Abstract
Parathyroid hormone-related protein (PTHrP) is an autocrine/paracrine factor produced by breast cancer cells that is speculated to play a major role in permitting breast cancer cells to grow into the bone microenvironment by stimulating the bone resorption axis. It has been previously shown that EGFR signaling induces the production of PTHrP in several primary and transformed epithelial cell types. Therefore, we investigated the relationship between EGFR and PTHrP gene expression in human breast cancer cells. Of a panel of 7 breast epithelial and cancer cell lines, the osteolytic, EGFR- positive lines (MDA-MB-231 and NS2T2A1) exhibited higher levels of PTHrP transcript expression. Amphiregulin mRNA levels in all lines were approximately 2 orders of magnitude higher than those of TGFα or HBEGF. In the EGFR bearing lines, the receptor was phosphorylated at tyrosine 992 under basal conditions, and the addition of 100 nM amphiregulin did not lead to the phosphorylation of other tyrosine residues typically phosphorylated by the prototypical ligand EGF. Treatment of the EGFR positive lines with the EGFR inhibitor PD153035 and amphiregulin-neutralizing antibodies reduced PTHrP mRNA levels by 50–70%. Stable EGFR expression in the MCF7 line failed to increase basal PTHrP mRNA levels; however, treatment of this cell line with exogenous EGF or amphiregulin increased PTHrP transcription 3-fold. Transient transfection analysis suggests that the MAPK pathway and ETS transcription factors mediate EGFR coupling to PTHrP gene expression. Taken together, it appears that autocrine stimulation of EGFR signaling by amphiregulin is coupled to PTHrP gene expression via EGFR Tyr992 and MAPK, and that this pathway may contribute to PTHrP expression by breast tumor cells.
Keywords: Amphiregulin, Breast cancer, Epidermal growth factor receptor (EGFR), Parathyroid hormone-related peptide (PTHrP)
Introduction
Signaling by parathyroid hormone-related protein (PTHrP) and the PTH/PTHrP receptor (PPR) has been recognized to play a major role in mammary gland development, physiology and neoplasia. PTHrP is expressed in the breast epithelia and induces glandular morphogenesis during early development, modulates branching morphogenesis during puberty and mobilizes calcium from the bone during lactation [1, 2]. Evidence from animal models of breast cancer metastasis to bone suggests that PTHrP-PPR signaling participates in a vicious cycle of tumor/bone cell interactions. PTHrP derived from tumor cells stimulates the PPR on cells of the osteoblast lineage. This leads to the recruitment and activation of osteoclasts that resorb bone and release bound growth factors such as TGFβ. The released growth factors stimulate the proliferation of tumor cells, as well as their expression of additional PTHrP [3].
Controversy has emerged regarding how PTHrP expression influences breast cancer progression. The earliest studies evaluating breast cancer metastases revealed a striking dichotomy of PTHrP expression in breast cancer tumors in the bone as compared to other sites. Over 90% of tumors in the bone expressed high levels of PTHrP antibody immunoreactivity or mRNA, whereas only 20% of metastases to visceral sites exhibited detectable PTHrP expression [4, 5]. Studies using protein detection methods have indicated that PTHrP is expressed in a large fraction (∼60–80%) of primary tumors, but a substantial subset of the tumors lack detectable levels of this factor [6, 7]. At this juncture, it is not clear whether primary tumors with high levels of PTHrP expression are more likely to metastasize to bone or whether PTHrP gene expression in the primary tumor is irrelevant because the PTHrP gene expression is activated in the bone microenvironment. Two retrospective studies using immunohistochemistry and RTPCR, concluded that primary tumors with higher PTHrP gene expression were more likely to produce bone metastasis [6, 7]. In contrast, a very large prospective study found that primary tumors with greater PTHrP immunoreactivity were associated with better prognosis, whereas the nonimmunoreactive tumors were more likely to generate metastases to all sites, including bone [8, 9]. This second set of studies raises the possibility that PTHrP expression may confer on the tumor cell a less invasive phenotype outside of the bone microenvironment [9]. A greater understanding of the factors that control the PTHrP gene expression might provide insight into the role of the peptide at different stages of breast cancer progression.
There is an emerging understanding of tumor cell regulation of PTHrP gene expression both within and outside the bone microenvironment. The human PTHrP gene is a complex transcriptional unit driven by 3 promoters and is alternatively spliced in the 5′ and 3′ regions of the transcript. Promoter activity appears to account for regulation of PTHrP gene expression in a great variety of normal and cancer cell types. However, PTHrP gene expression can also be regulated at the level of mRNA stability [10]. In both breast cancer tumors and cell lines, activity of the PTHrP-P3 promoter accounts for the bulk of the transcripts expressed [6, 11]. The activity of this promoter in all cell lines studied to date is dependent on an ETS binding site that lies ∼50 bp upstream of the P3 promoter [11-14]. Ets family transcription factors activate PTHrP gene expression in several breast epithelial and cancer cell lines and play a role in TGF-β mediated activation of the gene [11, 13, 15].
Growth factor signaling mediated through the mitogen activated protein kinase pathway (MAPK) leads to Ets factor mediated induction of PTHrP gene expression [16]. For example, the epidermal growth factor receptor (EGFR), whose coupling to MAPK has been extensively characterized, activated PTHrP gene expression via the Ets binding site upstream of the P3 promoter in human keratinocytes [14]. Peptide growth factor agonists for the EGFR include epidermal growth factor (EGF), transforming growth factor α (TGFα,) amphiregulin (AREG), heparin-binding EGF-like growth factor (HB-EGF), betacellulin (BTC), epiregulin (EPR) and epigen (EPG) [17]. Consequently, exogenous application of EGF and TGFα activated PTHrP gene expression in many types of epithelial cells [18-21]. Indeed, autocrine expression of AREG and consequent stimulation of EGFR signaling appears to account for the high levels of PTHrP produced by cultured keratinocytes [14]. EGFR is expressed in ∼60% of breast cancer tumors [22, 23] and breast tumor cells frequently express growth factors that stimulate EGFR signaling [22, 24-27]. Taken together these observations suggest ligand-induced EGFR signaling may regulate PTHrP gene expression in some breast cancers.
In this manuscript, we report that elevated levels of AREG and EGFR were observed in human breast epithelial and cancer cell lines that have a capacity to form lytic bone lesions in animal models. Moreover, inhibition of EGFR signaling reduced PTHrP expression and enhanced EGFR signaling increased PTHrP expression in these cell lines. Thus, these results suggest that AREG-induced EGFR signaling regulates PTHrP expression in breast cancers.
Materials and methods
Cell lines and cell culture
MDA-MB-231 and MCF7 cells were grown in DMEM/P (Sigma) supplemented with 10% FBS and 10 ng/ml insulin. The MCF/LXSN and MCF7/EGFR were grown in MEM (Mediatech) supplemented with 10% FBS, 10 ng/ml insulin, and 100 mg/ml G418 (Mediatech). The A1 and S1 cell lines were grown in a 50:50 mixture of RPMI and DMEM:F12 (Sigma) supplemented with 10% FBS. All cell lines were grown to confluence (plus one day) for the all RNA and protein expression experiments.
Pharmacologic reagents
Epidermal Growth Factor (EGF) and staurosporine was purchased from Sigma (St. Louis, Mo). Human recombinant AREG was purchased from R&D Systems. The EGFR tyrosine kinase inhibitor PD153035 was purchased from Tocris (United Kingdom). The anti-AREG goat antibody and the control, goat IgG, were all purchased from R&D Systems and used at a concenctration of 1 μg/ml.
AREG, HB-EGF, TGFα Elisa Assays
MDA, A1, and S1 cells were grown to confluency in a 12-well dish and then serum starved for 24 h. The conditioned media was collected and PMSF was added to a final concentration of 1 mM. Samples were cleared by centriguation for 10 min at 4°C. The AREG, HB-EGF, or TGFα concentrations were determined using the DuoSet ELISA kit and the instructions of the manufacturer (R&D Systems).
PTHrP Protein Analysis
S1 cells were grown to confluency in a 48-well dish and then serum starved for 16 h. 10 nM EGF or 100 nM AREG was added for 48 h. The culture medium was collected and frozen at −80 C. The cells were harvested by trypsinization. After staining the nonviable cells with trypan blue, the viable cells were counted on a hemacytometer. The PTHrP concentration in the culture medium was assayed according to the instructions of the manufacturer (Diagonstic Systems Laboratories, Inc.).
Analyses of EGFR phosphorylation and expression
We adapted previously published procedures for assaying ligand stimulation of EGFR tyrosine phosphorylation [28]. Briefly, cells were plated on a 100 mm dish and grown to confluence. The cells were incubated on ice for 30 min. The cells were then washed with ice cold PBS and treated with ligand for 7 min. The ligand containing media was aspirated and isotonic EBC (50 mM Tris pH 7.4, 120 mM NaCl, 0.5% NP40) lysis buffer was added. The cells were scraped into microcentrifuge tubes and were lysed on ice. The lysates were cleared by centrifugation and transferred to a fresh tube.
Following ligand stimulation, EGFR expression and tyrosine phosphorylation were analyzed essentially as described previously [28]. Briefly, concanavalin A-sepharose beads were used to precipitate glycoproteins (which include ErbB receptors) from cell lysates. The precipitates were resolved by SDS-PAGE on a 7.5% polyacrylamide gel and the resolved samples were electrotransferred to PVDF membrane (BioRad). The blots were probed using an anti-phosphotyrosine mouse monoclonal antibody (Upstate Biotechnology) or an anti-phosphospecific rabbit polyclonal antibodies (Cell Signaling Technology). Primary antibody binding was detected using a goat anti-mouse or goat anti-rabbit antibody conjugated to horseradish peroxidase (Kirkegarrd and Perry Labs Inc) and enhanced chemiluminescence (Santa Cruz). The immunoblots were then stripped and reprobed with an anti-EGFR rabbit polyclonal antibody (Santa Cruz) and detected as described above.
RNA isolation and quantitative real-time reverse transcription PCR (Q-RT-PCR)
Total RNA was prepared using the mini RNA isolation II kit from Zymo Research Corporation according to the manufacture's instructions. Reverse transcription (RT) of total RNA was performed in a final volume of 50 μl. Total RNA (1.5 μg) was treated with RQ1 DNase (Promega Corp.) in 1 × PCR puffer, 5 mM MgCl2, 1 mM dNTP, and 1 U RNasin inhibitor (Promega Corp.) at 37°C for 30 min prior to first strand cDNA synthesis. The DNase-treated total RNA was subjected to reverse transcription using 1 U Moloney murine leukemia virus (MuLV) reverse transcriptase and 2 μM random hexamers (Applied Biosystem) at 42°C for 30 min. The reverse transcriptase was inactivated by heating at 75°C for 5 min and cooling at 4°C for 5 min.
Quantitative real-time PCR (QRT-PCR) was performed using DyNAmo HS SYBR Green qPCR master mix (New England Biolabs) according to the manufacturer's instruction employing 2 μl cDNA. PCR reactions were performed in a DNA Engine Opticon System (MJ Research Inc.). The PCR cycling comprised an initial incubation at 95°C for 15 min to activate HotStartTaq DNA polymerase and 45 cycles of denaturation at 94°C for 15 s, annealing (temperature listed in [14]) for 30 s, and extension at 72°C for 30 s. To verify the specificity of the PCR products, a melting curve analysis was routinely performed in the range of temperature from 55 to 95°C. No-template control and no RT-treated samples were included as negative controls; they did not produce significant signals.
Plasmids and Plasmid Construction
The recombinant retrovirus expression vector pLXSN/ EGFR was used as a template for site-directed mutagenesis (Stratagene) reactions to generate a mutation in which a phenylalanine codon is substituted for the tyrosine 992 codon. The sequence for the primers used to make the mutant is available upon request.
Transient transfection
Transient transfections were performed using Lipofectamine (Sigma) according to the manufacturer's instructions. About 24-h after plating 1 × 105 cells in 6-well culture dishes, MCF7 cells were about 50% confluent. They were then transfected with 1 μg of the luciferase reporter construct and 1 μg of expression vectors. Each transfection mixture also contained 5 ng cytomegalovirus (CMV)-β-galactosidase expression vector. Exogenous ligand was added 24 h post transfection. After 48 h, transfected MCF7 cells were extracted with 200 μl luciferase lysis buffer (Promega). About 20 μl of cell extract were used in a reaction mixture that contained beetle luciferin (Promega) and luciferase activity was measured in a TD-20/20 luminometer (Turner Designs). The luciferase activity was normalized to β-galactosidase activity.
Results
The expression of PTHrP, ErbB receptors and EGFR ligands in a panel of human breast cancer and epithelial cell lines
To begin studying the relationship between EGFR and PTHrP gene expression, we assembled a panel of human breast cancer cell lines. MDA-MB-231 (MDA) is a highly tumorigenic human breast tumor cell line that forms rapidly growing lytic lesions in the bones of nude mice after intracardiac injections [29]. MCF7 is a human breast tumor cell line that generally is less aggressive than MDA and has been reported to form slow growing osteolytic or osteoblastic lesions after intracardiac injections [29, 30]. S1T3 (S1) is an SV40-immortalized human breast epithelial cell line that does not form tumors in nude mice [31]. NS2T2A1 (A1) is a highly tumorigenic clone selected by multiple in vivo and in vitro passages from an SV40-immortalized human breast epithelial line that had a marginal capacity to grow in nude mice [31]. A1 forms lytic lesions in bone after intracardiac injection into nude mice (unpublished data Z.B.). BC61, BC3A, and 8701 are clones derived from a parental cell line that came from a biopsy of a ductal infiltrating carcinoma of the breast [32, 33]. Furthermore, these cells have been reported to express PTHrP and the PTHrP receptor [34, 35]. Luparello and colleagues have also grouped these cells based on their ability to invade matrigel. The BC3A are considered noninvasive and are growth stimulated by PTHrP fragments. The BC61 are highly invasive and PTHrP fragments inhibit their proliferation. The 8701-cell line is the parental cell line that the other two were derived from [35].
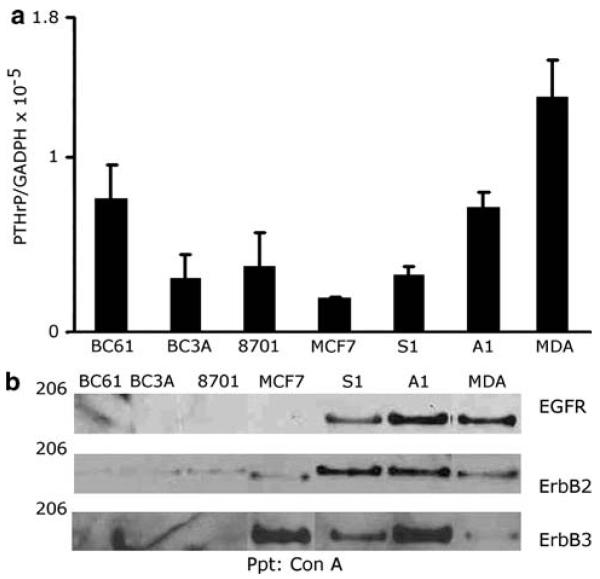
MDA cells express PTHrP mRNA and protein, which contributes to their ability to colonize bone [36]. Thus, we first examined PTHrP gene expression in our panel of cell lines in order to assess whether peptide mRNA expression correlated with the ability to form lytic lesions after intracardiac injection in nude mice. PTHrP gene expression was measured by quantitative real time reverse transcriptase polymerase chain reaction (QRTPCR). Every cell line in our panel contained PTHrP mRNA when grown under basal conditions (Fig. 1a). The osteolytic MDA and A1 cell lines exhibited slightly elevated PTHrP mRNA levels when compared to the BC3A, 8701, MCF7 and S1 cell lines. The BC61 line expressed levels of PTHrP mRNA similar to the MDA and A1 lines; however, this line does not appear to form osteolytic lesions after intracardiac injection suggesting PTHrP mRNA levels did not correlate with the growth of tumor cells in bone (JLG and JF unpublished data).
Fig. 1.
The expression of PTHrP and erbB receptors in a panel of human breast cancer cell lines. (a) Relative PTHrP mRNA levels in the various cell lines. The relative ratios of PTHrP mRNA to GADPH mRNA were expressed as a means ± s.d. of triplicate cultures from a single experiment. This experiment was repeated 3 times with similar results. (b) Cell extracts were prepared from unstimulated cells and quantified by a Bradford assay. Glyocoproteins from 500 μg of protein were precipitated with concanavalin A and analyzed. The immunoblots were probed with the corresponding antibodies. This experiment was repeated twice
Next, we evaluated the expression of EGFR, ErbB2, and ErbB3 in our panel of human breast tumor cell lines. We precipitated ErbB receptors from cell lysates using concanavalin A sepharose and we immunoblotted the resolved precipitates using specific antisera. A1 and MDA cells displayed greater EGFR expression than did the S1 cell line (Fig. 1b). In contrast, the MCF7 cell line lacked detectable EGFR expression. The two cell lines that displayed the highest amount of EGFR expression (MDA, A1) also exhibited robust PTHrP mRNA levels. Varying levels of ErbB2 and ErbB3 expression were observed in our panel of cell lines (Fig. 1b) and no correlation between PTHrP expression and the expression of these receptors could be discerned. However, it should be noted that all of the cell lines that expressed EGFR also expressed ErbB2, suggesting these receptors might possibly heterodimerize.
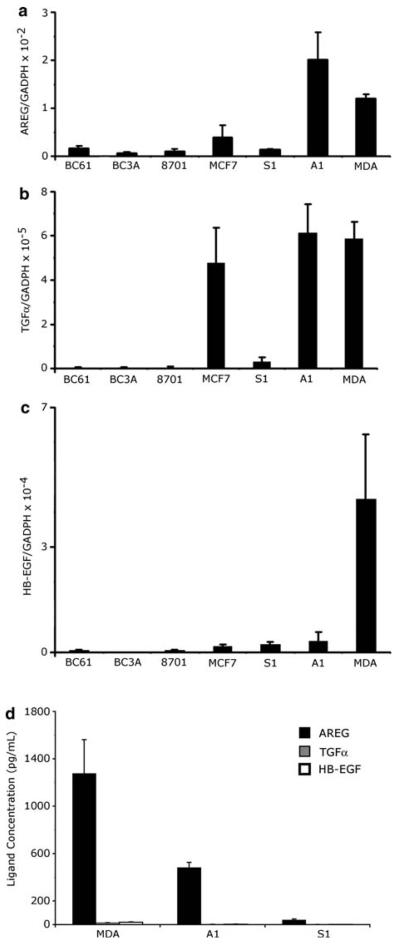
Using our panel of breast cell lines, we used QRTPCR to compare mRNA of the EGFR agonists HB-EGF, TGF-α, and AREG and ELISA assays compare the amount of secreted HB-EGF, TGF-α, and AREG. AREG transcripts in these cell lines were 100 to 1,000 times greater than HB-EGF or TGF-α transcripts and the A1 and MDA cells also contained higher levels of AREG transcripts than did the S1, MCF7, BC61 and BC3A cell lines (Fig. 2a, b, c). Moreover, the concentration of AREG secreted into the medium by the MDA, A1, and S1 cell lines was 100 to 1,000 times greater than HB-EGF or TGF-α (Fig. 2d, Table 1). Collectively, these data indicate that AREG appears to be the most abundant breast cancer associated EGFR agonist expressed by these cells.
Fig. 2.
EGF-ligand expression in a panel of human breast cancer cells. Relative AREG, TGF-α and HB-EGF mRNA levels in human breast cancer cells were measured by QRT-PCR. The relative ratios of the EGF-like ligands mRNA to GADPH mRNA levels were expressed as a means ± s.d. of triplicate cultures from a single experiment. Note the y-axis of panel (a) is 100 to a 1,000 times greater than panels (b) and (c). This experiment was repeated 3 times. (d) AREG, TGFα, and HB-EGF protein concentrations (pg/ml) were measured using sandwich Elisa assays on the media harvested from MDA, A1, and S1 cell lines and average concentrations were taken from 4 independent samples. These experiments were repeated twice
Table 1.
Protein concentration of EGFR ligands from conditioned media analyzed by ELISA
| Cell Line | AREG (pg/ml) | TGF-α (pg/ml) | HB-EGF (pg/ml) |
|---|---|---|---|
| MDA | 1276 ± 286.5 | 15 ± 1 | 20 ± 2.9 |
| A1 | 481 ± 44.2 | n.d. | n.d. |
| S1 | 51 ± 8.7 | n.d. | n.d. |
n.d.: not detectable, the detection limit is 12 pg/ml
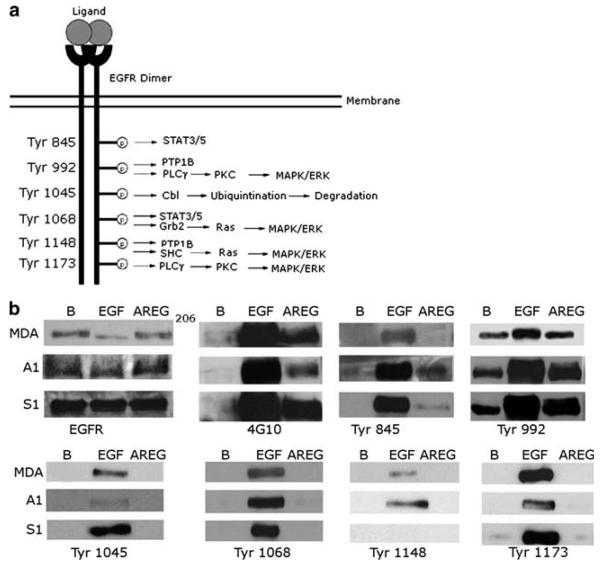
AREG stimulated a distinct pattern of EGFR tyrosine phosphorylation
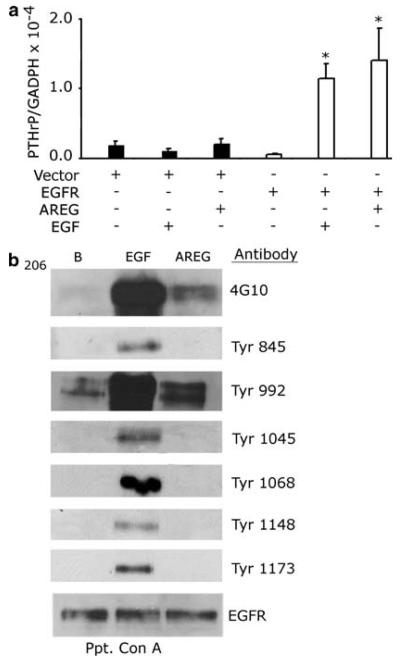
The S1, A1, and MDA cell lines exhibit EGFR expression as well as endogenous AREG expression. This suggests that these cells might display autocrine stimulation of EGFR signaling by AREG. Using phospho-specific anti-EGFR antibodies to a subset residues that are phosphorylated after EGF stimulation (Tyr845, Tyr992, Tyr1045, Tyr1068, Tyr1148 and Tyr1173), we have evaluated EGFR phosphorylation at specific tyrosine residues following mock stimulation or stimulation with EGF (10 nM) or AREG (100 nM). In the absence of ligand stimulation, EGFR phosphorylation is detected at Tyr992 in the MDA, S1 and A1 lines, despite the fact that global EGFR tyrosine phosphorylation cannot be detected using the generic anti-phosphotyrosine antibody 4G10 (Fig. 3b).
Fig. 3.
AREG stimulates a distinct pattern of EGFR tyrosine phosphorylation. The A1, S1 and MDA-MB-231 cells were stimulated with 10 nM of EGF, 100 nM of AREG, or PBS (B). Glycoproteins from the protein extracts were precipitated with concanavalin A sepharose and resolved by SDS-PAGE. The immunoblots were then probed with the corresponding antibody. This experiment was repeated three times
In general, EGF stimulated the EGFR phosphorylation at residues Tyr845, Tyr992, Tyr1045, Tyr1068, Tyr1148 and Tyr1173. Stimulation with 100 nM AREG resulted in a global increase in EGFR tyrosine phosphorylation, as detected using the generic anti-phosphotyrosine antibody 4G10 (Fig. 3b). The amount of stimulation is less that that caused by 10 nM EGF. This suggests that AREG stimulated EGFR phosphorylation on fewer EGFR tyrosine residues than does EGF. Indeed, unlike EGF, AREG did not stimulate a detectable increase in EGFR phosphorylation at Tyr1045, Tyr1068, Tyr1148, or Tyr1173 in the three cell lines. Furthermore, relative to EGF, AREG stimulated only a modest increase in EGFR phosphorylation at Tyr845 in the A1 and S1 cell lines. In contrast, AREG stimulated abundant EGFR phosphorylation of Tyr992 in the A1 and S1 cell lines and a modest increase in the MDA cell line. Thus, it is clear that EGF is more efficient at inducing phosphorylation of the EGFR tyrosine residues 845, 1045, 1068, 1148, 1173 than AREG, but both ligands induce phosphorylation of Tyr992.
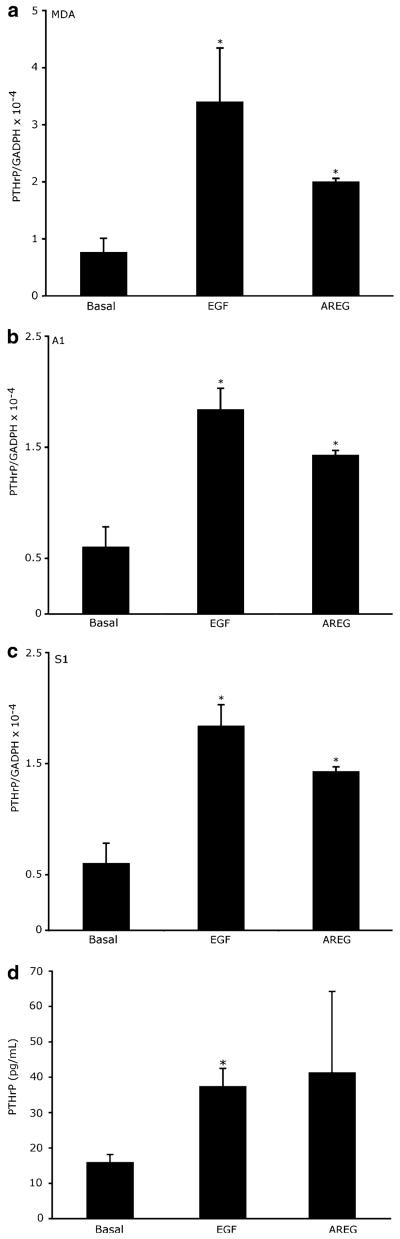
Activation of EGFR by AREG or EGF stimulates PTHrP mRNA and protein expression
Given that the S1, A1, and MDA cells display both EGFR and AREG expression, we hypothesized that treatment of these cells with the EGFR ligands AREG or EGF would stimulate PTHrP expression. As seen in Fig. 4, EGF and AREG stimulated increased PTHrP mRNA in all three cells lines. Furthermore, EGF and AREG stimulate an increase in PTHrP release into the medium by the S1 cell line (Fig. 4d).
Fig. 4.
Activation of EGFR by AREG or EGF stimulates PTHrP mRNA and protein expression. (a) MDA (b) A1, or (c) S1 cells were treated with 10 nM EGF, 100 nM AREG or PBS (Basal) for 6 h. The mRNA was harvested from three independent cultures and analyzed. This experiment was repeated 3 times. (d) S1 cells were serum starved for 16 h and then treated with 10 nM EGF, 100 nM AREG, or PBS for 48 h. The conditioned medium was collected from each culture and assayed for PTHrP concentration as described in the materials and methods. *P < 0.01 vs. basal
Inhibition of AREG and EGFR signaling decreased PTHrP mRNA expression
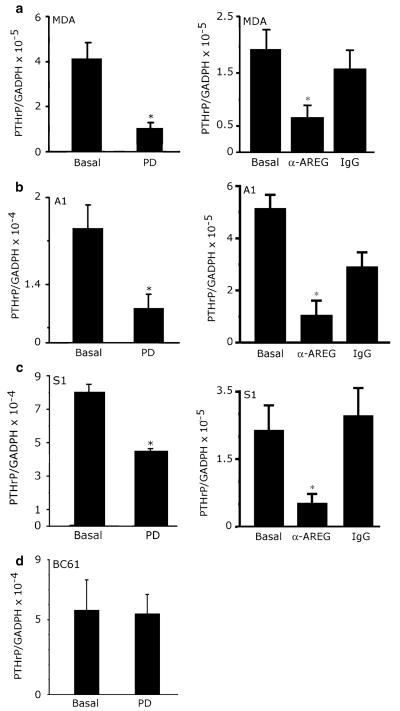
We speculated that stimulation of EGFR signaling in the A1, S1 and MDA cell lines by endogenous AREG accounts for the high levels of basal PTHrP expression displayed by these cells. We directly tested this hypothesis by examining the effect of the potent EGFR tyrosine kinase inhibitor PD153035 (PD) on PTHrP mRNA levels in the three cell lines that display EGFR expression (A1, S1, and MDA) and one cell line (BC61) that lacks EGFR expression. PD significantly and specifically inhibited expression of PTHrP mRNA in the A1, S1, and MDA cell lines (Fig. 5a, b). In contrast, PD failed to inhibit PTHrP expression in the BC61 cell line (Fig. 5d). Furthermore, a neutralizing anti-AREG antibody significantly and specifically inhibited PTHrP expression in the A1, S1, and MDA cell lines (Fig. 5a, b, c). Taken together these findings suggest that AREG induces PTHrP expression via the EGFR in the S1, A1 and MDA lines.
Fig. 5.
Inhibition of AREG and EGFR signaling decreases PTHrP expression. (a) MDA, (b) A1, (c) S1 or (d) BC61 cells were treated with 1 μM of PD for 6 h, 10 μg/ml of an AREG neutralizing antibody for 6 (a) or 24 h (b, c), or 10 μg/ml of goat IgG as a control. Messenger RNA was harvested from three independent cultures and analyzed. This experiment was repeated twice. *P < 0.01 vs. basal
Ectopic EGFR expression increased the expression of PTHrP mRNA
Because MCF7 cells display minimal EGFR expression, we hypothesized that ectopic EGFR expression in this cell line might be sufficient to induce PTHrP mRNA expression. We used infection with a recombinant retrovirus to stably express EGFR in MCF7 cells and assayed PTHrP mRNA expression in the absence of EGFR agonists or following treatment with EGF (10 nM) or AREG (100 nM). Ectopic EGFR expression was not sufficient to elevate PTHrP mRNA expression; however, AREG or EGF stimulated PTHrP expression in the MCF7/EGFR cell line but not in the vector control cell line (Fig. 6a). Moreover, the pattern of EGFR tyrosine phosphorylation displayed by the MCF7/EGFR cells after EGF and AREG stimulation was identical to the pattern observed in the A1, S1, and MDA cell lines. Specifically, EGF stimulated EGFR phosphorylation at Tyr845, Tyr992, Tyr1045, Tyr1068, Tyr1148 and Tyr1173, whereas AREG stimulated EGFR tyrosine phosphorylation primarily at Tyr992 (Fig. 6b). These findings suggested that EGFR expression in the absence of ligand stimulation is not sufficient to cause increased PTHrP mRNA expression, whereas EGFR signaling is sufficient to cause increased PTHrP mRNA expression.
Fig. 6.
Introduction of an EGFR signaling cascade increases the expression of PTHrP and phosphorylation of EGFR Tyr992. (a) MCF7 cells were infected with an LXSN vector control recombinant retrovirus of with the LXSN-EGFR recombinant retrovisur, resulting in the MCF7 vector control and MCF7/EGFR cell lines, respectively. The MCF7/EGFR and vector control cells were incubated with EGF (10 nM) or 100 nM AREG (AREG) for 6 h. The mRNA was harvested from three independent cultures. This experiment was repeated twice. (b) The cells were stimulated with 10 nM EGF or 100 nM AREG. The glycoproteins in protein extracts were precipitated with concanavalin A sepharose and resolved by SDS-PAGE. The immunoblots were then probed with the corresponding antibody. This experiment was repeated twice. *P < 0.01 vs. EGFR unstimulated
Signaling components downstream of the EGFR that activate PTHrP expression
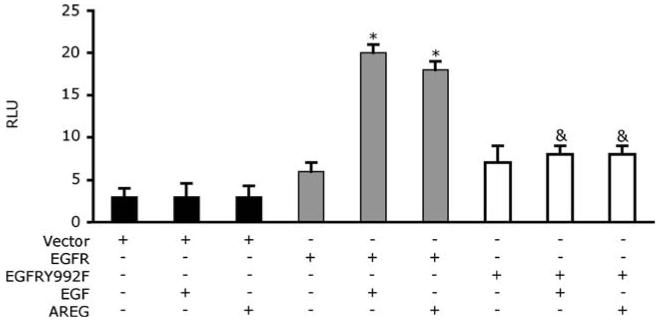
Phosphorylation of EGFR Tyr992 appears to correlate with PTHrP expression. We have examined the role of this site of EGFR phosphorylation, using an EGFR mutant in which Tyr992 is replaced with a phenylalanine residue (EGFRY992F). As shown in Fig. 7, the transient expression of wild-type EGFR in the MCF7 cell line permits AREG and EGF stimulation of the PTHrP-P3 promoter. However, transient expression of the EGFRY992F mutant failed to permit AREG or EGF stimulation of the PTHrP promoter, suggesting that phosphorylation of EGFR Tyr992 is indeed necessary for EGFR coupling to PTHrP transcription.
Fig. 7.
Mutation of Tyr residue at 992 reduces basal and stimulated PTHrP reporter gene expression. A transient transfection assay comparing the reporter activity of the vector, wild-type EGFR (EGFR), and EGFRY992F mutant transfected cells with and without the addition of AREG (100 nM) or EGF (10 nM). The luciferase activities were normalized to β-galactosidase levels and were expressed as means ± s.d. of triplicate cultures. RLU, relative luciferase units. *P < 0.01 vs. EGFR unstimulated &P < 0.01 vs. EGFR stimulated with AREG or EGF
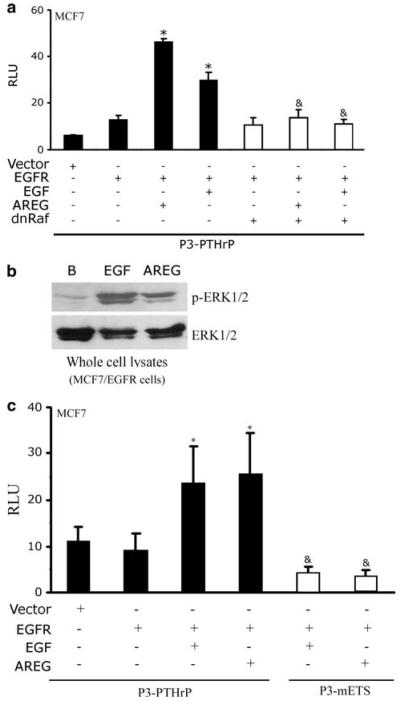
The phosphorylated Tyr992 residue has been reported to bind phospholipase Cγ (PLCγ), leading to increased PLCγ activity and activation of the MAPK pathway. Therefore, we investigated the importance of the MAPK pathway to EGFR coupling to PTHrP gene expression. As shown in Fig. 8a, transient expression of a dominant negative Raf (dnRaf) mutant [15, 37] blunted AREG or EGF stimulation of the PTHrP promoter in MCF7 cells.
Fig. 8.
Identification of signaling components downstream of EGFR that activate PTHrP gene expression. (a) and (c) Transient transfections assay comparing the reporter activity of the wild-type EGFR (WT), EGF (EGF) or AREG (AREG)-stimulated wild-type EGFR (WT) in the presence or absence of dnRaf or with a P3-mETS reporter. The luciferase activities were normalized to β-galactosidase levels and were expressed as means ± s.d. of triplicate cultures. RLU, relative luciferase units. (b) MCF7/EGFR cells were treated for 10 min with 10 nM EGF or 100 nM AREG. Whole cell lysates were harvested and resolved by SDS-PAGE. The immunoblots were then probed with the corresponding antibody. This experiment was repeated twice. *P < 0.01 vs. unstimulated EGFR &P < 0.01 vs. AREG or EGF stimulated EGFR
Signaling by the MAPK pathway is characterized by phosphorylation of ERK. Therefore, we tested whether EGFR coupling to PTHrP expression is accompanied by increased ERK phosphorylation. Stimulation of MCF7/EGFR cells with AREG (100 nM) or EGF (10 nM) resulted in increased phospho-Erk1/2 immunoreactivity detected by western blotting as compared to untreated cells (Fig. 8b).
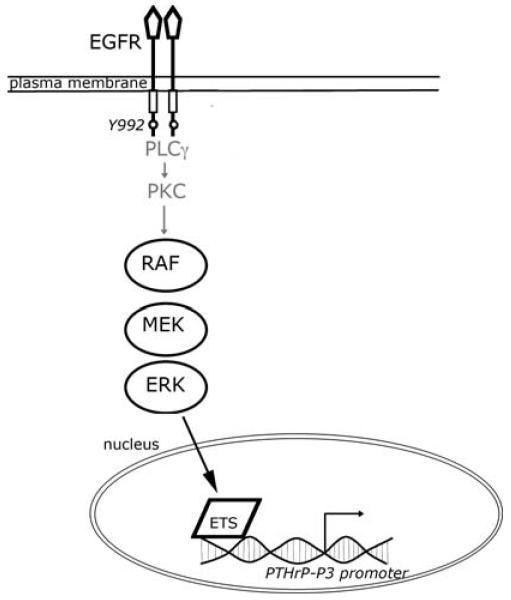
The MAPK pathway is coupled to ETS transcription factors, which in turn are critical regulators of PTHrP gene expression in breast epithelial and cancer cells [11, 15]. Mutation of the ETS site decreased basal activity of the PTHrP promoter and AREG did not stimulate transcription from this mutant promoter (Fig. 8c). Thus, an intact Ets site is required for EGFR couple to increased PTHrP gene expression. In Fig. 9, we present a model based on these findings that postulates that EGFR stimulates PTHrP gene expression through Tyr992, Raf, ERK1/2 and Ets pathway.
Fig. 9.
Proposed model of increase PTHrP expression in breast cancer cells. Depicted is a proposed model of how EGFR can stimulate PTHrP expression. Our data suggests that activation of EGFR on Tyr992 is necessary for PTHrP expression. Tyr992 has been reported to be coupled to the PLC γ and the PKC pathways but we did not evaluate this. PKC will then activated the MAPK pathway and eventually activate the Ets transcription factor. The Ets protein can then activate the transcription of the PTHrP-P3 promoter
Discussion
Our findings indicate that AREG stimulates EGFR coupling to PTHrP mRNA expression in breast epithelial and cancer cell lines. It appears that EGFR coupling is mediated by the MAPK pathway and activation of PTHrP gene expression requires the Ets site upstream of the PTHrP-P3 promoter. Thus, the EGFR-PTHrP signaling pathway in breast cells is similar to that characterized in cultured primary human keratinocytes [14]. Moreover, our results are consistent with the observation that MAPK signaling contributes to basal and TGF-β simulation of PTHrP expression in the MDA breast cancer line [38]. AREG and EGFR immuno-staining have been observed in a subset of primary breast cancers [22, 23, 27], indicating that some breast cancers may contain an autocrine signaling loop that could drive PTHrP gene expression outside the bone microenvironment. Unfortunately there have not been any systematic profiling of EGFR or AREG expression in breast cancer metastases in the bone, and this is probably due in part to infrequent resection of these tumors. However, the emerging analysis of breast cancer micro-metastases or disseminated tumor cells from the bone has indicates that ErbB2 [39] and possibly EGFR are expressed by these cells [40]. An issue that has not been clarified by these studies is whether EGFR dependent activation of PTHrP is the result of signaling from homodimers or heterodimers containing ErbB2. Collectively, these observations raise the possibility that autocrine ligand-induced-EGFR/ErbB signaling could contribute to PTHrP gene expression both within and outside of the bone microenvironment.
Here we demonstrate that endogenous and exogenous AREG stimulates a reduced level of phosphorylation of specific EGFR tyrosine residues as compared to the prototypical ligand, EGF. This observation is consistent with the concept that different ligands for the same ErbB family receptor produce differences in sites of receptor phosphorylation and this has been correlated with distinct functional outcomes. Recently, BTC has been reported to phosphorylate Y1068 residue and recruit the Grb2-MEKK1 complex to the plasma membrane, but EGF could not [41]. Chemicals that induce oxygen radicals have been reported to directly activate the EGFR resulting in phosphorylation of limited subset of tyrosine residues and receptor turnover was reduced as compared to the EGF-stimulated receptor [42]. There have been a number of reports that AREG stimulates unique biological outcomes as compared to other EGFR ligands. For example, transgenic mice that overexpress AREG in the skin display skin inflammation and articular cartilage erosion reminiscent of severe psoriasis [43], whereas analogous TGFα transgenic mice display only a modest epidermal hyperplasia [44]. Moreover, simultaneous disruption of TGFα, EGF, and AREG expression in mouse mammary glands results in less ductal growth than is observed in the mammary glands of mice lacking EGF and TGFα [45]. In MDCK cell line, AREG stimulates E-cadherin redistribution within the cell, as well as an epithelial to mesenchymal morphologic transition, neither of which occur following treatment with TGFα [46]. Finally, a recent study indicates that ectopic stimulation with AREG and endogenous AREG expression-stimulate increased motility in a mammary epithelial cell line, an effect that is not induced by ectopic EGF treatment [47]. It is tempting to speculate that differences in the sites of EGFR phosphorylation following stimulation with different EGFR agonists may account for the functional differences between these EGFR agonists.
Although we cannot rule out the possibility that AREG stimulates phosphorylation of other EGFR tyrosine residues such as Tyr974, Tyr1086, Tyr1101 and Tyr1114 [48, 49], our data highlights the importance of phosphorylation of EGFR Tyr992 in coupling EGFR to PTHrP gene expression. Phosphorylated Tyr992 binds to PLCγ, RasGAP, SHP2, and PTP1B [50-53]. PLCγ cleaves membrane phospholipids that generating bioactive fragments such as inositol triphosphate and diacylglycerol that stimulate downstream second messenger cascades [54]. RasGAP serves as an inhibitor of ras activation, but also may function as an adapter protein bind to EGFR complexes [47]. PTP1B and SHP2 are both protein tyrosine phosphatases reported to influence EGFR signaling in very different ways. PTP1B dephosphorylates EGFR and competes with PLCγ for binding to the phosphorylated EGFR [49]. Likewise, SHP2 competes with RasGAP for binding to phosphorylated EGFR and blocks the translocation of RasGAP to the membrane in A431 and COS-1 cells [51]. This results in prolonged Erk1/2 and Ras signal [51]. Thus, the phosphorylated EGFR Tyr992 residue binds multiple effector molecules that could influence PTHrP gene expression [55].
Our findings suggest that Erk1/2 and the Ets transcription factor are critical for EGFR coupling to PTHrP gene expression. This is consistent with the observation that inhibition of Raf or Ras or a mutated Ets transcription factor decreased the activity of the PTHrP-P3 transcriptional promoter in human keratinocytes [14]. Finally, xenografts of RWGT2 and HARA, human lung squamous cell carcinomas, display decreased EGFR and Erk1/2 phosphorylation and diminished plasma PTHrP expression following treatment with the EGFR tyrosine kinase inhibitor, gefitinib [56].
Several therapeutics targeted to the EGFR have been developed and tested in clinical trials involving advanced breast cancer patients [57, 58]. Unfortunately, the results of these studies have demonstrated no increase in median survival or decrease in progression of the disease [57, 58]. Nevertheless, an encouraging anecdotal observation has emerged from some of these studies. Gefitinib relieves bone pain in patients that were suffering from breast cancer metastases to bone [59, 60]. It has been proposed that anti-EGFR agents could provide relief for patients with metastasis by blocking EGFR-induced stimulation of bone resorptive factors RANKL and CSF-1in bone marrow stromal cells [61]. Intriguingly, PPR signaling has been reported to increase AREG and EGFR expression by bone marrow stromal cells and osteoblast-like cells [62, 63]. Combined with our findings, these data suggest that the intersection of AREG-EGFR signaling with the PTHrP-PPR pathway represents yet another point at which between breast cancer cells and cells of the bone microenvironment may be an ideal target for therapeutics designed to ameliorate the devastating consequences of bone metastases.
Acknowledgements
We wish to thank Dr. Irma Minafra for her generous gift of the BC61, BC3A, and 8701 cell lines. We are greatful for the support from National Institutes of Health; AR45585 DK067875, American Cancer Society Institutional Grant to Indiana University; IRG-84-002-15 and the Ruth Estrin Goldberg Memorial for Cancer Research to J. F., Walther Cancer Center Postdoctoral Grant to J.L.G., and Susan G. Komen Postdoctoral Award to J.L.G.
References
- 1.Wysolmerski JJ, Cormier S, Philbrick WM, et al. Absence of functional type 1 parathyroid hormone (PTH)/PTH-related protein receptors in humans is associated with abnormal breast development and tooth impaction. J Clin Endocrinol Metab. 2001;86:1788–1794. doi: 10.1210/jcem.86.4.7404. [DOI] [PubMed] [Google Scholar]
- 2.Dunbar ME, Dann P, Brown CW, et al. Temporally regulated overexpression of parathyroid hormone-related protein in the mammary gland reveals distinct fetal and pubertal phenotypes. J Endocrinol. 2001;171:403–416. doi: 10.1677/joe.0.1710403. [DOI] [PubMed] [Google Scholar]
- 3.Yin JJ, Selander K, Chirgwin JM, et al. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Southby J, Kissin MW, Danks JA, et al. Immunohisto-chemical localization of parathyroid hormone-related protein in human breast cancer. Cancer Res. 1990;50:7710–7716. [PubMed] [Google Scholar]
- 5.Vargas SJ, Gillespie MT, Powell GJ, et al. Localization of parathyroid hormone-related protein mRNA expression in breast cancer and metastatic lesions by in situ hybridization. J Bone Miner Res. 1992;7:971–979. doi: 10.1002/jbmr.5650070814. [DOI] [PubMed] [Google Scholar]
- 6.Bouizar Z, Spyratos F, De vernejoul MC. The parathyroid hormone-related protein (PTHrP) gene: use of downstream TATA promotor and PTHrP 1–139 coding pathways in primary breast cancers vary with the occurrence of bone metastasis. J Bone Miner Res. 1999;14:406–414. doi: 10.1359/jbmr.1999.14.3.406. [DOI] [PubMed] [Google Scholar]
- 7.Carron JA, Fraser WD, Gallagher JA. PTHrP and the PTH/PTHrP receptor are co-expressed in human breast and colon tumours. Br J Cancer. 1997;76:1095–1098. doi: 10.1038/bjc.1997.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson M, Danks J, Moseley J, et al. Parathyroid hormone-related protein production by breast cancers, improved survival, and reduced bone metastases. J Natl Cancer Inst. 2001;93:234–237. doi: 10.1093/jnci/93.3.234. [DOI] [PubMed] [Google Scholar]
- 9.Henderson MA, Danks JA, Slavin JL, et al. Parathyroid hormone-related protein localization in breast cancers predict improved prognosis. Cancer Res. 2006;66:2250–2256. doi: 10.1158/0008-5472.CAN-05-2814. [DOI] [PubMed] [Google Scholar]
- 10.Sellers RS, Capen CC, Rosol TJ. Messenger RNA stability of parathyroid hormone-related protein regulated by transforming growth factor-beta1. Mol Cell Endocrinol. 2002;188:37–46. doi: 10.1016/s0303-7207(01)00752-3. [DOI] [PubMed] [Google Scholar]
- 11.Cataisson C, Gordon J, Roussiere M, et al. Ets-1 activates parathyroid hormone-related protein gene expression in tumorigenic breast epithelial cells. Mol Cell Endocrinol. 2003;204:155–168. doi: 10.1016/s0303-7207(02)00298-8. [DOI] [PubMed] [Google Scholar]
- 12.Lindemann RK, Braig M, Ballschmieter P, et al. Protein kinase Calpha regulates Ets1 transcriptional activity in invasive breast cancer cells. Int J Oncol. 2003;22:799–805. [PubMed] [Google Scholar]
- 13.Lindemann RK, Braig M, Hauser CA, et al. Ets2 and protein kinase C epsilon are important regulators of parathyroid hormone-related protein expression in MCF-7 breast cancer cells. Biochem J. 2003;372:787–797. doi: 10.1042/BJ20030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho YM, Lewis DA, Koltz PF, et al. Regulation of parathyroid hormone-related protein gene expression by epidermal growth factor-family ligands in primary human keratinocytes. J Endocrinol. 2004;181:179–190. doi: 10.1677/joe.0.1810179. [DOI] [PubMed] [Google Scholar]
- 15.Lindemann RK, Ballschmieter P, Nordheim A, et al. Transforming growth factor beta regulates parathyroid hormone-related protein expression in MDA-MB-231 breast cancer cells through a novel Smad/Ets synergism. J Biol Chem. 2001;276:46661–46670. doi: 10.1074/jbc.M105816200. [DOI] [PubMed] [Google Scholar]
- 16.Watabe T, Yoshida K, Shindoh M, et al. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–137. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Roskoski R., Jr The ErbB/HER receptor protein-tyrosine kinases and cancer. Biochem Biophys Res Commun. 2004;319:1–11. doi: 10.1016/j.bbrc.2004.04.150. [DOI] [PubMed] [Google Scholar]
- 18.Burton PB, Knight DE. Parathyroid hormone-related peptide can regulate the growth of human lung cancer cells, and may form part of an autocrine TGF-alpha loop. FEBS Lett. 1992;305:228–232. doi: 10.1016/0014-5793(92)80674-6. [DOI] [PubMed] [Google Scholar]
- 19.Cramer SD, Peehl DM, Edgar MG, et al. Parathyroid hormone–related protein (PTHrP) is an epidermal growth factor-regulated secretory product of human prostatic epithelial cells. Prostate. 1996;29:20–29. doi: 10.1002/(SICI)1097-0045(199607)29:1<20::AID-PROS3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 20.Heath JK, Southby J, Fukumoto S, et al. Epidermal growth factor-stimulated parathyroid hormone-related protein expression involves increased gene transcription and mRNA stability. Biochem J. 1995;307(Pt 1):159–167. doi: 10.1042/bj3070159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrari SL, Rizzoli R, Bonjour JP. Effects of epidermal growth factor on parathyroid hormone-related protein production by mammary epithelial cells. J Bone Miner Res. 1994;9:639–644. doi: 10.1002/jbmr.5650090508. [DOI] [PubMed] [Google Scholar]
- 22.Ferrero JM, Ramaioli A, Largillier R, et al. Epidermal growth factor receptor expression in 780 breast cancer patients: a reappraisal of the prognostic value based on an eight-year median follow-up. Ann Oncol. 2001;12:841–846. doi: 10.1023/a:1011183421477. [DOI] [PubMed] [Google Scholar]
- 23.Tsutsui S, Ohno S, Murakami S, et al. Prognostic value of epidermal growth factor receptor (EGFR) and its relationship to the estrogen receptor status in 1029 patients with breast cancer. Breast Cancer Res Treat. 2002;71:67–75. doi: 10.1023/a:1013397232011. [DOI] [PubMed] [Google Scholar]
- 24.Suo Z, Risberg B, Karlsson MG, et al. The expression of EGFR family ligands in breast carcinomas. Int J Surg Pathol. 2002;10:91–99. doi: 10.1177/106689690201000202. [DOI] [PubMed] [Google Scholar]
- 25.Lodge AJ, Anderson JJ, Gullick WJ, et al. Type 1 growth factor receptor expression in node positive breast cancer: adverse prognostic significance of c-erbB-4. J Clin Pathol. 2003;56:300–304. doi: 10.1136/jcp.56.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomon DS, Normanno N, Ciardiello F, et al. The role of amphiregulin in breast cancer. Breast Cancer Res Treat. 1995;33:103–114. doi: 10.1007/BF00682718. [DOI] [PubMed] [Google Scholar]
- 27.Normanno N, Kim N, Wen D, et al. Expression of messenger RNA for amphiregulin, heregulin, and cripto-1, three new members of the epidermal growth factor family, in human breast carcinomas. Breast Cancer Res Treat. 1995;35:293–297. doi: 10.1007/BF00665981. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore JL, Riese DJ., II secErbB4-26/549 antagonizes ligand-induced ErbB4 tyrosine phosphorylation. Oncol Res. 2004;14:589–602. doi: 10.3727/0965040042707907. [DOI] [PubMed] [Google Scholar]
- 29.Guise TA, Yin JJ, Mohammad KS. Role of endothelin-1 in osteoblastic bone metastases. Cancer. 2003;97:779–784. doi: 10.1002/cncr.11129. [DOI] [PubMed] [Google Scholar]
- 30.Thomas RJ, Guise TA, Yin JJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140:4451–4458. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 31.Cataisson C, Lieberherr M, Cros M, et al. Parathyroid hormone-related peptide stimulates proliferation of highly tumorigenic human SV40-immortalized breast epithelial cells. J Bone Miner Res. 2000;15:2129–2139. doi: 10.1359/jbmr.2000.15.11.2129. [DOI] [PubMed] [Google Scholar]
- 32.Minafra S, Morello V, Glorioso F, et al. A new cell line (8701-BC) from primary ductal infiltrating carcinoma of human breast. Br J Cancer. 1989;60:185–192. doi: 10.1038/bjc.1989.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pucci-Minafra I, Minafra S, Alessandro R, et al. An ultrastructural evaluation of cell heterogeneity in invasive ductal carcinomas of the human breast. II. An in vitro study. J Submicrosc Cytol Pathol. 1989;21:489–499. [PubMed] [Google Scholar]
- 34.Luparello C, Burtis WJ, Raue F, et al. Parathyroid hormone-related peptide and 8701-BC breast cancer cell growth and invasion in vitro: evidence for growth-inhibiting and invasion-promoting effects. Mol Cell Endocrinol. 1995;111:225–232. doi: 10.1016/0303-7207(95)03577-t. [DOI] [PubMed] [Google Scholar]
- 35.Luparello C, Birch MA, Gallagher JA, et al. Clonal heterogeneity of the growth and invasive response of a human breast carcinoma cell line to parathyroid hormone-related peptide fragments. Carcinogenesis. 1997;18:23–29. doi: 10.1093/carcin/18.1.23. [DOI] [PubMed] [Google Scholar]
- 36.Guise TA, Yin JJ, Taylor SD, et al. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J Clin Invest. 1996;98:1544–1549. doi: 10.1172/JCI118947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brtva TR, Drugan JK, Ghosh S, et al. Two distinct Raf domains mediate interaction with Ras. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- 38.Kakonen SM, Selander KS, Chirgwin JM, et al. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J Biol Chem. 2002;277:24571–24578. doi: 10.1074/jbc.M202561200. [DOI] [PubMed] [Google Scholar]
- 39.Woelfle U, Breit E, Zafrakas K, et al. Bi-specific immunomagnetic enrichment of micrometastatic tumour cell clusters from bone marrow of cancer patients. J Immunol Methods. 2005;300:136–145. doi: 10.1016/j.jim.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13:1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 41.Saito T, Okada S, Ohshima K, et al. Differential activation of epidermal growth factor (EGF) receptor downstream signaling pathways by betacellulin and EGF. Endocrinology. 2004;145:4232–4243. doi: 10.1210/en.2004-0401. [DOI] [PubMed] [Google Scholar]
- 42.Goldkorn T, Balaban N, Matsukuma K, et al. EGF-Receptor phosphorylation and signaling are targeted by H2O2 redox stress. Am J Respir Cell Mol Biol. 1998;19:786–798. doi: 10.1165/ajrcmb.19.5.3249. [DOI] [PubMed] [Google Scholar]
- 43.Cook PW, Pittelkow MR, Piepkorn M. Overexpression of amphiregulin in the epidermis of transgenic mice induces a psoriasis-like cutaneous phenotype. J Invest Dermatol. 1999;113:860. doi: 10.1046/j.1523-1747.1999.00753.x. [DOI] [PubMed] [Google Scholar]
- 44.Vassar R, Hutton ME, Fuchs E. Transgenic overexpression of transforming growth factor alpha bypasses the need for c-Haras mutations in mouse skin tumorigenesis. Mol Cell Biol. 1992;12:4643–4653. doi: 10.1128/mcb.12.10.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luetteke NC, Qiu TH, Fenton SE, et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 46.Chung E, Graves-Deal R, Franklin JL, et al. Differential effects of amphiregulin and TGF-alpha on the morphology of MDCK cells. Exp Cell Res. 2005;309:149–160. doi: 10.1016/j.yexcr.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281:37728–37737. doi: 10.1074/jbc.M606532200. [DOI] [PubMed] [Google Scholar]
- 48.Wu SL, Kim J, Bandle RW, et al. Dynamic profiling of the post-translational modifications and interaction partners of epidermal growth factor receptor signaling after stimulation by epidermal growth factor using Extended Range Proteomic Analysis (ERPA) Mol Cell Proteomics. 2006;5:1610–1627. doi: 10.1074/mcp.M600105-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Jorissen RN, Walker F, Pouliot N, et al. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 50.Sturla LM, Amorino G, Alexander MS, et al. Requirement of Tyr-992 and Tyr-1173 in phosphorylation of the epidermal growth factor receptor by ionizing radiation and modulation by SHP2. J Biol Chem. 2005;280:14597–14604. doi: 10.1074/jbc.M413287200. [DOI] [PubMed] [Google Scholar]
- 51.Agazie YM, Hayman MJ. Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol. 2003;23:7875–7886. doi: 10.1128/MCB.23.21.7875-7886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rotin D, Margolis B, Mohammadi M, et al. SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C gamma. Embo J. 1992;11:559–567. doi: 10.1002/j.1460-2075.1992.tb05087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milarski KL, Zhu G, Pearl CG, et al. Sequence specificity in recognition of the epidermal growth factor receptor by protein tyrosine phosphatase 1B. J Biol Chem. 1993;268:23634–23639. [PubMed] [Google Scholar]
- 54.Hofmann J. Protein kinase C isozymes as potential targets for anticancer therapy. Curr Cancer Drug Targets. 2004;4:125–146. doi: 10.2174/1568009043481579. [DOI] [PubMed] [Google Scholar]
- 55.Richard V, Rosol TJ, Foley J. PTHrP gene expression in cancer: do all paths lead to Ets? Crit Rev Eukaryot Gene Expr. 2005;15:115–132. doi: 10.1615/critreveukaryotgeneexpr.v15.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorch G, Gilmore JL, Koltz PF, et al. Inhibition of epidermal growth factor receptor signalling reduces hypercalcaemia induced by human lung squamous-cell carcinoma in athymic mice. Br J Cancer. 2007;97:183–193. doi: 10.1038/sj.bjc.6603828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Normanno N, De Luca A, Maiello MR, et al. Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors in breast cancer: current status and future development. Front Biosci. 2005;10:2611–2617. doi: 10.2741/1725. [DOI] [PubMed] [Google Scholar]
- 58.Kim TE, Murren JR. Erlotinib OSI/Roche/Genentech. Curr Opin Investig Drugs. 2002;3:1385–1395. [PubMed] [Google Scholar]
- 59.Albain KS, Elledge R, Gradishar WJ, Hayes DF, et al. Open-label, phase II, multicenter trail of ZD1839 (‘Iressa’) in patients with advanced breast cancer. Breast Cancer Res Treatment. 2002;76:S33. Abstract. [Google Scholar]
- 60.von Minckwitz GJW, Beckmann M, De Bois A, et al. A multicenter phase II trial to evaluate gefitinib (‘Iressa’, ZD1839) (500 mg/day) in patients with metastatic breast cancer after previous chemotherapy treatment. Eur J Cancer. 2003;1 Abstract. [Google Scholar]
- 61.Normanno N, De Luca A, Aldinucci D, et al. Gefitinib inhibits the ability of human bone marrow stromal cells to induce osteoclast differentiation: implications for the pathogenesis and treatment of bone metastasis. Endocr Relat Cancer. 2005;12:471–482. doi: 10.1677/erc.1.00956. [DOI] [PubMed] [Google Scholar]
- 62.Qin L, Tamasi J, Raggatt L, et al. Amphiregulin is a Novel growth factor Involved in normal bone development and in the cellular response to parathyroid hormone stimulation. J Biol Chem. 2005;280:3974–3981. doi: 10.1074/jbc.M409807200. [DOI] [PubMed] [Google Scholar]
- 63.Qin L, Qiu P, Wang L, et al. Gene expression profiles and transcription factors involved in parathyroid hormone signaling in osteoblasts revealed by microarray and bioinformatics. J Biol Chem. 2003;278:19723–19731. doi: 10.1074/jbc.M212226200. [DOI] [PubMed] [Google Scholar]