Abstract
Formamidopyrimidine DNA glycosylase (Fpg) and endonuclease VIII (Nei) share an overall common three-dimensional structure and primary amino acid sequence in conserved structural motifs but have different substrate specificities, with bacterial Fpg proteins recognizing formamidopyrimidines, 8-oxoguanine (8-oxoG) and its oxidation products guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp) and bacterial Nei proteins recognizing primarily damaged pyrimidines. In addition to bacteria, Fpg has also been found in plants, while Nei is sparsely distributed among the prokaryotes and eukaryotes. Phylogenetic analysis of Fpg and Nei DNA glycosylases demonstrated, with 95% bootstrap support, a clade containing exclusively sequences from plants and fungi. Members of this clade exhibit sequence features closer to bacterial Fpg proteins than to any protein designated as Nei based on biochemical studies. The Candida albicans (Cal) Fpg DNA glycosylase and a previously studied Arabidopsis thaliana (Ath) Fpg DNA glycosylase were expressed, purified and characterized. In oligodeoxynucleotides, the preferred glycosylase substrates for both enzymes were Gh and Sp, the oxidation products of 8-oxoG, with the best substrate being a site of base loss. GC/MS analysis of bases released from γ-irradiated DNA show FapyAde and FapyGua to be excellent substrates as well. Studies carried out with oligodeoxynucleotide substrates demonstrate that both enzymes discriminated against A opposite the base lesion, characteristic of Fpg glycosylases. Single turnover kinetics with oligodeoxynucleotides showed that the plant and fungal glycosylases were most active on Gh and Sp, less active on oxidized pyrimidines and exhibited very little or no activity on 8-oxoG. Surprisingly, the activity of AthFpg1 on an AP site opposite a G was extremely robust with a kobs of over 2500 min−1.
Keywords: Base excision repair, DNA glycosylases, Phylogenetics, Formamidopyrimidine DNA glycosylase, Endonuclease VIII (Nei), Arabidopsis thaliana, Candida albicans, Guanidinohydantoin, Spiroiminodihydantoin
1. Introduction
Cellular DNA is under constant exposure to oxidative damage from endogenous and exogenous sources and these damages must be recognized and repaired quickly and with high fidelity. Endogenous damages include single-strand breaks, deaminations, depurinations, alkylations and oxidative modifications of the DNA bases [1]. Of the more than 25,000 DNA damages a human cell receives per day about 6000 are oxidatively induced base lesions. The consequences of these damages include mutations, cancer and/or cell death. Base excision repair (BER) has evolved to recognize and repair these damages with the first step in repair being recognition of a damaged base by a DNA glycosylase (for reviews see [2–4]). The DNA glycosylase binds to the DNA, cleaves the N-glycosyl bond releasing the damaged base resulting in an apurinic or apyrimidinic (AP) site. Most oxidative DNA glycosylases also cleave the phosphodiester backbone of DNA on the 3′ side of the AP site leaving either a α,β-unsaturated aldehyde (β-elimination) or a phosphate (β,δ-elimination). Since DNA polymerases require a 3′ hydroxyl group for synthesis the phosphodiesterase activity of an AP endonuclease is required to remove the α,β-unsaturated aldehyde or phosphate and leave a 3′ hydroxyl group. In mammalian cells, polynucleotide kinase is used to remove the phosphate generated by Fpg/Nei family glycosylases in order to leave an extendable 3′ end [5]. DNA polymerase then adds the cognate nucleotide and DNA ligase seals the gap. Eukaryotes use additional accessory proteins to maximize the efficiency of both recognition and repair (for reviews see [6,7]).
Candida albicans is the most prevalent human fungal pathogen. Normally it exists as a dimorphic commensal yeast found on humans, however in immune compromised individuals, such as AIDS patients and neonates, C. albicans can become an opportunistic pathogen. The human immune response recruits monocytes, dendritic cells and macrophages to the site of infection where genes involved with the production and release of reactive oxygen species (ROS) are induced [8]. Therefore C. albicans has to deal with endogenous ROS as well as the exogenous ROS produced by the host immune response. Arabidopsis thaliana is a vascular plant and as such it has to contend with the endogenous production of singlet oxygen from photosynthesis, internal cell signaling mediated by ROS as well as ROS from exposure to visible light [9]. Repair of oxidatively induced damage caused by treating reporter plasmid DNA with methylene blue and visible light has been demonstrated using Arabidopsis cell extracts [10].
The oxidative DNA glysosylases can be divided into two families based on structural and sequence homology (for reviews see [11,12]). The Nth superfamily contains Escherichia coli endonuclease III (EcoNth) and its yeast counterparts Ntg1 and Ntg2, that remove oxidized pyrimidines and formamidopyrimidines [4,6-diamino-5-formamidopyrimidine (FapyAde), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua)], MutY that removes A opposite 8-oxoG, Ogg1 that removes 8-oxoG and FapyGua opposite C, and AlkA that removes alkylated bases [2–4,13]. The Fpg/Nei family contains formamidopyrimidine DNA glycosylase (Fpg) also called MutM that removes 8-oxoG and FapyGua opposite C and FapyAde opposite T, and endonuclease VIII (Nei) that removes oxidized pyrimidines and FapyAde. With the exception of MutY and AlkA, which are monofunctional and only exhibit glysosylase activity, the glycosylases listed above are bifunctional and exhibit lyase activity in addition to glycosylase activity [2–4]. Members of the Nth superfamily are found throughout the three major kingdoms. Fpg proteins are primarily bacterial, while Nei and Nei-like glycosylases are found in only some of the γ-proteobacteria, actinobacteria, and metazoans [4] and we now place members of this family in acidobacteria, planctobacteria and cyanobacteria (see Fig. 1).
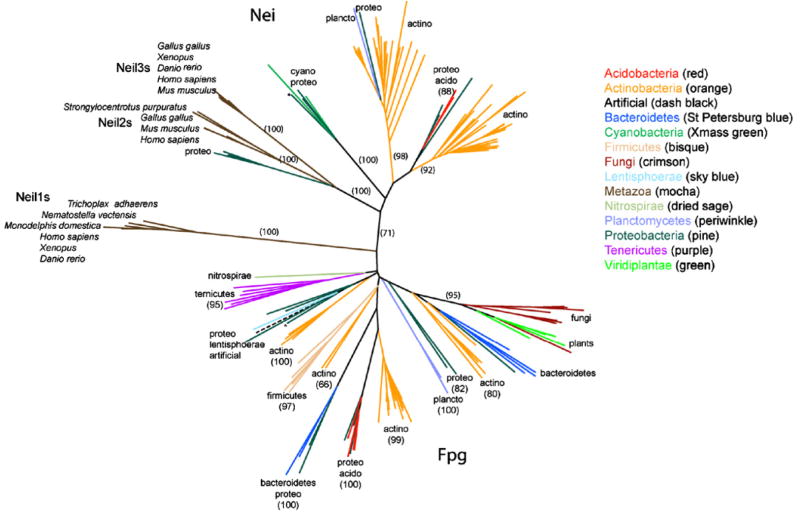
Fig. 1.
Unrooted phylogeny of the Fpg/Nei family. Nei sequences are in the top half of the tree and the bacterial Fpg are on the bottom half of the tree. The Neil1 clade falls between the Nei and Fpg clades. The bootstrap values are indicated where appropriate. * indicates the position of the E. coli Nei and Fpg sequences within the tree.
The BER system has redundancy with respect to overlapping substrate specificity such that the loss of a single DNA glycosylase will usually not result in a complete loss of repair for a specific oxidatively induced lesion. In addition to the glycosylases characterized in this study, C. albicans has the following but as yet uncharacterized putative proteins involved with the initial steps of BER: Ogg1, Ntg, Udg, AlkA and two AP endonucleases. Single mutants defective in Ogg1, Ntg1 or Apn1 are not sensitive to oxidizing agents, antifungal drugs nor are they altered in their response to macrophages [14]. A. thaliana has an Ogg1, MutY, MutT, Nth, six 3-methyadenine DNA glycosylases, three AP endonucleases in addition to potentially seven different forms of AthFpg that could be produced by alternative splicing [15]. We have characterized AthFpg1 in this study.
An unrooted phylogeny of the Fpg/Nei tree from a previous study from our laboratory [4] exhibited a long edge (having moderate, 52%, bootstrap support) that separated enzymes reported to have specificity for 8-oxoG from those reported to prefer oxidized pyrimidines. The phylogeny, then, supported a simple model according to which only one change in substrate preference occurred during Fpg/Nei family evolution and enzymes were thus parsimoniously named either “Fpg” or “Nei”. Here we show that the evolution of substrate specificity in this family is strikingly complex. The best substrates for CalFpg and AthFpg in oligodeoxynucleotides are Sp and Gh; CalFpg also recognizes oxidized pyrimidines. Both enzymes mainly release FapyAde and FapyGua and, to some extent, several pyrimidine-derived lesions from γ-irradiated DNA containing multiple lesions (with distinct preferences) but have little or no activity on 8-oxoG.
2. Materials and methods
2.1. Sequence identification and phylogeny
Fpg/Nei homologs were identified using the PFAM domain profile (pfam06831 [16]) through the CDD database [17]. CDTree [17] was used to organize the data. All the eukaryotic sequences were kept with the exception of redundant sequences, and sequences of clades for which the edges leading to them had a length greater than 2.0 (more than 1.5 substitutions per site on average). We used this criterion because the phylogenetic analysis software was unable to place such divergent sequences accurately. These sequences included two bacteroides, and the viral and archael sequences. We further discriminated Fpg from Nei by visually inspecting sequence alignments of putative Fpg/Nei proteins, and looking for conservation of key residues. T Coffee [18,19] was used to remove nearly identical sequences (98%), MAFFT [20] for high accuracy parameters for alignment and PFAAT [21] for visualization. Seaview [22] was used to remove phylogenetically uninformative sites and the phylogenetic tree was made with Phylip’s PROTDIST and NEIGHBOR [23] using SEQBOOT to create 100 bootstrap replicates.
2.2. Enzyme purification
C. albicans genomic DNA was used as a template to amplify CalFpg by the polymerase chain reaction (PCR). The gene is located on two exons but the vast majority of the gene is on one of the exons so that exon was amplified by PCR. The rest of the gene from the short exon was added with a long oligodeoxynucleotide primer of the appropriate sequence in a subsequent round of PCR. The CalFpg gene sequence contains three CUG codons, one at amino acid position 205, a second at position 375 and the last at position 377. The CUG codon in C. albicans is an exception to the universal codon code and codes for serine instead of leucine. Site directed mutagenesis using the Quikchange kit (Stratagene) was used to change these three codons so that E. coli would insert a serine at these positions into the CalFpg amino acid sequence. The CalFpg gene was cloned into pET22b with a C-terminal his tag and expressed in Rosetta DE3 plysS cells (Novagen). Transformed cells from a single colony were grown in 2 l LB under appropriate selection and expression was induced with 1 mM IPTG at an OD600 of 0.585 and grown at 16 °C overnight with shaking at 300 rpm. The cells were harvested and flash frozen in liquid nitrogen and stored at −80 °C. The cell pellet was resuspended in 50 ml of 50 mM sodium phosphate pH 8, 100 mM NaCl, 10 mM imidazole pH 8, 10% glycerol, 5 mM β-mercaptoethanol, 1 mM PMSF and 10 mM benzamidine. The cells were lysed by sonication six times for 2 min on ice. The lysate was clarified by centrifugation and the supernatant loaded onto a 5 ml HiTrap chelating HP column charged with 100 mM nickel sulfate. The protein was eluted at a flow rate of 2 ml/min with a linear gradient of 50 mM sodium phosphate pH 8, 100 mM NaCl, 500 mM imidazole pH 8, 10% glycerol and 5 mM β-mercaptoethanol. Fractions containing CalFpg, as determined by SDS-PAGE, were dialyzed against 20 mM HEPES pH 7.6, 150 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol. The protein was further purified utilizing a 5 ml HiTrap SP FF column. The protein was eluted at a flow rate of 2 ml/min with a linear gradient of 20 mM HEPES pH 7.6, 1 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol. Fractions containing CalFpg were combined and dialyzed overnight at 4 °C against 20 mM HEPES pH 7.6, 150 mM NaCl, 1 mM DTT and 25% glycerol. The following day the protein was dialyzed against 20 mM HEPES pH 7.6, 150 mM NaCl, 1 mM DTT and 50% glycerol and stored at −20 °C.
AthFpg1 in pET 28b with a C-terminal his tag was kindly provided by Terrence Murphy (University of California, Davis) [24]. There are an additional ten amino acids between the C-terminus of the gene and the hexa-histidine tag as a result of the cloning strategy employed. The TGA stop codon was mutated to TGC that codes for cysteine, therefore the additional amino acid sequence is CVDKLAAALE followed by the hexa-His tag. In addition, the first six codons that encode PELPEV were optimized for expression in E. coli. AthFpg1 is one of seven potential splice variants of AthFpg in A. thaliana. We will refer to AthFpg1 as AthFpg in the remainder of the paper. The procedure for expression of AthFpg was identical to that for CalFpg except for the following items. Expression was induced with 1 mM IPTG at an OD600 of 0.680. The protein was eluted from the 5 ml HiTrap chelating HP column with a linear gradient of 50 mM sodium phosphate pH 8, 150 mM NaCl, 500 mM imidazole pH 8, 10% glycerol, 5 mM β-mercaptoethanol. Protein concentrations were determined using the Bradford assay method (Bio-Rad). The fraction of active DNA glycosylase was determined by the molecular accessibility method of Blaisdell and Wallace [25] and all concentrations of protein used in this study were corrected for the fraction of active enzyme.
2.3. Substrates
Oligodeoxynucleotides (35-mers) with various oxidatively induced damages which were located 14 nucleotides from the 5′ end were purchased from Midland Certified Reagent Co. (Midland, Texas). The damage-containing oligodeoxynucleotide sequence is as follows: 5′-TGTCAATAGCAAGXGGAGAAGTCAATCGTGAGTCT-3′, where X denotes the oxidative damage: 7,8-dihydro-8-oxoguanine (8-oxoG), thymine glycol (Tg), 5-hydroxycytosine (5-OHC), 5-hydroxyuracil (5-OHU), 5,6-dihydrothymine (DHT) and 5,6-dihydrouracil (DHU). An oligodeoxynucleotide with a uracil at this position was used to create a site of base loss (AP site). Gh, Sp1 and Sp2 were synthesized as described previously [26] in the following sequence context: GCGTCCAZGTCTAC, where Z denotes a Gh, Sp1 or Sp2. The damage-containing strand, 1 pmol, was end-labeled for 15 min at 37 °C with [γ-32P] ATP by T4 polynucleotide kinase (New England Biolabs). The labeling reaction was terminated by the addition of EDTA and heat inactivation; the oligodeoxynucleotide was ethanol precipitated. The labeled oligodeoxynucleotide was suspended with 9 pmol of the appropriate damage, 10 pmol of the complementary strand to a final concentration of 250 nM in 10 mM Tris pH 8.0, 50 mm NaCl, heated to 95 °C and cooled to room temperature to create the double-stranded substrate. The complementary strand for the 35mer system had the sequence, 5′-AGACTCACGATTGACTTCTCC(N)CTTGCTATTGACA-3′, where N denotes the base opposite the damage. The complementary strand for the 14mer system had the sequence, GTAGAC(N)TGGACGC where N denotes the base opposite the damage. To create an AP site, the double-stranded oligodeoxynucleotide containing a uracil was treated with uracil DNA glycosylase (UDG-New England Biolabs) for 15 min at room temperature.
2.4. Glycosylase assays
The reaction conditions for the various DNA glycosylases used in this study were as follows: EcoFpg, and EcoNei (10 mM Tris–HCl (pH 8.0), 50 mM NaCl and 1 mM EDTA); hOGG1 (10 mM glycylglycine, 100 mM NaCl, 1 mM EDTA (pH 8.05); hNEIL1 (20 mM CHES–KOH (pH 9.5), 100 mM NaCl, and 1 mM EDTA); EcoNth (10 mM HEPES-KOH, 100 mM KCl, and 10 mM EDTA (pH 7.4) [Trevigen 1X REC Buffer 4]); CalFpg and AthFpg (10 mM Bis Tris Propane–HCl, 10 mM MgCl2, 1 mM DTT (pH 7.0) [New England Biolabs NEBuffer 1]); all reactions were supplemented with BSA 100 μg/ml. The E. coli glycosylase controls were used where appropriate such that EcoFpg was only used on damaged purines, while EcoNei and Nth were used on damaged pyrimidines with the caveat that EcoNei was also used on Gh and Sp. hOGG1 was only used on 8-oxoG while hNEIL1 was used on the damaged pyrimidines as well as Gh and Sp. Glycosylase assays were carried out at 37 °C except for AthFpg and CalFpg which were carried out at room temperature. Reactions were terminated with an equal volume of formamide stop buffer (98% formamide, 0.2N NaOH, 10 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanole). Single-stranded substrate reactions were carried out with an equimolar concentration of enzyme and substrate (25 nM) for 1 h. Double-stranded substrate (25 nM) reactions were carried out with AthFpg and CalFpg at 2.5, 25 and 250 nM and the appropriate DNA glycosylase controls (25 nM). The opposite base specificity was determined by incubating AthFpg (25 nM) or CalFpg (0.25 nM) with each lesion opposite all four bases (25 nM) for 1 h at room temperature.
2.5. Single-turnover kinetics
CalFpg or AthFpg (25 nM) was incubated with 3.1 nM substrate in 10 mM Bis Tris Propane–HCl, 10 mM MgCl2, 1 mM DTT (pH 7.0) [New England Biolabs NEBuffer 1] with 100 μg/ml BSA. Reactions were terminated at the appropriate time points with formamide stop buffer. The time points for DHU:G with CalFpg were 2, 4, 6, 8, 10, 15, 30 and 60 s since this substrate appeared to be the best, all other time points were 1, 2, 4, 8, 15, 30, 60 and in some cases 120 min. Kin-Tek Rapid Quench was used for the single turnover experiments with AthFpg and an AP:G pair since this reaction was complete in less than two seconds. A 1% solution of SDS was used as the quench to terminate this set of reactions. Terminated reactions were ethanol precipitated to remove the SDS and the DNA was suspended in 0.5× formamide stop buffer. The reaction products were separated on a 12% (w/v) polyacrylamide sequencing gel and quantitated with an isotope imaging system (Molecular Imaging System, Bio-Rad). Kinetic parameters were calculated using Prism 4 (GraphPad Software, Inc.).
2.6. GC/MS data analysis
Calf thymus DNA was γ-irradiated with 40 Gray in N2O saturated phosphate buffer solution to create the modified DNA bases as described previously [27–29]. Stable isotope-labeled analogs of modified bases were added to 50 μg aliquots of γ-irradiated DNA. The samples were then incubated with 2 μg of enzyme in 50 μl phosphate buffer (pH 7.4), 100 mM KCl, 1 mM EDTA and 0.1 mM dithiotreitol at 37 °C for 1 h. The rest of the analysis was performed as described [27–29]. The GC/MS data were collected from three independent experiments using three independently prepared DNA substrates.
The GC/MS data are represented by
where e indexes enzymes, p indexes products, Qepr is a GC/MS peak intensity, r indexes replicates, Rep is a set of replicates (e = 0 indicates no enzyme), and |Rep| is the number of replicates. We report Sep along with its sample standard deviation, obtained from triplicate reactions performed on the same day with the same enzyme and substrate preparations.
The data are also visualized after normalization based on
where
In contrast to the case in which enzymes react with substrates that are in distinct reaction mixtures,
under the Michaelis Menten model and under excess of substrate over enzyme, where fp is the fraction of oxidized bases that appear as product p. Therefore, contingent upon constancy of fp, S^ep serves as a single vector measure of substrate specificity.
3. Results
3.1. The Ath and Cal proteins are members of the Fpg/Nei family
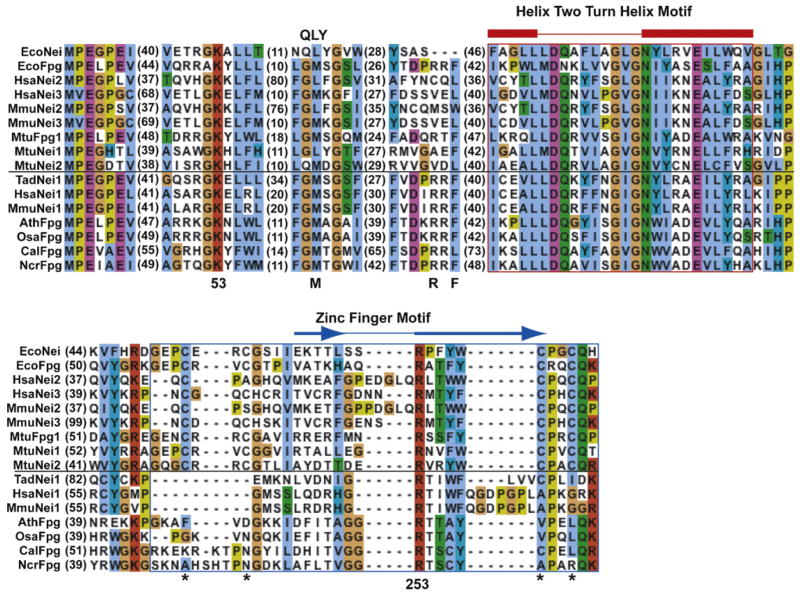
The Ath and Cal amino acid sequences were aligned with those of representative members of the Fpg/Nei family (Fig. 2). Searches of NCBI’s conserved domain database [PMID: 17135202] identified the Fpg/Nei family domain with high statistical significance (E < 10−15). Nearly all sequences from this family have an N-terminal proline (Pro2) with an α-amino group that functions as a nucleophile in the glycosylase/lyase reaction in EcoNei and EcoFpg. A valine is present at this position in hNEIL3 [30], mouse NEIL3 [31] and Mimivirus Nei2 [32] and has been proposed to function as the nucleophile. A glutamic acid (Glu3) is found in all sequences and is required for glycosylase activity in EcoFpg [33] and EcoNei [34]. Like other members of the Fpg/Nei family the Ath and Cal sequences have a lysine that aligns with position 53 in EcoNei, a residue that has been shown to be involved directly in catalysis [35], as well as the sequence signature of the helix-two-turn helix motif (position 154–177).
Fig. 2.
Sequence alignment of endonuclease VIII-like proteins with representative members of the Fpg/Nei family. Numbers in parenthesis represent the number of amino acids not shown between the regions. Residues important for catalytic activity or binding to substrate are indicated with a number that corresponds to the EcoNei sequence. The H2TH domain is outlined in red (α helices in red) and the zinc/zincless finger motif is outlined in blue, the helix and β strand schematics (blue arrows) are positioned according to the EcoNei sequence. The sequences below the horizontal black line have a ‘zincless finger’ while the sequences above the black line have a more typical zinc finger. The positions of the four cysteines in the zinc finger motif are denoted by an *. The position of the EcoNei QLY insertion loop is indicated on the top of the alignment and the position of the M74, R109 and P111 (position number based on EcoFpg sequence-will not match position number for EcoNei sequence) insertion loop are indicated on the bottom of the alignment. EcoNei: Escherichia coli Nei (gi|16128689); EcoFpg: Escherichia coli Fpg (gi|15804176); HsaNei2: Homo sapiens Neil2 (gi|21450800); HsaNei3: Homo sapiens Neil3 (gi|157388969); MmuNei2: Mus musculus Neil2 (gi|118130803); MmuNei3: Mus musculus Neil3 (gi|22122759); MtuFpg1: Mycobacterium tuberculosis Fpg (gi|148662769); MtuNei1: Mycobacterium tuberculosis Nei1 (gi|15609601); MtuNei2: Mycobacterium tuberculosis Nei2 (gi|15610433); TadNei1 Trichoplax adhaerens (gi|190586348); HsaNei1: Homo sapiens Neil1 (gi|119619664); AthFpg: Arabidopsis thalania Fpg1 (gi|18404050); OsaFpg: Oryza sativa Fpg (gi|115469160); CalFpg: Candida albicans (gi|3850130); NcrFpg: Neurospora crassa Fpg (gi|157072070).
3.2. Plant and fungal Fpg/Nei family members form a clade
An unrooted phylogeny of the Fpg/Nei family (Fig. 1) exhibits strong bootstrap support (95%) for an edge that separates plant and fungal sequences from all other sequences. Based on the similarity among plant and fungal sequences (the Ath and Cal sequences are over 50% identical) relative to other sequences in the phylogeny, the root of the phylogeny falls outside this branch. Thus the plant and fungal sequences share a common ancestor with each other that they do not share with other proteins in the tree. Notably this clade excludes metazoan sequences, in contrast to our expectation based on the eukaryotic family tree.
Like NEIL1 [37], AthFpg and CalFpg are missing the canonical zinc finger; AthFpg has no residual cysteine residues found in the typical CCCC Zn-finger while CalFpg has only the third cysteine. AthFpg and CalFpg have greater than 50% identity to each other in the putative Zn-less finger region (presumably their most recent common ancestor had no Zn-finger) but no detectable similarity with Fpg/Nei Zn-finger sequences or NEIL1 Zn-less finger sequences.
Enzymes are named based on either their function or phylogeny, options that are often consistent. To adhere to the usual practice of naming base excision repair enzymes based on phylogenetic relationships to previously named enzymes (that is, without regard for catalytic activities) and the previous designation of AthFpg [20], we refer to the C. albicans protein as CalFpg.
3.3. CalFpg and AthFpg are more similar in sequence to Fpg than Nei proteins
In the Fpg members of the family, Glu3 is followed almost exclusively by a conserved leucine (Leu4) in the bacterial, plant, and fungal sequences while a conserved glycine (Gly4) is observed in the Nei representatives (Fig. 2). The only exceptions are CalFpg, and NcrFpg which have a valine and isoleucine, respectively. AthFpg and CalFpg exhibit three amino acids (aligned with M74, R109, and F111 in the EcoFpg sequence) similar to those shown to play a role in opposite base interactions in Fpg, but are unlike those seen in Nei sequences.
Thus distinctive sequence features suggest that the fungal and plant sequences are more similar to those of Fpg than Nei, consistent with the distribution of sequence-sequence distances (not shown) and the existence of a long edge (Fig. 1) separating the bacterial Fpg and plant/fungal sequences from sequences designated as Nei.
3.4. An ancestor of extant metazoans possessed a NEIL1-like gene
Sequence similarity searches resulted in the identification of Fpg/Nei family members among early branching metazoans, including Trichoplax, sea anemone, and sea urchin. Like NEIL1, but unlike NEIL2/3, these sequences do not have a Zn-finger. Phylogenetic estimation (Fig. 1) suggests a clade (100% bootstrap support) consisting of these sequences and the vertebrate NEIL1 sequences.
3.5. AthFpg and CalFpg prefer Gh, Sp and oxidized pyrimidines over 8-oxoG in double-stranded oligodeoxynucleotides
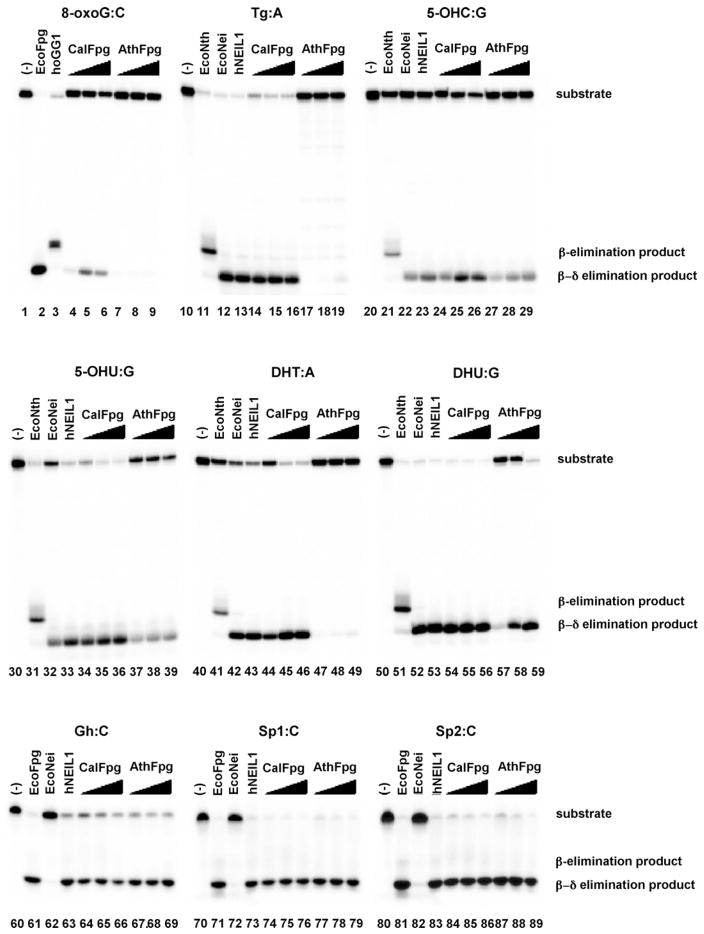
To see if the substrates for these Fpg-clade glycosylases included pyrimidines, we examined their activity on various oxidatively induced damages in double-stranded DNA as shown in Fig. 3 along with EcoFpg (lanes 2, 61, 71 and 81), hOGG1 (lane 3), EcoNth (lanes 11, 21, 31, 41 and 51), EcoNei (lanes 12, 22, 32, 42, 52, 62, 72 and 82) and hNEIL1 (lanes 13, 23, 33, 43, 53, 63, 73 and 83) as enzyme controls. No enzyme controls were also run (lanes 1, 10, 20, 30, 40, 50, 60, 70 and 80). The best oxidized pyrimidine substrates for CalFpg were DHU:G (lanes 54–56) and 5-OHU:G (lanes 34–36). The same oxidized pyrimidines were preferred by AthFpg, DHU:G (lanes 57–59) and 5-OHU:G (lanes 37–39). 8-oxoG:C was the worst substrate tested for CalFpg and AthFpg. Although the level of cleavage by AthFpg of the oligodeoxynucleotide containing 8-oxoG:C was similar to that previously observed [24] and led to its name as Fpg, no other substrates were tested. While 8-oxoG:C was a poor substrate for both CalFpg and AthFpg, the oxidation products of 8-oxoG namely, Gh:C, Sp1:C and Sp2:C (lanes 64–69, 74–79 and 84–89, respectively) were the best substrates for these enzymes. Also as shown in Fig. 3, CalFpg and AthFpg, like EcoFpg, EcoNei and NEIL1, are bifunctional glycosylases that leave a phosphate on the 3′ end of the single strand break indicative of a β,δ elimination reaction (Fig. 3).
Fig. 3.
DNA glycosylase/lyase activities of AthFpg and CalFpg. Double-stranded substrates (25 nM) were incubated with the appropriate control enzymes (25 nM) (EcoFpg and hOGG1 for 8oxoG:C, EcoNth, EcoNei and hNEIL1 for double-stranded pyrimidines, EcoFpg, EcoNei and hNEIL1 for Gh:C, Sp1:C and Sp2:C); 2.5, 25 or 250 nM AtFpg and CalFpg as described in Section 2.
3.6. Single-stranded DNA is also a substrate for CalFpg and AthFpg
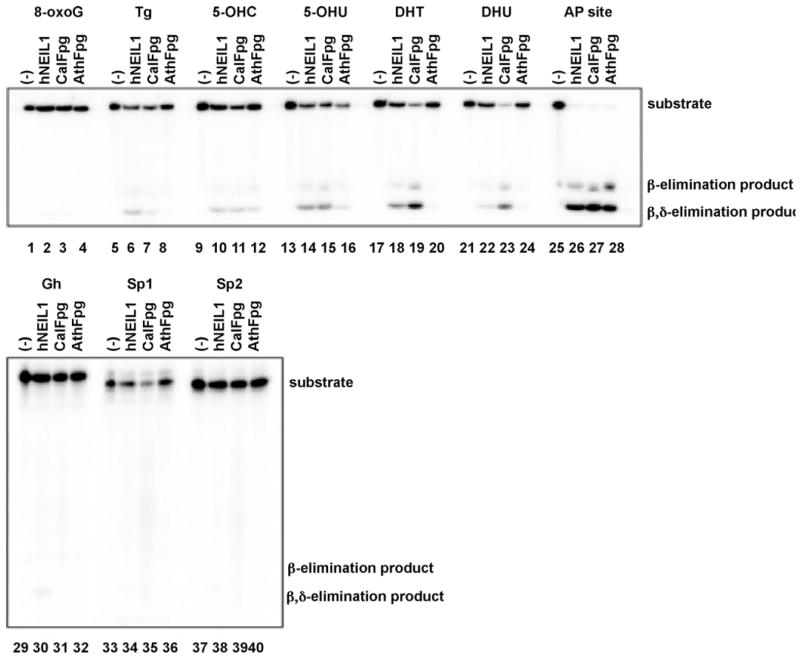
Because a number of Fpg/Nei family glycosylases recognize lesions in single stranded DNA [32,38,39], AthFpg and CalFpg were tested against single-stranded oligodeoxynucleotides containing oxidative damages or an AP site (Fig. 4). No activity against 8-oxoG (lanes 2–4) was detected for any of the glycosylases while essentially complete cleavage was observed with an AP site (lanes 26–28). As we [32,40] have previously observed, the substrates from best to worst for NEIL1 were AP followed by 5-OHU and DHT followed by Tg, 5-OHC, DHU. CalFpg also recognized these damages but preferred AP followed by DHU and DHT then 5-OHU, 5-OHC and Tg while AthFpg recognized AP the best, then 5-OHU and 5-OHC followed by DHU, DHT and Tg. The three glycosylases showed no activity on single-stranded DNA containing 8-oxoG. Interestingly, the best substrates in double stranded DNA, Gh (lanes 31, 32), Sp1 (lanes 35, 36) and Sp2 (lanes 39, 40), did not appear to be cleaved by CalFpg and AthFpg to any appreciable extent when present in single stranded DNA.
Fig. 4.
AthFpg and CalFpg recognize single-stranded substrates. Single-stranded substrates (25 nM) were incubated with hNEIL1, AthFpg and CalFpg (25 nM) as described in Section 2.
3.7. CalFpg and AthFpg discriminate against A opposite the lesion
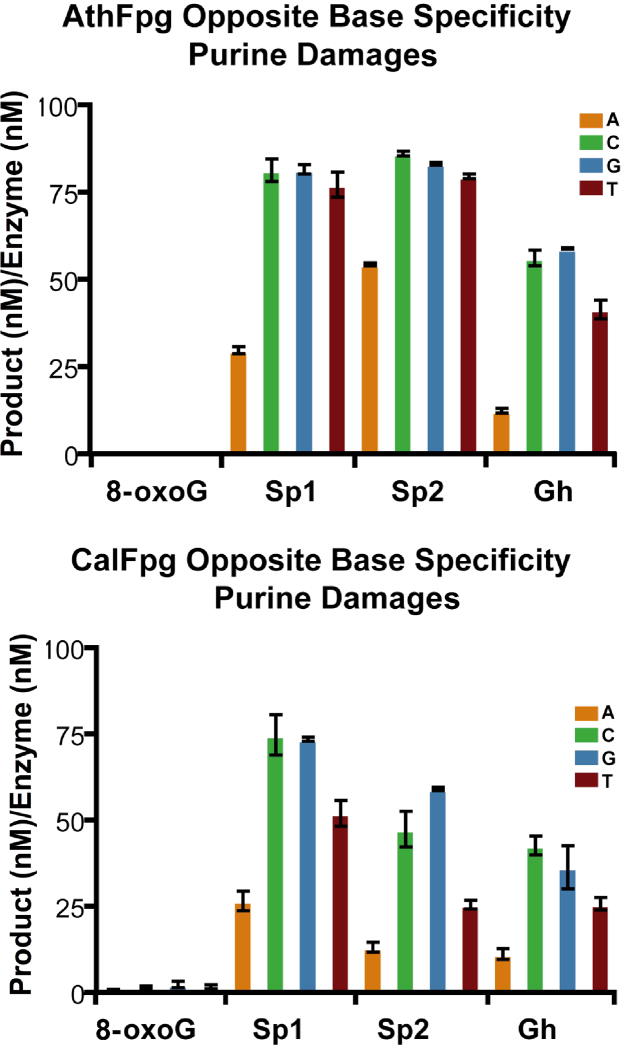
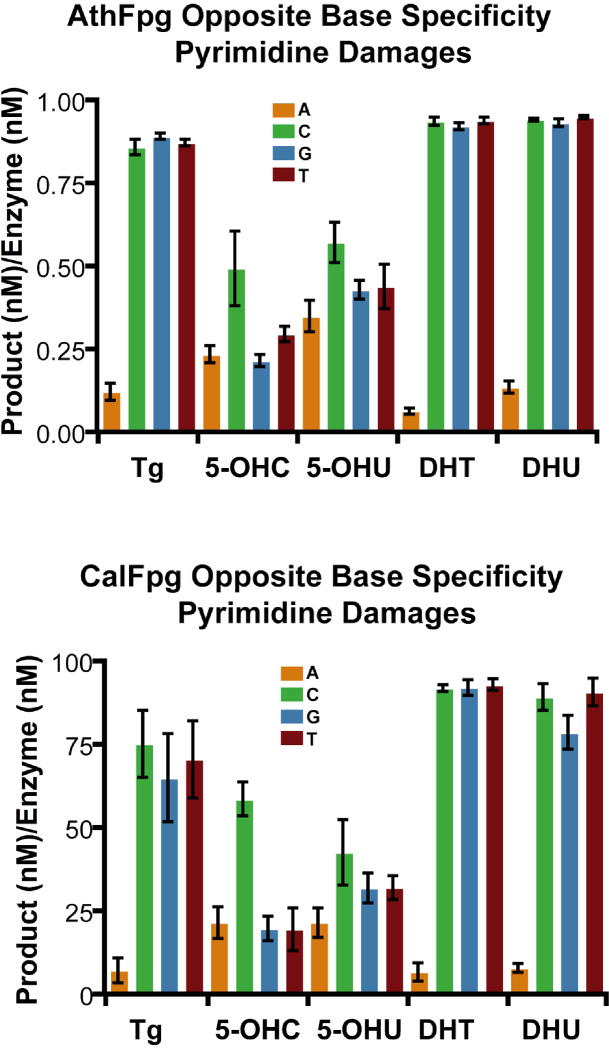
Because CalFpg and AthFpg were members of the Fpg clade, we looked at whether they retained the opposite base discrimination characteristic of Fpg proteins [41]. To do this, CalFpg and AthFpg were incubated with 25 nM labeled oligodeoxynucleotides containing an 8-oxoG, Gh, Sp1, Sp2, Tg, 5-OHC, 5-OHU, DHT and DHU, paired with an oligodeoxynucleotide such that an A, C, G or T was opposite the lesion (Figs. 5 and 6). CalFpg was used at 0.25 nM for all substrates. A 100-fold higher concentration of AthFpg, 25 nM, was required for the same amount of activity as CalFpg on 8-oxoG and the damaged pyrimidines but 0.25 nM was all that was required for Gh, Sp1 and Sp2. As shown in Figs. 5 and 6, CalFpg and AthFpg discriminate against A opposite the lesion. This is especially apparent for the best substrates, Gh, Sp1, Sp2, Tg, DHT and DHU. The only exception is that C is preferred over A, G or T opposite 5-OHU and 5-OHC for both enzymes. These results also clearly indicate that 8-oxoG is not a good substrate for CalFpg or AthFpg regardless of the base opposite the lesion (Fig. 5).
Fig. 5.
Opposite base specificity of AthFpg and CalFpg for purine damages. AthFpg and CalFpg (0.25 nM) were incubated with the substrates listed (25 nM) for 60 min at room temperature. AthFpg: 25 nM for 8oxoG, 0.25 nM for Gh, Sp1 and Sp2. Error bars indicate the standard error of the mean from three independent experiments. The uncertainties are standard errors.
Fig. 6.
Opposite base specificity of AthFpg and CalFpg for pyrimidine damages. AthFpg and CalFpg (0.25 nM) were incubated with the substrates listed (25 nM) for 60 min at room temperature. AthFpg: 25 nM for Tg, 5-OHC, 5-OHU, DHT and DHU. Error bars indicate the standard error of the mean from three independent experiments. The uncertainties are standard errors.
3.8. Single turnover kinetics analysis
Kinetic parameters for AthFpg and CalFpg were determined with pyrimidine lesions DHU and Tg opposite an A and G. DHU and Tg were chosen because they are the best pyrimidine substrates for AthFpg and CalFpg. Kinetic parameters with the oxidation products of 8-oxoG, Gh, Sp1 and Sp2 paired with a C and A were also determined. As shown in Table 1, the kobs for CalFpg with DHU (13.6 min−1) and AP sites (10.4/63 min−1) opposite G are reasonable and in the range of other Nei glycosylases [29,32,38,42–44] but are poor for DHU opposite A and Tg opposite A and G; 8-oxoG was such a poor substrate that kinetic values could not be determined for either glycosylase. The kobs for AthFpg on DHU and Tg opposite G were very low but measurable (0.05) while DHU:A and Tg:A were such poor substrates for AthFpg that kinetic parameters could not be determined. The kobs for the oxidation products of 8-oxoG for CalFpg was the highest with Sp1:C (39.94 min−1) while Gh and Sp2 opposite C were about the same (~2 min−1). The kobs for AthFpg with all three lesions were good with Sp1:C, 29.3 min−1, Sp2:C, 17.4 min−1, and Gh:C, 11.8 min−1. In every case the kobs for Gh, Sp1 and Sp2 opposite C was higher than the value for the same lesion opposite A. All of the single turnover kinetics fit a double exponential with the exception of CalFpg with Sp2:C. The kobs of CalFpg with an AP opposite a G fit a double exponential curve with a K1 value of 10.4 min−1 and a K2 of 63 min−1. The kobs of AthFpg with an AP opposite a G fit a double exponential curve with a K1 value of 240 min−1 and a K2 of 2700 min−1, values greater than 102–104 higher than with DHU or Tg. AthFpg appears to behave more like an AP endonuclease than a DNA glycosylase.
Table 1.
Kinetic rates (kobs) per minute of CalFpg and AthFpg with select double-strand substrates as determined by single turnover experiments. Two values in the box indicate double exponential kinetics (k1, k2). Nd—kinetic reaction did not go to completion.
| Lesion | Gh |
Sp1 |
Sp2 |
DHU |
Tg |
AP | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Opposite base | A | C | A | C | A | C | A | G | A | G | G |
| CalFpg | 0.165 | 0.1609 | 0.177 | 0.888 | 1.142 | 1.93 | 0.08 | 13.6 | 0.18 | 0.38 | 10.4 |
| 0.734 | 2.004 | 1.912 | 39.94 | 0.053 | 0.68 | 63 | |||||
| AthFpg | 0.069 | 0.17 | 0.156 | 1.66 | 0.2466 | 4.44 | Nd* | 0.05 | Nd* | 0.05 | 240 |
| 2.122 | 11.8 | 1.214 | 29.3 | 1.843 | 17.4 | 2700 | |||||
3.9. In γ-irradiated DNA CalFpg prefers FapyGua while AthFpg prefers FapyAde
The product spectrum of damages released from γ-irradiated DNA as determined by GC/MS after incubation with CalFpg or AthFpg exhibit some differences. Although both enzymes recognize and release the FapyAde and FapyGua, CalFpg releases predominantly FapyGua while AthFpg releases predominantly FapyAde (Table 2, Fig. 7). The CalFpg product spectrum is further distinguished from that of bacterial Nei and Nth by the small amount of Tg or 5-OHC released; AthFpg did not release either product. Unlike EcoFpg both enzymes release 5-hydroxy-5-methylhydantoin to some extent; however, only a small amount of 8-oxoG was released by CalFpg and none by AthFpg. Interestingly, 7,8-dihydro-8-oxoadenine (8-oxoA) was also a substrate of CalFpg similar to EcoFpg. On the other hand, only a small amount of 8-oxoA was released by AthFpg.
Table 2.
DNA glycosylase activity on γ-irradiated DNA. The numbers represent released damaged bases per 106 total bases from γ-irradiated DNA as measured by GC/MS.
| 5OHMH | 5-OHC | Tg | FapyAde | 8-oxoA | FapyGua | 8-oxoG | |
|---|---|---|---|---|---|---|---|
| EcoNth | 21.4 | 256.3 | 55.9 | 77 | 33.8 | 7.4 | 10.9 |
| EcoNei | 31.5 | 179.7 | 74.6 | 196.7 | 52.7 | 2.4 | −0.1 |
| EcoFpg | 3.2 | 0 | 0 | 200.3 | 50.3 | 689.6 | 514.3 |
| CalFpg | 24.3 | 24.7 | 4.3 | 197.3 | 63.7 | 532.3 | 5.6 |
| AthFpg | 10.4 | 0 | 0 | 133.3 | 7.2 | 57.8 | 0.15 |
Fig. 7.
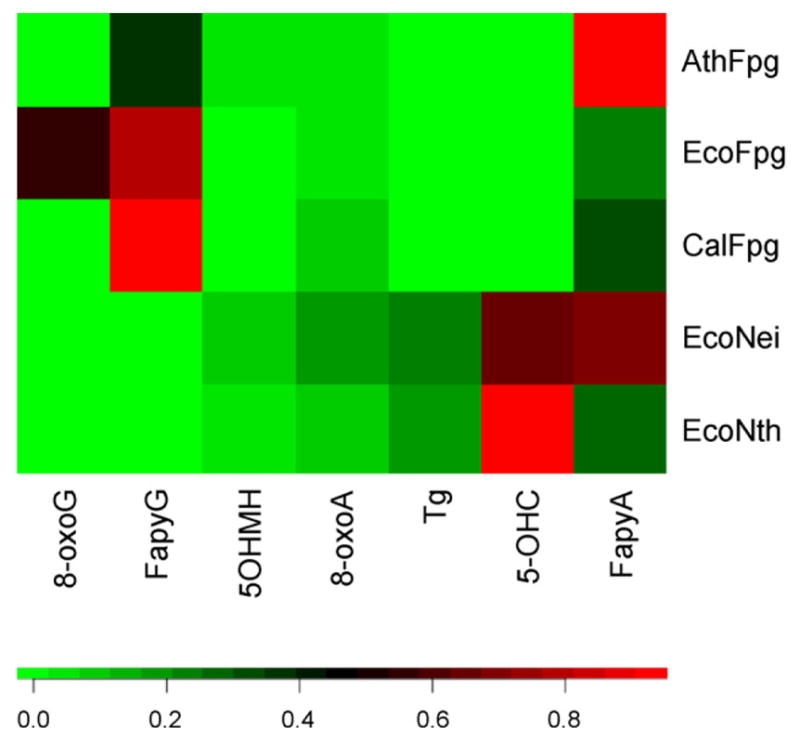
Heat map expressing GC/MS measurements. A color map (bottom) was used to associate normalized (see Section 2) GC/MS intensities (Table 2) with colors. Normalization adjusts in an ad hoc way for different overall differences in the amount of all products released. For example, comparison of the FapyGua and FapyAde cells associated with AthFpg with those associated with CalFpg clearly indicates the different substrate preferences of these two enzymes, even though the unnormalized intensities of the FapyAde are not dramatically different.
4. Discussion
4.1. Multiple absences of Fpg/Nei homologs among metazoans are due to gene loss
Earlier discussions of the Fpg/Nei phylogeny[4,30] emphasized the presence of family members in vertebrates, plants, and certain fungi in the context of absences from protists, Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster. The narrow phylogenetic distribution among eukaryotes raised the possibility that a vertebrate ancestor acquired a family member directly from bacteria, an event that is regarded as implausible based on the small number of carefully documented examples of such an event and on the requirement that the transfer reach germline cells. The results reported here clarify the phylogeny of eukaryotic proteins in this respect. The presence of Neil1 sequences additionally in Trichoplax adhaerens suggests that the ancestor of all known metazoans contained a family member, consistent with the presence of homologs in sea anemone, sea urchin, and fish. The absences of family members among ecdysozoa are therefore most likely due to gene loss. It seems unlikely, then, that the origin of all Fpg/Nei family members involved gene transfer to vertebrates.
In contrast, gene transfers to single celled or colonial eukaryotes seem plausible. Plants, fungi, and metazoans presumably each have single celled or colonial ancestors not shared with the other two. Absences of family members from protists can then be reasonably accounted for either by gene transfer or loss. The fungal phylogeny (not shown) suggests that absence from S. cerevisiae is most parsimoniously explained by a loss rather than horizontal transfer.
4.2. What is the origin of the Zn-finger in NEIL2 and NEIL3?
Metazoans contain Fpg/Nei homologs that can be confidently assigned to one of two classes, NEIL1 or NEIL2/3, based on overall sequence similarity or on certain slowly evolving sequence features that include a Zn-finger signature. The presence of at most one Fpg/Nei homolog in plants and fungi might suggest that the origin of the three vertebrate sequences was in gene duplication during metazoan evolution; it is common to see large families of homologous proteins in vertebrates. However, phylogenetic estimation places the NEIL2/NEIL3/bacterial Nei clade outside the NEIL1 clade. Although there is ample precedent for the gain of Zn fingers, which might occur through recombination or convergent evolution, such a possibility is inconsistent with the phylogeny shown in Fig. 1.
4.3. Actinobacteria exhibit multiple Fpg/Nei homologs
A remarkable six actinobacterial gene clades occur (Fig. 1): two among Nei, four dispersed throughout the Fpg clade. Thus actinobacteria alone represent well the diversity of the entire Fpg/Nei phylogeny. The above scenarios are consistent with actinobacteria either serving as the origin of the Nei sequences or simply being adept at scavenging sequences from other species. The large number of Fpg/Nei homologs among actinobacteria may result from selection or drift.
4.4. CalFpg and AthFpg are active against formamidopyrimidines and oxidation products of 8-oxoG but not against 8-oxoG
The experiments shown in Figs. 3 and 4 clearly demonstrate that 8-oxoG is a very poor substrate for CalFpg. The best oxidatively damaged oligodeoxynucleotide substrates for these enzymes are Gh:C, Sp1:C and Sp2:C as can be seen in Figs. 3 and 4. More detailed kinetics analysis (Table 1) demonstrates that the best purine substrate for CalFpg is Sp1:C; while Sp1:C is also the best purine substrate for AthFpg; this enzyme also recognizes Sp2:C and Gh:C. The results of the GC/MS experiments demonstrate that AthFpg and CalFpg are both active against FapyAde while CalFpg has an even more robust activity on FapyGua in DNA containing multiple radiation-induced lesions (Table 2 and Fig. 7). In terms of the lack of activity toward 8-oxoG, the results obtained with oligodeoxynucleotides are in excellent agreement with those obtained using γ-irradiated DNA. A. thaliana and C. albicans each have an Ogg DNA glycosylase for the recognition and removal of 8-oxoG and this may have allowed their respective Fpgs to evolve such that the oxidation products of 8-oxoG and formamidopyrimidines are their preferred substrates. Human and mouse NEIL1, like AthFpg and CalFpg, almost exclusively remove FapyAde and FapyGua from DNA containing multiple radiation-induced lesions [29,40,45]. EcoFpg, AthFpg and CalFpg all remove FapyAde and FapyGua but what distinguishes these enzymes from each other is that only EcoFpg removes 8-oxoG. Therefore, with respect to substrate preferences, AthFpg and CalFpg are more similar to mammalian NEIL1 than to EcoFpg [29,40,45].
4.5. The best substrate for AthFpg is an AP site
AthFpg was originally identified as a single copy gene comprised of 10 exons and two splice variants formed by differential splicing of exon 8 and called AtMMH, A. thaliana MutM homolog 1 and 2 [46] and at the same time by Murphy and Gao (1998) [47] who named them AtFpg-1 and 2. Polyclonal antibodies raised against the recombinant proteins identified a single 44 kDa polypeptide that co-migrated with the in vitro translation product of AthFpg in a Western blot analysis [46]. AthFpg expressed as an in vitro translation product was shown to cleave 8-oxoG in a double stranded oligodeoxynucleotide when opposite a cytosine and guanine but not opposite an adenine or thymine [46]. AtFpg1 and 2 expressed as recombinant proteins both cleave double stranded depurinated DNA; however, only AtFpg1 but not 2 cleaved 8-oxoG opposite C [24]. We have further characterized the activity of AthFpg1 on substrates containing 8-oxoG, FapyAde and FapyGua as well as damaged pyrimidines and an AP site and shown that oxidized pyrimidines FapyAde and FapyGua as well as AP sites were much better substrates than 8-oxoG (Figs. 3, 4 and 7; Tables 1 and 2). However, even with oxidized pyrimidines, AthFpg was an extremely poor glycosylase with a kobs on the two measurable base damage substrates being only 0.05 min−1 (Table 1). Interestingly, orthologs of AthFpg 1 and 2 in sugarcane, scMUTM1 and scMUTM2 were over-expressed and were able to complement E. coli deficient in Fpg and MutY decreasing the mutation frequency 10-fold compared to the Fpg and MutY mutants [48]. An AP site was by far the best substrate for AthFpg in both double-stranded and single-stranded DNA (Figs. 3 and 4) with the kobs being 2 orders of magnitude higher for an AP site than for a base lesion (Table 1). These data suggest that the AP lyase activity of AthFpg may perform a physiological role in Arabidopsis. There is precedence for this since in S. cerevisiae [49,50] and Schizosaccharomyces pombe [51,52] the lyase activity of endonuclease III cleaves the AP site left by 3-methyladenine DNA glycosylase. In fact, in S. pombe whole cell extracts, the Nth1 lyase is the major AP incision activity [51]. A similar suggestion has been made for NEIL1 in repair of alkylation damage [5] and in repair of the AP site left by hOGG1 [53]. It should be noted, however, that AthFpg is only expressed in flowers and roots [47]. Alternatively, for the glycosylase activity to be manifest, it may require interacting partners.
4.6. The Fpg insertion loop is present in CalFpg and AthFpg
The crystal structures of Fpg-DNA complexes from E. coli [54], Lactococcus lactus [55] and Bacilles stereothermophilus [35] have been solved. In all three structures, the void created by the vacated base is filled with amino acid residues homologous to Met74, Arg109 and Phe111 in E. coli which are conserved in Fpg glycosylases. From the E. coli Fpg-DNA Schiff base intermediate structure, Met74 occupies the space left by the vacated base, Arg109 and Phe111 play a role in unstacking the bases and kinking the DNA. Arg109 also contributes to opposite base specificity [54] by forming two hydrogen bonds with the orphaned cytosine as well as with thymine. G opposite 8-oxoG is also a good substrate for Fpg and assumes a syn conformation for hydrogen bonding with Arg109. Arg109 is among the most slowly evolving amino acids in the plant/fungi clade, clearly demonstrating that it is maintained by selective pressure. Such interactions are not possible with an orphaned adenine [35]. The eukaryotic functional homolog of bacterial Fpg, Ogg, exhibits more stringent specificity for C opposite 8-oxoG with members of its insertion loop forming five hydrogen bonds with the estranged C [36]. The crystal structure of EcoNei covalently crosslinked to substrate has also been solved and Gln69, Leu70 and Tyr71, the QLY loop, are inserted into the gap created by the everted base [56]. In the structure, Gln69 forms a single weak hydrogen bond with the A opposite the Tg and there is no discriminating element equivalent to Arg109 in Fpg.
AthFpg has Met74, Arg109 and Phe111, while CalFpg has Met74, Arg109 but Leu111 instead of Phe111. Thus the insertion loops of CalFpg and AthFpg are equivalent to the amino acids found in EcoFpg and very different from the amino acids in the QLY loop from EcoNei and accordingly, like Fpg, these enzymes discriminate against A opposite the lesion (Figs. 5 and 6).
NEIL1 also has the Fpg intercalation loop. Although the opposite base specificity of NEIL1 has not been extensively investigated, G is preferred over A opposite the Tg lesion [44]. Although the preference for G over A may be counterintuitive for the ring saturation products DHT, DHU (Fig. 4) and Tg (Fig. 4 and [44]), the latter authors have shown that Tg is the major oxidation product of 5-methylcytosine which would have G as its cognate partner. Interestingly, for most oxidized pyrimidines cleavage by EcoNei, which has the QLY insertion loop, exhibited no strong opposite base preference. However, when tested against DHU and cis-(5S,6R) Tg, less biologically relevant substrates, cleavage was lower when the lesion was opposite A [42]. The opposite base specificity of NEIL1 with respect to Gh and Sp is different insofar as T is the preferred opposite base [57]. This also differs from our result with CalFpg and AthFpg where the C or G are preferred bases opposite the Gh and Sp lesions. This is not surprising since the NEIL1 protein and the AthFpg and CalFpg proteins are found in different clades (Fig. 1) and may have evolved separately.
5. Summary
CalFpg and AthFpg fall into a plant and fungal clade that exhibits sequence features similar to bacterial Fpg, and the preferred substrates for these DNA glycosylases are Gh, Sp1, Sp2, FapyAde and FapyGua not 8-oxoG. Like NEIL1 these glycosylases have a zinc-less finger and can recognize oxidatively induced damage in single stranded DNA. AthFpg is especially interesting because the kobs with an AP site opposite a G is remarkably high for a DNA glycosylase. Clearly much needs to be done to ascertain the biological role of these unusual Fpg/Nei DNA glycosylases.
Acknowledgments
The authors would like to thank Wendy Cooper and Alicia Holmes for expression and purification of the proteins used in this study. We are grateful to Jeffrey Blaisdell for determining the active fraction of the enzymes and helpful discussion regarding kinetics. We would also like to thank Matthew Hogg for assistance with the KinTek Rapid Quench apparatus. Dr Douglas Johnson (University of Vermont) kindly provided genomic DNA from C. albicans that was used as the DNA template to amplify CalFpg. Terrence Murphy, University of California at Davis, graciously provided us with the vector containing the AthFpg construct used in this study. This research was supported in part by NIH grant P01 CA098993 awarded by the National Cancer Institute.
Certain commercial equipment or materials are identified in this paper in order to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Abbreviations
- Fpg
formamidopyrimidine DNA glycosylase
- Nei
endonuclease VIII
- Nth
endonuclease III
- 8-oxoG
8-oxo-7,8-dihydroguanine
- 8-oxoA
7,8-dihydro-8-oxoadenine
- Gh
guanidinohydantoin
- Sp
spiroiminodihydantoin
- CalFpg
Candida albicans (Cal) Fpg DNA glycosylase
- AthFpg
Arabidopsis thaliana (Ath) Fpg DNA glycosylase
- BER
base excision repair
- AP site
apurinic or apyrimidinic
- ROS
reactive oxygen species
- EcoNth
Escherichia coli endonuclease III
- EcoNei
Escherichia coli endonuclease VIII
- PCR
polymerase chain reaction
- Tg
thymine glycol
- 5-OHC
5-hydroxycytosine
- 5-OHU
5-hydroxyuracil
- DHT
5,6-dihydrothymine
- DHU
5,6-dihydrouracil
- UDG
uracil DNA glycosylase
- FapyAde
4,6-diamino-5-formamidopyrimidine
- FapyGua
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- 5OHMH
5-hydroxy-5-methylhydantoin
Footnotes
Conflict of interest statement
The above authors have no financial interests.
References
- 1.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 2.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson DM, III, Sofinowski TM, McNeill DR. Repair mechanisms for oxidative DNA damage. Front Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- 4.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 5.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 7.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 9.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 10.Li A, Schuermann D, Gallego F, Kovalchuk I, Tinland B. Repair of damaged DNA by Arabidopsis cell extract. Plant Cell. 2002;14:263–273. doi: 10.1105/tpc.010258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 14.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. Analysis of base excision and nucleotide excision repair in Candida albicans. Microbiology. 2008;154:2446–2456. doi: 10.1099/mic.0.2008/017616-0. [DOI] [PubMed] [Google Scholar]
- 15.Murphy TM, Gao MJ. Multiple forms of formamidopyrimidine-DNA glycosylase produced by alternative splicing in Arabidopsis thaliana. J Photochem Photobiol B. 2001;61:87–93. doi: 10.1016/s1011-1344(01)00172-5. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 19.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 20.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caffrey DR, Dana PH, Mathur V, Ocano M, Hong EJ, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC Bioinformatics. 2007;8:381. doi: 10.1186/1471-2105-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.6, Distributed by the author. Department of Genome Sciences, University of Washington; Seattle: 2005. [Google Scholar]
- 24.Gao MJ, Murphy TM. Alternative forms of formamidopyrimidine-DNA glycosylase from Arabidopsis thaliana. Photochem Photobiol. 2001;73:128–134. doi: 10.1562/0031-8655(2001)073<0128:afofdg>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Blaisdell JO, Wallace SS. Rapid determination of the active fraction of DNA repair glycosylases: a novel fluorescence assay for trapped intermediates. Nucleic Acids Res. 2007;35:1601–1611. doi: 10.1093/nar/gkm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 27.Dizdaroglu M. Chemical determination of oxidative DNA damage by gas chromatography–mass spectrometry. Methods Enzymol. 1994;234:3–16. doi: 10.1016/0076-6879(94)34072-2. [DOI] [PubMed] [Google Scholar]
- 28.Dizdaroglu M, Bauche C, Rodriguez H, Laval J. Novel substrates of Escherichia coli nth protein and its kinetics for excision of modified bases from DNA damaged by free radicals. Biochemistry. 2000;39:5586–5592. doi: 10.1021/bi9927787. [DOI] [PubMed] [Google Scholar]
- 29.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 30.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 31.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 32.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst) 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavrukhin OV, Lloyd RS. Involvement of phylogenetically conserved acidic amino acid residues in catalysis by an oxidative DNA damage enzyme formamidopyrimidine glycosylase. Biochemistry. 2000;39:15266–15271. doi: 10.1021/bi001587x. [DOI] [PubMed] [Google Scholar]
- 34.Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002;277:2938–2944. doi: 10.1074/jbc.M110499200. [DOI] [PubMed] [Google Scholar]
- 35.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 36.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 37.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 39.Neto JB, Gentil A, Cabral RE, Sarasin A. Mutation spectrum of heat-induced abasic sites on a single-stranded shuttle vector replicated in mammalian cells. J Biol Chem. 1992;267:19718–19723. [PubMed] [Google Scholar]
- 40.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- 42.Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006;45:12039–12049. doi: 10.1021/bi060663e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dizdaroglu M, Burgess SM, Jaruga P, Hazra TK, Rodriguez H, Lloyd RS. Substrate specificity and excision kinetics of Escherichia coli endonuclease VIII (Nei) for modified bases in DNA damaged by free radicals. Biochemistry. 2001;40:12150–12156. doi: 10.1021/bi015552o. [DOI] [PubMed] [Google Scholar]
- 44.Ocampo-Hafalla MT, Altamirano A, Basu AK, Chan MK, Ocampo JE, Cummings A, Jr, Boorstein RJ, Cunningham RP, Teebor GW. Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis–trans epimerization. DNA Repair (Amst) 2006;5:444–454. doi: 10.1016/j.dnarep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. Human polymorphic variants of the NEIL1 DNA glycosylase. J Biol Chem. 2007;282:15790–15798. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- 46.Ohtsubo T, Matsuda O, Iba K, Terashima I, Sekiguchi M, Nakabeppu Y. Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol Gen Genet. 1998;259:577–590. doi: 10.1007/s004380050851. [DOI] [PubMed] [Google Scholar]
- 47.Murphy TM, Gao MJ. Two cDNAs encoding Arabidopsis thaliana homologs of bacterial formamidopyrimidine-DNA glycosylase genes are produced by alternative processing (accession nos. AF099970 and AF099971) Plant Physiol. 1998;118:1535. [Google Scholar]
- 48.Scortecci KC, Lima AF, Carvalho FM, Silva UB, Agnez-Lima LF, Batistuzzo de Medeiros SR. A characterization of a MutM/Fpg ortholog in sugarcane—a monocot plant. Biochem Biophys Res Commun. 2007;361:1054–1060. doi: 10.1016/j.bbrc.2007.07.134. [DOI] [PubMed] [Google Scholar]
- 49.Hanna M, Chow BL, Morey NJ, Jinks-Robertson S, Doetsch PW, Xiao W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alseth I, Korvald H, Osman F, Seeberg E, Bjoras M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:5119–5125. doi: 10.1093/nar/gkh851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osman F, Bjoras M, Alseth I, Morland I, McCready S, Seeberg E, Tsaneva I. A new Schizosaccharomyces pombe base excision repair mutant, nth1, reveals overlapping pathways for repair of DNA base damage. Mol Microbiol. 2003;48:465–480. doi: 10.1046/j.1365-2958.2003.03440.x. [DOI] [PubMed] [Google Scholar]
- 53.Mokkapati SK, Wiederhold L, Hazra TK, Mitra S. Stimulation of DNA glycosylase activity of OGG1 by NEIL1: functional collaboration between two human DNA glycosylases. Biochemistry. 2004;43:11596–11604. doi: 10.1021/bi049097i. [DOI] [PubMed] [Google Scholar]
- 54.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 55.Serre L, Pereira de Jesus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21:2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]