Abstract
The small GTPase Rap1 affects cell adhesion and cell motility in numerous developmental contexts. Loss of Rap1 in the Drosophila wing epithelium disrupts adherens junction localization, causing mutant cells to disperse, and dramatically alters epithelial cell shape. While the adhesive consequences of Rap1 inactivation have been well described in this system, the effects on cell signaling, cell fate specification, and tissue differentiation are not known. Here we demonstrate that Egfr-dependent cell types are lost from Rap1 mutant tissue as an indirect consequence of DE-cadherin mis-localization. Cells lacking Rap1 in the developing wing and eye are capable of responding to an Egfr signal, indicating that Rap1 is not required for Egfr/Ras/MAPK signal transduction. Instead, Rap1 regulates adhesive contacts necessary for maintenance of Egfr signaling between cells, and differentiation of wing veins and photoreceptors. Rap1 is also necessary for planar cell polarity in these tissues. Wing hair alignment and ommatidial rotation, functional readouts of planar cell polarity in the wing and eye respectively, are both affected in Rap1 mutant tissue. Finally, we show that Rap1 acts through the effector Canoe to regulate these developmental processes.
Keywords: Rap1, adhesion, cadherin, planar cell polarity, canoe, wing vein, ommatidia
Introduction
Members of the Ras superfamily of small GTPases serve as molecular switches in many biological systems, translating environmental signals into specific cellular responses. GTPases of this type cycle between guanosine diphosphate (GDP)-bound and guanosine triphosphate (GTP)-bound states, typically adopting active conformations when associated with GTP (Bourne et al., 1990). GTP-bound Ras-like proteins bind downstream effectors, thereby activating signal transduction pathways. Among factors that regulate the activity of these proteins are GTPase exchange factors (GEFs) that promote the active GTP-bound state, and GTPase activating proteins (GAPs) that promote the inactive state (Vetter and Wittinghofer, 2001). Frequently, Ras-like proteins function as part of intercellular signaling pathways, acting downstream of transmembrane receptors to mediate cytosolic or transcriptional effects. The wide variety of biological effects mediated by these proteins includes cell proliferation, differentiation, cytoskeletal remodeling, and vesicular trafficking, to name just a small subset. In Drosophila, there are over one hundred genes in the Ras superfamily, roughly divided into the Ras/Rap/Ral, Rho, Rab, Ran and Arf/Sar families. Although a certain amount of crosstalk exists, each family is specialized for particular functions. For example, Rho GTPases generally control actin dynamics important during cell migration, while Rab GTPases are critical for intracellular membrane trafficking (Lundquist, 2006).
The gene Roughened (also called Rap1) encodes the Drosophila Rap1 homologue. Rap1 is the GTPase most highly related to Ras, and was initially identified based on its ability to antagonize Ras function in cell culture, restoring a malignant phenotype of K-Ras-transformed fibroblasts (Kitayama et al., 1989). Canonically, an Egf signal is conveyed through the Egfr/Ras/Raf/MEK/MAPK signal transduction pathway to affect the activity of transcription factors in the nucleus. It has been demonstrated in untransformed fibroblasts that activated Rap1 inhibits Ras signaling through MAP-Kinase (MAPK), potentially inhibiting Ras activation of Raf (Cook et al., 1993). More recent data, however, suggests that Rap1 does not always antagonize Ras signaling. In the Drosophila retina, data indicates that Ras and Rap1 function through independent pathways (Asha et al., 1999), while during embryogenesis, Rap1 has been shown to bind Raf and positively regulate the Raf/ERK/MAPK pathway in a Ras-independent fashion (Mishra et al., 2005). Exactly how Ras and Rap1 interact to mediate signaling downstream of receptor tyrosine kinases remains controversial.
A dominant theme emerging from the literature, however, is that Rap1 regulates adhesion between cells, affecting the localization and integrity of cell junctions. In particular, Rap1 has been shown to affect the inside-out activation of integrins (Bos et al., 2003; Caron, 2003; Han et al., 2006) and localization of the apico-lateral adherens junction complex (Hogan et al., 2004; Price et al., 2004; Wang et al., 2006). It is not surprising therefore, that Rap1 is a critical determinant of a cellular morphology and migration. This was first demonstrated when activation of Rap1 signaling, through misexpression of the Rap1-specific GEF C3G, increased cell spreading and attachment to the matrix in culture, while the Rap1-specific GAP Spa1 caused cell rounding and detachment from the matrix (Tsukamoto et al., 1999). Rap1 function has been most extensively characterized in the immune system where it plays a key role in the inside-out activation of integrins, controlling integrin receptor avidity (clustering), and ligand affinity in leucocytes (Kinashi and Katagiri, 2005). In addition, Rap1 polarizes leucocytes, generating a leading edge and stimulating transmigration through the vascular endothelium (Shimonaka et al., 2003).
Our understanding of Rap1 function is less complete in the context of cadherin-based cell adhesion. Cadherins are transmembrane proteins that mediate primarily homophilic cell interactions (Nose et al., 1988). While the extracellular cadherin repeat domains form contacts between neighboring cells, the cadherin intracellular domain binds β-catenin, which in turn complexes with the actin-binding protein α-catenin. In this way, cadherins serve as critical links between the cytoskeletons of adjacent cells. In polarized epithelia, cadherins are part of the adherens junction complex of proteins, localized in a ring around each cell at the boundary between the apical and basolateral membrane domains (apico-lateral). It was initially observed that Rap1 affects adherens junctions in the Drosophila wing epithelium (Knox and Brown, 2002). Wild-type wing cells maintain contact with their neighbors as they proliferate (Gibson et al., 2006), resulting in highly cohesive cell clones. In contrast, Rap1 mutant clones of cells disperse, intermingling with their wild-type neighbors. In these mutant cells, adherens junctions are not evenly distributed around the apical circumference, reducing adhesion to neighboring cells and causing fragmentation of the clone (Knox and Brown, 2002). This result indicates that Rap1 mediates an active process, which during the course of epithelial development maintains circumferential cell-to-cell contacts. A similar phenotype is seen in the Drosophila testes, where Rap1 is essential for stem cell anchoring to the niche (Wang et al., 2006). Since the initial observations in Drosophila, it has also been shown that Rap1 activity is required in mammalian cells to maintain E-cadherin-based cell-cell contacts (Hogan et al., 2004; Price et al., 2004).
Here we demonstrate that in developing epithelia the adhesive defects associated with Rap1 loss of function affect signaling between cells and tissue morphogenesis. In particular, Rap1 activity is critical for differentiation of Epidermal growth factor receptor (Egfr)-dependent cell types in the Drosophila wing and eye, as wing veins and photoreceptors are often lost from Rap1 mutant tissue. Planar cell polarity (PCP) is also impaired in the absence of Rap1, since wing hair formation and alignment, as well as ommatidial rotation are abnormal. Finally, we identify Canoe as a critical Rap1 effector in these developmental processes.
Materials and Methods
Genetics
The Rap1RB3 allele contains a point mutation, resulting in a premature stop codon near the N-terminus of the Rap1 protein (Hariharan et al., 1991). Rap1CD3 (Asha et al., 1999) and Rap1CD5 (Boettner et al., 2003) contain deletions that remove the entire Rap1 coding sequence. All three alleles behave as genetic nulls. Additional genetic reagents include FRT80B, Ubi-GFP (Xu et al., 1993), MARCM 80b (Lee and Luo, 2001), UAS-sSpitz, UAS-p35 (Zhou et al., 1997), GMR-p35 (Johnson et al., 2002), ey-flp (Newsome et al., 2000), NP2631-Gal4 (National Institute of Genetics, Japan), UAS-Rap1-IR (v20761 and v33437, Vienna Drosophila RNAi Center), UAS-Rap1N17 (Boettner et al., 2000), shg2 (Tepass et al., 1996), Actin>CD2>Gal4, UAS-GFP (Neufeld et al., 1998; Pignoni and Zipursky, 1997; Struhl and Basler, 1993), sev-Gal4 (K25) (Wilk et al., 1996), mδ0.5-Gal4 (Gaengel and Mlodzik, 2003), Star48-5 (Gaengel and Mlodzik, 2003), ptc-Gal4, stan192 (Rawls and Wolff, 2003), dbtar (Rothenfluh et al., 2000), mwh1 (Wong and Adler, 1993), UAS-canoe-IR (National Institute of Genetics, Japan), cno2 (Miyamoto et al., 1995), apGal4 (Calleja et al., 1996).
Mitotic recombination was induced using the Flp/FRT system (Xu et al., 1993). Larvae containing the hs-flp transgene were heat shocked for 45–60 minutes in a 37°C water bath at 72 hours after egg deposition (AED). Alternatively, ey-flp (Newsome et al., 2000) was used to generate clones in the eye. To generate Flp/Gal4 overexpressing clones (Neufeld et al., 1998; Pignoni and Zipursky, 1997; Struhl and Basler, 1993) larvae were heat shocked 8–12 minutes.
Immunohistochemistry
Larval and pupal tissues were fixed in 4% paraformaldehyde/PBS for 20 minutes at room temperature. Samples were placed in PBT (0.1% Triton-X/PBS) containing 4% Normal Goat Serum or 0.1% BSA, and incubated over-night at 4°C or one hour at room temperature. Samples were then incubated with primary antibodies over-night at 4°C. Primary antibodies used were anti-DE-cadherin (1:100, DSHB), anti-DSRF (1:500, Active Motif), anti-dpERK (1:500, Sigma), anti-dpERK (1:100, Cell Signaling Technology), anti-cleaved Caspase-3 (1:100, Cell Signaling Technology), anti-Canoe (1:00, gift from D. Yamamoto), anti-Dlg (1:100, DSHB), anti-DN-cadherin (1:100, DSHB), anti-Elav (1:200, DSHB), anti-Atonal (1:2000, gift from D. Marenda), anti-Senseless (1:1000, gift from H. Bellen), anti-Rough (1:100, DSHB), anti-Prospero (1:10, DSHB), anti-Cut (1:80, DSHB), anti-Stan (1:50, DSHB), anti-Mwh (1:500, gift from P. Adler). Alexa 488-, 568-, and 633-conjugated (Molecular Probes, 1:1500), or Fluorescein-, Rhodamine-, DyLight 649- and Cy5-conjugated (Jackson ImmunoResearch, 1:200) secondary antibodies were used. Rhodamine-conjugated phalloidin (Molecular Probes, 1:1500 in PBS) was used to stain F-actin. Nuclei were stained with Hoechst 33258 (Acros, 1:1500). Discs were mounted in Fluoroguard (Biorad) or 80% Glycerol containing 5% N-propyl gallate. Images were obtained on a Leica TCS SP confocal or a Zeiss Apotome microscope. Stacks of images were compiled and analyzed using Image J and Adobe Photoshop.
Western blots
Western blots were performed as described in Current Protocols in Molecular Biology. Briefly, 30 eye-antennal discs were dissected and lysed in 30 µL RIPA buffer (150mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris pH 8.0, 1mM phenyl methylsulfonyl fluoride). 30 µL 2x SDS-PAGE buffer (Bio-Rad) was added and samples were boiled 10 minutes to denature proteins. 20 µL of each sample were separated by SDS-PAGE and transferred to an Immobilon-P membrane (Millipore). Membranes were probed with anti-Rap1 (1:500, BD-Transduction Laboratories), washed and probed with horseradish peroxidase-conjugated antibodies (1:2,500, Jackson ImmunoResearch). Chemiluminescent detection was performed and anaylzed with a ChemiDoc XRX imaging system and Quantity One software (Bio-Rad). Membranes were stripped and re-probed with anti-Ras (1:500, Cell Signaling Technologies) and anti-Tubulin (1:500, Developmental Studies Hybridoma Bank) and analyzed as above.
Quantitative PCR
Thirty NP2631-Gal4, UAS-Rap1-IR/+ or NP2631-Gal4/+ eye-antennal imaginal discs were dissected from larvae grown at 25°C. PolyA+ RNA was isolated using a Dynabeads ®mRNA Direct kit™ (Invitrogen). Three independent mRNA isolations were performed for each genotype (biological replicates). The mRNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop). Reverse transcription reactions were performed on 20 ng of mRNA using an iScript™select cDNA synthesis kit (Biorad) following the manufacturer’s instructions. cDNA was then diluted 1:10. The PCR quantification was performed using 1µl cDNA per reaction with a ICycler IQ ™multicolored Real Time PCR Detection System (Biorad) using the IQ™ SYBR®Green Supermix (Biorad) as the detection dye. Each cDNA sample was run in triplicate and the average CT was used to calculate the fold change using the −2DDCT method (Livak and Schmittgen, 2001). Expression levels were normalized to rp49 expression. A Student’s two-tailed t-test was used to assess for statistical significance between control and sample fold change.
The following primers were used:
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Rap1 | CAGTGCATCTTCGTTGGAGAA | CGTGATCGAGTAGACCAGCA |
| Ras85D | AGAGGTGGCCAAACAGTACG | CGCACCAGTGTGTAAAATGC |
| Ric | CACGAAAACGAATCAAATGC | ACATGGACACGACACATTGC |
| Rab-RP3 | AGCGGTCTTCCTTTTCCAAT | AGGTCTCCATCACGAACAGG |
| Rap21 | GAAAGGAAGTCAGCCAGCAC | GAACGATGGTGGCGAATACT |
| Rp49 | CCAAGGACTTCATCCGCCACC | GCGGGTGCGCTTGTTCGATCC |
Adult eye imaging
Prior to sectioning, adult fly heads were fixed in 2% Glutaraldehyde, 2% OsO4 in phosphate buffer (0.1 M sodium phosphate pH 7.2) for 1 hour on ice, then transferred to 2% OsO4 in phosphate buffer for 2 hours on ice. Samples were serially dehydrated to 100% ethanol followed by two 10 minute washes in propylene oxide (Electron Microscopy Sciences). Samples were incubated overnight in 50% propylene oxide and Durcapan resin (Electron Microscopy Sciences), followed by 4 hours in 100% resin at room temperature. Samples were oriented in molds in 100% resin and incubated at 60°C overnight. Samples were tangentially sectioned (1µm) at the equatorial region on either a Leica UC6 or a Sorvall MT2 B Ultramicrotome. SEM adult eye images were obtained using a Hitachi TM1000 Table Top Scanning Electron Microscope.
PCP quantifications
Techniques were devised to quantify relative orientation and alignment of wing hairs and ommatidia. In the wing, patched-Gal4 was used to express a Rap1 RNAi transgene in a stripe between veins L3 and L4. An image of fixed size was taken immediately distal to the posterior cross-vein and anterior to vein L4. 250–300 wing hairs were contained in each image. Using Image J, a line was drawn over each hair delineating the hair orientation. Image J was used to determine an orientation angle for each hair (pointing distally equaled zero). Mean wing hair angle and wing hair angle variance were calculated for each wing with 5–10 wings quantified for each genotype. In wildtype wings, mean wing hair angle was expected to be near zero, and wing hair angle variance low, as hairs generally point distally and are aligned. Deviations in mean wing hair angle indicate a coordinated rotation of hairs away from the distal orientation (hairs could remain aligned). An increase in wing hair angle variance indicates hair misalignment. This calculation can be applied to other aligned epithelial structure as a measure of variation in planar polarity.
In the eye, an image of fixed size was taken at the R7 plane of a tangential section centered at the equator. 50–120 scorable ommatidia were contained in each image. Similar to the wing, using Image J an orientation angle was determined for each ommatidium, and angle means and variances calculated for each eye (3–7 eyes per genotype). The Wilcoxon Rank-Sum two-tailed test was used to determine the statistical significance of measurements.
Results
Rap1 affects epithelial cell adhesion and wing vein differentiation
As has been previously demonstrated, clones of cells lacking Rap1 activity in the Drosophila pupal wing had abnormal adhesive properties (Knox and Brown, 2002). Compared to wild-type controls at 36 hours after puparium formation (APF) (Fig.1A), Rap1 mutant cells dispersed, scattering into the adjacent wildtype epithelium (Fig.1B). Distribution of the homophilic cell adhesion molecule DE-cadherin (DE-cad) (encoded by the gene shotgun (shg) in Drosophila) was also affected. In Rap1 mutant cells DE-cad was abnormally concentrated at a single cell-cell interface (Knox and Brown, 2002); Fig.1C), creating asymmetric adhesive contacts and disrupting the regular hexagonal packing of wing epithelial cells. Throughout this analysis, three Rap1 alleles were analyzed (Rap1RB3, Rap1CD3, and Rap1CD5) resulting in equivalent phenotypes. These results confirm that Rap1 functions to maintain an even distribution of adherens junctions about the apical circumference of epithelial cells.
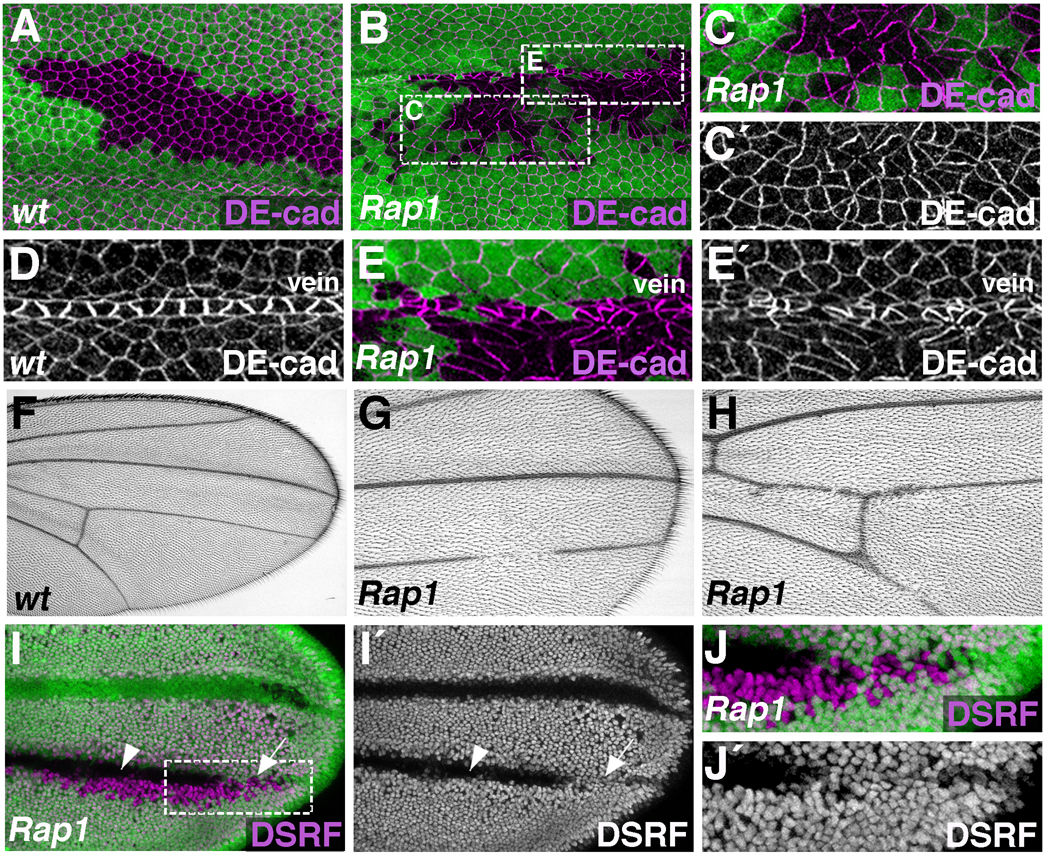
Figure 1. Rap1 regulates epithelial cell shape, DE-cad localization, and wing vein integrity.
(A–J) The Flp/FRT system was used to generate either wildtype (A,D,F), or Rap1 mutant (B,C,E,G–J) clones of cells. (A–E) Pupal wings (36 hours APF) stained for DE-cad. (A) In wildtype clones (GFP-negative), cells are generally hexagonal, and form a single cohesive unit. Green marks GFP-positive cells. (B) Rap1 mutant cells (GFP-negative) are irregularly shaped and intermingle with neighboring wildtype cells. In addition, DE-cad is asymmetrically distributed in Rap1 mutant cells. (C) Enlarged view of boxed region in B is shown. (D) Wildtype vein cells are morphologically distinct from surrounding intervein cells. (E) Loss of Rap1 severely disrupts vein cell morphology. Enlarged view of boxed region in B is shown. Compared to control adult wings (F), wings containing Rap1 mutant clones of cells have loss of wing vein material (G,H). (I) Pupal wing (36 hours APF) stained for DSRF, a marker of intervein cell fate. Rap1 mutant clones of cells (GFP-negative) are associated with ectopic DSRF expression (arrow), however many Rap1 mutant cells remain DSRF-negative (arrowhead). (J) Enlarged view of boxed region in I is shown.
While it is clear that Rap1 regulates cell adhesion in the wing, we next tested whether manipulations of Rap1 activity would affect differentiation of wing-specific cell types. In particular, the wing blade has a characteristic pattern of veins that add rigidity and are critical for flight (Fig.1F). Vein cell precursors are specified during larval stages (Sturtevant et al., 1993), and by 36 hours APF have adopted a distinct, non-hexagonal shape (Fig.1D). These cells express high levels of DE-cad (compared to surrounding intervein tissue), and this adhesive difference is important for their morphological differentiation (O'Keefe et al., 2007). When Rap1 loss-of-function clones overlapped vein territories, severe disruptions in vein cell morphology were observed (Fig.1E), suggesting that Rap1 is required for vein formation. In support of this hypothesis, adult wings containing Rap1 mutant clones of cells had vein defects never seen in control animals (Fig.1G,H). Wing vein discontinuities were frequently observed, although abnormalities in wing hair formation prevented us from identifying mutant cells in this context. To resolve this issue, pupal wings containing Rap1 mutant clones were dissected and stained for DSRF/Blistered, a marker of intervein cell fate (Montagne et al., 1996). Similar to the adult phenotype, vein discontinuities in the pupal wing were observed, as DSRF localization frequently expanded into vein territories (Fig.1I,J, arrow). This effect was weakly penetrant, however, as many Rap1 mutant cells maintained the vein cell fate (DSRF-negative) (Fig.1I, arrowhead).
Rap1 has been shown to affect both Egfr signal transduction (Cook et al., 1993; Mishra et al., 2005), and DE-cad-mediated cell adhesion (Knox and Brown, 2002), processes important for wing vein specification (Brunner et al., 1994; Clifford and Schupbach, 1989; Diaz-Benjumea and Hafen, 1994; Guichard et al., 1999; Karim and Rubin, 1998; Prober and Edgar, 2000; Sturtevant et al., 1993) and morphogenesis respectively. To better understand the role Rap1 plays during wing vein formation, we used the MARCM system (Lee and Luo, 2001) to activate Egfr signaling (via a secreted version of the ligand Spitz) in either wild-type or Rap1 mutant cells and measured the effect on vein cell fate and cell adhesion. In terms of cell fate, Rap1 did not affect the ability of Spitz to promote vein identity; DSRF levels were downregulated in the presence or absence of Rap1 function (Fig.2E,F, Supplemental Fig.S1C,D). A similar result was obtained when discs were stained for di-phospho-ERK (a more direct readout of MAPK activity in vein cells (Gabay et al., 1997)) (Supplemental Fig.S1A). In terms of cell adhesion, however, Rap1 significantly affected Spitz-expressing cells. Clones of cells expressing Spitz in the pupal wing (36 hours APF) had elevated levels of DE-cadherin, resulting in dramatic apical constriction (Fig.2A,C). Loss of Rap1 in this context did not affect DE-cad levels, nor DE-cad apical/basal localization (Fig.2B,D), however, asymmetric adhesive contacts were prevalent. Taken together these results indicate that in the developing wing epithelium Rap1 does not directly regulate the Egfr signaling cascade to affect vein/intervein identity. Instead, Rap1 affects adhesion between cells, and the effects on Egfr-dependent cell types are a consequence of these adhesive deficits. In Rap1 mutant clones of cells, therefore, vein cells are specified normally, but vein cell fate maintenance and vein differentiation are compromised.
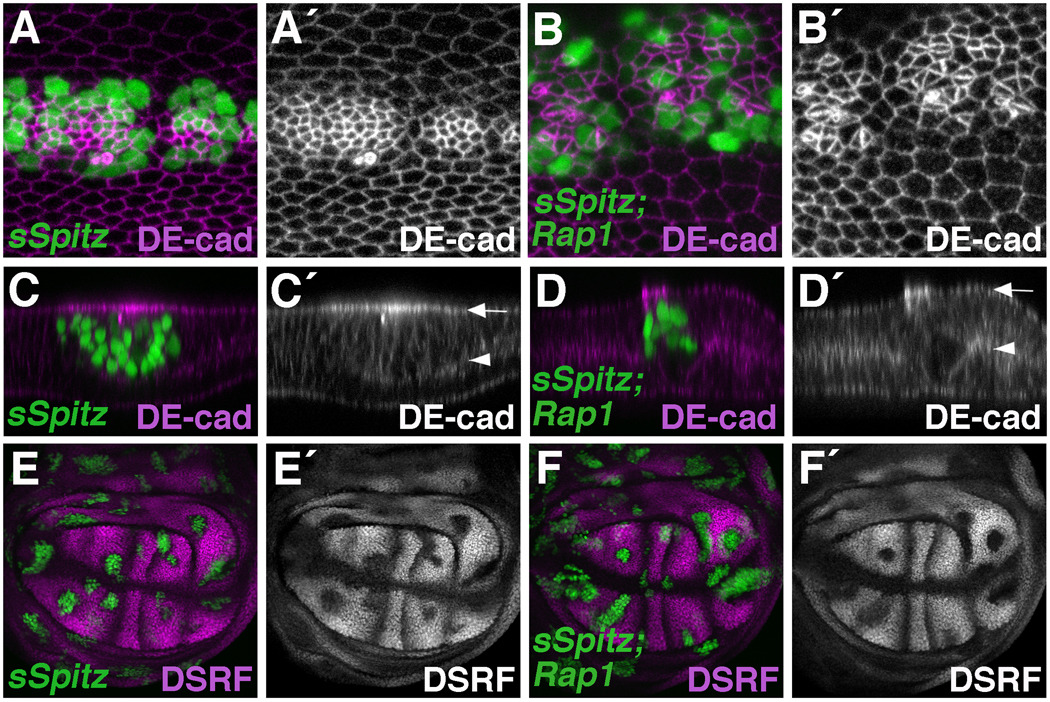
Figure 2. Rap1 and Egfr signaling affect different aspects of DE-cad localization.
(A–F) The MARCM system was used to express a secreted version of Spitz (sSpitz) in wildtype (A,C,E) or Rap1 mutant (B,D,F) cells. (A–D) Pupal wings (36 hours APF) stained for DE-cad are shown. (A) sSpitz expressing cells (GFP-positive) have high levels of DE-cad and are apically constricted compared to their wildtype neighbors. (B) Loss of Rap1 dramatically affects DE-cad localization in sSpitz expressing cells. (C,D) Optical cross-sections through the bi-layered wing epithelium are shown. Arrows and arrowheads indicate apical and basal surfaces of the dorsal wing epithelia respectively. (C) In sSpitz expressing cells, DE-cad is concentrated near the apical cell surface (apico-laterally). (D) Loss of Rap1 does not affect the apical/basal distribution of DE-cad in sSpitz expressing cells. (E,F) Wing imaginal discs stained for the intervein marker DSRF are shown. (E) sSpitz expressing cells downregulate DSRF indicating a switch to vein cell fate. (F) Loss of Rap1 does not affect the ability of sSpitz to downregulate DSRF.
Rap1 affects DE-cad localization and cell shape in the developing eye
To ask whether the cell adhesion phenotypes associated with Rap1 in the wing are seen in other developmental contexts, we disrupted Rap1 function in the Drosophila eye. Using an ey-flp transgene, clones of cells lacking Rap1 function were generated from the earliest stages of eye development. As was seen in the wing, DE-cad localization was affected by the loss of Rap1 activity. In third instar eye imaginal discs cells anterior to the morphogenetic furrow are undifferentiated and proliferating, while behind the furrow cells are exiting the cell cycle and differentiating (Wolff and Ready, 1991). At this stage of development, all epithelial cells in the eye imaginal disc express DE-cad, but DE-cad protein is enriched in cells of the morphogenetic furrow and at cell-cell contacts between photoreceptor precursors (Mirkovic and Mlodzik, 2006) (Fig.3A). DE-cad localization was not affected in Rap1 mutant clones anterior to the furrow, or in the furrow itself. Posterior to the furrow, however, a dramatic effect on DE-cad localization was observed. While many Elav-expressing photoreceptors were specified in the absence of Rap1 function, DE-cad enrichment between these cells was not observed (Fig.3A). Surprisingly, DN-cadherin (DN-cad) localization in the eye disc was not affected by Rap1 (Supplemental Fig.S2), indicating that Rap1 specifically affects DE-cad based adherens junctions at this stage of development.
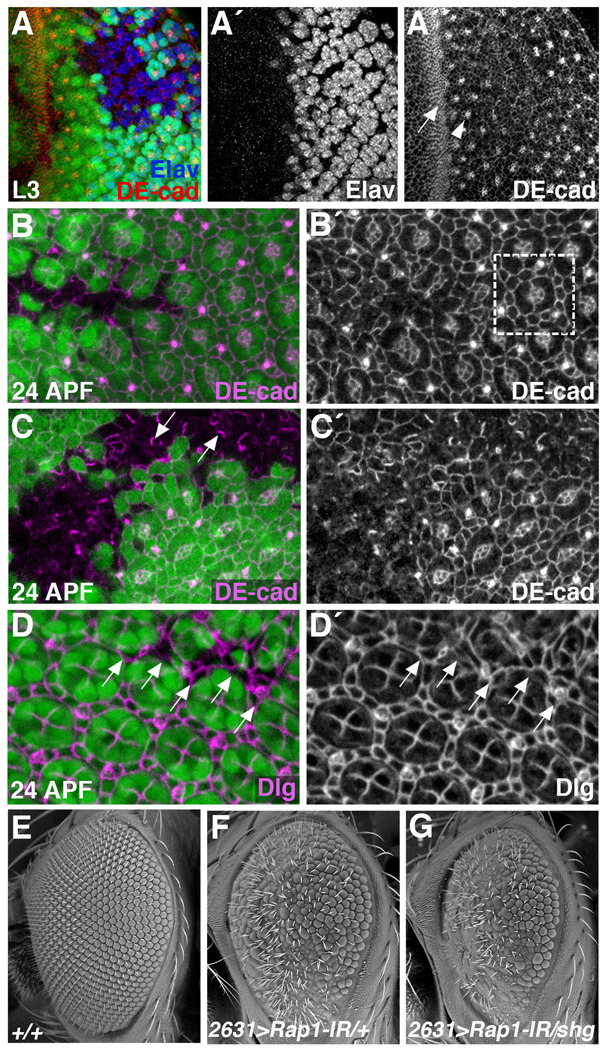
Figure 3. Rap1 affects cell adhesion and cell shape in the developing eye.
(A–D) Clones of cells lacking Rap1 (GFP negative) were generated using the Flp/FRT system. Eye tissue from late L3 larvae (A) or 24 hour APF pupae (B–) were stained for DE-cad (A–C) or Discs Large (Dlg) (D). (A) During larval stages, DE-cad levels are highest in the morphogenetic furrow (arrow) and at junctions between photoreceptor precursors (arrowhead). In Rap1 mutant cells, photoreceptors are still specified (A´), but the pattern of DE-cad localization is altered (A´´). (B,C) At 24 hours APF, DE-cad localization in the eye demarcates a regular lattice of cone and pigment cells. Boxed region in B´ indicates a single ommatidial unit. Loss of Rap1 disrupts DE-cad localization and the regular pattern of cell types. (C) Asymmetrical distributions of DE-cad are frequently observed in Rap1 mutant tissue (arrows). (D) Dlg localization similarly reveals a regular pattern of cone and pigment cells in wildtype 24 APF pupal eyes. Arrows point to Rap1 mutant cells, which are morphologically defective. (E–G) SEM images of adult eyes are shown. NP2631-Gal4, when crossed to a Rap1-RNAi transgene (Rap1-IR), knocks down Rap1 function in most eye cells. Compared to wildtype eyes (E), NP2631>Rap1-IR eyes are rough, typically indicating a defect in cell-type specification or differentiation (F). (G) The NP2631>Rap1-IR phenotype is significantly worsened when a single copy of the DE-cad/shg gene is eliminated.
At 24 hours APF, DE-cad localization in the wildtype pupal eye reveals a highly organized lattice of cell types (Fig.3B,C). At this stage, loss of Rap1 in the eye had similar, but more dramatic effects on DE-cad localization compared to those seen in the wing. Asymmetries in DE-cad were frequently observed within the clones (Fig.3C, arrows), but more frequent was a diffuse distribution of DE-cad. Cell shapes were also highly disorganized near Rap1 mutant tissue, with cells both within and immediately adjacent to the clone affected. However, since the loss of Rap1 so dramatically affected DE-cad localization, it was often difficult to delineate cell boundaries within the mutant clones. To circumvent this problem, we visualized Discs large (Dlg) in Rap1 mutant eye tissue. Dlg is a critical component of the septate junction (Woods et al., 1996), and its localization is not affected by Rap1 activity (Knox and Brown, 2002). At 24 hours APF, Dlg localization reveals a regular pattern of cone and interommatidial cells in wildtype eyes. This pattern was severely disrupted in and around Rap1 mutant tissue (Fig.3D). These results indicate that in the differentiating eye, Rap1 dramatically affects DE-cad localization and cell shape.
To demonstrate a functional relationship between Rap1 and DE-cad during eye development, we asked whether reduced levels of DE-cad could enhance a hypomorphic Rap1 phenotype. NP2631-Gal4 (NP2631) expresses in most cells of the third instar eye disc, both anterior and posterior to the morphogenetic furrow (Supplemental Fig.S3A). NP2631 was used to drive expression of a Rap1 RNAi transgene (UAS-Rap1-IR), knocking down Rap1 function in the developing eye (genotype: NP2631>Rap1-IR). Controls indicate that Rap1-IR specifically affects Rap1 expression and not other closely related GTPases (Supplemental Fig.S4). In wildtype adult eyes, ommatidia and bristles are arranged in a remarkably regular pattern (Fig.3E). In comparison, NP2631>Rap1-IR eyes were small and rough (Fig.3F). When one copy of the DE-cad/shg gene was removed from this genetic background (genotype: NP2631>Rap1-IR/shg2) the Rap1 phenotype was greatly enhanced (Fig.3G). As a control we determined that shg2 did not interact with the NP2631 driver alone (data not shown). This genetic interaction strongly suggests that Rap1 and DE-cad function in a common pathway to regulate differentiation and morphogenesis of the Drosophila eye.
Egfr-dependent cell types are lost from Rap1 mutant eye tissue
We next asked whether Rap1 activity is necessary for Egfr-dependent processes in the eye disc, as we had observed in the wing. Egfr signaling has several roles in eye development, but is first required in the morphogenetic furrow for the formation and spacing of ommatidial precursor clusters of cells (Baonza et al., 2001; Brown et al., 2006; Dominguez et al., 1998; Spencer et al., 1998; Yang and Baker, 2001). In differentiating cells posterior to the morphogenetic furrow Egfr signaling is required for proper spacing of the founding R8 photoreceptor, and for subsequent recruitment of all other ommatidial cell types. In the absence of Egfr signaling, therefore, an irregular pattern of R8 photoreceptors is specified, but all other ommatidial cell types are lost (Baonza et al., 2001; Dominguez et al., 1998; Lesokhin et al., 1999; Yang and Baker, 2001). Similarly, a partial decrease in Egfr signaling results in fewer ommatidial cells (Freeman, 1996). In addition, it has been demonstrated that Egfr activity is required for high levels of DE-cad in photoreceptors (Brown et al., 2006), suggesting that cell adhesion plays an important role in photoreceptor specification and differentiation (as in wing veins). Consistent with this hypothesis, DE-cad/shg mutants show loss of photoreceptors (Mirkovic and Mlodzik, 2006). As we described above, Rap1 also affects DE-cad levels in photoreceptor precursors (Fig. 2A), our first indication that Rap1 plays a role in Egfr-dependent processes in the eye.
To test whether Rap1 plays a role during the earliest stages of ommatidial development, we stained eye discs containing Rap1 mutant clones of cells for dpERK and Senseless. In the developing eye, Egfr/MAPK activity is highest in intermediate groups within the morphogenetic furrow (Baonza et al., 2001; Spencer et al., 1998) (Fig.4C, arrow), and loss of Rap1 did not affect dpERK levels, or the spacing of intermediate groups (Fig.4D, arrow). Similar results were obtained when levels and spacing of Atonal were examined (data not shown). Senseless localizes to a subset of cells within each intermediate group (the R8 equivalence group) (Frankfort et al., 2001) (Fig.4E), and within the furrow its localization was normal in Rap1 mutant clones (Fig.4F). These results indicate that Rap1 is not required for the patterning events that drive formation and spacing of the intermediate clusters or the R8 equivalence groups. In addition, Rap1 does not prevent the activation of the Egfr signaling pathway, as judged by the normal dpERK levels in Rap1 mutant tissue.
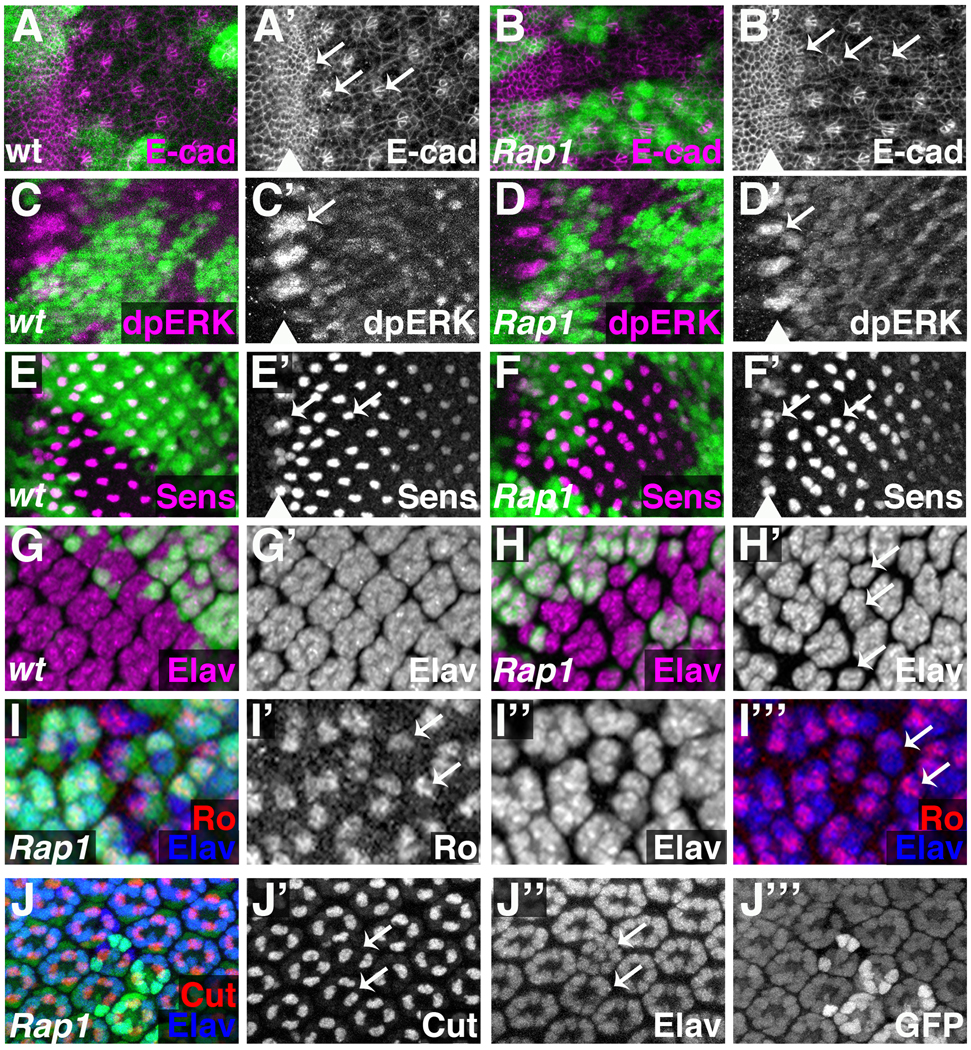
Figure 4. Rap1 is required for proper ommatidial cell development.
Wildtype (A,C,E,G) or Rap1 mutant (B,D,F,H–J) clones of cells (GFP negative) were generated using the Flp/FRT system. An ey-flp transgene was used to generate clones from the earliest stages of eye development. Larval eye imaginal discs were stained for E-cadherin (E-cad) (A,B), dpERK (C,D), Sens (E,F), Elav (G,H), Ro/Elav (I). 40-hour APF pupal disc was stained for Cut/Elav (J). An arrowhead marks the position of the morphogenetic furrow (A’–F’). (A) E-cadherin is expressed at high levels in the furrow. Cells in the furrow form rosettes (left arrow), which reorganize into arcs (middle arrow); the arcs close to form clusters, each of which is the precursor of an ommatidium. E-cadherin is maintained at high levels in arcs and clusters, but not in intervening cells. (B) E-cadherin staining reveals that cells in Rap1- tissue form morphologically abnormal arcs and clusters (arrows). (C) In wild-type eye discs, high levels of di-phosphorylated MAPK (dpERK) are present in intermediate groups, with lower levels behind the furrow. (D) dpERK levels and spacing intermediate groups is not affected by Rap1- clones. (E) Senseless is expressed in R8 equivalence groups (left arrow) and at high levels in R8 (an Egfr-independent cell type). (F) Senseless expression in R8 equivalence groups is normal, and Senseless-expressing R8 cells are present in Rap1 mutant clones, although their spacing is altered. (G) Elav marks post-mitotic neurons, normally found in clusters posterior to the morphogenetic furrow. (H) In Rap1 mutant clones, Elav clusters are often irregularly shaped (arrows), suggesting a loss of photoreceptor precursors. Rough most strongly labels R2 and R5, and ommatidia containing a single Rough positive cell are frequently found in and around Rap1 mutant tissue (I, arrows). Cut localizes to cone cells, which are frequently lost from Rap1 clones (J), including from ommatidia containing a full complement of photoreceptors (arrows).
As ommatidial pre-clusters emerge from the furrow, however, Rap1 phenotypes become apparent. Posterior to the furrow, Senseless-expressing R8 cells were present in Rap1 clones, although they were irregularly spaced (Fig. 4F, arrows). This is consistent with a reduction in Egfr function. The spacing phenotype is not due to cell apoptosis, as expression of the cell death inhibitor p35 (Hay et al., 1994) in Rap1 mutant cells did not restore normal spacing to ommatidia (Supplemental Fig.S5). To examine in more detail why R8 cells were affected, we stained these discs for DE-cad to visualize ommatidial precursors at this stage. In wild-type eye discs, cells in the furrow express high levels of DE-cad and form rosettes, which rearrange to form arcs, which close to form clusters of photoreceptor precursors (Wolff and Ready, 1993) (Fig 4A, arrows). DE-cad is maintained at high levels in clusters, while being downregulated in surrounding cells. Rap1 does not affect DE-cad localization in the furrow (as mentioned previously). However, although arcs and clusters within Rap1 mutant clones attempt to form, they are structurally abnormal and fail to maintain high levels of DE-cad (Fig 4B, arrows). These early morphological defects, which are likely to result from the effects of Rap1 on cell adhesion, may account for the irregular spacing of R8 cells. Taken together, our results suggest that the morphological changes characteristic of ommatidial precursors as they exit the furrow are regulated by Rap1, but that Rap1 is dispensable for the patterning events that determine and position the rosettes, arcs and clusters.
With R8 present, we next asked whether each ommatidium contained the regular complement of photoreceptor precursors. Elav is expressed by post-mitotic neurons, and posterior to the furrow labels regularly spaced clusters of photoreceptors (Fig.4G). In Rap1 mutant clones the Elav clusters were variable in size, suggesting a loss of photoreceptor cell types (Fig.4H, arrows). To determine which cells were affected we first examined Rough (R2/R5) and Prospero (R7/cone) expression in Rap1 mosaic eyes. Both Rough and Prospero cell types were frequently absent from Rap1 mutant ommatidial clusters (Fig.4I and data not shown). Cone cells (labeled by the Cut protein (Blochlinger et al., 1993)) require Egfr signaling for their specification and were often lost from eye discs containing Rap1 mutant clones (data not shown), revealing another cell type that requires Rap1 function.
We also examined Cut localization in pupal eyes (40 hours APF). In some cases, ommatidia associated with Rap1 mutant cells contained a full complement of photoreceptors, but were missing cone cells (Fig.4J). This indicates that cone cell loss is not a secondary consequence of missing photoreceptors. Interestingly, the arrangement of photoreceptors in the pupal eye can also be affected by Rap1 loss-of-function clones, as demonstrated by the ommatidium with one photoreceptor surrounded by the seven others (all eight photoreceptors are normally arranged in a circle). This is consistent with improper maintenance of adhesive contacts in ommatidia containing Rap1 mutant cells.
Effects of Rap1- clones on photoreceptor development do not result from apoptosis
During the course of the experiments described above we noticed occasional Elav-positive nuclei in ectopic locations. As photoreceptors are specified their nuclei migrate to the apical surface of the eye epithelium (Wolff and Ready, 1993). Optical sections through Rap1 mutant clones, however, revealed Elav nuclei at the basal surface. They were consistently located well behind the furrow in a region where developing ommatidia are fairly mature. Whether these are cells that are being eliminated from the epithelium, or simply cells with mis-positioned nuclei remains to be determined. It is important to point out that Rap1 mutant ommatidia with photoreceptor loss are often detected in regions where basal nuclei are not present. Thus, despite these basal nuclei, the primary effect of Rap1 on photoreceptors is likely to be a failure of recruitment.
Nevertheless, to confirm that Rap1 is necessary for photoreceptor recruitment, and that photoreceptor loss from Rap1 mutant clones does not primarily result from apoptosis, we expressed the apoptosis inhibitor p35 in Rap1 mutant cells using the MARCM system or GMR-p35 (Johnson et al., 2002). Acridine orange stainings were used to confirm that p35 inhibited apoptosis (not shown). In both cases we still observed loss of Elav-positive cells from developing ommatidia, and basally localized Elav-positive nuclei (Supplemental Fig.S5). These results indicate that the loss of photoreceptors observed in Rap1 loss-of-function clones does not result from apoptosis.
In summary, Rap1 has no effect on early patterning events that occur ahead of and in the furrow. Instead, Rap1is required for proper morphology of ommatidial preclusters, for proper spacing of R8 precursors, and for recruitment of photoreceptors and cone cells after R8 specification. The fact that Rap1 clones do not affect dpERK levels indicates that, as in the wing, Rap1 is not required for Egfr signaling per se. Rather, our results are consistent with the idea that the role of Rap1 in Egfr-dependent developmental events results from effects of Rap1 on cell adhesion.
Rap1 function is required for ommatidial rotation
In addition to apical/basal polarity, cells within a developing tissue are often polarized within the plane of the epithelium. Termed planar cell polarity (PCP), this process allows for a large number of cells to coordinately differentiate asymmetric structures. In the Drosophila retina each ommatidium is polarized since the R3 and R4 photoreceptors occupy asymmetric positions. Within each ommatidium, the PCP network of proteins (Frizzled and Dishevelled for example) determines which cell adopts the R3 versus R4 fate; thus determining ommatidial chirality (Adler, 2002; Mlodzik, 1999; Strutt and Strutt, 1999). Following specification of R3/R4, each ommatidial precluster undergoes a 90-degree rotation toward the midline of the developing eye field. This rotation involves a complex rearrangement of cell-cell contacts, and requires input from planar cell polarity, although the mechanisms linking the PCP complex of proteins to the rotation process remain unclear (Seifert and Mlodzik, 2007; Wolff et al., 2007). In this way, ommatidial rotation is a readout of PCP signaling in the eye. It has been demonstrated that Egfr activity and cadherin-based cell adhesion both affect ommatidial rotation. Hypomorphic mutations in the Egfr pathway that allow for normal specification of ommatidial cell types frequently have rotation phenotypes, but proper chirality (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003; Strutt and Strutt, 2003), and DE-cad has been shown to promote ommatidial rotation (Mirkovic and Mlodzik, 2006). Since Rap1 affects Egfr-dependent processes and cadherin-based cell adhesion, we asked whether Rap1 was necessary for ommatidial rotation in the developing eye.
The protein Rough is strongly expressed in photoreceptors R2 and R5 that, under wildtype circumstances, are located on opposite sides of each ommatidium. This pattern allows one to track the degree of rotation for ommatidia in the larval eye disc (Fig.5A). Using an ey-flp transgene, clones of cells lacking Rap1 function were generated and late third instar discs were stained for Rough. In these discs, the ommatidial rotation pattern was disrupted, as adjacent ommatidia were frequently misaligned (Fig.5B); only complete ommatidia were included in the analysis. Based on the scattered nature of the clones, it was not possible to determine whether this was a cell-autonomous effect. We therefore examined sections through adult eyes containing Rap1 mutant clones. In these retinas, most mutant cells did not survive, leaving behind scar-like tissue devoid of recognizable cell types (Fig.5C). A few mutant cells did survive, however, and were typically found at the edges of clones in incomplete or fused ommatidia. These phenotypes have been described before (Asha et al., 1999). When ommatidial orientation was scored in these eyes (again, only complete ommatidia were included), those containing Rap1 mutant cells were sometimes misaligned (Fig.5C, white arrow). In addition, ommatidia adjacent to Rap1 mutant territories, but containing entirely wildtype photoreceptors, were also affected (Fig.5C, black arrows). This is not surprising since the rotation process involves the coordinated making and breaking of cell-cell contacts, so a cell lacking normal adhesive properties would likely affect its neighbors. Rap1 therefore plays an important role in ommatidial rotation in the eye.
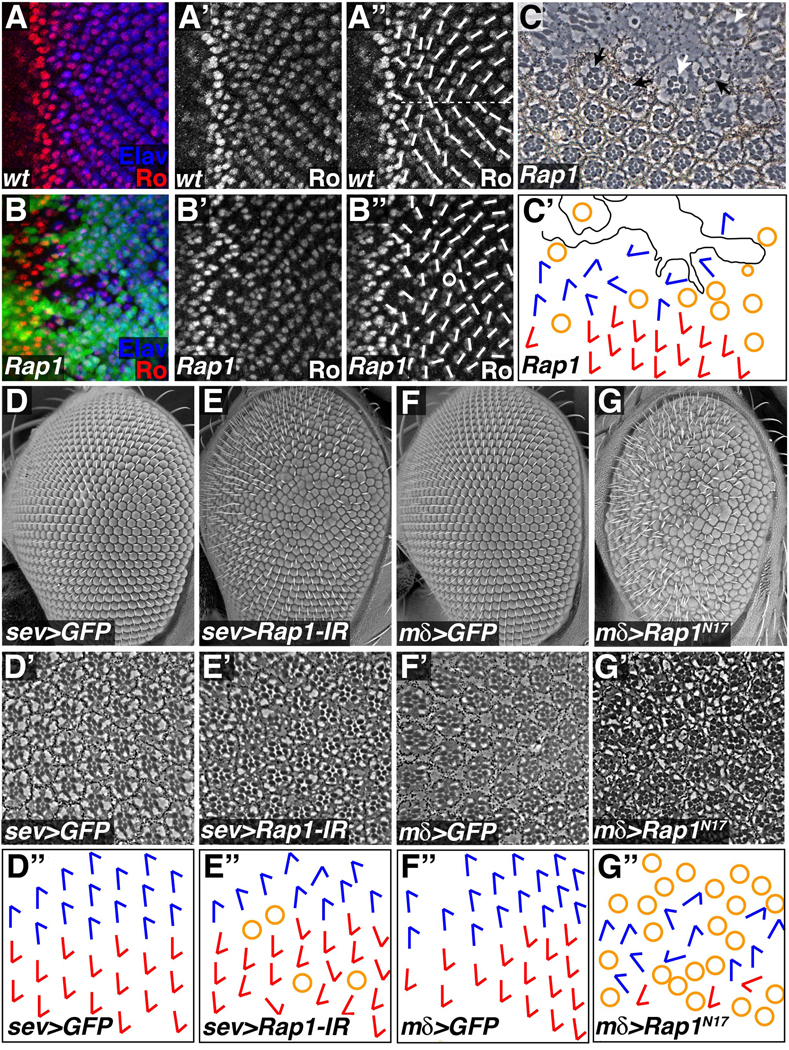
Figure 5. Rap1 affects ommatidial rotation in the developing eye.
(A,B) Eye imaginal discs were stained for Rough (Ro) and Elav. Short white lines indicate ommatidial orientation, while the horizontal dotted line marks the equator. (A) In wildtype discs a regular pattern of ommatidial orientation is apparent. (B,C) Clones of cells lacking Rap1 (GFP negative) were generated using the Flp/FRT system. (B) Rap1 mutant cells disrupt the overall pattern of ommatidial rotation. Circle denotes an ommatidium with abnormal numbers of photoreceptors. (C) In tangential sections of adult eyes Rap1 mutant cells lack pigment. Scar-like tissue often formed in Rap1 mutant clones. At the edges of clones misaligned ommatidia containing Rap1 mutant cells (white arrow), or entirely wildtype cells (black arrows) could be found. White arrowhead indicates an ommatidium containing Rap1 mutant cells that has a normal alignment. (C´) Schematic representation of the section in C is shown. Blue and red arrows indicate ommatidial orientation; yellow circles indicate incomplete ommatidia; black line demarcates scar tissue. (D–G) Loss and gain of Rap1 function in subsets of cells also disrupts ommatidial rotation. SEM micrographs (D–G), tangential sections (D´–G´), and schematic representations of ommatidial orientation (D´´–G´´) are shown for each genotype. (E) sev-Gal4 in combination with Rap1-IR was used to knock down Rap1 function in a subset of differentiating photoreceptors (R3, R4, R7) and cone cells. (G) Similarly, mδ0.5-Gal4 was used to express Rap1N17 in R4 and weakly in R3 and R7. In both cases, reduction of Rap1 activity significantly increased ommatidial misalignment when compared to controls (D and F respectively).
In order to generate adult retinas in which Rap1 function had been compromised to a lesser degree, sevenless-Gal4 (K25) was used to drive expression of Rap1-IR, knocking down Rap1 gene function in a subset of retinal cell types (R3, R4, R7 and cone cells, Supplemental Fig.S6). At the gross level, a rough eye phenotype was obtained (compare Fig.5D and 5E). When sections through the sev-Gal4>Rap1-IR adult eye were examined, a small number of ommatidia contained an irregular complement of photoreceptors, similar to the Rap1 loss-of-function result (Fig.5E). Ommatidial orientation was scored in these eyes, examining only those ommatidia containing a wildtype set of photoreceptors. Compared to controls, Rap1-IR expressing eyes had significant defects in ommatidial rotation. In order to quantify this phenotype, an orientation angle (with respect to the dorsal/ventral axis) was measured for each ommatidium (normal orientation equals 90 degrees). Mean orientation angle and angle variance was then calculated for each eye, and these values were used to compare between genotypes. In control eyes, ommatidia were essentially aligned resulting in a low orientation variance (mean angle=91.9; median angle variance 17.6, n=3). Knocking down Rap1 function with sev-Gal4 had little effect on orientation angle, but significantly (p<.05) increased angle variance (mean angle=92.4; median angle variance 282.2, n=6), indicating that Rap1 activity is essential for proper ommatidial rotation.
Similar albeit stronger results were obtained when the dominant negative Rap1N17 was expressed in the eye. The phenotype obtained with sev-Gal4 was too severe for analysis, so a weaker driver, mδ0.5-Gal4 was used. mδ0.5-Gal4 is expressed at highest levels in R4, but also more weakly in R3 and R7 (Supplemental Fig.S3). Reduction of Rap1 signaling in this subset of cells resulted in a rough eye phenotype, many ommatidia with abnormal numbers of cells, and severe effects on ommatidial rotation (Fig.5G). Thus, results from three independent methods of reducing Rap1 function demonstrate that Rap1 has a role in ommatidial rotation.
Rap1 genetically interacts with Egfr pathway components in the eye
To more firmly establish a role for Rap1 in ommatidial rotation, we asked whether Rap1 genetically interacts with genes known to affect this process. Star is a transmembrane protein located in the endoplasmic reticulum that facilitates the trafficking and activation of Egf ligands, positively regulating signaling through the Egfr pathway (Lee et al., 2001). It has been demonstrated that Star is haploinsufficient in the eye (Heberlein et al., 1993), and Star heterozygotes have subtle effects on ommatidial formation and rotation (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003) (Fig. 6A,D). The adult eyes of Star heterozygous flies (Star48-5/+) are mildly rough with misarranged ommatidia, and occasional ommatidial fusions (Fig. 6A). We first asked whether reduction of Star activity could affect the NP2631>Rap1-IR rough eye phenotype (Fig. 6B) by generating NP2631>Rap1-IR/Star48-5 animals. Adult eyes from these individuals demonstrated a clear genetic interaction between Rap1 and Star48-5 in this context (Fig.6C), but the dramatic nature of the phenotype precluded an analysis of ommatidial rotation.
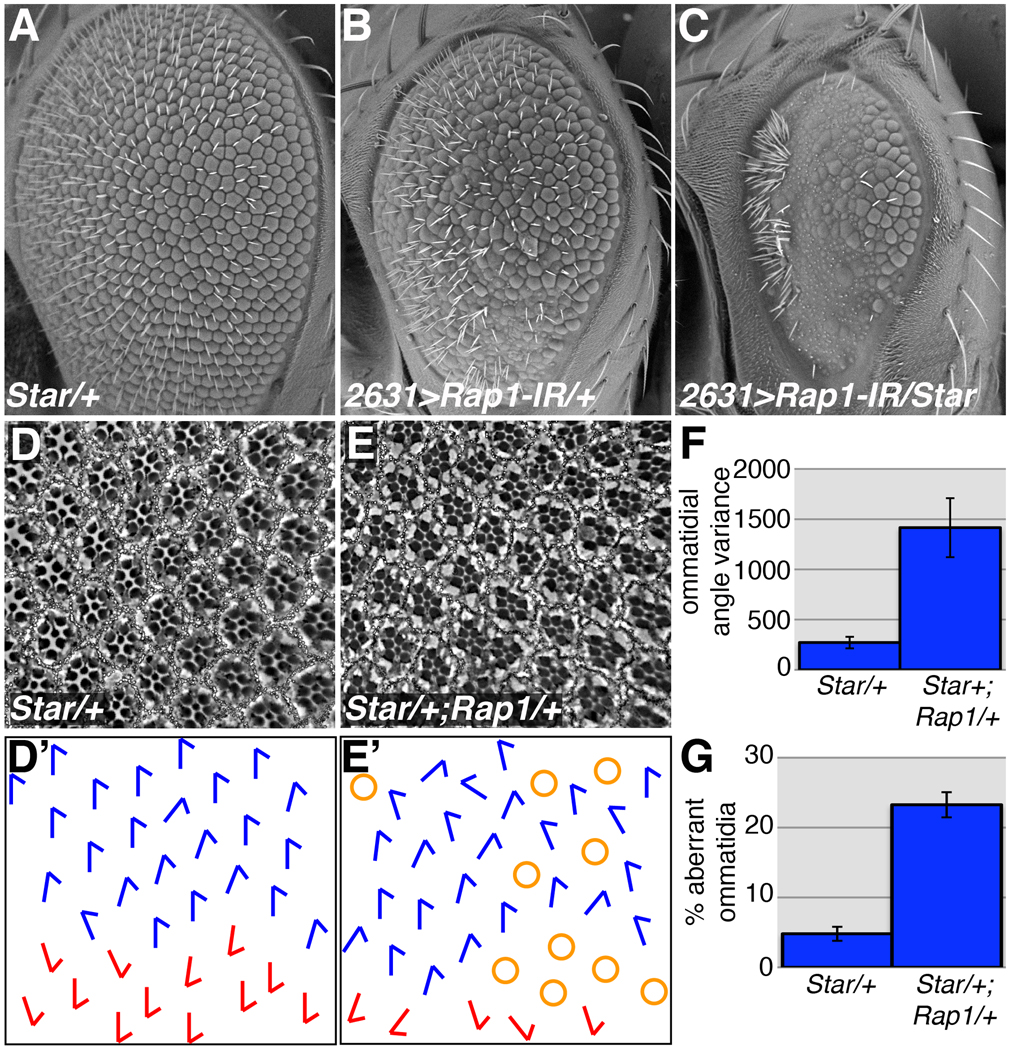
Figure 6. Rap1 and Star genetically interact.
Star is a positive regulator of Egf signaling, and Star48-5 heterozygotes have a subtle effect on adult eye architecture (A). The rough eye phenotype associated with NP2631>Rap1-IR (B) is dramatically worsened when a single copy of Star48-5 is eliminated (C). (D) When sections through adult Star48-5/+ eyes are examined, ommatidia are slightly misaligned compared to wildtype. Loss of one copy of Rap1 significantly enhances the Star48-5 adult eye phenotype (E). Tangential sections through adult eyes (D,E) and schematic diagrams of ommatidial orientation (D´,E´) are shown for each genotype. (F,G) Quantifications of the Rap1, Star genetic interaction are shown. Loss of one copy of Rap1 significantly increases ommatidial misalignment (F), and the percentage of aberrant ommatidia associated with Star48-5 heterozygotes (p<0.05).
To generate a subtler phenotype involving Rap1 and Star, double heterozygotes were generated (genotype: Star48-5/+; Rap1RB3/+). As mentioned above, when tangential sections of Star/+ adult eyes were examined, mild ommatidial rotation defects (mean angle=85.0; median angle variance=262.4) and occasional loss of photoreceptors (median % aberrant=5.2%) were observed (Fig.6D). Importantly, Rap1/+ animals had normal ommatidial rotation (mean angle=89.6; median angle variance=48.1), and very few ommatidia with loss or gain of photoreceptors (median % aberrant=1.1%) (Supplemental Fig.S6). Star/+;Rap1/+ doubly heterozygous flies showed a statistically significant (p<0.05) increase in ommatidial misalignment (mean angle=101.3; median angle variance=1657.2) (Fig.6E,F), and in the number of ommatidia containing aberrant numbers of photoreceptors (median % aberrant=22.2%) (Fig.6G).
We also tested whether sensitized genotypes involving other Egfr pathway members could be modified by Rap1 loss-of-function alleles. Kekkon 1 (Kek) is a transmembrane protein that negatively regulates Egfr activity (Ghiglione et al., 1999). Expression of kek under sev-Gal4 control resulted in weak rotation defects (mean angle=94.4; median angle variance=55.3) and some photoreceptor loss (median % aberrant=13.7%), (Supplemental Fig.S6). Removing a single copy of Rap1 significantly (p<0.05) enhanced the effects of sev>kek on both ommatidial rotation and photoreceptor loss (mean angle=89.9; median angle variance=293.1; median % aberrant=25.5%) (Supplemental Fig.S6). Expression of GFP under sev-Gal4 control had no effect on rotation (mean angle=92.0; median angle variance=17.5), and no photoreceptor loss/gain was detected.
Expression of an activated Egfr (λ-top) (Queenan et al., 1997) under sev-Gal4 control also resulted in ommatidial rotation and photoreceptor defects (mean angle=94.8; median angle variance=645.2; median % aberrant=2.2%) (Supplemental Fig.S6). Although Rap1 appears to have no effect on the sev-λ-top photoreceptor gain/loss phenotype (median % aberrant=1.0%), it slightly but significantly (p<0.05) suppresses the ommatidial rotation defects (mean angle=95.5; median angle variance=503.3) (Supplemental Fig.S6). Thus, with respect to ommatidial rotation, reducing Rap1 levels by a single gene copy dominantly enhanced the effects of Egfr loss (Star and sev>kek), and dominantly suppressed the effects of Egfr overactivation (sep>λ-top). These results demonstrate a clear role for Rap1 in ommatidial rotation, an Egfr/DE-cad-dependent process.
Rap1 is required for PCP signaling in the wing
To explore in more detail whether Rap1 affects PCP signaling (as our data in the eye suggested) we returned to the wing. Each cell in the wing blade generates a single hair located at its distal edge that points distally. PCP signaling is required for the proper formation and orientation of these wing hairs. Adult wings containing Rap1 loss of function clones had patches of disorganized wing hairs that were never seen in control wings (Fig.7A). These hairs were not distally oriented and often more than one hair formed from a single wing cell (Fig.7A’, arrow).
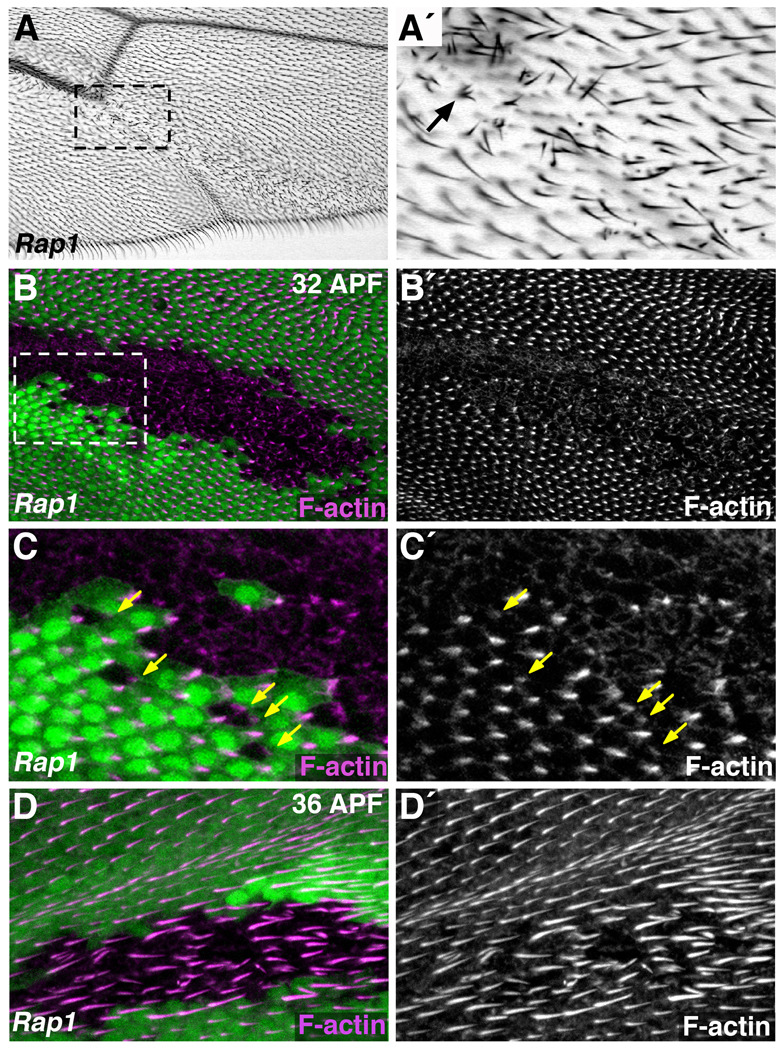
Figure 7. Rap1 is necessary for the polarized formation of wing hairs.
(A–D) The Flp/FRT system was used to generate Rap1 mutant clones of cells. (A) Adult wings containing Rap1 mutant clones of cells often have patches of disorganized wing hairs. These hairs do not point distally, as in wildtype cells, and multiple hairs seem to form from single wing cells (arrow, A´). (A´) Magnified view of boxed region in A is shown. (B–D) Fluorescently labeled phalloidin was used to visualize F-actin (highly enriched in wing hairs) during pupal stages. (B) At 32 hours APF Rap1 mutant cells (GFP-negative) are defective in pre-hair formation. (C) Magnified view of boxed region in B is shown. Wildtype cells adjacent to Rap1 mutant clones form normal prehairs, indicating a cell autonomous role for Rap1. Arrows indicate defective prehairs of Rap1 mutant cells. (D) At 36 hours APF irregular wing hair structures are associated with Rap1 mutant cells (GFP-negative).
To determine whether this phenotype was associated with Rap1 mutant cells, we examined pupal wings of the same genotype. At 32 hours APF, F-actin staining reveals the position of the wing pre-hair. Compared to the surrounding wild-type tissue, Rap1 mutant cells had an abnormal pattern of F-actin distribution (Fig.7B,C, Supplemental Fig.S7A). Pre-hair formation was clearly delayed in these cells, as F-actin foci were less defined, or absent altogether. When prehairs were evident, however, they were found at the distal edge of Rap1 mutant cells (arrows Fig.7C), suggesting that these cells were polarized appropriately, but unable to coordinate changes in the actin cytoskeleton necessary for wing hair formation. Consistent with this interpretation, localization of the PCP protein Starry Night (Stan) was essentially normal (concentrated at proximal/distal cell junctions) in Rap1 mutant cells (Supplemental Fig.7C). These experiments also indicated that Rap1 acts cell-autonomously in this process, as the development of wing hairs was unaffected in wild-type cells immediately adjacent to, or surrounded by, Rap1 mutant tissue (Fig.7C). By 36 hours APF, most Rap1 mutant cells had generated a wing hair, although they were disorganized compared to surrounding wild-type tissue (Fig.7D). Based on these results, we conclude that Rap1 is not involved in propagation of the PCP signal through the wing epithelium, but instead functions as a cell-autonomous PCP effector, organizing the polarized formation of wing hairs.
Genes involved in PCP signaling worsen a mild Rap1 phenotype
To understand more clearly the role played by Rap1 during PCP signaling in the wing, we asked whether known PCP genes could modify a mild Rap1 phenotype. ptc-Gal4 was used to drive expression of Rap1-IR in the wing, knocking down Rap1 function in a stripe of cells between veins L3 and L4 (Supplemental Fig.S3D). At 25°C wing hair alignment in this area of the wing was severely disrupted (compare Fig.8A and 8B). To quantify this effect, an orientation angle (with respect to the proximal/distal axis) was calculated for each wing hair in the affected region. Mean hair angle (zero degrees points distally) and angle variance were then calculated for each wing, and these values were used to compare between genotypes. In control wings, the hairs uniformly pointed distally resulting in low mean angle and low hair angle variance (mean angle=−1.1; median angle variance=36.6) (Fig.8G,H). Rap1-IR expression resulted in a slight posterior deflection of wing hairs (mean angle=−5.4), and more random wing hair orientation, and therefore, a significantly larger wing hair angle variance (median angle variance=545.8). To generate a more moderate phenotype for use in genetic interaction experiments, ptc>Rap1-IR animals were raised at 18°C to reduce Rap1-IR expression (Fig.8C), lowering wing hair angle variance (median angle variance=80.0). For these wings, and all subsequent genotypes, wing hairs were deflected posteriorly a few degrees (Fig.8G), so our analysis will focus on angle variance.
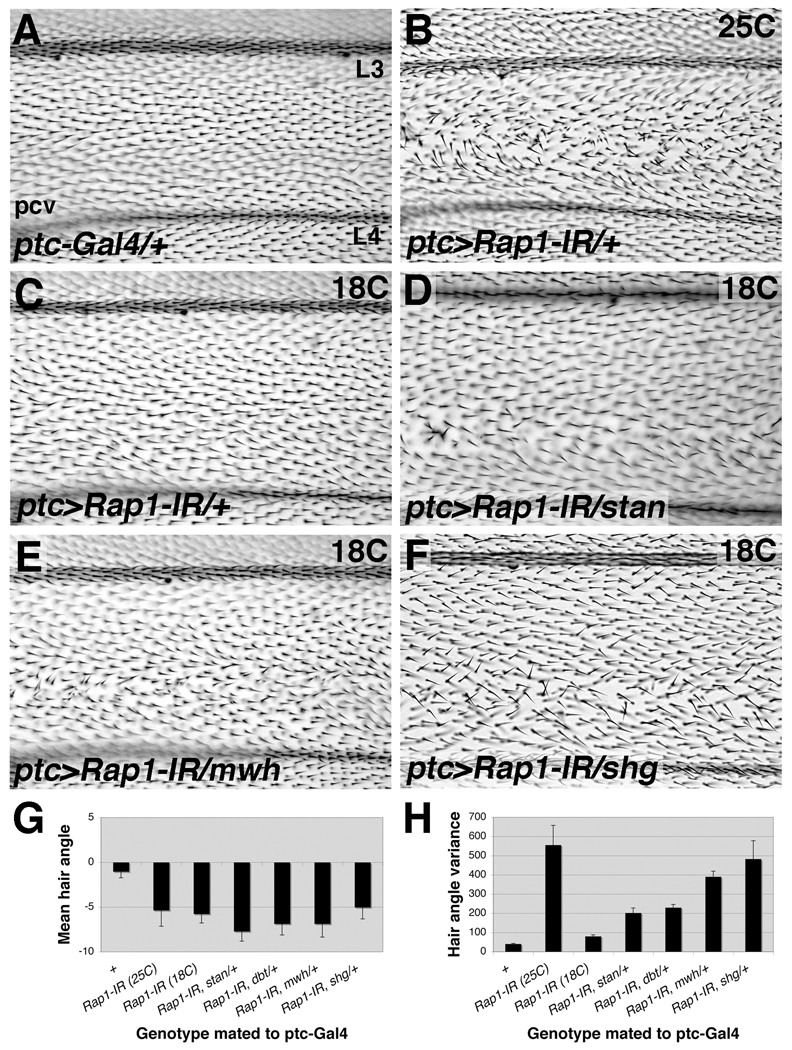
Figure 8. Genes involved in PCP signaling worsen a mild Rap1 phenotype.
Compared to control wings (A) expression of Rap1-IR via ptc-Gal4 at 25°C results in wing hair misalignment between veins L3 and L4 (B). (C) At 18°C ptc>Rap1-IR results in a mild misalignment phenotype. Wing hair orientation becomes significantly more variable if one copy of stan (D), mwh (E), or DE-cad (encoded by the gene shg) is removed (F). For each image vein L3 (top), vein L4 and the posterior cross vein (pcv) are shown. (G) Mean wing hair angle quantification is shown. Zero degrees points to the right (distally), while negative values point down (posteriorly). (H) Quantification of wing hair angle variance is shown. Error bars indicate SEM.
starry night (stan), also called Flamingo, encodes for an atypical cadherin that functions as a core PCP gene (Chae et al., 1999). When a loss-of-function allele of stan192 was crossed into the ptc>Rap1-IR background, wing hair angle variance was significantly more affected (median angle variance=204.4) (Fig.8D,H). Similarly, a loss-of-function allele of double time (dbtar) genetically interacts with the ptc>Rap1-IR phenotype (median angle variance=220.4) (Fig.8H). dbt is not a core PCP gene, but instead regulates the activity of the core component Dishevelled (Cong et al., 2004; Klein et al., 2006; Strutt et al., 2006; Tsai et al., 2007). Finally, the gene multiple wing hairs (mwh1) severely affected wing hair alignment in combination with reduced Rap1 function (median angle variance=380.1) (Fig.8E,H). mwh encodes a novel GTPase-binding domain/formin homology 3 (GBD/FH3) domain protein, and functions as a cell-autonomous, downstream effector of the PCP signaling pathway (Peyer and Hadorn, 1965; Strutt and Warrington, 2008; Yan et al., 2008). Importantly, ptc-Gal4 did not interact with stan, dbt, or mwh on its own (data not shown). Rap1 genetically interacts, therefore, with genes at every level of the PCP hierarchy: core components, regulators, and effectors. The strong interaction with mwh, however, combined with the nature of the Rap1 phenotype, suggests that Rap1 functions together with mwh as a PCP effector in the wing; coordinating cytoskeletal components to differentiate a single, polarized wing hair.
We examined whether Mwh localization was altered in Rap1 mutant clones of cells. At 32 hours APF, Mwh is concentrated at growing wing hairs (Strutt and Warrington, 2008; Yan et al., 2008). Loss of Rap1 had a dramatic effect on Mwh localization, but the pattern resembled that seen with F-actin: delay in prehair formation and loss of focused Mwh accumulation, but correct localization to the distal edge of Rap1 mutant cells (compare Supplemental Fig.S7A and B). This suggests that in the absence of Rap1, Mwh still localizes appropriately to F-actin foci at this timepoint. Whether Rap1 affects Mwh activity, however, remains an open question.
We also asked whether DE-cad/shg could modify the ptc>Rap1-IR phenotype (genotype: ptc>Rap1-IR;shgR69/+). DE-cad regulates ommatidial rotation in the eye (Mirkovic and Mlodzik, 2006), as mentioned previously, and evidence suggests that PCP genes regulate DE-cad localization during the hexagonal packing of wing epithelial cells (Classen et al., 2005). Wing hairs were significantly more disorganized in these animals (median angle variance= 496.64) (Fig.8F). The fact that this interaction is stronger than with stan, dbt or mwh confirms that the critical link is between Rap1 and DE-cad during this process, as it is in other contexts.
The Rap1 effector Canoe affects PCP signaling, cell adhesion and epithelial cell shape in the developing wing
To understand the molecular mechanisms underlying Rap1 function in the wing, we asked whether the GTPase effector Canoe plays a role in this developmental context. Canoe is a scaffolding protein containing several protein-protein interacting motifs, and has been shown to act downstream of both Ras and Rap1 (Boettner et al., 2003; Carmena et al., 2006; Gaengel and Mlodzik, 2003). In the eye, canoe phenotypes include photoreceptor loss and ommatidial rotation defects (Gaengel and Mlodzik, 2003; Matsuo et al., 1997; Miyamoto et al., 1995); effects very similar to loss of Rap1. In the wing epithelium it has been demonstrated that loss of Rap1 results in Canoe mislocalization (similar to DE-cad) (Knox and Brown, 2002).
The FRT/Gal4 system was used to generate clones of cells expressing a canoe RNAi transgene (cno-IR), and adult or pupal wings analyzed. Adult wings containing cno-IR clones of cells frequently had patches of disorganized wing hairs (Fig.9A) and wing vein discontinuities (arrow Fig.9A´). When pupal wings were examined at 32 hours APF, prehair formation (revealed by F-actin staining) was defective in cno-IR expressing cells (Fig.9C). The effects were subtler than those seen in Rap1 loss of function cells (prehair formation was not delayed), but cno-IR expression likely results in a hypomorphic phenotype. In addition, compared to control GFP-expressing clones, cno-IR clones were fragmented. cno-IR clones did not form a single cohesive unit, but instead intermingled with surrounding wild-type cells (Fig.9C,D). This phenotype is remarkably similar to that observed with Rap1 null clones.
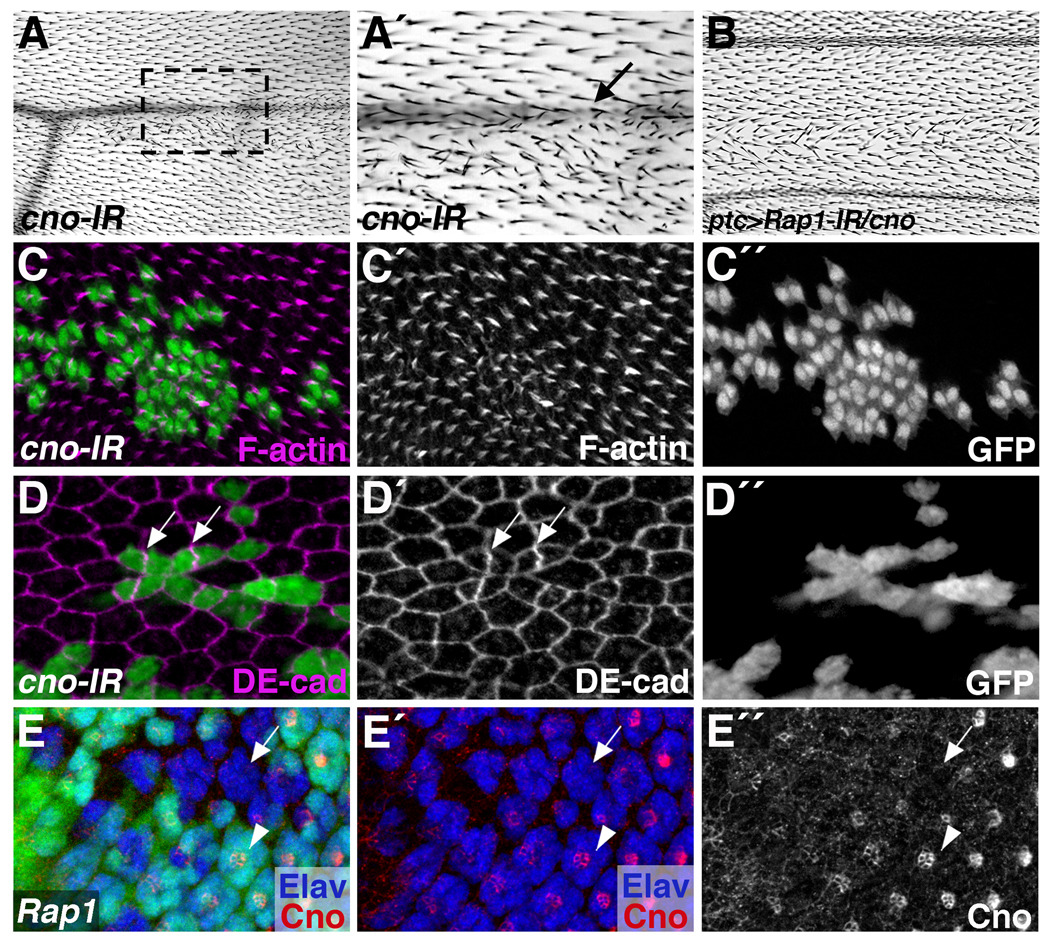
Figure 9. canoe exhibits a Rap1-like phenotype in the wing.
(A,C,D) The FRT/Gal4 system was used to express a canoe-RNAi transgene (cno-IR) in clones of cells. (A) Adult wings lacking canoe in clones of cells often have patches of disorganized wing hairs, and loss of wing vein material (arrow, A´). (A´) Magnified view of the boxed region in A is shown. (B) canoe genetically interacts with Rap1. A subtle PCP phenotype associated with ptc>Rap1-IR is exacerbated when one copy of canoe is removed. (C,D) In pupal wings, clones of cells expressing cno-IR are fragmented. (C) Wing prehair formation is abnormal in cno-IR cells (32 hours APF). (D) cno-IR expressing cells exhibit cell shape and DE-cad mislocalization phenotypes similar to Rap1 loss of function (36 hours APF). Arrows indicate high levels of DE-cad found at the interface between two cno-IR expressing cells. (E) The Flp/FRT system was used to generate Rap1 mutant clones of cells (GFP-negative) in the larval eye. Cno localization is disrupted in Rap1 mutant cells. Arrow points to an ommatidium lacking Rap1, while arrowhead indicates a wildtype ommatidium.
To analyze Canoe’s effect on epithelial cell shape and adherens junction localization, 36 hour APF wings containing cno-IR clones were dissected and stained for DE-cad. In these wings, DE-cad asymmetries were frequently observed, with abnormally high levels of DE-cad found between pairs of cells (Fig.9D, arrows). The regular hexagonal packing of wing epithelial cells was also disrupted by cno-IR expression in these wings. Based on these experiments, it is clear that in this developmental context canoe and Rap1 phenocopy one another to a high degree. Since this combination of phenotypes is so unique (clone dispersal and DE-cad asymmetry in particular), this strongly suggests that Canoe functions together with Rap1 to control cell adhesion in the wing epithelium.
To demonstrate a more direct role for Canoe in Rap1 developmental processes we next asked whether reducing canoe function could worsen a hypomorphic Rap1 phenotype. When a single copy of canoe was removed from the ptc>Rap1-IR genetic background, wing hair misalignment significantly increased (median angle variance=333.1) (compare Fig.8A and Fig.9B). canoe did not interact with the Gal4 driver alone (data not shown).
Although Canoe has been previously linked to Ras in cone cell development and ommatidial rotation (Gaengel and Mlodzik, 2003; Matsuo et al., 1997), our results led us to ask whether Canoe could also be linked to Rap1 in the eye. We therefore stained discs containing Rap1- clones of cells with anti-Canoe. In wildtype ommatidia Canoe is enriched at junctions between photoreceptors in a pattern similar to that of DE-cad. When Rap1 mutant clones were generated in the developing eye disc, Canoe enrichment at these cell junctions was eliminated (Fig.9E). Taken together, these data suggest that Canoe functions as a critical effector of Rap1 signaling in both the Drosophila wing and eye to regulate epithelial cell shape, cellular differentiation and tissue morphogenesis.
Discussion
Rap1 affects Egfr-dependent developmental processes
In Drosophila imaginal discs, low levels of Egfr signaling are required for growth and survival of developing epithelial cells (Clifford and Schupbach, 1989; Diaz-Benjumea and Hafen, 1994; Prober and Edgar, 2000), while high pathway activity is found in spatially restricted patterns and directs the differentiation of specific cell types (Freeman, 1996; Guichard et al., 1999; Kumar et al., 1998; Martin-Blanco et al., 1999; Yang and Baker, 2001). In the experiments presented here, we have demonstrated that Rap1 loss-of-function phenotypes resemble Egfr hypomorphic mutations in both the wing and eye. Following these initial observations, we investigated in greater depth the nature of these defects, asking first whether Rap1 was directly affecting Egfr signal transduction. Our data indicates that Rap1 mutant cells are fully capable of responding to an Egf signal, as Spitz expression in Rap1 clones results in high Egfr/Ras/MAPK pathway activity (as measured by dpERK levels, and loss of DSRF expression). Moreover, in wing and eye imaginal discs, loss of Rap1 does not affect the pattern of dpERK localization. At this early stage of development, therefore, vein cells and ommatidial precursors in the furrow (intermediate groups) are specified appropriately in the absence of Rap1 function. Interestingly, in these contexts Rap1 does not have dramatic effects on cell adhesion; Rap1 clones in the wing disc do not scatter, and DE-cad localization is unaffected in the eye furrow. At later stages of development, however (posterior to the furrow, and the pupal wing), adhesive deficits associated with Rap1 loss-of-function are manifest and Egfr-dependent cell types are lost. In the pupal wing, Rap1 mutant cells scatter, the Egfr signal is not maintained, and wing veins fail to differentiate. Similarly in the eye, abnormal adhesive contacts between ommatidial precursors are observed as they exit the furrow. As a result, cells within a developing ommatidium likely fail to maintain their relative positions, photoreceptor/cone cell recruitment is impaired, and an incomplete ommatidium differentiates. In these contexts, therefore, Rap1 does not activate the Egfr/Ras/MAPK signaling cascade to specify cell fate, but instead regulates and/or maintains adhesive contacts necessary for the continued receipt of an Egfr signal.
Why are Egfr-dependent cell types particularly sensitive to Rap1 loss? In several developmental contexts, including the Drosophila wing, eye and trachea, it has been demonstrated that Egfr/Ras signaling up-regulates DE-cad levels (both transcriptionally and post-translationally) (Brown et al., 2006; Cela and Llimargas, 2006; O'Keefe et al., 2007). Loss of Egfr signaling reduces DE-cad levels, often leading to the loss of epithelial integrity. When DE-cad levels are manipulated, Egfr-dependent cell types are often affected (even when epithelial integrity is maintained). In particular, reduction of DE-cad results in photoreceptor loss, ommatidial rotation defects (Mirkovic and Mlodzik, 2006), and wing vein discontinuities (O'Keefe et al., 2007). These data suggest that DE-cad functions to stabilize or maintain Egfr/Ras/MAPK signaling between cells. In the Drosophila eye this makes intuitive sense as cells with highest levels of MAPK activity (ommatidial precursors) form tightly adhesive clusters of cells, segregating themselves from the rest of the epithelium as differentiation proceeds. In Rap1 mutant cells the ability of Egfr/Ras/MAPK signaling to positively affect DE-cad-mediated adhesion is clearly compromised. As a result, Egfr signaling between cells is weakened, and/or not maintained, and Egfr-dependent cell types are lost. In this way, Ras and Rap1 act in parallel to regulate DE-cad-mediated adhesion, facilitating consistent responses to developmental signals, and the appropriate differentiation of cell types within an epithelium.
Rap1 is necessary for planar cell polarity
PCP signaling orients large groups of cells within the plane of an epithelium allowing for the coordinated differentiation of asymmetric structures. For example, in the wing epithelium each cell generates a single hair at its distal edge that points distally, and mutations in the planar cell polarity network of proteins result in wing hair misalignment, and often problems in wing hair formation. Briefly, a core set of PCP proteins (including Frizzled, Disheveled, Prickle, Strabismus, Starry night and Diego) becomes asymmetrically distributed along the proximal/distal axis of each wing cell. While the precise mechanisms by which these protein asymmetries are established remain unclear, a number of effector proteins have been identified that translate the asymmetric PCP signal into the polarized formation of actin-based wing hairs (Klein and Mlodzik, 2005; Seifert and Mlodzik, 2007; Zallen, 2007). These same genes direct ommatidial alignment in the eye.
Here we identify Rap1 as a novel effector of the PCP signaling pathway in Drosophila. In the eye, PCP signaling determines chirality (based on R3/R4 cell fate specification) as well as the direction of ommatidial rotation. Rap1 mutant cells cause very subtle defects in chirality (data not shown), but dramatically affect ommatidial rotation. In other words, transmission of the PCP signal through the eye epithelium is essentially unaffected in Rap1 mutant tissue, but the adhesive/cytoskeletal response to PCP is compromised. In the eye, therefore, Rap1 primarily acts as a PCP effector. What are the mechanisms by which Rap1 affects PCP in the eye? Current evidence suggests that additional signaling inputs are required downstream of the PCP genes to mechanically drive the rotation process (nemo for example) (Choi and Benzer, 1994). One signaling pathway that acts downstream of PCP (or in parallel) to control ommatidial rotation is the Egfr pathway, as Egfr signaling has previously been shown to affect ommatidial rotation (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003; Strutt and Strutt, 2003). It is possible that Rap1 mutant cells mis-rotate because Egfr signaling is compromised, however this is not the only possibility. Rap1 also regulates DE-cad/ adherens junction localization in the developing eye and it has been demonstrated that DE-cad promotes ommatidial rotation (Mirkovic and Mlodzik, 2006). It is most likely, therefore, that Rap1 maintains/regulates DE-cad-mediated adhesive contacts necessary for ommatidial rotation, and in this way effects PCP in the eye.
The mechanisms by which Rap1 affects PCP in the wing, however, are less clear, as Egfr (data not shown) and DE-cad do not play clear roles in this process. Adult wings containing Rap1 mutant cells often contained disorganized patches of wing hairs, and when pupal wings were examined, prehair formation was disrupted. It has been previously shown that many PCP genes (including the core components) have non-autonomous effects on wing hair formation, however this was not the case for Rap1. In the pupal wing, wildtype cells immediately adjacent to Rap1 mutant clones had normal prehairs. Loss of Rap1 also did not affect localization of the core PCP protein Stan, and when prehairs formed in Rap1 mutant cells, they were found at the distal edge. These data indicate that Rap1 mutant cells are polarized appropriately. As in the eye, therefore, Rap1 does not affect propagation of the PCP signal through the wing epithelium, but instead acts as a local, cell-autonomous PCP effector, translating cell polarity into a coordinated cytoskeletal response.
We demonstrated genetic interactions between a hypomorphic Rap1 phenotype in the wing (ptc>Rap1-IR) and several PCP genes. Loss of a single copy of starry night (a core PCP gene), double time (a PCP regulator), or multiple wing hairs (a PCP effector) all exacerbated wing hair misalignments associated with the ptc>Rap1-IR genotype. Among these factors, the strongest interaction was seen between Rap1 and multiple wing hairs (mwh), consistent with Rap1 acting as a PCP effector. In adult and pupal wings containing Rap1 mutant clones, multiple wing hairs were frequently observed (similar to the mwh phenotype). Interestingly, mwh has recently been cloned (Strutt and Warrington, 2008; Yan et al., 2008), and found to encode a GTPase-binding domain/formin homology 3 (GBD/FH3) domain containing protein, functioning to inhibit actin filament formation. Although our data suggests that Rap1 does not directly affect Mwh localization, it will be interesting to see whether Rap1 regulates Mwh activity to control actin dynamics essential for the polarized formation of wing hairs. Ommatidial rotation is also perturbed in mutants for the Drosophila Myosin II, Zipper (Fiehler and Wolff, 2007), also implicating actin dynamics in this process. Thus, the effects of Rap1 on PCP readouts in both the wing and eye could be connected at the level of the actin cytoskeleton.
The most striking genetic interaction with ptc>Rap1-IR, however, was seen with DE-cad/shg, suggesting that the adhesive and cell shape phenotypes associated with Rap1 disrupt PCP signaling in the wing. In fact, it has recently been demonstrated that subtle defects in epithelial cell packing (associated with the posterior crossvein, or loss of PTEN) exacerbate polarity phenotypes associated with fat mutant clones (Ma et al., 2008). It is not surprising, therefore, that extreme cell shape defects associated with Rap1 loss-of-function clones affect wing hair alignment. Rap1 PCP phenotypes are more severe than those described by Ma et al., however, as wing hairs associated with Rap1 mutant cells are not swirled, but instead are randomly oriented.
Deciphering the mechanisms by which Rap1 regulates DE-cad localization, therefore, is critical to understanding Rap1 function in this context and others (cancer cell metastasis in particular). An intriguing link between Rap1 and components of the exocyst has recently been established (Frische et al., 2007). Since the exocyst is involved in recycling E-cad from endosomes to the plasma membrane (Langevin et al., 2005), it is possible that Rap1 interacts with exocyst components to control recycling of DE-cad-based adherens junctions, promoting the making and breaking of cell contacts necessary for hexagonal packing of wing epithelial cells and ommatidial rotation. In the pupal wing, evidence suggests that PCP factors regulate the polarized trafficking of DE-cad-containing exocyst vesicles to coordinate epithelial cell shape (Classen et al., 2005). Therefore, Rap1 may serve as the critical link between PCP factors and the exocyst in this developmental context, a hypothesis to be tested in the future.
Canoe is a Rap1 effector in the Drosophila wing and eye
The protein encoded by AF-6/Canoe includes two N-terminal Ras association (RA) domains, a phospho-peptidebinding Forkhead-Associated domain (FHA), a DIL domain found in myosins that likely mediates protein-protein interactions, and a PDZ domain involved in protein-protein interactions. In developing epithelia, it has been demonstrated that AF-6/Canoe functions as a scaffolding protein (linking proteins together via its multiple protein-protein interaction domains) to regulate adhesion between cells and the actin cytoskeleton. Both AF-6 and Canoe bind the junctional protein ZO-1/Polychaetoid, and localize in vivo to tight or adherens junctions (Takahashi et al., 1998; Yamamoto et al., 1997). Consistent with this role, mouse embryos lacking AF-6 are not viable due to severe defects in cell adhesion and cell polarity (Zhadanov et al., 1999).
It has been demonstrated that localization of the AF-6/ZO-1 complex to cell junctions is a regulated process. The GTP-bound form of Ras binds the RA domains of AF-6 (Kuriyama et al., 1996), causing dissociation of the AF-6/ZO-1 complex, and loss of both proteins from cell junctions (Boettner et al., 2000; Yamamoto et al., 1997). In this way, AF-6 acts as an effector of the Egfr/Ras signaling pathway at points of cell contact. This hypothesis is supported in vivo as Canoe plays a critical role in Egfr/Ras-dependent developmental processes in the Drosophila eye (Gaengel and Mlodzik, 2003; Matsuo et al., 1997). While this data suggests that AF-6/Canoe acts downstream of Ras, it has subsequently been demonstrated that AF-6/Canoe binds more effectively to Rap1 (Boettner et al., 2000; Boettner et al., 2003; Linnemann et al., 1999; Su et al., 2003). In several contexts, including dorsal closure of the Drosophila embryo (Boettner et al., 2003), activity-dependent remodeling of dendritic spines in neurons (Xie et al., 2005), and integrin-based adhesion in cultured cells (Su et al., 2003), functional connections between Rap1 and AF-6/Canoe have been demonstrated. It seems, therefore, that AF-6/Canoe functions downstream of multiple GTPases, to control cell adhesion. Finally, GTPases such as Ras and Rap1 are not the only signal transducers to affect AF-6/Canoe. In various developmental systems it has been shown that Canoe interacts with the Notch, JNK and Wg signaling pathways as well (Carmena et al., 2006; Miyamoto et al., 1995; Takahashi et al., 1998). These results suggest that AF-6/Canoe functions as a critical scaffolding protein at cell junctions, binding multiple signal transduction effectors and integrating a complex array of signals to affect adhesion and actin dynamics of developing cells.
In the studies described here, we identify two novel roles for canoe in the Drosophila wing epithelium, both of which involve Rap1. First, canoe loss-of-function cells had adhesive defects. As opposed to wildtype cells that maintain adhesive contacts with their neighbors as they grow and divide, canoe mutant cells dispersed, resulting in highly fragmented clones of cells. The adherens junctions of these cells were not evenly distributed, but instead concentrated at one cell-cell interface. In this way, pairs of canoe mutant cells adhered tightly to one another, scattered into the adjacent wildtype epithelium, and disrupt the characteristic, hexagonal packing of wing epithelial cells. This phenotype is very unusual and phenocopies Rap1 in this context. These results strongly suggest that Canoe acts as a Rap1 effector in the developing wing, controlling the even distribution of adherens junctions about the apical circumference of epithelial cells. Secondly, we demonstrate a role for canoe in the planar cell polarity pathway. Signaling through the planar cell polarity pathway is necessary for appropriate wing hair formation and alignment. Adult wings containing canoe mutant clones of cells often had disorganized patches of wing hairs, and at pupal stages, canoe mutant cells were defective in actin-rich prehair formation. In this context Rap1 and canoe genetically interacted, as removal of a single copy of canoe dramatically worsened wing hair alignment in a hypomorphic Rap1 background. Canoe, therefore, acts together with Rap1 as an effector of the planar cell polarity pathway, likely affecting actin dynamics essential for wing hair formation and alignment.
Supplementary Material
Acknowledgements
We would like to thank Iswar Hariharan, Marek Mlodzik, Daniel Marenda, Hugo Bellen, Benjamin Boettner, Paul Adler, David Strutt, Daisuke Yamamoto, Ryu Ueda and his colleagues at The National Institute of Genetics, The Drosophila Genetic Resource Center, The Bloomington Drosophila Stock Center, and The Developmental Studies Hybridoma Bank for reagents. Thanks to Yan Liu and Kathryn Hanley for patient help with statistical analysis, to Ivy Yu for help with quantitative PCR, and Julio Vasquez for assistance with microscopy. Finally, thanks to Dino, Crash and the entire Edgar and Curtiss labs for advice, support and entertainment. This work was supported by NCI 5 U56 CA096288 and NCI 1 U54 CA132381 to FHCRC; NCI 5 U56 CA096286 and NCI 1 U54 CA132383 to NMSU; NMSU MARC NIH GM07667-31; and NMSU HHMI-USE 52005881.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler PN. Planar signaling and morphogenesis in Drosophila. Dev Cell. 2002;2:525–535. doi: 10.1016/s1534-5807(02)00176-4. [DOI] [PubMed] [Google Scholar]
- Asha H, de Ruiter ND, Wang MG, Hariharan IK. The Rap1 GTPase functions as a regulator of morphogenesis in vivo. Embo J. 1999;18:605–615. doi: 10.1093/emboj/18.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Jan LY, Jan YN. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc Natl Acad Sci U S A. 2000;97:9064–9069. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, Gaul U. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics. 2003;165:159–169. doi: 10.1093/genetics/165.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, de Bruyn K, Enserink J, Kuiperij B, Rangarajan S, Rehmann H, Riedl J, de Rooij J, van Mansfeld F, Zwartkruis F. The role of Rap1 in integrin-mediated cell adhesion. Biochem Soc Trans. 2003;31:83–86. doi: 10.1042/bst0310083. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baonza A, Freeman M. Epithelial cell adhesion in the developing Drosophila retina is regulated by Atonal and the EGF receptor pathway. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–5412. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Carmena A, Speicher S, Baylies M. The PDZ protein Canoe/AF-6 links Ras-MAPK, Notch and Wingless/Wnt signaling pathways by directly interacting with Ras, Notch and Dishevelled. PLoS ONE. 2006;1:e66. doi: 10.1371/journal.pone.0000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J Cell Sci. 2003;116:435–440. doi: 10.1242/jcs.00238. [DOI] [PubMed] [Google Scholar]
- Cela C, Llimargas M. Egfr is essential for maintaining epithelial integrity during tracheal remodelling in Drosophila. Development. 2006;133:3115–3125. doi: 10.1242/dev.02482. [DOI] [PubMed] [Google Scholar]
- Chae J, Kim MJ, Goo JH, Collier S, Gubb D, Charlton J, Adler PN, Park WJ. The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development. 1999;126:5421–5429. doi: 10.1242/dev.126.23.5421. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Classen AK, Anderson KI, Marois E, Eaton S. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell. 2005;9:805–817. doi: 10.1016/j.devcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Clifford RJ, Schupbach T. Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics. 1989;123:771–787. doi: 10.1093/genetics/123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H. Casein kinase Iepsilon modulates the signaling specificities of dishevelled. Mol Cell Biol. 2004;24:2000–2011. doi: 10.1128/MCB.24.5.2000-2011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Rubinfeld B, Albert I, McCormick F. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. Embo J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Hafen E. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310:348–362. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Frische EW, Pellis-van Berkel W, van Haaften G, Cuppen E, Plasterk RH, Tijsterman M, Bos JL, Zwartkruis FJ. RAP-1 and the RAL-1/exocyst pathway coordinate hypodermal cell organization in Caenorhabditis elegans. Embo J. 2007;26:5083–5092. doi: 10.1038/sj.emboj.7601922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–5423. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Carraway KL, 3rd, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96:847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Patel AB, Nagpal R, Perrimon N. The emergence of geometric order in proliferating metazoan epithelia. Nature. 2006;442:1038–1041. doi: 10.1038/nature05014. [DOI] [PubMed] [Google Scholar]
- Guichard A, Biehs B, Sturtevant MA, Wickline L, Chacko J, Howard K, Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development. 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH.Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3 Curr Biol 2006161796–1806. f [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Carthew RW, Rubin GM. The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell. 1991;67:717–722. doi: 10.1016/0092-8674(91)90066-8. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Hariharan IK, Rubin GM. Star is required for neuronal differentiation in the Drosophila retina and displays dosage-sensitive interactions with Ras1. Dev Biol. 1993;160:51–63. doi: 10.1006/dbio.1993.1285. [DOI] [PubMed] [Google Scholar]
- Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, Braga VM, Birchmeier W, Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Grawe F, Grzeschik N, Knust E. Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr Biol. 2002;12:1675–1680. doi: 10.1016/s0960-9822(02)01180-6. [DOI] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- Kinashi T, Katagiri K. Regulation of immune cell adhesion and migration by regulator of adhesion and cell polarization enriched in lymphoid tissues. Immunology. 2005;116:164–171. doi: 10.1111/j.1365-2567.2005.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- Klein TJ, Jenny A, Djiane A, Mlodzik M. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr Biol. 2006;16:1337–1343. doi: 10.1016/j.cub.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Klein TJ, Mlodzik M. Planar cell polarization: an emerging model points in the right direction. Annu Rev Cell Dev Biol. 2005;21:155–176. doi: 10.1146/annurev.cellbio.21.012704.132806. [DOI] [PubMed] [Google Scholar]
- Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science. 2002;295:1285–1288. doi: 10.1126/science.1067549. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Kuriyama M, Harada N, Kuroda S, Yamamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C, Canaani E, Kaibuchi K. Identification of AF-6 and canoe as putative targets for Ras. J Biol Chem. 1996;271:607–610. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- Langevin J, Morgan MJ, Sibarita JB, Aresta S, Murthy M, Schwarz T, Camonis J, Bellaiche Y. Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev Cell. 2005;9:365–376. doi: 10.1016/j.devcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lesokhin AM, Yu SY, Katz J, Baker NE. Several levels of EGF receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev Biol. 1999;205:129–144. doi: 10.1006/dbio.1998.9121. [DOI] [PubMed] [Google Scholar]
- Linnemann T, Geyer M, Jaitner BK, Block C, Kalbitzer HR, Wittinghofer A, Herrmann C. Thermodynamic and kinetic characterization of the interaction between the Ras binding domain of AF6 and members of the Ras subfamily. J Biol Chem. 1999;274:13556–13562. doi: 10.1074/jbc.274.19.13556. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lundquist EA. Small GTPases. WormBook. 2006:1–18. doi: 10.1895/wormbook.1.67.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Amonlirdviman K, Raffard RL, Abate A, Tomlin CJ, Axelrod JD. Cell packing influences planar cell polarity signaling. Proc Natl Acad Sci U S A. 2008;105:18800–18805. doi: 10.1073/pnas.0808868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E, Roch F, Noll E, Baonza A, Duffy JB, Perrimon N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development. 1999;126:5739–5747. doi: 10.1242/dev.126.24.5739. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Takahashi K, Kondo S, Kaibuchi K, Yamamoto D. Regulation of cone cell formation by Canoe and Ras in the developing Drosophila eye. Development. 1997;124:2671–2680. doi: 10.1242/dev.124.14.2671. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Mlodzik M. Cooperative activities of drosophila DE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–3293. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- Mishra S, Smolik SM, Forte MA, Stork PJ. Ras-independent activation of ERK signaling via the torso receptor tyrosine kinase is mediated by Rap1. Curr Biol. 2005;15:366–370. doi: 10.1016/j.cub.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Nihonmatsu I, Kondo S, Ueda R, Togashi S, Hirata K, Ikegami Y, Yamamoto D. canoe encodes a novel protein containing a GLGF/DHR motif and functions with Notch and scabrous in common developmental pathways in Drosophila. Genes Dev. 1995;9:612–625. doi: 10.1101/gad.9.5.612. [DOI] [PubMed] [Google Scholar]
- Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. Embo J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J, Groppe J, Guillemin K, Krasnow MA, Gehring WJ, Affolter M. The Drosophila Serum Response Factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development. 1996;122:2589–2597. doi: 10.1242/dev.122.9.2589. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- O'Keefe DD, Prober DA, Moyle PS, Rickoll WL, Edgar BA. Egfr/Ras signaling regulates DE-cadherin/Shotgun localization to control vein morphogenesis in the Drosophila wing. Dev Biol. 2007;311:25–39. doi: 10.1016/j.ydbio.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyer B, Hadorn E. [The manifestation pattern of the mutant "multiple wing hairs" (mwh) of Drosophila melanogaster] Arch Julius Klaus Stift Vererbungsforsch Sozialanthropol Rassenhyg. 1965;40:19–26. [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Price LS, Hajdo-Milasinovic A, Zhao J, Zwartkruis FJ, Collard JG, Bos JL. Rap1 regulates E-cadherin-mediated cell-cell adhesion. J Biol Chem. 2004;279:35127–35132. doi: 10.1074/jbc.M404917200. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Ras1 promotes cellular growth in the Drosophila wing. Cell. 2000;100:435–446. doi: 10.1016/s0092-8674(00)80679-0. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Rawls AS, Wolff T. Strabismus requires Flamingo and Prickle function to regulate tissue polarity in the Drosophila eye. Development. 2003;130:1877–1887. doi: 10.1242/dev.00411. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Abodeely M, Young MW. Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr Biol. 2000;10:1399–1402. doi: 10.1016/s0960-9822(00)00786-7. [DOI] [PubMed] [Google Scholar]
- Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- Shimonaka M, Katagiri K, Nakayama T, Fujita N, Tsuruo T, Yoshie O, Kinashi T. Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J Cell Biol. 2003;161:417–427. doi: 10.1083/jcb.200301133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Strutt D, Warrington SJ. Planar polarity genes in the Drosophila wing regulate the localisation of the FH3-domain protein Multiple Wing Hairs to control the site of hair production. Development. 2008;135:3103–3111. doi: 10.1242/dev.025205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Price MA, Strutt D. Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila. Curr Biol. 2006;16:1329–1336. doi: 10.1016/j.cub.2006.04.041. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. Polarity determination in the Drosophila eye. Curr Opin Genet Dev. 1999;9:442–446. doi: 10.1016/S0959-437X(99)80067-7. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–1457. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, Bier E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 1993;7:961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- Su L, Hattori M, Moriyama M, Murata N, Harazaki M, Kaibuchi K, Minato N. AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem. 2003;278:15232–15238. doi: 10.1074/jbc.M211888200. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Matsuo T, Katsube T, Ueda R, Yamamoto D. Direct binding between two PDZ domain proteins Canoe and ZO-1 and their roles in regulation of the jun N-terminal kinase pathway in Drosophila morphogenesis. Mech Dev. 1998;78:97–111. doi: 10.1016/s0925-4773(98)00151-8. [DOI] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Tsai IC, Amack JD, Gao ZH, Band V, Yost HJ, Virshup DM. A Wnt-CKIvarepsilon-Rap1 pathway regulates gastrulation by modulating SIPA1L1, a Rap GTPase activating protein. Dev Cell. 2007;12:335–347. doi: 10.1016/j.devcel.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto N, Hattori M, Yang H, Bos JL, Minato N. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J Biol Chem. 1999;274:18463–18469. doi: 10.1074/jbc.274.26.18463. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Wang H, Singh SR, Zheng Z, Oh SW, Chen X, Edwards K, Hou SX. Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell. 2006;10:117–126. doi: 10.1016/j.devcel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Wilk R, Weizman I, Shilo BZ. trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- Wolff T, Guinto JB, Rawls AS. Screen for Genetic Modifiers of stbm Reveals that Photoreceptor Fate and Rotation Can Be Genetically Uncoupled in the Drosophila Eye. PLoS ONE. 2007;2:e453. doi: 10.1371/journal.pone.0000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Martinez-Arias MBA, editor. The development of Drosophila melanogaster. Cold Spring Harbor: Cold Spring Harbor Press; 1993. pp. 1277–1326. [Google Scholar]
- Wong LL, Adler PN. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J Cell Biol. 1993;123:209–221. doi: 10.1083/jcb.123.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Huganir RL, Penzes P. Activity-dependent dendritic spine structural plasticity is regulated by small GTPase Rap1 and its target AF-6. Neuron. 2005;48:605–618. doi: 10.1016/j.neuron.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM, Greenwood S, Struhl G. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. The Ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Huen D, Morely T, Johnson G, Gubb D, Roote J, Adler PN. The multiple-wing-hairs gene encodes a novel GBD-FH3 domain-containing protein that functions both prior to and after wing hair initiation. Genetics. 2008;180:219–228. doi: 10.1534/genetics.108.091314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- Zallen JA. Planar polarity and tissue morphogenesis. Cell. 2007;129:1051–1063. doi: 10.1016/j.cell.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Zhadanov AB, Provance DW, Jr, Speer CA, Coffin JD, Goss D, Blixt JA, Reichert CM, Mercer JA. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr Biol. 1999;9:880–888. doi: 10.1016/s0960-9822(99)80392-3. [DOI] [PubMed] [Google Scholar]
- Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc Natl Acad Sci U S A. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.