DOI] [PubMed] [Google Scholar]
- 27.Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol. 2004;5:685–692. doi: 10.1038/ni1088. [DOI] [PubMed] [Google Scholar]
- 28.Cresswell P. Invariant chain structure and MHC class II function. Cell. 1996;84:505–507. doi: 10.1016/s0092-8674(00)81025-9. [DOI] [PubMed] [Google Scholar]
- 29.Sant AJ, Miller J. MHC class II antigen processing: biology of invariant chain. Curr Opin Immunol. 1994;6:57–63. doi: 10.1016/0952-7915(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 30.Maric MA, Taylor MD, Blum JS. Endosomal aspartic proteinases are required for invariant-chain processing. Proc Natl Acad Sci U S A. 1994;91:2171–2175. doi: 10.1073/pnas.91.6.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riese RJ, Wolf PR, Bromme D, Natkin LR, Villadangos JA, Ploegh HL, Chapman HA. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity. 1996;4:357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- 32.Watts C. Antigen processing in the endocytic compartment. Curr Opin Immunol. 2001;13:26–31. doi: 10.1016/s0952-7915(00)00177-1. [DOI] [PubMed] [Google Scholar]
- 33.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 34.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 35.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 36.Pieters J. MHC class II compartments: specialized organelles of the endocytic pathway in antigen presenting cells. Biol Chem. 1997;378:751–758. [PubMed] [Google Scholar]
- 37.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crotzer VL, Blum JS. Cytosol to lysosome transport of intracellular antigens during immune surveillance. Traffic. 2008;9:10–16. doi: 10.1111/j.1600-0854.2007.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, Gregg JL, Wang N, Zhou D, O'Donnell P, Blum JS, Crotzer VL. Compartmentalization of class II antigen presentation: contribution of cytoplasmic and endosomal processing. Immunol Rev. 2005;207:206–217. doi: 10.1111/j.0105-2896.2005.00297.x. [DOI] [PubMed] [Google Scholar]
- 41.Lunemann JD, Munz C. Autophagy in CD4(+) T-cell immunity and tolerance. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.113. [DOI] [PubMed] [Google Scholar]
- 42.Strawbridge AB, Blum JS. Autophagy in MHC class II antigen processing. Curr Opin Immunol. 2007;19:87–92. doi: 10.1016/j.coi.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 43.Dul JL, Davis DP, Williamson EK, Stevens FJ, Argon Y. Hsp70 and antifibrillogenic peptides promote degradation and inhibit intracellular aggregation of amyloidogenic light chains. J Cell Biol. 2001;152:705–716. doi: 10.1083/jcb.152.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riedel A, Nimmerjahn F, Burdach S, Behrends U, Bornkamm GW, Mautner J. Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur J Immunol. 2008;38:2090–2095. doi: 10.1002/eji.200737900. [DOI] [PubMed] [Google Scholar]
- 46.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 47.Dorfel D, Appel S, Grunebach F, Weck MM, Muller MR, Heine A, Brossart P. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005;105:3199–3205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 48.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 49.Tewari MK, Sinnathamby G, Rajagopal D, Eisenlohr LC. A cytosolic pathway for MHC class II-restricted antigen processing that is proteasome and TAP dependent. Nat Immunol. 2005;6:287–294. doi: 10.1038/ni1171. [DOI] [PubMed] [Google Scholar]
- 50.Lich JD, Elliott JF, Blum JS. Cytoplasmic processing is a prerequisite for presentation of an endogenous antigen by major histocompatibility complex class II proteins. J Exp Med. 2000;191:1513–1524. doi: 10.1084/jem.191.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malnati MS, Marti M, LaVaute T, Jaraquemada D, Biddison W, DeMars R, Long EO. Processing pathways for presentation of cytosolic antigen to MHC class II-restricted T cells. Nature. 1992;357:702–704. doi: 10.1038/357702a0. [DOI] [PubMed] [Google Scholar]
- 52.Dissanayake SK, Tuera N, Ostrand-Rosenberg S. Presentation of endogenously synthesized MHC class II-restricted epitopes by MHC class II cancer vaccines is independent of transporter associated with Ag processing and the proteasome. J Immunol. 2005;174:1811–1819. doi: 10.4049/jimmunol.174.4.1811. [DOI] [PubMed] [Google Scholar]
- 53.Dani A, Chaudhry A, Mukherjee P, Rajagopal D, Bhatia S, George A, Bal V, Rath S, Mayor S. The pathway for MHCII-mediated presentation of endogenous proteins involves peptide transport to the endo-lysosomal compartment. J Cell Sci. 2004;117:4219–4230. doi: 10.1242/jcs.01288. [DOI] [PubMed] [Google Scholar]
- 54.Taylor GS, Long HM, Haigh TA, Larsen M, Brooks J, Rickinson AB. A Role for Intercellular Antigen Transfer in the Recognition of EBV-Transformed B Cell Lines by EBV Nuclear Antigen-Specific CD4+ T Cells. J Immunol. 2006;177:3746–3756. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 55.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 56.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HK, Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20:23–29. doi: 10.1016/j.coi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Talloczy Z, Virgin HWt, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 61.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 62.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 63.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 65.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 66.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 67.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. Embo J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, Browning D, Rawson J, Smith SB, Barrett JT, Schoenlein PV. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7:2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 69.Lleo A, Invernizzi P, Selmi C, Coppel RL, Alpini G, Podda M, Mackay IR, Gershwin ME. Autophagy: highlighting a novel player in the autoimmunity scenario. J Autoimmun. 2007;29:61–68. doi: 10.1016/j.jaut.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 72.Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A, Mansfield JC, Lewis CM, Schreiber S, Mathew CG. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 73.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]