Abstract
Receptors for immunoglobulins (Fc receptors) play a central role during an immune response, as they mediate the specific recognition of antigens of almost infinite diversity by leucocytes, thereby linking the humoral and cellular components of immunity. Indeed, engagement of Fc receptors by immunoglobulins initiates a range of immunoregulatory processes that might also play a role in disease pathogenesis. In the circulation, five main types of immunoglobulins (Ig) exist – namely IgG, IgA, IgE, IgM and IgD and receptors with the ability to recognize and bind to IgG (Fcγ receptor family), IgE (FcεRI and CD23), IgA (CD89; Fcα/µR) and IgM (Fcα/µR) have been identified and characterized. However, it is astonishing that nearly all the known human Fc receptors display extensive genetic variation with clear implications for their function, thus representing a substantial genetic risk factor for the pathogenesis of a range of chronic inflammatory disorders.
Keywords: Fc receptors, immunoglobulins, polymorphisms, copy number variation, chronic inflammatory diseases
The Fcγ receptor family
Immunoglobulin G (IgG) is the most abundant Ig class in serum, constituting over 75% of circulating immunoglobulin. It mediates key effector functions through interaction with Fcγ receptors, which are encoded by eight different genes, each with multiple transcriptional isoforms and located in a locus on the long arm of chromosome 1 (1q21–23). Fcγ receptors are related structurally and belong to the Ig protein superfamily with multiple Ig-like domains. Fcγ receptors are divided generally into three main classes: FcγRI (CD64), FcγRII (CD32) and FcγRIII (CD16), each with distinct structural and functional properties (Fig. 1).
Fig. 1.
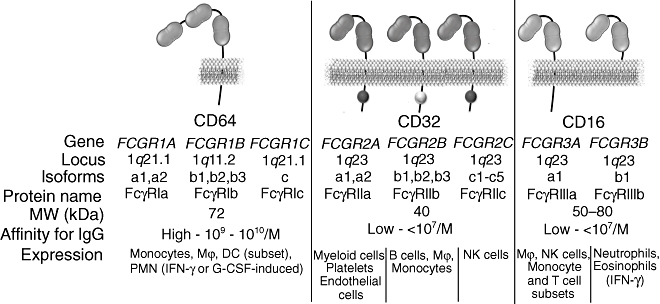
Overview of the Fcγ receptor family. Mϕ, macrophages; DC, dendritic cells; PMN, polymorphonuclear leucocytes; IFN-γ, interferon-γ; G-CSF, granulocyte colony-stimulating factor; NK, natural killer.
FcγRI is a high-affinity receptor for monomeric IgG (Ka: 109–1010/M) with three extracellular Ig-like domains expressed constitutively by monocytes and macrophages, as well as by many myeloid progenitor cells. Three genes coding for FcγRI have been characterized: FCGR1A, FCGR1B and FCGR1C[1]. However, it is generally accepted that only FcγRIa is capable of IgG binding, whereas FcγRIb and FcγRIc possibly represent truncated or soluble forms of the receptor, with poorly characterized function. The α ligand-binding chain of FcγRI associates with a disulphide-bonded dimer of FcR γ chain, a signal transducing polypeptide described originally as a component of the high affinity IgE receptor that associates with various Fc receptors. The γ chain carries an activatory signalling motif (immunoreceptor tyrosine-based activation motif: ITAM), which mediates signal transduction upon FcγRI engagement.
In contrast to FcγRI, the other two classes of Fcγ receptor, FcγRII and FcγRIII, display low affinity for monomeric IgG. They are capable of binding to aggregated IgG through multimeric low-affinity, high-avidity interactions, which are particularly important in the recognition and binding of antibody–antigen complexes during an immune response. IgG binding to low-affinity FcγR can trigger a range of effector and immunoregulatory functions, including degranulation, phagocytosis and regulation of antibody production [2]. Examination of the genomic organization of the FcγRII and FcγRIII locus suggests that these receptors arose as a consequence of multiple gene duplication and recombination processes, followed by gain-of-function mutations. FcγRII is encoded by three genes: FCGR2A, FCGR2B and FCGR2C. All the members of the FcγRII class share a characteristic structure unique to FcγRII, that includes functional signalling motifs in their cytoplasmic domains, namely ITAM for FcγRIIa and FcγRIIc and immunoreceptor tyrosine-based inhibition motif (ITIM) for FcγRIIb. FcγRII is expressed by diverse cell types: FcγRIIa by myeloid cells, including polymorphonuclear leucocytes, monocytes, macrophages, platelets and certain types of endothelial cells; FcγRIIb by B cells, monocytes and macrophages; while FcγRIIc expression is restricted solely to natural killer (NK) cells.
The other low-affinity Fcγ receptor, FcγRIII, is encoded by genes FCGR3A and FCGR3B. Although FcγRIIIa and FcγRIIIb share high levels of sequence homology, they exhibit distinct structural differences. FcγRIIIa is a transmembrane protein that associates with the FcRγ chain, whereas FcγRIIIb is processed post-translationally as a glycosylphosphatidylinositol (GPI)-anchored protein, lacking transmembrane and intracellular domains. This difference is thought to be the result of an original gene duplication of the ancestral gene followed by point mutation in the membrane proximal region of the extracellular domain that created a GPI-anchor signal sequence (serine at position 203 rather than the phenylalanine present in the transmembrane FcγRIIIa). The FcγRIIIa isoform is expressed widely by several leucocyte cell types, including macrophages, NK cells and subsets of T cells and monocytes, while FcγRIIIb is expressed constitutively only by neutrophils [2].
The Fcε receptor family
Although normally present in serum at very low levels, IgE may be up-regulated in certain individuals where it associated with atopy, and in immune responses to certain parasites where it is believed to play a protective role. Unlike Fcγ receptors, there are no structural similarities shared by the two classes of IgE receptors: FcεRI, representing the high-affinity receptor for IgE, and FcεRII (CD23), a lower-affinity receptor. FcεRI is a heterotetrameric structure that is comprised of one α subunit, one β subunit and a homodimer of the FcRγ subunit, encoded by FCER1A, MS4A2 and FCER1G genes, respectively. The α subunit mediates IgE binding and its structure resembles closely those of the Fcγ receptors, as it consists of two extracellular Ig-like domains with a transmembrane and a cytoplasmic region. The β subunit of FcεRI is a 4-α helix membrane-spanning protein with tyrosine phosphorylation motifs in its C-terminal cytoplasmic region. Recent studies have demonstrated that it exists in two splice variant forms, the β and βT, the latter having a role in the cellular targeting of FcεRIa [3]. Finally, the γ subunit together with the β subunit facilitates signalling following engagement of FcεRIa. FcεRI is expressed constitutively by mast cells and basophils, whereas monocytes, eosinophils and dendritic cells express FcεRI, but only in its trimeric form (α,γ-γ), lacking the β subunit [3].
FcεRII (CD23) is the low-affinity receptor for IgE, having an affinity of approximately 107/M. It is expressed constitutively by B cells and is involved in the regulation of IgE production by B cells – a role related to that of FcγRIIb. In addition, it has been demonstrated that several cell types, including eosinophils, neutrophils, macrophages, monocytes and T cells, up-regulate FcεRII expression upon treatment with interleukin (IL)-4 [4]. FcεRII can also bind and interact with other molecules, such as CD21, CD11b and CD11c, although the functional significance of these interactions is still being defined. FcεRII displays susceptibility to proteolytic cleavage by A disintegrin and metalloproteinase (ADAM) sheddases and soluble FcεRII has been reported to have mitogenic properties [5,6].
Receptors for IgM and IgA
A number of receptors specific for IgA and IgM have been characterized, including the polymeric immunoglobulin receptor (pIgR) and Fcα/µR. These two receptors bind to IgA and IgM with intermediate affinity and are encoded by the PIGR and FCAMR genes present at the same locus on chromosome 1 (1q32). Fcα/µR is expressed by mature B cells and macrophages, as well as in secondary lymphoid organs such as lymph nodes, intestine and appendix, and it is therefore anticipated that it possesses a range of immunoregulatory roles [7]. In contrast, pIgR is expressed predominantly on the basolateral surface of epithelial cells and is involved in the transport of mucosal IgA and IgM across the epithelia [8].
Although pIgR and Fcα/µR, along with several other molecules, can bind specifically to IgA, the only ‘classical’ Fc receptor specific for IgA is FcαRI (CD89) [9]. While the ligand-binding (α) chain of FcαRI is related structurally to those of FcγR and FcεRI it is a more distantly related member of the family, and the FCAR gene maps to chromosome 19, alongside genes for leucocyte Ig-like receptors and natural killer cell receptors (KIRs). Like many of the FcγR and FcεRI, the FcαRI a chain associates with a homodimer, the FcRγ chain, although it is often expressed in the absence of FcRγ chain pairing.
Fc receptor genetic variation – implications for function
Single nucleotide polymorphisms (SNPs)
The vast majority of the Fc receptor encoding genes display genetic variation either in the form of SNPs or alteration in their copy number. Although many SNPs have been identified for Fc receptors, for most of them their precise impact upon receptor function remains unknown. Functionally relevant genetically determined SNPs can be categorized into three main types, based on the effect they have on receptor function: (i) augmenting the affinity of Fcγ receptors for particular IgG subclasses; (ii) altering the receptor function and consequently downstream effector events; and (iii) affecting transcriptional promoter activity or mechanisms that alter the levels of receptor expression (Table 1).
Table 1.
Functional effects of genetic variants of Fc receptors.
| Receptor | Gene | Variant | Effect | Reference |
|---|---|---|---|---|
| FcγRIIa | FCGR2A | R131H | H131: higher affinity for IgG2 | [12] |
| FcγRIIb | FCGR2B | I232T | T232: lower affinity for lipid rafts/decreased inhibitory activity | 17,18 |
| −386G > C | −386C: higher promoter activity | [23] | ||
| −343G > C | −343C: loss of AP-1 binding site/transcriptionally repressed | [24] | ||
| −120T > A | −120A: higher promoter activity | [23] | ||
| FcγRIIc | FCGR2C | Q57X | Truncated non-functional protein | [21] |
| −386G > C | −386C: increased promoter activity | [21] | ||
| −120C > A | −120A: linked with higher transcriptional activity | [21] | ||
| CNV | [21] | |||
| FcγRIIIa | FCGR3A | V158F | V158: higher affinity for IgG1 and 3, binds IgG4 | [14] |
| CNV | [35] | |||
| FcγRIIIb | FCGR3B | NA1/2 | NA1: higher affinity for IgG1 and 3 | [15] |
| SH/A78D | Unknown – linked to the NA2 allele | [16] | ||
| CNV | 33,108 | |||
| FcεRI α chain | FCER1A | −66T > C | −66T: higher promoter activity – additional GATA-1 binding site | [26] |
| −315C > T | −315T: increased transcriptional activity – Sp1 site | [27] | ||
| −335T > C | −335C: increased expression? | [109] | ||
| FcεRI β chain | MS4A2 | −426T > C | −426C: increased promoter activity | [31] |
| −654C > T | −654T: increased promoter activity-YY-1 binding | 28,31 | ||
| −109C > T | −109T: unknown/higher receptor expression | [28] | ||
| E237G | G237: associated with higher expression | [88] | ||
| I181L | L181: unknown/higher expression? | [88] | ||
| V183L | L183: unknown/higher expression? | [88] | ||
| FcεRII | FCER2 | R62W | W62: resistant to proteolytic cleavage | [5] |
| pIgR | PIGR | A580V | V580: near endoproteolytic cleavage site/reduced efficiency of IgA release? | [8] |
| FcαRI | FCAR | S248G | G248: enhanced IgA-mediated responses; increased cytokine release | [19] |
Ig: immunoglobulin.
One of the first functional SNP identified for Fc receptors was the R131H allelic variant for the low-affinity Fcγ receptor FcγRIIa. This point mutation (519G > A) results in an amino acid substitution [arginine (R) to histidine (H)] at position 131, which is located in the membrane proximal Ig-like domain of the extracellular region [10]. Recent crystallographic studies, along with previous mutational analyses, indicated that this region is involved in the receptor interface interacting with the Fc portion of IgG [11] and the R131H variant determines the affinity of FcγRIIa for human IgG2. In particular, while the R131 variant is not able to interact with hIgG2, H131 has been shown previously to bind and enable phagocytosis of hIgG2-coated particles [12]. The H131 variant might be particularly important in conditions characterized by high IgG2 antibody responses, where it may confer enhanced leucocyte activation by IgG2 and increased capacity for clearance of circulating IgG2 complexes.
SNPs affecting the binding affinity for IgG subclasses have also been characterized for FcγRIII. There are two co-dominantly expressed allelic variants of FcγRIIIa having either a valine (V) or a phenylalanine (F) at position 158. This single amino acid substitution has been demonstrated to increase the affinity of the V158 allotype for IgG1 and IgG3 compared to F158 and induce capacity for IgG4 binding [13,14]. Furthermore, IgG-induced NK cell activity has been found to be increased significantly in 158V/V rather than the 158F/F individuals.
FcγRIIIb is characterized by the presence of the human neutrophil antigen (HNA or NA), a polymorphic variant that comprises four non-synonymous and one synonymous mutation within the membrane distal Ig-like domain of the receptor. The four amino acid differences between the two NA allotypes (NA1 or HNA-1a and NA2 or HNA-1b) have an impact on the N-linked glycosylation of the receptor, and as a consequence affect the affinity for IgG subclasses. In particular, the NA1 allotype displays increased binding and phagocytosis of IgG1- and IgG3-coated particles and it has been shown to exhibit higher affinity for IgG3 compared to the NA2 allotype [15]. Apart from NA-1 and NA-2, the SH polymorphism (also termed as HNA-1c) has been added to the NA family of FcγRIIIb polymorphisms [16]. The SH allele determines the substitution of alanine at position 78 to aspartic acid (A78D) and is linked mainly to the NA2 allele; however, the precise role of this polymorphism in IgG binding is unknown.
In addition to the polymorphisms augmenting receptor affinity for IgG subclasses, SNPs that have a profound impact on the functional effector responses of Fc receptors following immunoglobulin binding have been described. For example, in the case of the inhibitory Fcγ receptor FcγRIIb, it has been demonstrated that a change of a non-polar isoleucine to a polar threonine at position 232 (I232T) within the transmembrane region could affect receptor activity. The T232 variant has been shown to be translocated less efficiently to membrane domains rich in cholesterol and sphingolipids, termed as lipid rafts [17,18]. As a consequence, reduced inhibitory activity was observed for the T232, due possibly to impaired interaction with protein kinases that reside preferentially in lipid raft membrane domains.
A recently identified functional polymorphism in FcεRII is the arginine (R) to tryptophan (W) substitution at position 62 (R62W). In contrast to the R62 variant, the W62 variant is resistant to proteolytic shedding following treatment with a broad range of different proteases, which might influence receptor function and mitogenic role [5]. Finally, a relatively common polymorphism identified in the coding region of FcαRI is associated with impaired capacity to trigger mechanisms such as cytokine release [19]. This polymorphism (844A > G) involves the change of serine 248 to glycine (S248G) within the cytoplasmic domain of the receptor's α chain [20]. IgA-mediated cross-linking of FcαRI on neutrophils from individuals homozygous for the G248 variant triggered significantly more IL-6 release than equivalent cross-linking of receptor on neutrophils from donors homozygous for the S248 variant. In fact, the G248 form, unlike the S248 variant, is capable of inducing cytokine release in the absence of the FcRγ chain. This capacity is presumed to be due, at least in part, to its ability to interact directly with the Src family member Lyn, an important component of the FcαRI signalling cascade.
A number of SNPs have been characterized that play a regulatory role in the expression of particular Fcγ and Fcε receptors. For example, within exon 3 of the FCGR2C gene, a single nucleotide substitution at position 202 results in the change of a glutamine residue to a stop codon at position 57 (Q57X), resulting in the production of a non-functional, truncated protein. Furthermore, NK cells from heterozygous 57Q/X donors displayed reduced expression compared to 57Q/Q donors, while homozygous 57X/X donors were negative for NK cell surface receptor expression [21].
Another determinant of the levels of receptor expression are SNPs within the upstream promoter sequences that may affect the transcriptional activity of the promoter. For example, two SNPs, −386G > C and −120T > A, have been identified within the almost identical promoter regions of the genes encoding the FcγRIIb and FcγRIIc receptors [21,22]. Using in vitro promoter activity assays, it has been shown that the less frequent −386C and −120A haplotypes exhibit enhanced transcriptional activity compared to the −386G and −120T ones, due mainly to the differential binding of GATA-4 and Yin-Yang 1 (YY-1) transcription factors [22,23]. In addition, genetic linkage was observed between the −386C and −120A alleles. Furthermore, an additional polymorphic site (G to C substitution at the –343 position; −343G > C) within the promoter of the FCGR2B gene enables the interaction of the YY-1 transcription factor, leading to transcriptional repression of FCGR2B via competition for binding with the c-Jun/AP-1 transcription complex [24,25]. Given the high sequence similarity between the FCGR2B and the FCGR2C gene promoters, the −343G > C polymorphism would also be predicted to be present within the FCGR2C promoter.
Similarly, within the promoter of the gene coding for the α chain of the FcεRI (FCER1A), a number of SNPs, including –66T > C, −315C > T and −335C > T, have been shown to regulate receptor expression through differential binding and transactivation of transcription regulatory factors. For example, the –66T variant displayed increased in vitro transcriptional activity compared to the −66C form, because the former has an additional GATA-1 binding motif [26]. Similarly, the C to T substitution at position −315 (also referred as −344) was associated with significantly higher promoter activity, an effect that was attributed to the binding of Sp1 transcriptional regulator [27]. Similar SNPs have been identified in the promoter regions of the gene encoding for the β chain of the FcεRI receptor [28–30]. Increased binding of YY-1 and consequently higher promoter activity has been observed for the −426C and −654T haplotypes, compared with the −426T and −654C haplotypes [31].
Copy number variation
Apart from SNPs, a number of recent studies have demonstrated that genes coding for Fcγ receptors exhibit variation in their copy numbers. Indeed, whole genome scans revealed that over 12% of the human genome is covered by copy number variation, accounting for a great proportion of genetic diversity between individuals (reviewed in [32]). In addition, astonishingly high variation has been noted within the Fcγ receptor locus (Fig. 2), but not within the loci where the genes coding for other Fc receptors are mapped. Copy number variation has been demonstrated for FCGR3B, FCGR2C and FCGR3A genes, but not for FCGR2A or FCGR2B. For FCGR3B, Willcocks and co-workers [33] have demonstrated recently an association between gene copy number and surface expression of FcγRIIIb in neutrophils. In addition, neutrophils isolated from donors with more than two gene copies displayed enhanced IgG-induced effector responses as well as increased cell adherence in IgG-coated surfaces compared with those from donors with less than two [33]. Similarly, NK cells from individuals with two or three copies of FCGR3A tend to express higher levels of receptor and exhibit greater antibody-dependent killing capacity than those from individuals with one copy of the gene [21]. Based on these findings, it is anticipated that higher copy numbers of the FcγRIIc gene may be associated with increased levels of surface receptor expression and potentiation of responses following stimulation with IgG. Although a number of studies have made use of well-validated complementary techniques for the assessment of copy number variation, there is controversy on the accuracy and sensitivity of some of these techniques, as they are still at an early stage of technical development.
Fig. 2.

Copy number variation of the Fcγ receptor locus revealed by genome-wide analysis. Array comparative genome hybridization data from the Whole Genome TilePath (WGTP) project of the Sanger Institute (based on Redon et al.[106], available at http://www.sanger.ac.uk/humgen/cnv/data/). Log intensity ratios from 270 HapMap samples for nine probes within the 1q23 region of chromosome 1, where the Fcγ receptor locus is mapped. Contrary to other probes, distinct clusters of intensity ratios within the Fcγ receptor probe (8H4) reveal extensive gene copy variation ranging from zero to more than three copies. Similar analyses of the same experimental dataset for the loci where other Fc receptor genes are mapped revealed minimal variation in intensity ratios among the HapMap individuals (data not shown). A notable exception was the FcαR locus (19q13) that displays significant variation, due probably to the close proximity of the FCAR gene with the KIR family genes, which have been described previously to exhibit extensive copy-number variation [107].
Fc receptor genetic variants as risk factors for chronic inflammatory diseases
The link between Fc receptor genetic variants and disease pathogenesis has been the subject for intensive investigation for a number of decades and it is accepted widely that they play a crucial role in the pathogenesis of a range of chronic inflammatory diseases, constituting significant genetic risk factors for disease development and prognosis. Based on the functional implications of the Fcγ receptor polymorphic and copy number variants, they can be categorized into either low- or high-responder variants. Low-responder polymorphic variants of Fcγ receptors, for example R131, F158 and NA2 for FCGR2A, FCGR3A and FCGR3B, respectively, as well as a low number of genomic copies, are associated usually with autoimmune pathologies that are characterized by the presence of circulating IgG complexes [34–36]. Reduced efficiency of IgG–Fc interactions with these low-responder variants may compromise clearance of IgG complexes from circulation, leading to their deposition in peripheral tissues; a process that initiates or exacerbates inflammatory processes with detrimental effects. Alternatively, high-responder variants are linked to chronic inflammatory disorders characterized by excessive or inappropriate leucocyte activation [37–39]. These variants may result in more efficient and prolonged Fc–IgG interactions. Reduced threshold for IgG-mediated cellular effector responses could promote leucocyte infiltration into tissues together with the release of histotoxic and cytotoxic compounds that amplify inflammatory cell-mediated tissue damage [40]. A number of chronic inflammatory diseases (summarized in Table 2) have been shown to be associated with Fcγ receptor genetic variants and include (but are not limited to) autoimmune pathologies, such as systemic lupus erythematosus (SLE) [14,17,22,24,25,36,41–60], rheumatoid arthritis [35,61–66], myasthenia gravis [67,68], certain neuropathies[69–72], acute allograft rejection [73] and vascular inflammatory and thrombotic disorders, such as coronary artery stenosis, peripheral atherosclerosis and vasculitis [37,38,74–81].
Table 2.
Association of Fcγ receptor variants with chronic inflammatory diseases.
| Gene | Variant | Disease | Reference |
|---|---|---|---|
| FCGR2A | H131 | GBS | [72] |
| R131 | Acute renal allograft rejection, APS, giant cell arteritis, HIT, ITP, IgA nephropathy, lupus nephritis, MG severity, peripheral atherosclerosis, RA severity, RF, SLE, WG | [34,36,38,39,41–44,46,47,49,52,57,64,66,68, 73,74,77,80–82,110–118] | |
| FCGR2B | −120A | SLE, CIDP | [22,71] |
| −343C | SLE | [24,25] | |
| −386C | SLE, CIDP | [22,71] | |
| T232 | SLE | [17,53,57–60] | |
| FCGR2C | High CNV | ITP | [21] |
| FCGR3A | F158 | Coronary artery stenosis, giant cell arteritis, lupus nephritis, SLE, WG | [14,37–39,46,50–53,82,83,119] |
| V158 | ACPA-positive RA, allergic rhinitis, bronchial asthma, HIT, ITP, IgA nephropathy severity, rheumatoid factor production, RA | [35,61–63,65,75,78,110,120,121] | |
| FCGR3B | NA1 | ANCA vasculitis, ITP, MG severity | [67,76,79] |
| NA2 | GBS severity, lupus nephritis, SLE | [55–57,69,70] | |
| High CNV | ANCA-positive vasculitis | [33] | |
| Low CNV | SLE, lupus nephritis | [33,86,108] |
ACPA: anti-citrullinated protein/peptide antibodies; ANCA: anti-neutrophil cytoplasmic antibodies; APS: anti-phospholipid syndrome; CIDP: chronic inflammatory demyelinating polyneuropathy; CNV: copy number variation; GBS: Guillain–Barré syndrome; HIT: heparin-induced thrombocytopenia; Ig: immunoglobulin; ITP: idiopathic thrombocytopenia purpura; MG: myasthenia gravis; NA: neutrophil antigen; RA: rheumatoid arthritis; RF: rheumatic fever; SLE: systemic lupus erythematosus; WG: Wegener's granulomatosis.
Although there is substantial evidence for the role of Fcγ receptor variants with susceptibility to all these diseases, SLE represents a prototype, multi-organ, antibody-mediated autoimmune disorder, characterized classically by elevated circulating IgG complexes. SLE has been shown to be associated strongly with almost all known Fcγ receptor polymorphic and copy number variants. Several groups have reported an increased frequency of the low-responder allele of FcγRIIIa, F158 among SLE patients [14,46,50–53,82,83]. Furthermore, this allele displays preferential segregation with affected individuals from multiplex SLE families, indicating clearly that it represents a significant risk factor for SLE susceptibility [84]. For FcγRIIa, the R131 allele has been shown to be associated with SLE in several ethnic groups, an observation that was confirmed further by recent meta-analyses [36,49]. Similarly, increased frequency among SLE patients of the low-responder allele of FcγRIIIb, NA2 has been reported widely [55–57]. It should be noted that there have been several reports that describe the lack of association of these polymorphisms with SLE, due possibly to the high genetic variation of these polymorphisms in populations of different origin [50,51,56,85]. In addition, although autoantibodies of all three major subclasses (IgG1, IgG2 and IgG3) can be detected in SLE patients, heterogeneity of the antibody subclass responses and differential clinical exacerbations among these patients could also constitute an additional determinant that accounts for the lack of association between particular Fcγ receptor polymorphisms with disease susceptibility.
A number of polymorphisms within the FCGR2B coding and promoter regions that are linked to the development of SLE have been identified. Many of these variants exhibit decreased activity or expression of FcγRIIb that would probably affect B cell function and antibody production, as well as the activity of other cell types such as monocytes and macrophages. A notable example is the transmembrane polymorphism T232, which displays lower affinity for lipid rafts, and as a consequence it exhibits decreased inhibitory activity and has been shown to be associated with SLE, at least in Asian populations [17,53,57–60]. Similarly, the −343C allele that displayed repressed FCGR2B promoter activity was enriched among SLE patients, compared to disease-free controls [24,25]. Other SNPs within the promoter of FCGR2B are −386G > C and −120T > A, which exhibit genetic linkage. Interestingly, Su and colleagues reported that the frequency of the −386C and −120A alleles, which exhibit enhanced transcriptional activity, was increased in Caucasian patients with SLE compared to ethnically matched controls [22]. The functional relevance of this finding remains to be determined, but one possibility is that these polymorphisms (−386G > C, −120T > A) might be linked genetically to other as-yet unidentified polymorphisms associated with SLE, or their over-representation in the SLE cohort to be due to their low haplotype frequency in the tested population. Finally, variation in the copy number of FCGR3B has been identified as an additional determinant for the development of SLE. In two independent studies, low copy numbers of the FCGR3B gene was associated strongly with SLE, as the percentage of individuals with less than two genomic copies of FCGR3B was significantly higher in the SLE compared to the control study cohort [33,86]. Reduced surface expression of FcγRIIIb in these individuals may compromise the clearance of IgG complexes, increasing the risk for the development of SLE.
Apart from Fcγ receptors, polymorphism within FcαRI has been associated with SLE, in that the proinflammatory G248 allele has been found to be enriched in SLE populations [19] (Table 3). In addition, several Fcε receptor polymorphisms have been linked with the development of allergy-related chronic inflammatory diseases, and new associations continue to be reported [87](Table 3). In particular, polymorphisms within the gene coding for the β subunit of the FcεRI (MS4A2) have been shown to be associated strongly with atopy, asthma, airway hyperresponsiveness, allergic rhinitis, serum IgE levels, atopic asthma and atopic dermatitis [28–30,88–97]. The extent of the impact of these polymorphisms on receptor function and properties is under investigation. Many atopy-associated SNPs within the promoter of FCER1A have been reported to determine transcriptional activity and subsequently receptor expression. In particular, the −66T, −315T and −335C alleles, which differ in their frequency between atopic and non-atopic subjects, at least in some populations, were associated strongly with increased serum IgE [26,27,98]. As the levels of IgE in circulation are correlated greatly with the surface expression of FcεRI, these polymorphisms might affect receptor expression directly or indirectly and consequently the effectiveness of IgE-mediated cellular responses.
Table 3.
Role of IgE, IgA and IgM receptor SNPs in disease pathogenesis.
| Receptor | SNP | Disease | Reference |
|---|---|---|---|
| FcεRI α chain | −66T > C | Atopy, high IgE levels in asthma | [26] |
| −315C > T | Aspirin-intolerant chronic urticaria, high IgE levels | [98] | |
| −335T > C | High IgE levels | [109] | |
| FcεRI β chain | E237G | Atopic asthma, high IgE levels, atopy, airway hyperresponsiveness, allergic rhinitis, asthma | [88,90–97,122] |
| I181L | Atopy, asthma | [88,89] | |
| −109C > T | High IgE levels | [28–30,95] | |
| FcαRI | S248G | SLE | [19] |
| pIgR | A580V | IgA nephropathy | [8] |
Ig: immunoglobulin; SLE: systemic lupus erythematosus; SNP: single nucleotide polymorphism.
In summary, Fc receptors have a key role in the regulation of immune cell function and the polymorphic and copy number variants identified so far undoubtedly contribute to the development of a number of chronic inflammatory diseases. Apart from these diseases, Fc receptor polymorphisms have also been shown to be associated strongly with susceptibility to pathogens, constituting a major genetic risk factor for a number of infectious diseases[99–103]. Furthermore, recent advances in the therapeutic use of intravenous immunoglobulins have highlighted the role of Fc receptor polymorphisms in the clinical outcome and therapeutic responsiveness [104,105]. One of the future challenges is to determine the precise role of the different classes of human Fc receptors in the control of innate and acquired immune responses and define how the observed genetic variation contributes to disease pathogenesis.
Acknowledgments
The support of the British Heart Foundation (FS/05/119/19568) and the Medical Research Council is gratefully acknowledged.
Disclosure
The authors declare no competing financial interests.
References
- 1.Ernst LK, Duchemin AM, Miller KL, Anderson CL. Molecular characterization of six variant Fcγ receptor class I (CD64) transcripts. Mol Immunol. 1998;35:943–54. doi: 10.1016/s0161-5890(98)00079-0. [DOI] [PubMed] [Google Scholar]
- 2.Nimmerjahn F, Ravetch JV. Fcγ receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 3.Donnadieu E, Jouvin MH, Rana S, et al. Competing functions encoded in the allergy-associated F(c)εRIβ gene. Immunity. 2003;18:665–74. doi: 10.1016/s1074-7613(03)00115-8. [DOI] [PubMed] [Google Scholar]
- 4.Yokota A, Kikutani H, Tanaka T, et al. Two species of human Fcε receptor II (FcεRII/CD23): tissue-specific and IL-4-specific regulation of gene expression. Cell. 1988;55:611–18. doi: 10.1016/0092-8674(88)90219-x. [DOI] [PubMed] [Google Scholar]
- 5.Meng JF, McFall C, Rosenwasser LJ. Polymorphism R62W results in resistance of CD23 to enzymatic cleavage in cultured cells. Genes Immun. 2007;8:215–23. doi: 10.1038/sj.gene.6364376. [DOI] [PubMed] [Google Scholar]
- 6.Liu YJ, Cairns JA, Holder MJ, et al. Recombinant 25-kDa CD23 and interleukin 1 α promote the survival of germinal center B cells: evidence for bifurcation in the development of centrocytes rescued from apoptosis. Eur J Immunol. 1991;21:1107–14. doi: 10.1002/eji.1830210504. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto N, Shibuya K, Shimizu Y, et al. A novel Fc receptor for IgA and IgM is expressed on both hematopoietic and non-hematopoietic tissues. Eur J Immunol. 2001;31:1310–16. doi: 10.1002/1521-4141(200105)31:5<1310::AID-IMMU1310>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Obara W, Iida A, Suzuki Y, et al. Association of single-nucleotide polymorphisms in the polymeric immunoglobulin receptor gene with immunoglobulin A nephropathy (IgAN) in Japanese patients. J Hum Genet. 2003;48:293–9. doi: 10.1007/s10038-003-0027-1. [DOI] [PubMed] [Google Scholar]
- 9.Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J Pathol. 2006;208:270–82. doi: 10.1002/path.1877. [DOI] [PubMed] [Google Scholar]
- 10.Clark MR, Clarkson SB, Ory PA, Stollman N, Goldstein IM. Molecular basis for a polymorphism involving Fc receptor II on human monocytes. J Immunol. 1989;143:1731–4. [PubMed] [Google Scholar]
- 11.Maxwell KF, Powell MS, Hulett MD, et al. Crystal structure of the human leukocyte Fc receptor, FcγRIIa. Nat Struct Biol. 1999;6:437–42. doi: 10.1038/8241. [DOI] [PubMed] [Google Scholar]
- 12.Warmerdam PA, Van de Winkel JG, Gosselin EJ, Capel PJ. Molecular basis for a polymorphism of human Fcγ receptor II (CD32) J Exp Med. 1990;172:19–25. doi: 10.1084/jem.172.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koene HR, Kleijer M, Algra J, Roos D, Von dem Borne AE, De Haas M. FcγRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell FcγRIIIa, independently of the FcγRIIIa-48L/R/H phenotype. Blood. 1997;90:1109–14. [PubMed] [Google Scholar]
- 14.Wu J, Edberg JC, Redecha PB, et al. A novel polymorphism of FcγRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest. 1997;100:1059–70. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmon JE, Edberg JC, Kimberly RP. Fcγ receptor III on human neutrophils. Allelic variants have functionally distinct capacities. J Clin Invest. 1990;85:1287–95. doi: 10.1172/JCI114566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bux J, Stein EL, Bierling P, et al. Characterization of a new alloantigen (SH) on the human neutrophil Fcγ receptor IIIb. Blood. 1997;89:1027–34. [PubMed] [Google Scholar]
- 17.Kono H, Kyogoku C, Suzuki T, et al. FcγRIIB Ile232Thr transmembrane polymorphism associated with human systemic lupus erythematosus decreases affinity to lipid rafts and attenuates inhibitory effects on B cell receptor signaling. Hum Mol Genet. 2005;14:2881–92. doi: 10.1093/hmg/ddi320. [DOI] [PubMed] [Google Scholar]
- 18.Floto R, Clatworthy M, Heilbronn K, et al. Loss of function of a lupus-associated FcγRIIb polymorphism through exclusion from lipid rafts. Nat Med. 2005;11:1056–8. doi: 10.1038/nm1288. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Ji C, Xie F, et al. FcαRI (CD89) alleles determine the proinflammatory potential of serum IgA. J Immunol. 2007;178:3973–82. doi: 10.4049/jimmunol.178.6.3973. [DOI] [PubMed] [Google Scholar]
- 20.Jasek M, Manczak M, Sawaryn A, et al. A novel polymorphism in the cytoplasmic region of the human immunoglobulin A Fc receptor gene. Eur J Immunogenet. 2004;31:59–62. doi: 10.1111/j.1365-2370.2004.00445.x. [DOI] [PubMed] [Google Scholar]
- 21.Breunis WB, van Mirre E, Bruin M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111:1029–38. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- 22.Su K, Wu J, Edberg JC, et al. A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcγRIIb alters receptor expression and associates with autoimmunity. I. Regulatory FCGR2B polymorphisms and their association with systemic lupus erythematosus. J Immunol. 2004;172:7186–91. doi: 10.4049/jimmunol.172.11.7186. [DOI] [PubMed] [Google Scholar]
- 23.Su K, Li X, Edberg JC, Wu J, Ferguson P, Kimberly RP. A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcγRIIb alters receptor expression and associates with autoimmunity. II. Differential binding of GATA4 and Yin-Yang1 transcription factors and correlated receptor expression and function. J Immunol. 2004;172:7192–9. doi: 10.4049/jimmunol.172.11.7192. [DOI] [PubMed] [Google Scholar]
- 24.Blank MC, Stefanescu RN, Masuda E, et al. Decreased transcription of the human FCGR2B gene mediated by the −343 G/C promoter polymorphism and association with systemic lupus erythematosus. Hum Genet. 2005;117:220–7. doi: 10.1007/s00439-005-1302-3. [DOI] [PubMed] [Google Scholar]
- 25.Olferiev M, Masuda E, Tanaka S, Blank MC, Pricop L. The role of activating protein 1 in the transcriptional regulation of the human FCGR2B promoter mediated by the −343 G −> C polymorphism associated with systemic lupus erythematosus. J Biol Chem. 2007;282:1738–46. doi: 10.1074/jbc.M605808200. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa M, Nishiyama C, Nishiyama M, et al. A novel −66T/C polymorphism in FcεRI α-chain promoter affecting the transcription activity: possible relationship to allergic diseases. J Immunol. 2003;171:1927–33. doi: 10.4049/jimmunol.171.4.1927. [DOI] [PubMed] [Google Scholar]
- 27.Kanada S, Nakano N, Potaczek DP, et al. Two different transcription factors discriminate the −315C > T polymorphism of the FcεRIα gene: binding of Sp1 to −315C and of a high mobility group-related molecule to −315T. J Immunol. 2008;180:8204–10. doi: 10.4049/jimmunol.180.12.8204. [DOI] [PubMed] [Google Scholar]
- 28.Hizawa N, Maeda Y, Konno S, Fukui Y, Takahashi D, Nishimura M. Genetic polymorphisms at FCER1B and PAI-1 and asthma susceptibility. Clin Exp Allergy. 2006;36:872–6. doi: 10.1111/j.1365-2222.2006.02413.x. [DOI] [PubMed] [Google Scholar]
- 29.Hizawa N, Yamaguchi E, Jinushi E, Kawakami Y. A common FCER1B gene promoter polymorphism influences total serum IgE levels in a Japanese population. Am J Respir Crit Care Med. 2000;161:906–9. doi: 10.1164/ajrccm.161.3.9903128. [DOI] [PubMed] [Google Scholar]
- 30.Hizawa N, Yamaguchi E, Jinushi E, Konno S, Kawakami Y, Nishimura M. Increased total serum IgE levels in patients with asthma and promoter polymorphisms at CTLA4 and FCER1B. J Allergy Clin Immunol. 2001;108:74–9. doi: 10.1067/mai.2001.116119. [DOI] [PubMed] [Google Scholar]
- 31.Nishiyama C, Akizawa Y, Nishiyama M, et al. Polymorphisms in the FcεRIβ promoter region affecting transcription activity: a possible promoter-dependent mechanism for association between FcεRIβ and atopy. J Immunol. 2004;173:6458–64. doi: 10.4049/jimmunol.173.10.6458. [DOI] [PubMed] [Google Scholar]
- 32.Schaschl H, Aitman TJ, Vyse TJ. Copy number variation in the human genome and its implication in autoimmunity. Clin Exp Immunol. 2009;156:12–16. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willcocks LC, Lyons PA, Clatworthy M, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–82. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karassa FB, Bijl M, Davies KA, et al. Role of the Fcγ receptor IIA polymorphism in the antiphospholipid syndrome: an international meta-analysis. Arthritis Rheum. 2003;48:1930–8. doi: 10.1002/art.11059. [DOI] [PubMed] [Google Scholar]
- 35.Thabet MM, Huizinga TW, Marques RB, et al. The contribution of Fcγ receptor IIIA gene 158V/F polymorphism and copy number variation to the risk of ACPA positive rheumatoid arthritis. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.099309. in press. doi: 10.1136/ard.2008.099309. [DOI] [PubMed] [Google Scholar]
- 36.Karassa FB, Trikalinos TA, Ioannidis JP, FcγRIIa-SLE Meta-Analysis Investigators Role of the Fcγ receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum. 2002;46:1563–71. doi: 10.1002/art.10306. [DOI] [PubMed] [Google Scholar]
- 37.Gavasso S, Nygård O, Pedersen ER, et al. Fcγ receptor IIIA polymorphism as a risk-factor for coronary artery disease. Atherosclerosis. 2005;180:277–82. doi: 10.1016/j.atherosclerosis.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Morgan AW, Robinson JI, Barrett JH, et al. Association of FCGR2A and FCGR2A–FCGR3A haplotypes with susceptibility to giant cell arteritis. Arthritis Res Ther. 2006;8:R109. doi: 10.1186/ar1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dijstelbloem HM, Scheepers RH, Oost WW, et al. Fcγ receptor polymorphisms in Wegener's granulomatosis: risk factors for disease relapse. Arthritis Rheum. 1999;42:1823–7. doi: 10.1002/1529-0131(199909)42:9<1823::AID-ANR5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 40.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–82. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 41.Sato H, Iwano M, Akai Y, et al. FcγRIIa polymorphism in Japanese patients with systemic lupus erythematosus. Lupus. 2001;10:97–101. doi: 10.1191/096120301677569675. [DOI] [PubMed] [Google Scholar]
- 42.Norsworthy P, Theodoridis E, Botto M, et al. Overrepresentation of the Fcγ receptor type IIA R131/R131 genotype in caucasoid systemic lupus erythematosus patients with autoantibodies to C1q and glomerulonephritis. Arthritis Rheum. 1999;42:1828–32. doi: 10.1002/1529-0131(199909)42:9<1828::AID-ANR6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Song YW, Han CW, Kang SW, et al. Abnormal distribution of Fcγ receptor type IIa polymorphisms in Korean patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41:421–6. doi: 10.1002/1529-0131(199803)41:3<421::AID-ART7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 44.Khoa PD, Sugiyama T, Yokochi T. Fcγ receptor II polymorphism in Vietnamese patients with systemic lupus erythematosus. Lupus. 2003;12:704–6. doi: 10.1191/0961203303lu454sr. [DOI] [PubMed] [Google Scholar]
- 45.Dijstelbloem HM, Bijl M, Fijnheer R, et al. Fcγ receptor polymorphisms in systemic lupus erythematosus: association with disease and in vivo clearance of immune complexes. Arthritis Rheum. 2000;43:2793–800. doi: 10.1002/1529-0131(200012)43:12<2793::AID-ANR20>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 46.Magnusson V, Johanneson B, Lima G, et al. Both risk alleles for FcγRIIA and FcγRIIIA are susceptibility factors for SLE: a unifying hypothesis. Genes Immun. 2004;5:130–7. doi: 10.1038/sj.gene.6364052. [DOI] [PubMed] [Google Scholar]
- 47.Balada E, Villarreal-Tolchinsky J, Ordi-Ros J, et al. Multiplex family-based study in systemic lupus erythematosus: association between the R620W polymorphism of PTPN22 and the FcγRIIa (CD32A) R131 allele. Tissue Antigens. 2006;68:432–8. doi: 10.1111/j.1399-0039.2006.00695.x. [DOI] [PubMed] [Google Scholar]
- 48.Lee EB, Lee YJ, Baek HJ, et al. Fcγ receptor IIIA polymorphism in Korean patients with systemic lupus erythematosus. Rheumatol Int. 2002;21:222–6. doi: 10.1007/s00296-001-0171-x. [DOI] [PubMed] [Google Scholar]
- 49.Yuan H, Pan HF, Li LH, et al. Meta analysis on the association between FcγRIIa-R/H131 polymorphisms and systemic lupus erythematosus. Mol Biol Rep. 2009;36:1053–8. doi: 10.1007/s11033-008-9280-x. [DOI] [PubMed] [Google Scholar]
- 50.Jönsen A, Gunnarsson I, Gullstrand B, et al. Association between SLE nephritis and polymorphic variants of the CRP and FcγRIIIa genes. Rheumatology. 2007;46:1417–21. doi: 10.1093/rheumatology/kem167. [DOI] [PubMed] [Google Scholar]
- 51.Koene HR, Kleijer M, Swaak AJ, et al. The FcγRIIIA-158F allele is a risk factor for systemic lupus erythematosus. Arthritis Rheum. 1998;41:1813–18. doi: 10.1002/1529-0131(199810)41:10<1813::AID-ART13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 52.Zuñiga R, Ng S, Peterson MG, et al. Low-binding alleles of Fcγ receptor types IIA and IIIA are inherited independently and are associated with systemic lupus erythematosus in Hispanic patients. Arthritis Rheum. 2001;44:361–7. doi: 10.1002/1529-0131(200102)44:2<361::AID-ANR54>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 53.Chu ZT, Tsuchiya N, Kyogoku C, et al. Association of Fcγ receptor IIb polymorphism with susceptibility to systemic lupus erythematosus in Chinese: a common susceptibility gene in the Asian populations. Tissue Antigens. 2004;63:21–7. doi: 10.1111/j.1399-0039.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 54.Kyogoku C, Tsuchiya N, Wu H, Tsao BP, Tokunaga K. Association of Fcγ receptor IIA, but not IIB and IIIA, polymorphisms with systemic lupus erythematosus: a family-based association study in Caucasians. Arthritis Rheum. 2004;50:671–3. doi: 10.1002/art.20029. [DOI] [PubMed] [Google Scholar]
- 55.Hatta Y, Tsuchiya N, Ohashi J, et al. Association of Fcγ receptor IIIB, but not of Fcγ receptor IIA and IIIA polymorphisms with systemic lupus erythematosus in Japanese. Genes Immun. 1999;1:53–60. doi: 10.1038/sj.gene.6363639. [DOI] [PubMed] [Google Scholar]
- 56.González-Escribano MF, Aguilar F, Sánchez-Román J, Núñez-Roldán A. FcγRIIA, FcγRIIIA and FcγRIIIB polymorphisms in Spanish patients with systemic lupus erythematosus. Eur J Immunogenet. 2002;29:301–6. doi: 10.1046/j.1365-2370.2002.00324.x. [DOI] [PubMed] [Google Scholar]
- 57.Siriboonrit U, Tsuchiya N, Sirikong M, et al. Association of Fcγ receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens. 2003;61:374–83. doi: 10.1034/j.1399-0039.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 58.Kyogoku C, Dijstelbloem HM, Tsuchiya N, et al. Fcγ receptor gene polymorphisms in Japanese patients with systemic lupus erythematosus: contribution of FCGR2B to genetic susceptibility. Arthritis Rheum. 2002;46:1242–54. doi: 10.1002/art.10257. [DOI] [PubMed] [Google Scholar]
- 59.Chen JY, Wang CM, Ma CC, et al. Association of a transmembrane polymorphism of Fcγ receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum. 2006;54:3908–17. doi: 10.1002/art.22220. [DOI] [PubMed] [Google Scholar]
- 60.Tsuchiya N, Kyogoku C. Role of Fcγ receptor IIb polymorphism in the genetic background of systemic lupus erythematosus: insights from Asia. Autoimmunity. 2005;38:347–52. doi: 10.1080/08916930500123926. [DOI] [PubMed] [Google Scholar]
- 61.Kastbom A, Ahmadi A, Söderkvist P, Skogh T. The 158V polymorphism of Fcγ receptor type IIIA in early rheumatoid arthritis: increased susceptibility and severity in male patients (the Swedish TIRA project) Rheumatology. 2005;44:1294–8. doi: 10.1093/rheumatology/kei010. [DOI] [PubMed] [Google Scholar]
- 62.Lee YH, Ji JD, Song GG. Associations between FCGR3A polymorphisms and susceptibility to rheumatoid arthritis: a metaanalysis. J Rheumatol. 2008;35:2129–35. doi: 10.3899/jrheum.080186. [DOI] [PubMed] [Google Scholar]
- 63.Morgan AW, Griffiths B, Ponchel F, et al. Fcγ receptor type IIIA is associated with rheumatoid arthritis in two distinct ethnic groups. Arthritis Rheum. 2000;43:2328–34. doi: 10.1002/1529-0131(200010)43:10<2328::AID-ANR21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 64.Brun JG, Madland TM, Vedeler C. Immunoglobulin G fc-receptor (FcγR) IIA, IIIA, and IIIB polymorphisms related to disease severity in rheumatoid arthritis. J Rheumatol. 2002;29:1135–40. [PubMed] [Google Scholar]
- 65.Chen JY, Wang CM, Wu JM, Ho HH, Luo SF. Association of rheumatoid factor production with FcγRIIIa polymorphism in Taiwanese rheumatoid arthritis. Clin Exp Immunol. 2006;144:10–16. doi: 10.1111/j.1365-2249.2006.03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berdeli A, Celik HA, Ozyürek R, Aydin HH. Involvement of immunoglobulin FcγRIIA and FcγRIIIB gene polymorphisms in susceptibility to rheumatic fever. Clin Biochem. 2004;37:925–9. doi: 10.1016/j.clinbiochem.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Raknes G, Skeie GO, Gilhus NE, Aadland S, Vedeler C. FcγRIIA and FcγRIIIB polymorphisms in myasthenia gravis. J Neuroimmunol. 1998;81:173–6. doi: 10.1016/s0165-5728(97)00174-4. [DOI] [PubMed] [Google Scholar]
- 68.van der Pol WL, Jansen MD, Kuks JB, et al. Association of the Fcγ receptor IIA-R/R131 genotype with myasthenia gravis in Dutch patients. J Neuroimmunol. 2003;144:143–7. doi: 10.1016/j.jneuroim.2003.08.043. [DOI] [PubMed] [Google Scholar]
- 69.van Sorge NM, van der Pol WL, Jansen MD, et al. Severity of Guillain–Barré syndrome is associated with Fcγ Receptor III polymorphisms. J Neuroimmunol. 2005;162:157–64. doi: 10.1016/j.jneuroim.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 70.Vedeler CA, Raknes G, Myhr KM, Nyland H. IgG Fc-receptor polymorphisms in Guillain–Barré syndrome. Neurology. 2000;55:705–7. doi: 10.1212/wnl.55.5.705. [DOI] [PubMed] [Google Scholar]
- 71.Tackenberg B, Jelcic I, Baerenwaldt A, et al. Impaired inhibitory Fcγ receptor IIB expression on B cells in chronic inflammatory demyelinating polyneuropathy. Proc Natl Acad Sci USA. 2009;106:4788–92. doi: 10.1073/pnas.0807319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Pol WL, van den Berg LH, Scheepers RH, et al. IgG receptor IIa alleles determine susceptibility and severity of Guillain–Barré syndrome. Neurology. 2000;54:1661–5. doi: 10.1212/wnl.54.8.1661. [DOI] [PubMed] [Google Scholar]
- 73.Yuan FF, Watson N, Sullivan JS, et al. Association of Fcγ receptor IIA polymorphisms with acute renal-allograft rejection. Transplantation. 2004;78:766–9. doi: 10.1097/01.tp.0000132560.77496.cb. [DOI] [PubMed] [Google Scholar]
- 74.Williams Y, Lynch S, McCann S, Smith O, Feighery C, Whelan A. Correlation of platelet FcγRIIA polymorphism in refractory idiopathic (immune) thrombocytopenic purpura. Br J Haematol. 1998;101:779–82. doi: 10.1046/j.1365-2141.1998.00802.x. [DOI] [PubMed] [Google Scholar]
- 75.Fujimoto TT, Inoue M, Shimomura T, Fujimura K. Involvement of Fcγ receptor polymorphism in the therapeutic response of idiopathic thrombocytopenic purpura. Br J Haematol. 2001;115:125–30. doi: 10.1046/j.1365-2141.2001.03109.x. [DOI] [PubMed] [Google Scholar]
- 76.Foster CB, Zhu S, Erichsen HC, et al. Polymorphisms in inflammatory cytokines and Fcγ receptors in childhood chronic immune thrombocytopenic purpura: a pilot study. Br J Haematol. 2001;113:596–9. doi: 10.1046/j.1365-2141.2001.02807.x. [DOI] [PubMed] [Google Scholar]
- 77.Carlsson LE, Santoso S, Baurichter G, et al. Heparin-induced thrombocytopenia: new insights into the impact of the FcγRIIa-R-H131 polymorphism. Blood. 1998;92:1526–31. [PubMed] [Google Scholar]
- 78.Gruel Y, Pouplard C, Lasne D, Magdelaine-Beuzelin C, Charroing C, Watier H. The homozygous FcγRIIIa-158V genotype is a risk factor for heparin-induced thrombocytopenia in patients with antibodies to heparin-platelet factor 4 complexes. Blood. 2004;104:2791–3. doi: 10.1182/blood-2004-01-0058. [DOI] [PubMed] [Google Scholar]
- 79.Tse WY, Abadeh S, Jefferis R, Savage CO, Adu D. Neutrophil FcγRIIIb allelic polymorphism in anti-neutrophil cytoplasmic antibody (ANCA)-positive systemic vasculitis. Clin Exp Immunol. 2000;119:574–7. doi: 10.1046/j.1365-2249.2000.01182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Der Meer I, Witteman JC, Hofman A, Kluft C, de Maat MP. Genetic variation in Fcγ receptor IIa protects against advanced peripheral atherosclerosis. The Rotterdam Study. Thromb Haemost. 2004;92:1273–6. doi: 10.1160/TH04-05-0268. [DOI] [PubMed] [Google Scholar]
- 81.Raaz D, Herrmann M, Ekici AB, et al. FcγRIIa genotype is associated with acute coronary syndromes as first manifestation of coronary artery disease. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2009.01.013. in press. doi: 10.1016/j.atherosclerosis.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 82.Yun HR, Koh HK, Kim SS, et al. FcγRIIa/IIIa polymorphism and its association with clinical manifestations in Korean lupus patients. Lupus. 2001;10:466–72. doi: 10.1191/096120301678416015. [DOI] [PubMed] [Google Scholar]
- 83.Seligman VA, Suarez C, Lum R, et al. The Fcγ receptor IIIA-158F allele is a major risk factor for the development of lupus nephritis among Caucasians but not non-Caucasians. Arthritis Rheum. 2001;44:618–25. doi: 10.1002/1529-0131(200103)44:3<618::AID-ANR110>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 84.Edberg JC, Langefeld CD, Wu J, et al. Genetic linkage and association of Fcγ receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis Rheum. 2002;46:2132–40. doi: 10.1002/art.10438. [DOI] [PubMed] [Google Scholar]
- 85.Botto M, Theodoridis E, Thompson EM, et al. FcγRIIa polymorphism in systemic lupus erythematosus (SLE): no association with disease. Clin Exp Immunol. 1996;104:264–8. doi: 10.1046/j.1365-2249.1996.33740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fanciulli M, Norsworthy PJ, Petretto E, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–3. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weidinger S, Gieger C, Rodriguez E, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Green SL, Gaillard MC, Song E, Dewar JB, Halkas A. Polymorphisms of the β chain of the high-affinity immunoglobulin E receptor (FcεRI-β) in South African black and white asthmatic and nonasthmatic individuals. Am J Respir Crit Care Med. 1998;158:1487–92. doi: 10.1164/ajrccm.158.5.9707099. [DOI] [PubMed] [Google Scholar]
- 89.Li A, Hopkin JM. Atopy phenotype in subjects with variants of the β subunit of the high affinity IgE receptor. Thorax. 1997;52:654–5. doi: 10.1136/thx.52.7.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shirakawa T, Mao XQ, Sasaki S, et al. Association between atopic asthma and a coding variant of FcεRIβ in a Japanese population. Hum Mol Genet. 1996;5:1129–30. doi: 10.1093/hmg/5.8.1129. [DOI] [PubMed] [Google Scholar]
- 91.Zhang X, Zhang W, Qiu D, Sandford A, Tan WC. The E237G polymorphism of the high-affinity IgE receptor β chain and asthma. Ann Allergy Asthma Immunol. 2004;93:499–503. doi: 10.1016/s1081-1206(10)61419-6. [DOI] [PubMed] [Google Scholar]
- 92.Rigoli L, Di Bella C, Procopio V, et al. Molecular analysis of sequence variants in the Fcε receptor Iβ gene and IL-4 gene promoter in Italian atopic families. Allergy. 2004;59:213–18. doi: 10.1046/j.1398-9995.2003.00385.x. [DOI] [PubMed] [Google Scholar]
- 93.Nagata H, Mutoh H, Kumahara K, et al. Association between nasal allergy and a coding variant of the FcεRIβ gene Glu237Gly in a Japanese population. Hum Genet. 2001;109:262–6. doi: 10.1007/s004390100561. [DOI] [PubMed] [Google Scholar]
- 94.Laprise C, Boulet LP, Morissette J, Winstall E, Raymond V. Evidence for association and linkage between atopy, airway hyper-responsiveness, and the β subunit Glu237Gly variant of the high-affinity receptor for immunoglobulin E in the French-Canadian population. Immunogenetics. 2000;51:695–702. doi: 10.1007/s002510000185. [DOI] [PubMed] [Google Scholar]
- 95.Cui T, Wang L, Wu J, Xie J. The association analysis of FcεRIβ with allergic asthma in a Chinese population. Chin Med J. 2003;116:1875–8. [PubMed] [Google Scholar]
- 96.Kim YK, Park HW, Yang JS, et al. Association and functional relevance of E237G, a polymorphism of the high-affinity immunoglobulin E-receptor β chain gene, to airway hyper-responsiveness. Clin Exp Allergy. 2007;37:592–8. doi: 10.1111/j.1365-2222.2007.02680.x. [DOI] [PubMed] [Google Scholar]
- 97.Hill MR, Cookson WO. A new variant of the β subunit of the high-affinity receptor for immunoglobulin E (FcεRI-β E237G): associations with measures of atopy and bronchial hyper-responsiveness. Hum Mol Genet. 1996;5:959–62. doi: 10.1093/hmg/5.7.959. [DOI] [PubMed] [Google Scholar]
- 98.Bae JS, Kim SH, Ye YM, et al. Significant association of FcεRIα promoter polymorphisms with aspirin-intolerant chronic urticaria. J Allergy Clin Immunol. 2007;119:449–56. doi: 10.1016/j.jaci.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 99.Cooke GS, Aucan C, Walley AJ, et al. Association of Fcγ receptor IIa (CD32) polymorphism with severe malaria in West Africa. Am J Trop Med Hyg. 2003;69:565–8. [PubMed] [Google Scholar]
- 100.Platonov AE, Kuijper EJ, Vershinina IV, et al. Meningococcal disease and polymorphism of FcγRIIa (CD32) in late complement component-deficient individuals. Clin Exp Immunol. 1998;111:97–101. doi: 10.1046/j.1365-2249.1998.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshie H, Kobayashi T, Tai H, Galicia JC. The role of genetic polymorphisms in periodontitis. Periodontol 2000. 2007;43:102–32. doi: 10.1111/j.1600-0757.2006.00164.x. [DOI] [PubMed] [Google Scholar]
- 102.van der Pol WL, Huizinga TW, Vidarsson G, et al. Relevance of Fcγ receptor and interleukin-10 polymorphisms for meningococcal disease. J Infect Dis. 2001;184:1548–55. doi: 10.1086/324662. [DOI] [PubMed] [Google Scholar]
- 103.Forthal DN, Landucci G, Bream J, Jacobson LP, Phan TB, Montoya B. FcγRIIa genotype predicts progression of HIV infection. J Immunol. 2007;179:7916–23. doi: 10.4049/jimmunol.179.11.7916. [DOI] [PubMed] [Google Scholar]
- 104.Nimmerjahn F, Ravetch JV. Anti-inflammatory actions of intravenous immunoglobulin. Annu Rev Immunol. 2008;26:513–33. doi: 10.1146/annurev.immunol.26.021607.090232. [DOI] [PubMed] [Google Scholar]
- 105.Binstadt BA, Geha RS, Bonilla FA. IgG Fc receptor polymorphisms in human disease: implications for intravenous immunoglobulin therapy. J Allergy Clin Immunol. 2003;111:697–703. doi: 10.1067/mai.2003.1380. [DOI] [PubMed] [Google Scholar]
- 106.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–5. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 108.Aitman T, Dong R, Vyse T, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–5. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 109.Shikanai T, Silverman ES, Morse BW, Lilly CM, Inoue H, Drazen JM. Sequence variants in the FcεRI α chain gene. J Appl Physiol. 2002;93:37–41. doi: 10.1152/japplphysiol.00993.2001. [DOI] [PubMed] [Google Scholar]
- 110.Tanaka Y, Suzuki Y, Tsuge T, et al. FcγRIIa-131R allele and FcγRIIIa-176V/V genotype are risk factors for progression of IgA nephropathy. Nephrol Dial Transplant. 2005;20:2439–45. doi: 10.1093/ndt/gfi043. [DOI] [PubMed] [Google Scholar]
- 111.Bazilio AP, Viana VS, Toledo R, Woronik V, Bonfá E, Monteiro RC. FcγRIIa polymorphism: a susceptibility factor for immune complex-mediated lupus nephritis in Brazilian patients. Nephrol Dial Transplant. 2004;19:1427–31. doi: 10.1093/ndt/gfh121. [DOI] [PubMed] [Google Scholar]
- 112.Salmon JE, Millard S, Schachter LA, et al. FcγRIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest. 1996;97:1348–54. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duits AJ, Bootsma H, Derksen RH, et al. Skewed distribution of IgG Fc receptor IIa (CD32) polymorphism is associated with renal disease in systemic lupus erythematosus patients. Arthritis Rheum. 1995;38:1832–6. doi: 10.1002/art.1780381217. [DOI] [PubMed] [Google Scholar]
- 114.Gelmetti AP, Freitas AC, Woronik V, Barros RT, Bonfá E, Monteiro RC. Polymorphism of the FcγRIIα IgG receptor in patients with lupus nephritis and glomerulopathy. J Rheumatol. 2006;33:523–30. [PubMed] [Google Scholar]
- 115.Zuniga R, Markowitz GS, Arkachaisri T, Imperatore EA, D'Agati VD, Salmon JE. Identification of IgG subclasses and C-reactive protein in lupus nephritis: the relationship between the composition of immune deposits and Fcγ receptor type IIA alleles. Arthritis Rheum. 2003;48:460–70. doi: 10.1002/art.10930. [DOI] [PubMed] [Google Scholar]
- 116.Lee HS, Chung YH, Kim TG, et al. Independent association of HLA-DR and Fcγ receptor polymorphisms in Korean patients with systemic lupus erythematosus. Rheumatology. 2003;42:1501–7. doi: 10.1093/rheumatology/keg404. [DOI] [PubMed] [Google Scholar]
- 117.Dijstelbloem HM, Hepkema BG, Kallenberg CG, et al. The R-H polymorphism of Fcγ receptor IIa as a risk factor for systemic lupus erythematosus is independent of single-nucleotide polymorphisms in the interleukin-10 gene promoter. Arthritis Rheum. 2002;46:1125–6. doi: 10.1002/art.518. [DOI] [PubMed] [Google Scholar]
- 118.Jönsen A, Bengtsson AA, Sturfelt G, Truedsson L. Analysis of HLA DR, HLA DQ, C4A, FcγRIIa, FcγRIIIa, MBL, and IL-1Ra allelic variants in Caucasian systemic lupus erythematosus patients suggests an effect of the combined FcγRIIa R/R and IL-1Ra 2/2 genotypes on disease susceptibility. Arthritis Res Ther. 2004;6:R557–62. doi: 10.1186/ar1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kyogoku C, Tsuchiya N, Matsuta K, Tokunaga K. Studies on the association of Fcγ receptor IIA, IIB, IIIA and IIIB polymorphisms with rheumatoid arthritis in the Japanese: evidence for a genetic interaction between HLA-DRB1 and FCGR3A. Genes Immun. 2002;3:488–93. doi: 10.1038/sj.gene.6363921. [DOI] [PubMed] [Google Scholar]
- 120.Morgan AW, Barrett JH, Griffiths B, et al. Analysis of Fcγ receptor haplotypes in rheumatoid arthritis: FCGR3A remains a major susceptibility gene at this locus, with an additional contribution from FCGR3B. Arthritis Res Ther. 2006;8:R5. doi: 10.1186/ar1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zeyrek D, Tanac R, Altinoz S, et al. FcγRIIIa-V/F 158 polymorphism in Turkish children with asthma bronchiale and allergic rhinitis. Pediatr Allergy Immunol. 2008;19:20–4. doi: 10.1111/j.1399-3038.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- 122.Lee YL, Gilliland FD, Wang JY, Lee YC, Guo YL. Associations of FcεRIβ E237G polymorphism with wheezing in Taiwanese schoolchildren. Clin Exp Allergy. 2008;38:413–20. doi: 10.1111/j.1365-2222.2007.02916.x. [DOI] [PubMed] [Google Scholar]


