Abstract
Background
The importance of genetic variation to the etiology of neuropsychiatric disorders is well established and is currently being examined for diagnosis and treatment. The most popular method of obtaining material for genotype analysis, high-yielding DNA extraction from blood, has several limitations, including invasiveness, need for skilled individuals to collect material, and requirement for cold storage. Saliva sampling is non-invasive and trained personnel are less necessary, but it still requires a relatively high level of subject compliance. Buccal mucosa cells sampling is almost completely non-invasive, reducing compliance issues significantly. Samples collected have been shown to produce usable DNA after shipment through conventional mail. However, the DNA produced by rapid elution of these swabs in chaotropic buffers is of limited quality and low purity.
Objective
Our aim was to develop a rapid, economical, high-yielding and environmentally-safe method for extraction of high quality genomic DNA, which can be used to determine clinically important genotypes from trace quantity samples.
Methods
We developed a method of extracting high-quality genomic DNA from buccal swab, which we termed the `Rapid Method for Swab' (RMS). We compared RMS with two established procedures, specifically the original Rapid Method (RM) [Lahiri et al. J Biochem Biophys Methods. 25, 193-205 (1992)] and the commercially available BuccalAmp (Epicentre Biotechnologies, Madison, WI) method. We assessed the generated genomic DNAs by their i) quality, ii) quantity, iii) restriction enzyme digestibility and v) PCR-based genotyping in addition to time, cost and environmental impact of the procedures.
Main Results
DNA generated by RMS was of higher purity than that by BuccalAmp. RMS is non-enzymatic and does not use strong chaotropic salts or extreme pH. We also demonstrated the suitability of RMS-DNA for LA/LG genotyping as generated by PCR using 7-deaza dGTP.
Conclusion
The RMS procedure is novel, efficient, safe, and high-yielding, and produces DNA of high quality from a single human buccal swab. RMS is a non-invasive technique and particularly suitable for children and older subjects and in field collection settings.
Keywords: DNA purification, HTTLPR, genotyping, DNA, nucleic acid, genomic DNA, 5-HTT, large sample size, swab, buccal mucosa
Introduction
The importance of genetic variation to the etiology of neuropsychiatric disorders has been well established for research and is being examined for diagnosis and potential treatment purposes. Examples include a variable-length repeat polymorphism, `HTTLPR', and a further LA/LG `sub-variant' in a single repeat (Hu et al., 2006; Zalsman et al., 2006), in the promoter of the serotonin transporter gene (SLC6A4, 5-HTT) in depressive disorders (Cervilla et al., 2006);, the apolipoprotein E (APOE) ε4 genotype and late-onset AD (LOAD) (Lahiri et al., 2004); promoter polymorphisms in the APOE gene and LOAD (Bullido et al., 1998); and a VAL/MET polymorphism in the catechol-O-methyltransferase gene and schizophrenia (Li et al., 1996). In addition, genetic variation has been found to determine responses to psychopharmacologic therapy in diverse conditions, including but not limited to the HTTLPR polymorphism altering response to selective serotonin reuptake inhibitors (Eichhammer et al., 2003; Putzhammer et al., 2005) and APOE genotype effects on response to tacrine (Farlow et al., 1996).
Beyond direct medical applications, discovery of pathological associations based on genetic variation is stronger with larger sample sizes. Of 20 recent studies chosen at random from a Medline search of the keywords `HTTLPR' and `genotype', 16 used DNA prepared from whole blood or white blood cells (Cervilla et al., 2006; Finger et al., 2006; Kim et al., 2006; Little et al., 2006; Mannelli et al., 2006; Marsh et al., 2006; Roiser et al., 2006; Saunders et al., 2006; Segal et al., 2006; Smeraldi et al., 2006; Zalsman et al., 2006; Becker et al., 2007; Gerra et al., 2007; Heiser et al., 2007; Mergen et al., 2007; Nilsson et al., 2007); Lahiri and Nurnberger, 1991), 4 used saliva (Cervilla et al., 2006; Taylor et al., 2006; Covault et al., 2007; Kaufman et al., 2007), and 2 used buccal mucosa (Brocke et al., 2006; Zalsman et al., 2006). Some studies used more than one collection method.
The most popular method, high-yielding DNA extraction from blood, has several research advantages, specifically that currently accepted methods, including kits (e. g. `MasterPure', Epicentre, Madison, WI) or non-enzymatic methods (Lahiri and Nurnberger, 1991; (Lahiri et al., 1992) are rapid and reliable, and the DNA derived is of high quality and suitable for multiple uses and purposes for research and clinical setting. Unfortunately, blood collection has several limitations. Its invasiveness reduces the likelihood of compliance, especially among children, the elderly, and some neuropsychiatric patients. If venous blood is used, skilled individuals are needed to collect material, increasing cost. Such samples also require cold storage, further increasing cost. Some of these issues have been partially addressed by use of dry blood spots on paper-based collection media, used in one of the aforementioned studies (Gerra et al., 2007). However, this method reduces the DNA obtained from a given sample, limiting its direct use for multiple assays.
In contrast, saliva sampling is non-invasive and trained personnel are less necessary for sampling from the majority of subjects, but it still requires a relatively high level of subject compliance, necessary to produce 2-4 ml volumes of saliva currently required by current commercial methods (Rogers et al., 2007; Viet et al., 2007). Buccal mucosa cells sampling is almost completely non-invasive and painless, reducing compliance issues significantly. Samples collected have been shown to generate usable DNA after shipment through conventional mail (Ilveskoski et al., 1998). Sample collection does not require highly-trained personnel nor an established `cold chain' to preserve samples until DNA is extracted. For example, in a recent study of the HTTLPR polymorphism and temperamental fearfulness in children, our group used buccal mucosa samples collected either by parents in the children's homes or by technicians in a clinical setting (Hayden et al., 2007). For that study, a kit employing pH10 high-salt buffers (BuccalAmp, Epicentre, Madison, WI) was used to produce `DNA' that amounted to a crude cell lysate containing various proteins and nucleic acids. A total of 134 samples were collected, of which 95 produced a result via PCR (Table 1). Of the 134 samples 16 were not processed due to visible contamination of the swabs. Test preparations of such swabs with the BuccalAmp kit indicated no likelihood of successful genotyping. A further 29 did not produce discernable signal from genotyping PCR.
Table 1.
Success, failure and rejection of buccal tissue samples prepared by BuccalAmp method (Hayden et al., 2007).
| Group | Swabs | Rejected | Failed | Success | Success Rate |
|---|---|---|---|---|---|
| Parental Collectiona | 52 | 15 | 19 | 18 | 49% |
| Technician Collectiona | 82 | 1 | 10 | 71 | 88% |
| Total | 134 | 16 | 29 | 89 | 75% |
Parental collected samples were taken at home, with the parent administering the swab. Swabs were stored in home freezers or refrigerators and delivered to a laboratory at Stony Brook University (Stony Brook, NY, USA). Technician-collected samples were collected by clinical technicians at Stony Brook University.
In response to these difficulties we investigated an efficient and economical method to produce higher-quality DNA from very small quantity of human biological fluids, preferably obtained by a non-invasive and painless technique. We have developed a rapid and simple non-enzymatic method of DNA purification from buccal mucosa that does not require the use of enzymatic digest or proprietary reagents. DNA generated using this approach is of high purity and quality and suitable for use in genotyping. This method was based on our previous well-established rapid method (RM) for purifying DNA from blood samples (Lahiri and Nurnberger, 1991), but it required less time, fewer steps, and generated up to 4μg of DNA from a single buccal swab.
In comparison with DNA from buccal swab produced via the commercial BuccalAmp kit, our method produced DNA of equal amount but higher purity and at least comparable applicability for PCR-based genotyping. This method did not require the purchase of proprietary reagents and could be performed in any facility with common molecular biology reagents and equipment limited to a microcentrifuge, a vortexer, and water baths capable of 55°C and 65°C incubation. The rapid method for swab (RMS) used buccal swabs for its source material and was suitable for use in a variety of laboratories and potentially in field conditions.
Materials and Methods
Collection of samples from human subjects
All samples collected from human subjects, both buccal swab and whole blood, were collected per Institution Review Board (IRB)-approved protocols.
Collection of buccal swab samples
Polyester fiber-tipped swabs (Fisher Scientific, Pittsburgh, PA) were used for collection of cheek epithelial cells from eight laboratory volunteers. One swab was used per cheek. Volunteers were instructed to rub the inside of a cheek, swirling the swab at least ten times while so doing. Volunteers reported no discomfort when queried. Swab samples were cut off to length and stored in 15ml conical bottom tubes until extracted (overnight) at room temperature.
Collection of blood samples
Three volunteers had blood drawn by clinical personnel into EDTA Vacutainers (purple stoppered). Samples of between 0.5 and 5 ml were drawn. One volunteer required three attempts to draw blood, and total draw from that volunteer was less than 1ml. Blood samples were aliquoted and stored at -20°C until extracted.
Preparation of DNA from blood samples
The RM of DNA extraction developed for use in blood by Lahiri and colleagues (Lahiri et al., 1992) was employed and all steps performed at room temperature unless otherwise specified. Briefly, 500μl of whole blood was mixed with 500μl of TKM buffer (Tris-HCl pH 7.6 10mM, KCl 10mM, MgCl2 10mM, EDTA 2mM) in a microcentrifuge tube. To this, 1 ml of TKM + 2.5% Triton X-100 was added to disrupt the cell membrane without damaging nuclei. The tube was mixed by inversion (10 times) and then centrifuged at 1000g for 8 minutes at 4°C. The cytoplasmic supernatant was discarded and the nuclear pellet was resuspended in 1 ml of TKM by pipetting. Cells were again centrifuged at 1000g for 8 minutes and washed with TKM once more. The final pellet was resuspended in 200μl TKM, after which 15μl of 10% SDS was added to lyse nuclei and mixed by inversion. Material was incubated for 5 minutes at 55°C and 75μl of saturated NaCl solution was added. The tube was mixed by inversion and centrifuged at maximum speed (12,600g) in an Eppendorf microcentrifuge for 5 minutes at room temperature. Supernatant was transferred to a fresh tube and 0.7 volume of isopropanol was added. The sample was mixed by inversion and centrifuged for 10 minutes at maximum speed in a microcentrifuge. Pellet was washed in cold 70% ethanol and permitted to air dry for 15 minutes. DNA material was resuspended in 100μl TE at 65°C for one hour.
Preparation of DNA from buccal cell swabs by different methods
All methods were repeated at least three times. The RM was modified for use with buccal swab samples as follows: Swabs were swirled directly in 500μl of TKM buffer (Tris-HCl pH 7.6 10mM, KCl 10mM, MgCl2 10mM, EDTA 2mM). To this, 500μl of TKM + 2.5% Triton X-100 was added. The tube was mixed by inversion (10 times) and then centrifuged at 1000g for 8 minutes at 4°C. The supernatant was discarded and the pellet was resuspended in 1 ml of TKM by pipetting. Cells were again centrifuged at 1000g for 8 minutes and washed with TKM once more. The final pellet was resuspended in 200μl TKM, after which 15μl of 10% SDS was added and mixed by inversion. Material was incubated for 5 minutes at 55°C and 75μl of saturated NaCl solution was added. The tube was mixed by inversion and centrifuged at maximum speed in an Eppendorf microcentrifuge for 5 minutes at room temperature. Supernatant was transferred to a fresh tube and 0.7 volume of isopropanol was added. The sample was mixed by inversion and centrifuged for 10 minutes at maximum speed in a microcentrifuge. Pellet was washed in cold 70% ethanol and permitted to air dry for 15 minutes. DNA material was resuspended in 100μl TE at 65°C for one hour.
A modified rapid method for swab (RMS) was also employed. Briefly, the three washing steps were dispensed with and cells were directly suspended in 500μl of TKM + 1.25% Triton X-100. To this was added 37.5μl of 10% SDS. Samples were incubated at 55°C for 5 minutes and briefly vortexed. After the addition of 187.5μl of saturated NaCl, samples were mixed by inversion. Samples were then centrifuged and 0.7 volumes of isopropanol were added to the clear supernatant. The sample was mixed by inversion and centrifuged for 10 minutes at maximum speed in a microcentrifuge. Pellet was washed in cold 70% ethanol and permitted to air dry for 15 minutes. Material was resuspended in 100μl TE at 65°C for one hour. To optimize the method, the RMS was repeated under variant conditions, specifically Tris-HCl of pH 5.4 and pH 10.0, and in TKM + 50mM NaOH. The NaOH treatment was followed by neutralization with 50μl 1M Tris pH 7.6 for half of the NaOH treated samples. These pH conditions were chosen based on the pH of the QuickExtract buffer (pH 10) and on a conventional acidic pH for acetate salts used for precipitating DNA. The NaOH was included based on the long-standing use of NaOH-based lysis methods for collection of DNA from buccal swabs (Walker et al., 1999). For some samples, QuickExtract (Epicentre Biotechnologies, Madison, WI) buffer was used instead of TKM + Triton X-100, otherwise following the RMS protocol. In some RMS preparations, a phase boundary occurred after the NaCl precipitation/centrifugation step. Both upper and lower phases were collected and analyzed separately.
Cells were also extracted using QuickExtract buffer and the BuccalAmp method (Epicentre) according to the manufacturer's instructions. In addition, cells were suspended in TKM and the BuccalAmp method was applied to the suspension.
Evaluation of quantity and quality of DNA from buccal samples prepared by different methods via absorbance and fluorescence
DNA samples were evaluated by measurement of optical density at 260 nm (OD260) and at 280 nm (OD280) using a Tecan Genios microplate reader (Phenix Research Products, Asheville, NC). Quantitative DNA yield was assessed by OD260 for RM and all RMS methods, and by Hoechst 2258 dye for the `Epi' and `TKM/Epi' methods due to very low OD260/280 ratios for those particular samples. A volume of 25 μl of each sample was diluted with either 75 μl of TE (for RM, all RMS variants, and QuickExtract buffer followed by RMS method), by QuickExtract buffer (for BuccalAmp method) or by TKM (for suspension in TKM followed by BuccalAmp method). A Beckman DU-70 (Beckman-Coulter, Fullerton, CA) spectrophotometer was used to measure spectrum response at wavelengths from 220-300 nm. Those samples diluted in TE were zeroed with TE. The QuickExtract/BuccalAmp sample was zeroed against QuickExtract buffer. The sample diluted with TKM was zeroed against TKM. In addition, a 50ng sample of commercially available DNA (Roche, Indianapolis, USA) was diluted to 100μl total volume in TE and subject to spectrum analysis.
Semi-quantitative and qualitative evaluation of DNA from buccal samples prepared by different methods via agarose gel and restriction enzyme digest
A volume of 20μl of material prepared by each method was digested with EcoRI (Roche, Indianapolis) at 37°C for 60 minutes. Samples were run on 1% agarose gel alongside an equal volume of undigested material and visualized by ethidium bromide staining.
Genotyping PCR of DNA from samples prepared by different methods from buccal cells and whole blood
To test the genotyping suitability of DNA we generated in this study, we selected the HTTLPR length polymorphism of the serotonin transporter 6A4 protein. The HTTLPR polymorphism is a repeat-number polymorphism that begins approximately 1.4 kb upstream of the +1 transcription start site (TSS) of the SLC6A gene. The most common polymorphisms are 14- and 16- subunit repeats, usually termed `s' and `l'. Genotyping of the HTTLPR polymorphism was chosen to investigate the utility of various DNA samples for PCR-based protocols. Briefly, the oligomers HTTLPR-F (5'-GGCGTTGCCGCTCTGAATGC) and HTTLPR-R (5'-GAGGGACTGAGCTGGACAACCAC) were commercially synthesized (Invitrogen, Carlsbad, CA) and used in an HTTLPR genotyping reaction. Briefly, 21μl of HTTLPR-F, 0.5μM; HTTLPR-R, 0.5μM; dATP, 200μM; dCTP, 200μM; dTTP, 100μM, dGTP, 100μM; 7-deaza-dGTP (all dNTP from Roche); Taq polymerase (Roche), 2 units; was added to 9 μl of each different DNA preparation. The mixtures were hot-start denatured at 95°C for 5 minutes and subject to 35 cycles of 95°C, 30sec; 61.5°, 30sec; 72°, 45sec. Samples were then held at 4°C until analyzed by native Tris-acetate EDTA (TAE) 4.5% polyacrylamide gel. Alleles running at approximately 530bp length were designated `l' while those running at approximately 480bp length were designated `s'.
We also evaluated suitability of DNA generated by the specific PCR method we chose for LA/LG genotyping. To this end, we digested PCR product with HpaII. Incorporation of 7-deaza dGTP into a DNA strand has been shown to interfere with some but not all restriction enzymes that incorporate `G' into their recognition sequence (Grime et al., 1991).
An additional band of approximately 900bp in length was observed (see `HTTLPR genotyping PCR from whole blood and buccal cell DNA' in Results) when running heterozygote HTTLPR sample on TAE-polyacrylamide gel electrophoresis (PAGE). We repeated HTTLPR PCR on this sample and ran on 5% TAE-PAGE and 2.5% TAE-agarose to determine if the sample was a novel PCR-generated DNA fragment or a conformational complex between the smaller fragments.
Results
Comparison of quantity of DNA samples prepared by different preparation methods
DNA generated was subject to quality and quantity measures, the results of which are summarized in Table 2. Briefly, yields from all methods were comparable, 1.0-4.0 μg per subject, except for the RMS NaOH/N and TKM/epi methods, which had no measurable yield. Nucleic acid purity, as assessed by OD260/280 ratio was high (1.7-2.0 for the RM and RMS pH7.6 methods) (Table 2). The RMS pH5.4, pH10.0, and NaOH methods had greater OD260/280 ratio variability. Lower-phase RMS subsamples had no nucleic acid by any measure and were excluded from further analysis.
Table 2.
Summary of DNA Extraction Methods
| Method | Extraction Buffer | pH | Major steps |
Product | Yield, μg | A260/280 | Quality (gel) |
Time | Restr. Enzyme |
PCR- HTTLPR |
|---|---|---|---|---|---|---|---|---|---|---|
| RM | TKM+Triton X-100 | 7.6 | RM | DNA | 1.0-4.0 | 1.7-2.0 | high | 45 minc | yes | yes |
| RMS pH5.4 | TKM+Triton X-100. | 5.4 | RMS | DNA | 1.0-4.0 | 1.5-2.0 | mod | 20 minc | yes | yes |
| RMS pH7.4 | TKM+Triton X-100 | 7.6 | RMS | DNA | 1.0-4.0 | 1.7-2.0 | high | 20 minc | yes | yes |
| RMS pH10 | TKM+Triton X-100 | 10.0 | RMS | DNA | 1.0-4.0 | 1.5-2.0 | high | 20 minc | yes | yes |
| RMS NaOH | TKM+Triton X-100+NaOH | 13.4 | RMS | DNA | 1.0-4.0 | 1.2-2.0 | varies | 20 minc | no | yes |
| RMS NaOH/N | TKM+Triton X-100+NaOH Tris-neutralized | 8.3 | RMS | DNA | — | 3+ | not vis | 20 minc | no | no |
| Epi. | QuickExtract | ~10 | BuccalAmp | Crude lysate | 1.0-4.0b | 0.7-1.1 | varies | 5 min | no | yes |
| TKM/epi | TKM | 7.6 | BuccalAmp | Crude lysate | — | 1.2-1.3 | not vis | 5 min | no | no |
| Epi/RM | QuickExtract | ~10 | RMS | DNA | 0.3-0.4 | 1.5-2.0 | high | 5 min | - | yes |
aNo signal above background.
Yield was measured by Hoechst fluorescence dye due to very low OD260/OD280 ratio.
Time does not include a 60 minute incubation at 65°C to resuspend washed DNA pellet.
Determination of quality by restriction digestibility of DNA samples prepared by different methods
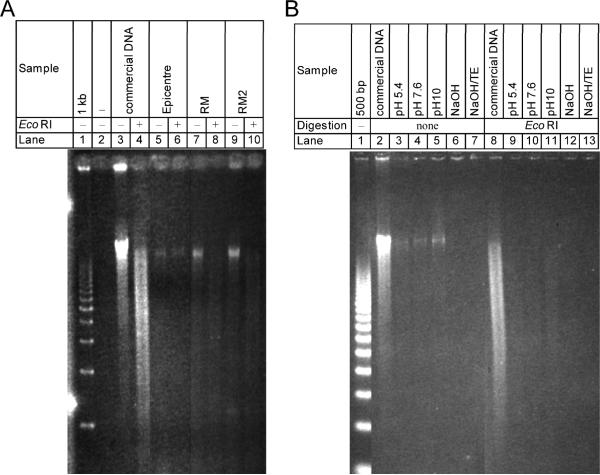
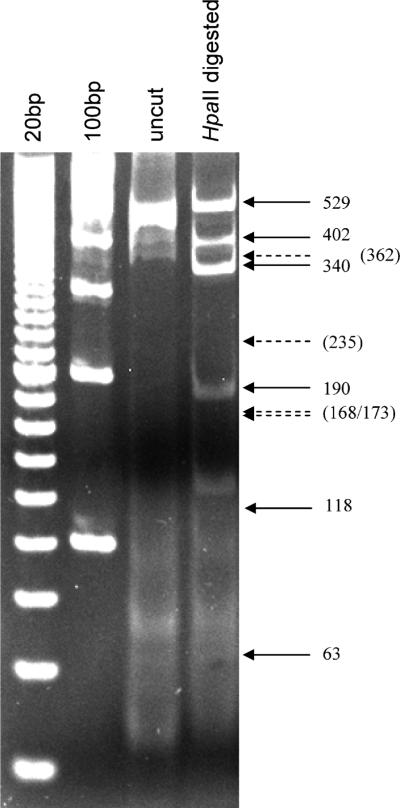
The BuccalAmp, RM and RMS methods all produced sufficient DNA that a 20μl sample was visible as a single large (> 12 kb) band on 1% agarose gel (Fig. 1A, lanes 5, 7, 9, respectively). However, DNA produced by the BuccalAmp method was not digestible by EcoRI restriction enzyme (lane 6) and remained intact, indicating the presence of significant amounts of contamination interfering with the reaction in the material. Digestion by EcoRI of both the RM and RMS samples (lanes 8 and 10) was successful. When 20μl samples of DNA prepared by variations of the RMS method were run on 1% agarose (Fig. 1B), only material from the Tris pH-modified samples had sufficient DNA to be visible as a single band (lanes 3-5) This material was digestible with EcoRI (lanes 9-11). Material from NaOH-modified RMS and neutralized NaOH-modified RMS was not visible (lanes 6, 7, 12, 13).
Fig. 1. Assessment of the quality of DNA generated by different extraction methods.
A. A volume of 20 μl of prepared DNA was run on 1% agarose gel, either uncut (lanes 3, 5, 7, 9) or after digestion with EcoRI (lanes 4, 6, 8, 10). Lane 1: commercial 1kb DNA ladder; lanes 3-4: commercially purchased DNA. lanes 5-6: DNA prepared by BuccalAmp kit; lanes 7-8: DNA prepared by the rapid method (RM); lanes 9-10; DNA prepared by the modified rapid method (RMS) B. samples of DNA purified by RM or by RMS with different buffer conditions were run on 1% agarose gel or digested with EcoRI and run on agarose gel. Lane 1: 500bp DNA ladder; lanes 2-7: undigested DNA; lanes 8-13: DNA digested with EcoRI. DNA was visible for commercial DNA (lanes 2, 8), and for RMS DNA prepared at buffer pH of 5.4 (lanes 3, 9), 7.6 (lanes 4, 10), and 10 (lanes 5, 11). Preparations from the RMS method adjusted by NaOH (lanes 6, 7, 12, 13) were also run.
Assessment of `contaminants' and purity by spectrum analysis of samples prepared by different methods
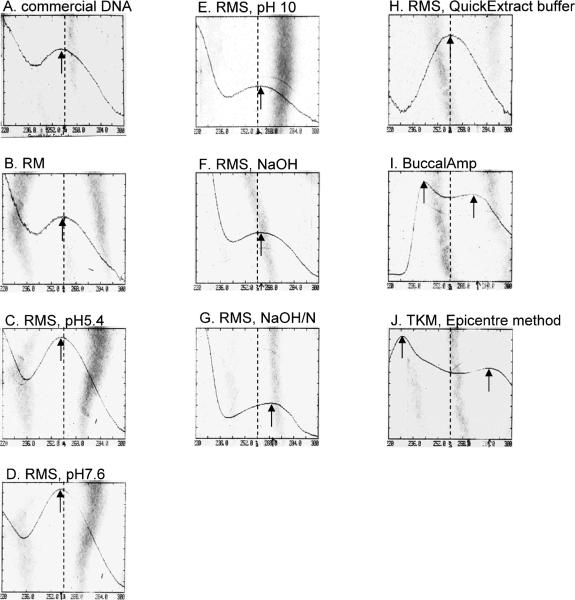
Spectra were run from 220nm to 300nm wavelength on commercially purchased DNA (Fig. 2A) and samples of 50μl that were taken from DNA prepared by either RM (Fig. 2B), a modification of RMS (Fig. 2C-G), the RM method applied to buccal cells suspended in QuickExtract buffer (Fig. 2H), by complete BuccalAmp method (Fig. 2I) or cells suspended in TKM and prepared by the BuccalAmp method (Fig. 2J). For most samples prepared by RM or a variant of RMS, the spectrum peak was at or near 260nm (Fig. 2A-F, H). However, the peak for the Tris-neutralized NaOH modification of RMS was closer to 270nm than to 260nm (Fig. 2G), indicating the presence of some contamination. Material prepared using the BuccalAmp method, regardless of initial suspension buffer, did not have a typical nucleic acid spectrum, which is suggestive of a heterogeneous sample (Fig. 2I, J).
Fig. 2. Spectral analysis of DNA extracted by different methods.
DNA was extracted from buccal swabs by ten different methods as described in the text. A spectrum for 100μl of each was generated at 220nm to 300 nm with a Beckman DU-70 spectrophotometer. On each spectrum, the position of OD260 is indicated with a dashed line. The major peak is indicated with an arrow. Samples analyzed were as follows: A. Commercial DNA sample, 100ng diluted into 100μl. B. Rapid Method (RM), C-H. Modified rapid method (RMS) with different buffer conditions, specifically pH 5.4 (C), pH 7.6 (D), pH 10 (E), 50mM NaOH (F), and 50mM NaOH equilibrated with 1M Tris pH 7.6 (G), Epicentre buffer used with RMS (H). I. The conventional Epicentre BuccalAmp method. J. TKM buffer used with the BuccalAmp `heat and vortex' method.
HTTLPR genotyping PCR from whole blood and buccal cell DNA
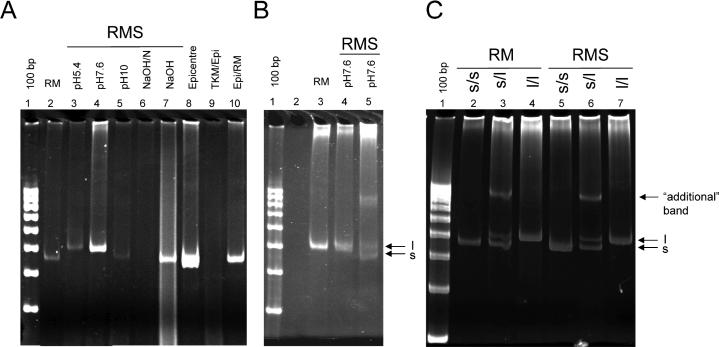
Conventional HTTLPR genotyping PCR was performed using 9μl of each of eight differently-prepared DNA samples and 10ng of commercially-obtained human DNA (Fig. 3A). Genotyping was possible for all methods of DNA preparation except for Tris-neutralized NaOH modified RMS (lane 8) and DNA from cells suspended in TKM and prepared by the BuccalAmp method (lane 11). The strongest signals from a 9μl sample were apparent in BuccalAmp prepared DNA (lane 10), followed by RMS DNA in pH7.6 buffer (lane 5), DNA from cells suspended in Epicentre buffer and prepared by RMS (lane 10), and NaOH modified RMS (lane 9), although significant smearing was seen with this last sample. The first group of laboratory volunteers appear to have all been either homozygous `s/s' or `l/l'. Additional volunteers were recruited, DNA prepared by RMS (pH7.6) and subject to HTTLPR genotyping to confirm that preparation methods had not been introducing PCR artifacts that would resemble only homozygous DNA. One additional volunteer's DNA was heterozygous `s/l' (Fig. 3B, lane 5). Finally, to compare the results of the RMS to more commonly-used DNA derived from blood samples, we performed HTTLPR genotyping PCR on three volunteer samples, each previously identified as l/l, s/s, or s/l genotype (Fig. 3C). In all three cases, as expected, results from buccal DNA were similar to those had from blood DNA.
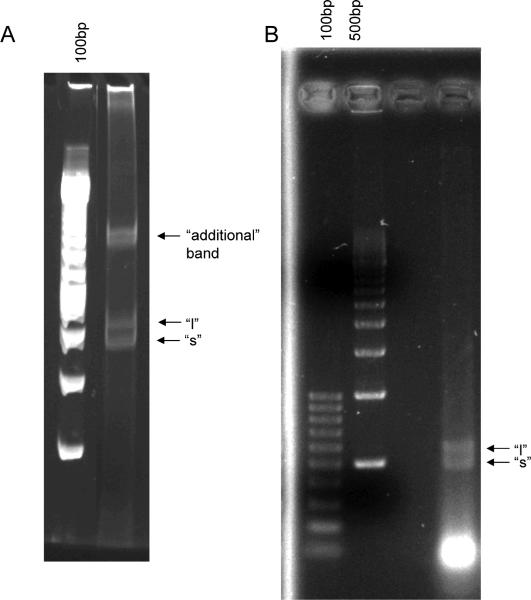
Fig. 3. HTTLPR genotyping PCR of DNA generated by different extraction methods.
HTTLPR genotyping PCR was carried out on DNA samples generated by different extraction/purification methods as described in the text. A volume of 9μl of each sample was used. DNA bands associated with the `s' and `l' alleles and the `additional' band associated with the `s/l' heterozygote are indicated. A: Lane 1: 100bp ladder; lanes 2-10: DNA prepared by the RM (2), RMS (3-7), Epicentre BuccalAmp (8), Epicentre buffer followed by RMS (9), or TKM followed by BuccalAmp heat and vortex method (10). B: Genotyping PCR carried out on additional lab samples to verify heterozygote in RMS DNA. Lane 1: 100bp ladder; lane 2 empty; lane 3: DNA prepared by RM; lanes 4-5: DNA prepared by RMS, pH 7.6 buffer. Positions of the `s' and `l' allele bands are indicated. C: Genotyping PCR carried out in parallel on DNA extracted from swabs via RMS and from whole blood via RM. Lane 1: 100bp ladder; 2, 5: DNA from volunteer `A'; 3, 6: DNA from volunteer `B'; 7, 9: DNA from volunteer `C'. Lanes 2-4 were prepared from buccal swab by RMS. Lanes 5-7 were prepared from whole blood by RM. Genotypes are indicated.
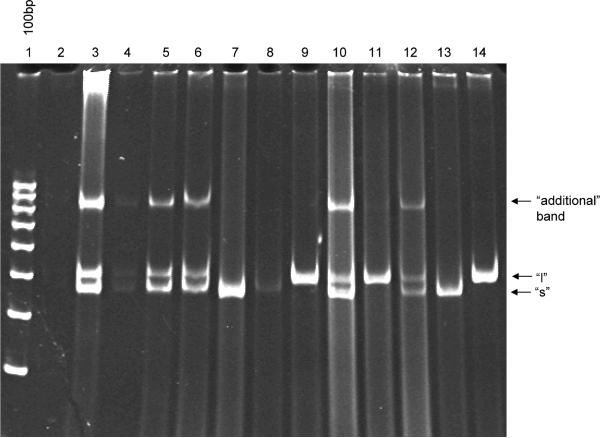
A slow-migrating `additional band' in the heterozygote DNA (approximately 900bp-1kb) appears to be a conformational DNA state composed of a mixture of `s' and `l' bands, and it has appeared whenever heterozygote genotypes show up for other HTTLPR studies by our group, whether the DNA is prepared by either RMS (Fig. 3B lane 5 and 3C lane 6), by RM (Fig. 3C lane 3), or in our previous work (Hayden et al., 2007) by the BuccalAmp method (Fig. 4, lanes 3, 4, 5, 6, 10, 12). To confirm that this `additional' band was conformational and not due to generation of an additional linear PCR fragment, genotyping PCR was performed on confirmed heterozygous (prepared by RMS) and subsamples were run on 5% TAE-PAGE and 2.5% TAE-agarose. The `additional' band only was visible on the TAE-PAGE gel (Fig. 5A) and appears to have resolved into `s' and `l' components on agarose (Fig. 5B) gel.
Fig. 4. Presence of `additional' (~900bp) band in TAE-PAGE of HTTLPR heterozygotes prepared by BuccalAmp method.
Multiple different human DNA samples were prepared by BuccalAmp method as described previously (Hayden et al., 2007). Samples were run on 4.5% native TAE-PAGE. Genotypes of `s/s', `l/l', and `s/l' appeared in the sample. An `additional' band of approximately 900bp appeared in all and was unique to `s/l' samples.
Fig. 5. Determination of the nature of a high molecular weight `band' by native acrylamide and agarose electrophoresis.
Heterozygous DNA was used for HTTLPR genotyping and subject to electrophoresis on `A'; nondenaturing TAE-PAGE, `B'; TAE agarose. An approximately 900bp band appeared only on the native TAE-PAGE gel. This band did not appear on the agarose gel.
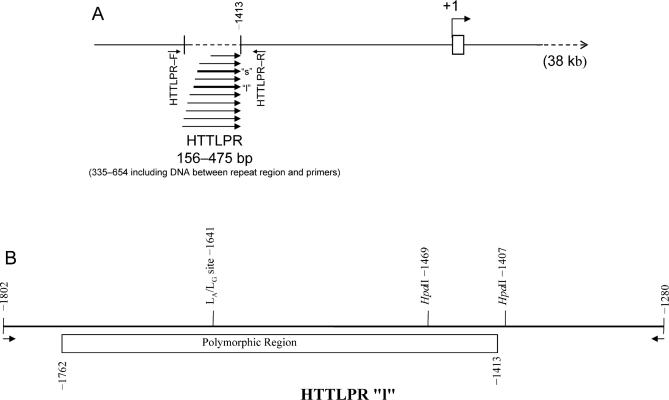
We also evaluated suitability of DNA generated by PCR using 7-deaza dGTP for LA/LG genotyping. To this end, we digested PCR product with HpaII. The LG genotype generates an additional `GGCC' recognition sequence for this restriction enzyme (Fig. 6). Potential full and partial digestion bands are summarized in Table 3. We determined, from the presence of several partial digestion bands, that 7-deaza dGTP interferes with HpaII digestion; however, PCR was carried out with a mixture of dGTP and 7-deaza dGTP, permitting sufficient digestion of products (Fig. 7) to determine that our example did not have the `LG' genotype.
Fig. 6. HTTLPR `l' and variant illustrating location of HpaII sites and the LA/LG site.
A. Location of the HTTLPR polymorphic region in relationship to the +1 transcription start site of the SLC6A4 gene, diagram based on published sequences (Lesch et al., 1994) and Nakamura et al (Nakamura et al., 2000). Sequences of multiple `l' variant alleles (Nakamura et al., 2000) were analyzed for the presence of HpaII sites. The LG polymorphic site (which creates an additional HpaII site) is also indicated.
Table 3.
DNA sizes resulting from HpaII digestion of HTTLPR genotyping PCR with 7-deaza dGTP.
Indicates a partial digestion product.
Fig. 7. Use of RMS-prepared DNA for HTTLPR LA/LG screening.
HTTLPR genotyping PCR was carried out on DNA samples generated by RMS as described in the text. A volume of 9μl of sample previously determined to be l/l genotype was used. PCR was followed by overnight (16hr) digestion of reaction products with HpaII restriction enzyme. Both full and partial digestion products were present. Sample was LA/LA, since none of the potential LG digestion products (dashed-line arrows) appeared.
Discussion
Establishing the effects of genetic variation on etiology of neuropsychiatric disorders often requires hundreds or thousands of subjects. This is due in part to the complexity of linkage between genetic variation and development of a neuropsychiatric disorder. For example, the influence of the HTTLPR polymorphism on affective disorders has been both supported (Cervilla et al., 2006) and rejected (Willis-Owen et al., 2005), and the field recognizes the need for larger sample sizes to make clinically valid determinations regarding the importance of this polymorphism (Stein et al., 2006). In addition, some potentially important genetic polymorphisms may be sufficiently rare that significant effects can only be determined with large samples. A need for larger sample sizes would be compounded further if interaction of multiple polymorphisms provides a specific risk, such as interaction between promoter polymorphisms in the APOE gene and the APOEε4 allele (Parker et al., 2005). With large samples, reducing the cost of individual DNA preparations while preserving quality and utility is an important consideration.
Collection of original materials (swab or whole blood) under good conditions is important for high quality DNA extraction. It is also important to emphasize that stability and integrity of genomic DNA depend on the storage conditions, such as temperature, freeze-thaw cycles and buffer composition. Although we have not studied the various parameters affecting stability of DNA prepared by the RMS procedure, we have previously tested different conditions of storage and stability for DNA prepared by RM (Lahiri and Schnabel, 1993). In short, DNA samples remained intact and undegraded for a long period of time when DNA was dissolved in higher concentrations of EDTA.
Ideally, tissue sampling should be non-invasive and painless. These concerns are especially critical in certain populations, such children and psychiatric groups. Preparation of DNA from these samples also needs to be rapid, reliable and consistent. In any large scale genetic association project, per-unit cost of DNA preparation becomes a significant issue. A kit such as the BuccalAmp extraction kit has a retail cost of $2.48 per preparation (based on information available at http://www.epicentre.com/ as of January 18, 2007). Time investment is approximately five minutes per preparation batch, with an added technical demand of vortexing to specific times.
We have determined that our RMS (pH 7.6 buffer) produces DNA of greater purity as measured by spectrophotometry than that produced by BuccalAmp. We chose spectrophotometry over a method such as direct DNA sequencing because optical measurement is a direct measurement of the constituents of a sample, and impure DNA will produce a markedly different overall spectrum than does high-quality material. While it is true that sequencing is sensitive to contaminants, it is also true that samples such traditional alkaline lysis miniprep DNA can be sequenced, indicating some tolerance for impurities in the process. While sequencing might indicate a `good enough' DNA sample for most purposes, it still would not provide as precise an indication of sample purity as would an optical spectrum.
DNA produced by the RMS is useful for experimental, and potentially clinical, genotyping, including for both the HTTLPR `s' and `l' genotype and the LA/LG genotype via PCR or PCR followed by restriction enzyme digest, respectively. While other methods for genotyping, such as Illumina, Affymetrix SNP chips, or TaqMan assays are effective high-throughput methods, we intentionally restricted our analysis to lower front-end cost methods such as repeat-length PCR and PCR followed by restriction digest. In addition, methods such as Affymetrix SNP chip do not discern the HTTLPR polymorphism. Moreover, the DNA sample generated by RMS was high molecular weight. Should a specific application require a `slightly degraded' sample, random shearing could be induced by simple (and low-cost) vortexing.
The RMS requires a sterile swab ($0.24 each, Fisher Scientific) and approximately $0.05 worth of common laboratory chemicals (Tris-HCl, EDTA, MgCl2, KCl, NaCl, Triton X-100, and SDS). Time requirements are estimated at 20 minutes per preparation batch, not including a 60-minute incubation at 65°C, during which time personnel could pay attention to other tasks. In addition to HTTLPR genotyping, DNA isolated by the RMS procedure can be utilized to study genetic polymorphism of other important genes, such as APOE and the β-amyloid precursor protein (APP).
Acknowledgements
This work was supported by grants from the Alzheimer's association and NIH (AG18379 and AG18884) to DKL.
Abbreviations
- SLC6A4
serotonin transporter gene
- 5-HTT
serotonin transporter gene (alternate abbreviation)
- LOAD
late-onset Alzheimer's disease
- AD
Alzheimer's disease
- APOE
apolipoprotein E gene
- RM
rapid method for DNA purification from blood
- RMS
rapid method for swab
- IRB
institution review board
- TSS
transcription start site
- TAE
tris-acetate EDTA
- TBE
tris-borate EDTA
- PAGE
polyacrylamide gel electrophoresis
REFERENCES
- Becker K, El-Faddagh M, Schmidt MH, Laucht M. Is the serotonin transporter polymorphism (5-HTTLPR) associated with harm avoidance and internalising problems in childhood and adolescence? J Neural Transm. 2007;114:395–402. doi: 10.1007/s00702-006-0577-4. [DOI] [PubMed] [Google Scholar]
- Brocke B, Armbruster D, Muller J, Hensch T, Jacob CP, Lesch KP, et al. Serotonin transporter gene variation impacts innate fear processing: acoustic startle response and emotional startle. Mol Psychiatry. 2006;11:1106–1112. doi: 10.1038/sj.mp.4001908. [DOI] [PubMed] [Google Scholar]
- Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, et al. A polymorphism in the regulatory region of APOE associated with risk for Alzheimer's dementia. Nat Genet. 1998;18:69–71. doi: 10.1038/ng0198-69. [DOI] [PubMed] [Google Scholar]
- Cervilla JA, Rivera M, Molina E, Torres-González F, Bellón JA, Moreno B, et al. The 5-HTTLPR s/s genotype at the serotonin transporter gene (SLC6A4) increases the risk for depression in a large cohort of primary care attendees: The PREDICT-gene study. Am J Med Genet B Neuropsych Genet. 2006;141B:912–917. doi: 10.1002/ajmg.b.30455. [DOI] [PubMed] [Google Scholar]
- Covault J, Tennen H, Armeli S, Conner TS, Herman AI, Cillessen AH, et al. Interactive effects of the serotonin transporter 5-HTTLPR polymorphism and stressful life events on college student drinking and drug use. Biol Psychiatry. 2007;61:609–616. doi: 10.1016/j.biopsych.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Eichhammer P, Langguth B, Wiegand R, Kharraz A, Frick U, Hajak G. Allelic variation in the serotonin transporter promoter affects neuromodulatory effects of a selective serotonin transporter reuptake inhibitor (SSRI) Psychopharmacology (Berl) 2003;166:294–297. doi: 10.1007/s00213-002-1370-1. [DOI] [PubMed] [Google Scholar]
- Farlow MR, Lahiri DK, Poirier J, Davignon J, Hui S. Apolipoprotein E genotype and gender influence response to tacrine therapy. Ann N Y Acad Sci. 1996;802:101–110. doi: 10.1111/j.1749-6632.1996.tb32603.x. [DOI] [PubMed] [Google Scholar]
- Finger EC, Marsh AA, Buzas B, Kamel N, Rhodes R, Vythilingham M, et al. The Impact of Tryptophan Depletion and 5-HTTLPR Genotype on Passive Avoidance and Response Reversal Instrumental Learning Tasks. Neuropsychopharmacology. 2006;32:206–215. doi: 10.1038/sj.npp.1301182. [DOI] [PubMed] [Google Scholar]
- Gerra G, Zaimovic A, Garofano L, Ciusa F, Moi G, Avanzini P, et al. Perceived parenting behavior in the childhood of cocaine users: Relationship with genotype and personality traits. Am J Med Genet B Neuropsych Genet. 2007;144B:52–57. doi: 10.1002/ajmg.b.30388. [DOI] [PubMed] [Google Scholar]
- Grime SK, Martin RL, Holaway BL. Inhibition of restriction enzyme cleavage of DNA modified with 7-deaza-dGTP. Nucleic Acids Res. 1991;19:2791. doi: 10.1093/nar/19.10.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EP, Dougherty LR, Maloney B, Durbin CE, Olino TM, Nurnburger JI, et al. Temperamental fearfulness in childhood and the serotonin transporter promoter region polymorphism: a multimethod association study. Psychiatr Genet. 2007;17:135–142. doi: 10.1097/YPG.0b013e3280147847. [DOI] [PubMed] [Google Scholar]
- Heiser P, Dempfle A, Friedel S, Konrad K, Hinney A, Kiefl H, et al. Family-based association study of serotonergic candidate genes and attention-deficit/hyperactivity disorder in a German sample. J Neural Transm. 2007;114:513–521. doi: 10.1007/s00702-006-0584-5. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilveskoski E, Lehtimaki T, Erkinjuntti T, Koivula T, Karhunen PJ. Rapid apolipoprotein E genotyping from mailed buccal swabs. J Neurosci Methods. 1998;79:5–8. doi: 10.1016/s0165-0270(97)00157-x. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Yang BZ, Douglas-Palumberi H, Crouse-Artus M, Lipschitz D, Krystal JH, et al. Genetic and environmental predictors of early alcohol use. Biol Psychiatry. 2007;61:1228–1234. doi: 10.1016/j.biopsych.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Kim H, Lim S-W, Kim S, Kim J, Chang YH, Carroll BJ, et al. Monoamine Transporter Gene Polymorphisms and Antidepressant Response in Koreans With Late-Life Depression. JAMA. 2006;296:1609–1618. doi: 10.1001/jama.296.13.1609. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Bye S, Nurnberger JI, Jr., Hodes ME, Crisp M. A non-organic and non-enzymatic extraction method gives higher yields of genomic DNA from whole-blood samples than do nine other methods tested. J Biochem Biophys Methods. 1992;25:193–205. doi: 10.1016/0165-022x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Nurnberger JI., Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Sambamurti K, Bennett DA. Apolipoprotein gene and its interaction with the environmentally driven risk factors: molecular, genetic and epidemiological studies of Alzheimer's disease. Neurobiol Aging. 2004;25:651–660. doi: 10.1016/j.neurobiolaging.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Schnabel B. DNA isolation by a rapid method from human blood samples: effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochem Genet. 1993;31:321–328. doi: 10.1007/BF02401826. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Balling U, Gross J, Strauss K, Wolozin BL, Murphy DL, et al. Organization of the human serotonin transporter gene. J Neural Transm Gen Sect. 1994;95:157–162. doi: 10.1007/BF01276434. [DOI] [PubMed] [Google Scholar]
- Li T, Sham PC, Vallada H, Xie T, Tang X, Murray RM, et al. Preferential transmission of the high activity allele of COMT in schizophrenia. Psychiatr Genet. 1996;6:131–133. doi: 10.1097/00041444-199623000-00005. [DOI] [PubMed] [Google Scholar]
- Little KY, Zhang L, Cook E. Fluoxetine-induced alterations in human platelet serotonin transporter expression: serotonin transporter polymorphism effects. J Psychiatry Neurosci. 2006;31:333–339. [PMC free article] [PubMed] [Google Scholar]
- Mannelli P, Patkar AA, Peindl K, Tharwani H, Gopalakrishnan R, Hill KP, et al. Polymorphism in the serotonin transporter gene and moderators of prolactin response to meta-chlorophenylpiperazine in African-American cocaine abusers and controls. Psychiatry Res. 2006;144:99–108. doi: 10.1016/j.psychres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Marsh AA, Finger EC, Buzas B, Soliman N, Richell RA, Vythilingham M, et al. Impaired recognition of fear facial expressions in 5-HTTLPR S-polymorphism carriers following tryptophan depletion. Psychopharmacology (Berl) 2006;189:387–394. doi: 10.1007/s00213-006-0581-2. [DOI] [PubMed] [Google Scholar]
- Mergen H, Karaaslan C, Mergen M, Deniz Ozsoy E, Ozata M. LEPR, ADBR3, IRS-1 and 5-HTT genes polymorphisms do not associate with obesity. Endocr J. 2007;54:89–94. doi: 10.1507/endocrj.k06-023. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ueno S, Sano A, Tanabe H. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol Psychiatry. 2000;5:32–38. doi: 10.1038/sj.mp.4000698. [DOI] [PubMed] [Google Scholar]
- Nilsson KW, Damberg M, Ohrvik J, Leppert J, Lindstrom L, Anckarsater H, et al. Genes encoding for AP-2[beta] and the Serotonin Transporter are associated with the Personality Character Spiritual Acceptance. Neurosci Lett. 2007;411:233–237. doi: 10.1016/j.neulet.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Parker GR, Cathcart HM, Huang R, Lanham IS, Corder EH, Poduslo SE. Apolipoprotein gene E4 allele promoter polymorphisms as risk factors for Alzheimer's disease. Psychiatr Genet. 2005;15:271–275. doi: 10.1097/00041444-200512000-00009. [DOI] [PubMed] [Google Scholar]
- Putzhammer A, Schoeler A, Rohrmeier T, Sand P, Hajak G, Eichhammer P. Evidence of a role for the 5-HTTLPR genotype in the modulation of motor response to antidepressant treatment. Psychopharmacology (Berl) 2005;178:303–308. doi: 10.1007/s00213-004-1995-3. [DOI] [PubMed] [Google Scholar]
- Rogers NL, Cole SA, Lan HC, Crossa A, Demerath EW. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol. 2007;19:319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiser JP, Rogers RD, Cook LJ, Sahakian BJ. The effect of polymorphism at the serotonin transporter gene on decision-making, memory and executive function in ecstasy users and controls. Psychopharmacology (Berl) 2006;188:213–227. doi: 10.1007/s00213-006-0495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CJ, de Milander L, Hew-Butler T, Xenophontos SL, Cariolou MA, Anastassiades LC, et al. Dipsogenic genes associated with weight changes during Ironman Triathlons. Hum Mol Genet. 2006;15:2980–2987. doi: 10.1093/hmg/ddl240. [DOI] [PubMed] [Google Scholar]
- Segal J, Pujol C, Birck A, Gus Manfro G, Leistner-Segal S. Association between suicide attempts in south Brazilian depressed patients with the serotonin transporter polymorphism. Psychiatry Res. 2006;143:289–291. doi: 10.1016/j.psychres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Smeraldi E, Serretti A, Artioli P, Loranzi C, Catalano M. Serotonin transporter genelinked polymorphic region: possible pharmacogenetic implications of rare variants. Psychiatr Genet. 2006;16:153–158. doi: 10.1097/01.ypg.0000218611.53064.a0. [DOI] [PubMed] [Google Scholar]
- Stein MB, Seedat S, Gelernter J. Serotonin transporter gene promoter polymorphism predicts SSRI response in generalized social anxiety disorder. Psychopharmacology (Berl) 2006;187:68–72. doi: 10.1007/s00213-006-0349-8. [DOI] [PubMed] [Google Scholar]
- Taylor SE, Way BM, Welch WT, Hilmert CJ, Lehman BJ, Eisenberger NI. Early Family Environment, Current Adversity, the Serotonin Transporter Promoter Polymorphism, and Depressive Symptomatology. Biological Psychiatry. 2006;60:671–676. doi: 10.1016/j.biopsych.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Viet CT, Jordan RC, Schmidt BL. DNA promoter hypermethylation in saliva for the early diagnosis of oral cancer. J Calif Dent Assoc. 2007;35:844–849. [PubMed] [Google Scholar]
- Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect. 1999;107:517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis-Owen SA, Turri MG, Munafo MR, Surtees PG, Wainwright NW, Brixey RD, et al. The serotonin transporter length polymorphism, neuroticism, and depression: a comprehensive assessment of association. Biol Psychiatry. 2005;58:451–456. doi: 10.1016/j.biopsych.2005.04.050. [DOI] [PubMed] [Google Scholar]
- Zalsman G, Huang Y-y, Oquendo MA, Burke AK, Hu X-z, Brent DA, et al. Association of a Triallelic Serotonin Transporter Gene Promoter Region (5-HTTLPR) Polymorphism With Stressful Life Events and Severity of Depression. Am J Psychiatry. 2006;163:1588–1593. doi: 10.1176/ajp.2006.163.9.1588. [DOI] [PubMed] [Google Scholar]