Abstract
Objective
Transcription regulatory complexes comprising myocardin and serum response factor (SRF) are critical for the transcriptional regulation of many smooth muscle-specific genes. However, little is known about the epigenetic mechanisms that regulate the activity of these complexes. In the current study, we investigated the role of SWI/SNF ATP-dependent chromatin remodeling enzymes in regulating the myogenic activity of myocardin.
Methods and Results
We found that both Brg1 and Brm are required for maintaining expression of several smooth muscle-specific genes in primary cultures of aortic smooth muscle cells. Furthermore, the ability of myocardin to induce expression of smooth muscle-specific genes is abrogated in cells expressing dominant negative Brg1. In SW13 cells, that lack endogenous Brg1 and Brm1, myocardin is unable to induce expression of smooth muscle-specific genes. Whereas, reconstitution of wild type, or bromodomain mutant forms Brg1 or Brm1, into SW13 cells restored their responsiveness to myocardin. SWI/SNF complexes were found to be required for myocardin to increase SRF binding to the promoters of smooth muscle-specific genes. Brg1 and Brm directly bind to the N-terminus of myocardin, in vitro, through their ATPase domains and Brg1 forms a complex with SRF and myocardin in vivo in smooth muscle cells.
Conclusion
These data demonstrate that the ability of myocardin to induce smooth muscle-specific gene expression is dependent on its interaction with SWI/SNF ATP-dependent chromatin remodeling complexes.
Keywords: Brg1, Brm, telokin, calponin, SRF
Introduction
Smooth muscle cells are important contractile components of the cardiovascular system that regulate blood pressure and flow. Vascular smooth muscle cells modulate their phenotype in response to extracellular cues during the development and progression of a variety of diseases including atherosclerosis and hypertension. These diseases are associated with decreased expression of proteins required for the normal contractile function of smooth muscle cells1. Understanding the mechanisms that control expression of contractile and regulatory proteins in smooth muscle cells is, therefore, an essential step toward determining how these processes are altered in pathological conditions.
The interaction of serum response factor (SRF) with the co-activator myocardin is a critical determinant of vascular smooth muscle development2, 3. Myocardin null mice lack differentiated smooth muscle cells in the dosal aorta and placental vasculature and die around E102. Myocardin is thus critically required for the differentiation of these populations of vascular smooth muscle cells. Myocardin does not bind directly to DNA, but interacts with genes via its binding to SRF through a basic domain and polyglutamine-rich (poly Q) domain in myocardin. Myocardin-bound SRF binds to CArG elements within the promoters of many smooth muscle-specific genes4 and myocardin activates transcription of these genes through a strong transcriptional activation domain at its C-terminus5. However, although myocardin is a potent activator of CArG box-containing cardiac and smooth muscle-specific genes, it poorly activates SRF-dependent skeletal muscle-specific genes or early response genes such as c-fos or Egr-13, yet how myocardin distinguishes smooth muscle-restricted genes from other SRF-dependent genes still remains elusive.
The dependence of myocardin on promoter bound SRF poses an interesting problem as there is little SRF bound to the promoters of smooth muscle-specific genes in nonmuscle cells6, yet myocardin can induce expression of smooth muscle-specific genes in these cells3. Myocardin also increases SRF binding to the promoters of smooth muscle-specific genes within intact chromatin6, although the mechanism underlying this phenomena is unknown. We hypothesized that ATP-dependent chromatin remodeling may be required for these functions of myocardin, allowing it to increase SRF binding to the promoters of smooth muscle-specific genes within chromatin. Recent studies of ATP-dependent remodeling enzymes have highlighted crucial roles for these proteins in diverse developmental processes7. Mammalian ATP-dependent chromatin-remodeling enzymes belong to the SNF2 family of DNA-dependent ATPases, which all have a helicase-like ATPase domain. The best-characterized mammalian ATP-dependent chromatin-remodeling complex is the SWI/SNF complex7. Brahma (Brm) or Brahma-like gene 1 (Brg1) are the ATPase subunits of the SWI/SNF complex. These proteins have been shown to activate or repress expression of genes during myeloid differentiation, erythropoiesis, adipogenesis, skeletal muscle myogenesis, liver development and gliogenesis (reviewed in7). Recently we have also shown that Brg1 and Brm are required for myocardin related transcription factor A (MRTFA) to induce smooth muscle-specific genes in nonmuscle cells8. However, as MRTFA knockout mice do not exhibit any major vascular defects9 the importance of MRTFA-SWI/SNF interactions in vascular smooth muscle cells is not clear.
In the current study we found that Brg1 and Brm are essential for maintaining expression of smooth muscle-specific genes in vascular smooth muscle cells. Brg1/Brm are required for myocardin’s ability to induce expression of smooth muscle-specific genes and to increase SRF binding to the promoters of these genes. We found that Brg1 forms a complex with myocardin and SRF in vivo and directly binds to myocardin in vitro through its ATPase domain. Together, our data demonstrate that SWI/SNF ATP-dependent chromatin remodeling complexes are required for differentiation of vascular smooth muscle cells.
Materials And Methods
A detailed methods section is included in the supplemental material (please see www.ahajournal.org). For adenoviral expression, Brg1 cDNAs were cloned into pShuttle (Clontech). All promoter reporter genes were constructed by cloning fragments of promoters into the pGL2B luciferase vector (Promega, Madison, WI) as described previously 10, 11. Primary mouse aortic smooth muscle cells were prepared from aorta dissected from 4-week-old mice essentially as described previously 8. For all experiments primary cells were replated at 7 × 104 per well in 12-well plates. 12 h after plating, cells were transfected with pre-designed Dharmacon siRNA pools targeting Brg1 or Brm, as well as a control siRNA pool at final concentration of 50nM using Lipofactamine 2000 (Invitrogen). 36 h after transfection, mRNA was harvested and gene expression measured by quantitative real time RT-PCR with respective gene-specific primers (See Supplemental Table I). Chromatin immunoprecipitation assays, protein co-immunoprecipitation and GST pull-down assays were performed as described previously 8, 12.
Results
Depletion of endogenous Brg1 and Brm in aortic SMCs attenuates expression of smooth muscle-specific genes
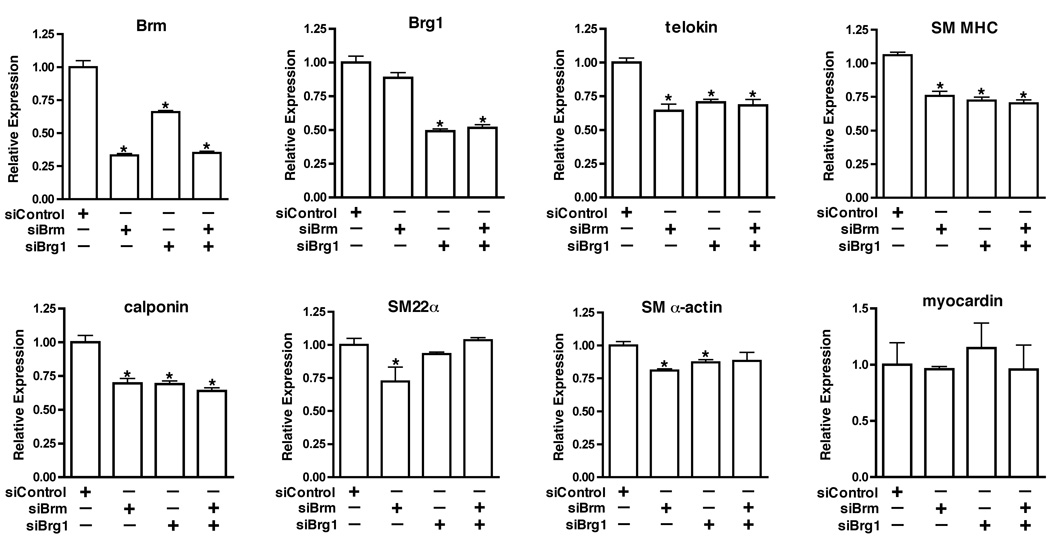
Previously we have shown that the ATP-dependent chromatin remodeling enzymes, Brahma-Related Gene 1 (Brg1) and Brahma (Brm) play an important role in balancing the ability of MRTFA to regulate expression of SRF-dependent smooth muscle-specific genes and immediate early genes 8. However, as MRTFA knockout mice have not been reported to exhibit any vascular defects9 the importance of MRTFA-SWI/SNF interactions in vascular SMCs is not clear. We therefore examined the functional role SWI/SNF in vascular SMCs. siRNA-mediated knockdown of Brg1 or Brm in primary mouse aortic SMCs attenuated expression of telokin, calponin and smooth muscle myosin heavy chain (SM MHC), late markers of smooth muscle differentiation, by approximately 40%. In contrast, knockdown of Brg1 or Brm had a lesser effect on expression of the early markers of smooth muscle differentiation, SM22α or SM α-actin (Figure 1). Silencing Brg1 also led to a 30% reduction in expression of endogenous Brm although Brm knockdown did not affect Brg1 expression. This is not a result of cross reactivity of siRNA molecules as similar results were obtained with multiple siRNA duplexes, with DN-Brg1 and in Brg1 knockout cells (data not shown), suggesting that Brm expression is at least partially dependent on Brg1 in smooth muscle cells. Surprisingly, knockdown of both Brg1 and Brm together did not result in any further attenuation of smooth muscle-specific genes, as compared to knockdown of either protein alone (Figure 1). These data suggest that Brg1 and/or Brm are important for maintaining expression of genes that are late differentiation markers of vascular SMCs.
Figure 1. Effects of depletion Brg1 or Brm on expression of endogenous smooth muscle-specific genes.
Mouse aortic smooth muscle cells were transfected with pre-designed Dharmacon siRNA smart pools targeting Brg1 or Brm, as well as a control siRNA, at final concentration of 50nM. 36 h after transfection, mRNA was harvested and the levels of Brg1, Brm and smooth muscle marker genes were measured by quantitative real time RT-PCR as indicated in each panel. Data presented are the mean±SEM of 6 samples from 2 independent experiments and expression levels were normalized to an hprt internal control and are expressed relative to levels in siRNA control samples (set to 1). * Indicates statistical significance as determined by a student T test (P<0.05).
DN-Brg1 represses activation of smooth muscle-specific genes by myocardin
As myocardin has been shown to be critical for vascular smooth muscle differentiation we next sought to determine if myocardin requires SWI/SNF to induce expression of smooth muscle-specific genes. For these experiments we utilized 10T1/2 embryonic fibroblasts and primary cultures of aortic smooth muscle cells, two well established systems in which myocardin has been shown to induce expression of smooth muscle-specific genes. To inhibit SWI/SNF activity, cells were transduced with adenovirus expressing an ATPase deficient mutant of Brg1 (K798R) that acts as a dominant negative 13. Consistent with previous reports, adenoviral-mediated expression of myocardin in 10T1/2 and primary aortic SMC cells resulted in induction of endogenous telokin, smMHC, calponin, SM22α and SM α-actin mRNA (Figure 2A, B). Expression of dominant negative Brg1 significantly attenuated the induction of telokin, smMHC, calponin, and SM22α by myocardin in 10T1/2 cells and aortic SMCs (Figure 2). In contrast, DN-Brg1 slightly augmented myocardin’s ability to activate SRF itself and did not significantly affect the activation of c-fos in aortic SMC (Figure 2B). Similar results were also seen in NIH3T3 cells induced to express a dominant negative Brg1 (Supplemental figure I). Interestingly DN-Brg1 also attenuated the ability of myocardin to induce the expression of the cardiac-specific ANF and cardiac α-actin genes in 10T1/2 cells (Figure 2A).
Figure 2. DN-Brg1 abrogates the induction of smooth muscle-specific genes by myocardin.
A. 10T1/2 cells were transduced with DN-Brg1 or control YFP adenovirus together with myocardin (solid bars) or YFP (open bars) adenovirus. 48 hrs following transduction total RNA was harvested and analyzed by qRT-PCR. Transcript levels was firstly normalized to hprt internal loading control and then normalized to their respective YFP control group. RQ=2−ΔΔCt and ΔΔCt = (Ct experimental - Ct hprt)- (Ct control –Ct hprt). Data presented are the mean±SEM of 6 samples obtained from 2 independent experiments. B. Primary mouse aortic smooth muscle cells were transduced by DN-Brg1 or YFP control adenovirus with or without myocardin adenovirus as described in “A” (transduction efficiency of these cells was approximately 50–60%). 36–48 hours after transduction, cells were lysed, mRNA harvested and transcript levels analyzed as described above. Data presented are the mean±SEM of 3 samples obtained from one experiment. Similar results were obtained in a replicate experiment.
Previous reports have shown that transient over-expression of wild-type Brg1 can increase the activity of SWI/SNF-dependent reporter genes, while expression of a dominant negative Brg1 decreases the activity of these reporter genes through formation of inactive SWI/SNF complexes14, 15. We therefore determined if DN-Brg1 could directly affect myocardin’s ability to activate the telokin and SM22α promoters. Luciferase reporter assays revealed that over-expression of DN-Brg1 in NIH3T3 cells attenuated myocardin’s activation of telokin and SM22α promoter reporter constructs (Supplemental figure I). Taken together, data presented here demonstrate that dominant negative Brg1 can attenuate myocardin’s ability to induce expression of many smooth muscle-specific genes in both nonmuscle and smooth muscle cells. In contrast, dominant negative Brg1 either did not affect or augmented myocardin’s ability to activate other SRF target genes such as SRF itself or c-fos.
Induction of endogenous smooth muscle-specific genes by myocardin requires Brg1/Brm ATPase activity but does not require their bromodomains
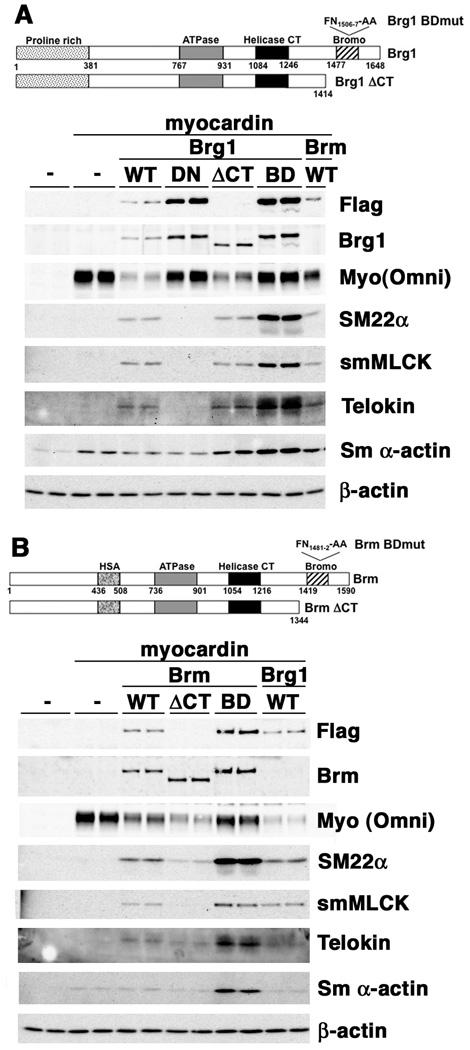
To formally confirm that the ATPase activity of Brg1 and Brm is required to support myocardin’s myogenic activity we utilized SW13 cells, an adenocarcinoma cell line that expresses no endogenous Brg1 or Brm16 (Figure 3). In these cells myocardin fails to induce expression of most smooth muscle-specific proteins (Figure 3A, B, left four lanes). Only SM α-actin induction could be detected. Co-transfection of SW13 cells with myocardin together with wild type Brg1 or Brm expression plasmids restored myocardin’s ability to induce expression of smooth muscle-specific proteins such as SM22α, telokin, and smMLCK (Figure 3). In contrast, the dominant negative, ATPase deficient Brg1 could not restore myocardin’s myogenic activity (Figure 3A, ‘DN’ lanes). Both Brg1 and Brm contain a bromodomain toward their c-termini that has been shown to be able to interact with acetylated histones17. To determine if Brg1/Brm-acteylated histone interactions are important for supporting myocardin’s myogenic activity, mutant Brg1/Brm molecules were generated that either completely lacked the c-terminus, including the bromodomains or that harbored two specific amino-acid mutations within the bromodomain (Brg1 FN1506-7 – AA, Brm FN1481-2 -AA). Mutation of these residues has been previously shown to inhibit the binding of Brg1 to acetylated histones17. These mutants were able to support myocardin’s myogenic activity similar to the wild type molecules, indicating that the bromodomains of Brg1 or Brm are not required for this activity (Figure 3). These data also demonstrate that both Brm and Brg1 have similar abilities to support myocardin’s myogenic activity (Figure 3B, compare the Brg1 WT lanes to the Brm WT lanes). Although identical amounts of myocardin plasmid were used in all co-transfections the resultant myocardin expression levels varied depending on which Brg1/Brm plasmid was co-transfected into the cells (Figure 3). This likely reflects differences in transfection efficiencies of specific plasmid mixtures. In general, the extent of induction of smooth muscle-specific genes by myocardin reflected the relative levels of myocardin expression. However, in cells transfected with the DN-Brg1 even high levels of myocardin failed to induce expression of smooth muscle-specific genes (Figure 3A). This latter finding further demonstrates that the ATPase activity of Brg1 is required to support myocardin’s myogenic activity.
Figure 3. Re-introduction of wild type Brg1 or Brm, but not an ATPase deficient mutant, into SW13 cells restores myocardin’s ability to induce expression of smooth muscle-specific genes.
SW13 cells grown in 6-well plates were co-transfected with expression plasmids (1µg) encoding either wild type Brg1 (WT) and Brm, DN-Brg1 (DN), CT-truncated Brg1 or Brm (ΔCT) or bromodomain mutants (BD) of Brg1 and Brm, together with myocardin expression plasmid (1µg, except in the absence of the Brg/Brm plasmid where 2µg were used), as indicated at the top of the panel (-, empty expression plasmid). 36 hours after transfection protein expression was analyzed by Western blotting with the indicated antibodies. Exogenous Brg1 and Brm were detected with anti-Flag antibodies (Flag) and exogenous smooth muscle myocardin was detected with an omni-epitope tag antibody (Myo(Omni)). 30µg of protein were loaded in each lane. The blots shown are representative of data obtained from 2 separate experiments. The Brm WT sample shown in panel ‘A’ is the same sample as shown in the first WT lane of panel ‘B’. Similarly the WT Brg1 samples shown in panel ‘B’ are the same samples as those shown in panel ‘A’.
DN-Brg1 inhibits myocardin’s ability to increase SRF binding to the promoters of smooth muscle-specific genes
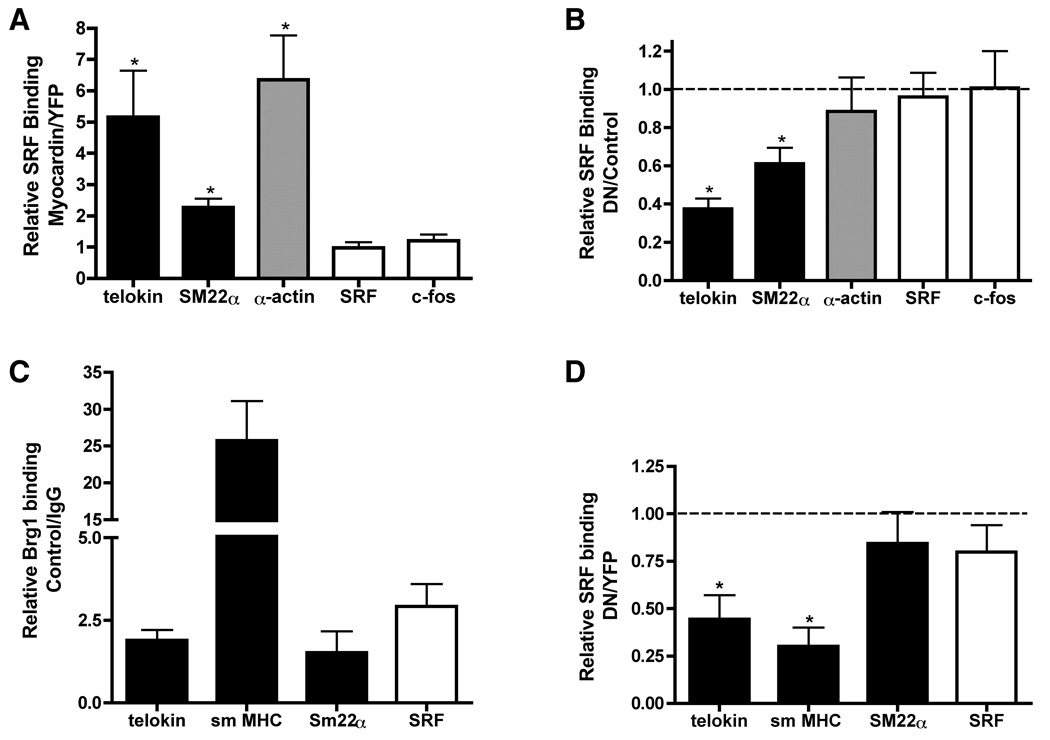
Previous studies have shown that in nonmuscle cells there is little SRF bound to the promoters of smooth muscle-specific genes but introduction of myocardin into these cells leads to increased SRF binding6. Similarly we have previously shown that MRTFA increases SRF binding to the promoters of smooth muscle-specific genes and this process is attenuated by DN-Brg18. We thus sought to determine if Brg1 is also required for myocardin to increase SRF binding to the promoters of smooth muscle-specific genes within intact chromatin. In agreement with previous reports6, using quantitative ChIP assays we observed enhanced SRF binding to the telokin, SM22α and SM α-actin promoters but not the c-fos and SRF promoters following myocardin transduction into NIH 3T3 cells (Figure 4A). Expression of DN-Brg1 attenuated this myocardin-induced increase in SRF binding to the telokin and SM22α promoters without significantly affecting SRF binding to the SM α-actin, SRF and c-fos promoters (Figure 4B). ChIP assays in differentiated primary SMCs also revealed Brg1 binding to the promoters of smooth muscle-specific genes under control conditions (Figure 4C). In addition, expression of DN-Brg1 in differentiated SMCs significantly inhibited the binding of endogenous SRF to the promoters of the telokin and SM MHC gene but not the SRF or SM22α genes (Figure 4D). These data are consistent with expression data shown in figure 1 that demonstrate that the later markers of smooth muscle differentiation such as telokin and smMHC are most dependent on Brg1/Brm.
Figure 4. DN-Brg1 blocks the ability of myocardin to increase SRF binding to the promoters of smooth muscle-specific genes within chromatin.
A–B. B22 cells (an NIH3T3 cells line that inducibly express DN-Brg1 in response to tetracycline withdrawal13) grown in the presence (control) or absence (induced DN-Brg1) of tetracycline were transduced with myocardin or YFP control adenovirus. After 30 hrs, cells were fixed and harvested for chromatin immunoprecipitation assays. Chromatin was precipitated using an antibody against SRF or using IgG negative control. The precipitated genomic DNA was purified and the presence of the promoters of SRF-dependent genes measured by real time PCR using gene specific primers8. A. The increase in SRF binding in samples transduced with myocardin is indicated relative to those transduced with YFP. These data were calculated and normalized to input levels as follows: Relative SRF binding=2−ΔΔCt, with ΔΔCt= (Ct myocardin-Ct input)-(CtYFP-Ct input). B. The relative inhibition of myocardin induced SRF binding by DN-Brg1 is shown. This was calculated as follows: Relative SRF binding= 2−ΔΔCt, with ΔΔCt= (Ct DN-Brg1+Myocardin -Ct input)-(Ct YFP+Myocardin -Ct input). Data shown in panels ‘A’ and ‘B’ are the mean±SEM of 7 samples obtained from 3 independent experiments. A one-way t-test was performed and the asterisks indicate the results that are statistically different from 1 (P<0.05). C–D. Primary colon smooth muscle cells were prepared from 4 week old mice. The cells were transduced by DN-Brg1 or YFP control adenovirus. After 36 hours, cells were fixed and harvested for chromatin immunoprecipitation assays as above. C. The relative Brg1 binding to several SRF dependent genes in control primary smooth muscle cells is shown. The Brg1 binding to promoters were normalized to IgG control. The relative Brg1 binding was calculated as RQ=2−ΔCt, with ΔCt= Ct Brg1-Ct IgG. D. The relative inhibition of SRF binding by DN-Brg1 is shown. The inhibition of SRF binding by DN-Brg1 is calculated as, RQ=2−ΔΔCt, with ΔΔCt= (Ct DN-Brg1-Ct input)-(Ct YFP-Ct input). Data presented are the mean ±SEM of 4 samples.
Brg1, myocardin and SRF form a complex in vivo
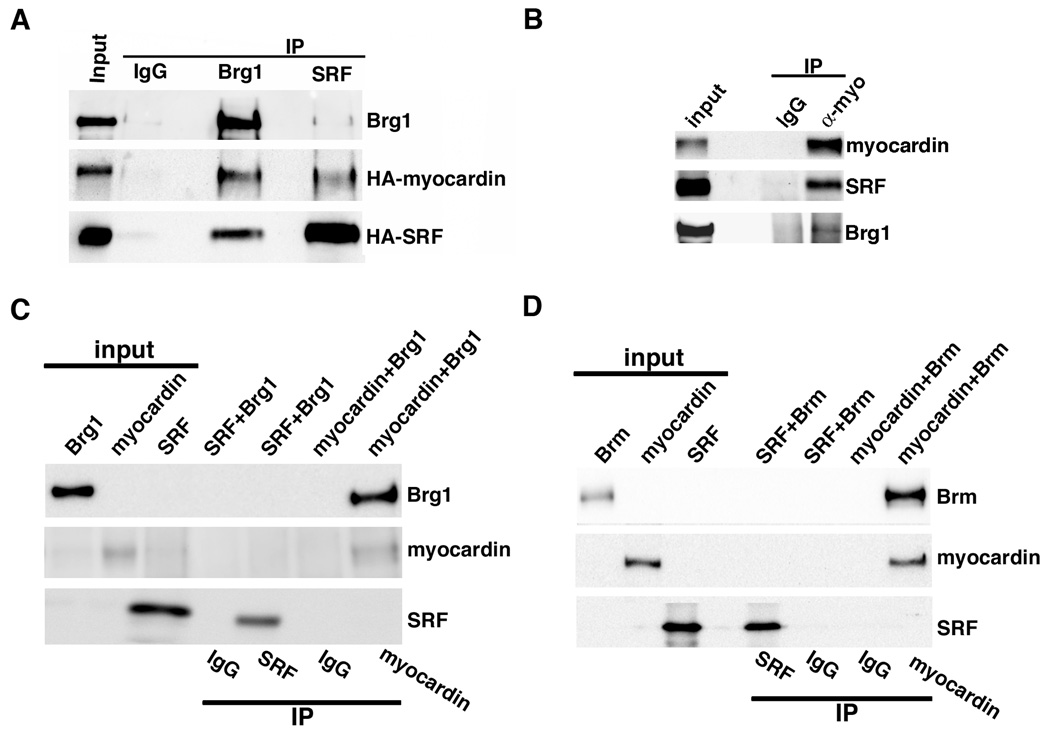
To determine if Brg1 interacts with myocardin/SRF complexes in vivo, immunoprecipitation assays were performed from COS cells transduced with adenoviruses encoding myocardin and SRF (Figure 5A). Brg1 immunoprecipitates were found to also contain myocardin and SRF. Similarly, SRF immunoprecipitates also contained Brg1 and myocardin. In rat aortic A10 SMCs, myocardin immunoprecipitates also contained SRF and Brg1 (Figure 5B). These data suggest that, either Brg1, myocardin and SRF exist in a single complex in vivo or that Brg1 binds to both myocardin and SRF. To distinguish these possibilities co-immunoprecipitation assays were performed using proteins generated in vitro. Results from these experiments demonstrated that Brg1 and Brm directly bind to myocardin but not to SRF (Figure 5C,D). When Brg1 or Brm were incubated with myocardin they could be co-immunoprecipitated with myocardin. In contrast, when Brg1 or Brm were incubated with SRF they were not co-immunoprecipitated with SRF. No endogenous SRF, myocardin and Brg1 were detected in the in vitro translation reaction system (data not shown).
Figure 5. Myocardin, SRF and Brg1 form a complex in vivo and Brg1 binds directly to myocardin in vitro.
A. COS cells were transduced with HA-tagged myocardin and HA-tagged SRF adenovirus. After 24 hours, nuclear protein was harvested and proteins were immunoprecipitated using Brg1, SRF or control IgG antibodies, as indicated. Immunoprecipitated proteins were identified by Western blotting using antibodies against Brg1 or the HA-epitope tags on myocardin and SRF as indicated at the right of the blot. B. A10 SMCs were transduced with omni-tagged myocardin for 24 hours. Nuclear protein was harvested and subsequently immunoprecipitated using myocardin or control IgG antibody. C. SRF, myocardin, and Brg1 or Brm (D) were transcribed and translated in vitro, the expressed proteins were then incubated together (SRF+Brg1/Brm or myocardin +Brg1/Brm) as indicated at the top of the blots. Protein mixtures were immunoprecipitated with myocardin, SRF or IgG control antibodies, as indicated below the blots. The precipitated proteins were analyzed by Western blotting, using antibodies indicated at the right of the blots. On all blots ‘input’ lanes represent 10% of the inputs that were mixed together and used for immunoprecipitates.
The ATPase domain of Brg1 binds to N-terminus of myocardin
Co-immunoprecipitation assays using in vitro transcribed and translated myocardin and fragments of Brg1 revealed that the region from amino acids 837 to 1446 of Brg1 binds to myocardin (Supplemental figure II). To further resolve the myocardin binding site within this region, an additional series of Brg1 deletion mutants were generated, expressed in bacteria and used in GST-pulldown assays with the N-terminus of myocardin as bait (Supplemental figure II). These assays demonstrate that the ATPase domain of Brg1 extending from amino acids 767–931 is sufficient to bind to myocardin. Interestingly this region is 94% identical between Brg1 and Brm, consistent with our observations that both of these molecules can bind to myocardin (Figure 5C,D). GST-pulldown assays were also performed to determine which portion of myocardin interacts with Brg1 (Supplemental figure II). Data from these experiments demonstrated that the Brg1 interacts with the N-terminal portion of myocardin, with the region spanning the basic and poly Q domains (myo Δ2-GST in supplemental figure IIe) having the highest apparent affinity. The GST-pulldown assays also confirm that Brg1 does not directly bind to SRF, while Barx2 can be readily detected bound to GST-SRF fusion proteins, as reported previously12.
Discussion
Our data demonstrate that the ability of myocardin to induce expression of most smooth muscle-specific genes is regulated by the activity of the SWI/SNF ATP-dependent chromatin remodeling complex. We suggest a model in which myocardin associates with the SWI/SNF complex through direct binding to the Brg1 or Brm ATPase subunit. This association is required in order for myocardin to increase SRF binding to the promoters of smooth muscle-specific genes within intact chromatin, thereby leading to activation of these genes during differentiation of vascular smooth muscle cells.
Although Brg1/Brm containing SWI/SNF complexes are required for myocardin to induce expression of many smooth muscle-specific genes the induction of SM α-actin by myocardin was largely independent of SWI/SNF (Figure 1–Figure 3 and supplementary figure 1). In addition, in SW13 cells that lack Brg1 and Brm, myocardin was still able to induce SM α-actin expression (Figure 3A). This correlates with the relatively high basal levels of SM α-actin expression in many of these cells in the absence of added myocardin. Thus it is likely that the SM α-actin promoter is already in an active transcriptionally favorable conformation in the absence of myocardin. This may suggest that the SWI/SNF complex is dispensable for myocardin-induced activation of genes, such as SM α-actin, in cells in which these genes are already transcriptionally active. In contrast, SWI/SNF activity is required for myocardin to induce expression of genes that are otherwise transcriptionally silent in a given cell type. Similar to SM α-actin expression of SM22α in primary smooth muscle cells was found to be largely independent of Brg1. This may also reflect the contribution of myocardin independent pathways in driving chromatin remodeling and transcription of the SM22α locus in smooth muscle cells. For example, both SM α-actin and SM22α can be induced by TGFβ in 10T1/2 cells by a myocardin independent pathway 18, 19. This model is, however, likely to be an over-simplification, as in primary cultures of mouse aortic smooth muscle cells we observed that the myocardin-induced increase in expression of smooth muscle-specific genes was at least partially attenuated by DN-Brg1 (Figure 2B). As all of the genes examined were expressed prior to myocardin over-expression, this may suggest that even if a gene is transcriptionally active, SWI/SNF induced changes in chromatin can further augment myocardin’s myogenic activity.
Domain mapping experiments suggest that Brg1 and Brm interact with the region of myocardin that spans the SRF interaction domain (basic and poly Q region). Despite the overlapping binding sites, co-immunoprecipitation studies show that myocardin is present in a complex that includes both Brg1 and SRF within intact cells. The Brg1 binding site in myocardin is present in both the long, cardiac-selective isoform (1–935) and the shorter, smooth muscle-selective isoform (80–935). These results suggest that both cardiac- and smooth muscle-selective myocardin isoforms will interact with and be regulated by SWI/SNF. In support of this proposal, the myogenic activity of both the cardiac myocardin (Supplemental figure I), and the smooth muscle myocardin (figure 2–figure 3) was attenuated by dominant negative Brg1. In addition, the ability of myocardin to induce expression of cardiac-specific genes in 10T1/2 cells was also attenuated by DN-Brg1. These observations suggest that the myogenic activity of myocardin both in the heart and in vascular SMCs is regulated by SWI/SNF.
Results from experiments in which we reconstituted Brg1 or Brm expression in SW13 cells suggest that either wild-type Brg1 or Brm containing SWI/SNF complexes are equally effective at supporting myocardin’s myogenic activity (Figure 3). In addition, knockdown of either Brg1 or Brm in aortic SMCs attenuated expression of smooth muscle-specific genes (Figure 1) and both Brg1 and Brm can directly bind to myocardin in vitro (Figure 5C and D). These data suggest that Brg1 or Brm containing SWI/SNF complexes may both play important roles in smooth muscle cells. In contrast to myocardin, the LIM domain protein CRP2 has recently been shown to interact specifically with Brg1, but not Brm, in order to induce expression of smooth muscle-specific genes in cardiomyocytes20. Despite the ability of either Brg1 or Brm to support myocardin’s myogenic activity in SW13 cells, knocking-down both Brg1 and Brm in smooth muscle cells did not result in any further attenuation of smooth muscle-specific genes as compared to knockout of either protein alone (Figure 1). The lack of an additive effect of the double knockdown, may suggest that Brg1- and Brm-containing SWI/SNF complexes act together in smooth muscle cells to regulate myocardin. This must however, be interpreted with caution as knockdown of Brg1 also attenuated expression of Brm. Further studies analyzing tissue specific single or double knockouts of Brg1 and Brm, in vivo, will be required to clarify the role of individual SWI/SNF complexes in regulating smooth muscle differentiation.
In addition to ATP-dependent chromatin remodeling complexes, enzymes that covalently modify histones are important to mediate myocardin activation of smooth muscle-specific genes21, 22. For example, myocardin has been shown to bind to p300 and to promote acetylation of histones associated with the promoters of smooth muscle-specific genes6, 21. As the bromodomains of Brg1 and Brm are known to bind to acetylated histones we initially speculated that myocardin-recruited HAT activity may help recruit SWI/SNF to promote transcriptional activation of genes in smooth muscle cells. However, data showing that the bromodomains of Brg1 and Brm are not important for supporting myocardin’s myogenic function argue against this proposal (Figure 3). Similarly, it is unlikely that direct DNA binding by the AT-hook domain of Brg1 or Brm is required to recruit myocardin to chromatin, as the C-terminal truncation of Brm that we analyzed also lacks this domain23 yet was still able to support myocardin’s myogenic activity (Figure 3).
In skeletal muscle, weak binding of MyoD to the myogenin promoter via MyoD interactions with Pbx, facilitates SWI-SNF recruitment through direct binding of MyoD and Brg1 24, 25. Chromatin remodeling by SWI/SNF then facilitates tight binding of MyoD to the E box within the myogenin promoter, facilitating promoter activation and skeletal muscle cell differentiation. By analogy we propose a model in which in undifferentiated SMC or in nonmuscle cells, SRF has a low binding affinity for CArG box elements in the promoters of smooth muscle-specific genes within intact chromatin. Little transcription activity of smooth muscle-specific genes such as telokin and SM-MHC, thus occurs in these cells. To induce smooth muscle differentiation, myocardin complexed with p300 and SWI/SNF, interacts with SRF weakly bound to the promoters of smooth muscle-specific genes. SWI/SNF binding to the promoter regions then leads to ATP-dependent chromatin remodeling and rearrangement of the nucleosomes that facilitates tight binding of SRF. This may also allow binding of additional activators to the adjacent DNA segments. These activators, together with the SRF/myocardin/p300 complex can then further modify chromatin to facilitate recruitment of general transcriptional factors, including RNA polymerase II, resulting in transcriptional activation of smooth muscle-specific genes.
Supplementary Material
Acknowledgements
We are grateful to April Hoggatt for expert technical assistance.
Sources of Funding
This work was supported by NIH HL58571, DK61130 and DK65644 and a Biomedical Research Grant from IUSOM to B.P.H., by GM56244 to A.N.I., by an American Heart Association Fellowship and Scientist Development Grant to J. Z. and by fellowships from the Fortune Fry Foundation and American Heart Association to M.Z.
Footnotes
Disclosures
None
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 4.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 6.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282:25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herring BP, Smith AF. Telokin expression is mediated by a smooth muscle cell-specific promoter. Am J Physiol. 1996;270:C1656–C1665. doi: 10.1152/ajpcell.1996.270.6.C1656. [DOI] [PubMed] [Google Scholar]
- 11.Hoggatt AM, Simon GM, Herring BP. Cell-specific regulatory modules control expression of genes in vascular and visceral smooth muscle tissues. Circ Res. 2002;91:1151–1159. doi: 10.1161/01.res.0000047508.30800.4f. [DOI] [PubMed] [Google Scholar]
- 12.Herring BP, Kriegel AM, Hoggatt AM. Identification of Barx2b, a serum response factor-associated homeodomain protein. J Biol Chem. 2001;276:14482–14489. doi: 10.1074/jbc.M011585200. [DOI] [PubMed] [Google Scholar]
- 13.de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 16.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, Wu J, Shi Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 18.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Kulik M, Lechleider RJ. Smad proteins regulate transcriptional induction of the SM22alpha gene by TGF-beta. Nucleic Acids Res. 2003;31:1302–1310. doi: 10.1093/nar/gkg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang DF, Belaguli NS, Chang J, Schwartz RJ. LIM-only protein, CRP2, switched on smooth muscle gene activity in adult cardiac myocytes. Proc Natl Acad Sci U S A. 2007;104:157–162. doi: 10.1073/pnas.0605635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S, Owens GK, Olson EN. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourachot B, Yaniv M, Muchardt C. The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol Cell Biol. 1999;19:3931–3939. doi: 10.1128/mcb.19.6.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.